Introduction
Raoultella species are aerobic, Gram-negative, non-motile, capsulated bacilli belonging to the family Enterobacteriaceae, that are closely related to Klebsiella spp.1, 2 They were initially included in the genus Klebsiella until the late 1990s.3 Many studies have shown that 0.2–19.0% of isolates initially identified as Klebsiella spp. were Raoultella spp. by 16S rRNA analysis.2 Raoultella spp. are known to occur in aquatic and soil environment. They are a rare cause of human infections and are known to cause soft tissue and bloodstream infections.1, 4 The most common species implicated in human infections are Raoultella ornithinolytica, Raoultella planticola and Raoultella terrigena.2 We report a rare case of post-operative sub-hepatic space infection caused by extensively drug resistant R. ornithinolytica.
Case report
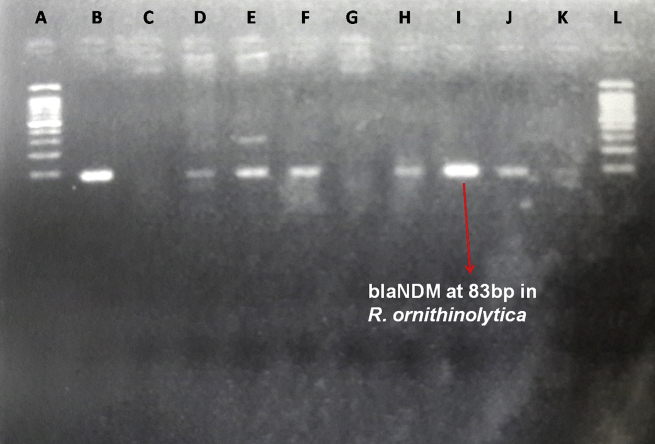
A 75 year old male diabetic and hypertensive patient presented with fever with chills and jaundice. On investigation, he was found to have a mass in the head of pancreas causing obstructive jaundice for which he underwent Whipple's pancreaticoduodenectomy. On 13th and 14th post-op day, fluid from the sub-hepatic drain was sent to microbiology laboratory for culture and sensitivity testing. Both the drain fluid specimens were processed according to standard protocol followed in the microbiology laboratory. After 24 h of incubation at 37 °C, large, lactose fermenting, glistening moist colonies were seen on MacConkey agar. Gram stain revealed Gram-negative stout rods resembling Klebsiella spp. The organism was catalase positive and oxidase negative. Primary biochemical reactions differentiated it from Klebsiella pneumoniae as the isolate was Methyl red test positive and produced indole. Differentiation from other species in the genus Raoultella was made by the ornithine decarboxylase test which is positive in case of R. ornithinolytica and negative in R. planticola and R. terrigena. Antimicrobial susceptibility was performed on Mueller-Hinton agar plates by the standard Kirby–Bauer disk diffusion method as per the Clinical and Laboratory Standards Institute (CLSI) guidelines 2014. The isolate was found to be resistant to all antibiotics tested. The isolate was also tested by Vitek-2 automated identification and antimicrobial susceptibility testing system, which confirmed it as R. ornithinolytica and the antimicrobial resistance profile was identical to the Kirby–Bauer method. In addition, Vitek-2 showed that the isolate was susceptible only to colistin and tigecycline. Screening for metallo-β-lactamase (MBL) production was carried out by the MBL (IP/IPI) E-test method (AB Biodisk, Solna, Sweden), as per the manufacturer's instructions, and the isolate was found to be positive. Polymerase chain reaction (PCR) was performed to detect the presence of New Delhi metallo-β-lactamase gene (blaNDM). The forward and reverse primers used in the PCR were procured from Integrated DNA Technologies (IDT), Inc., as described by Van der Zee, et al5 (Table 1). For a single sample reaction of 50 μl volume, various reagents were used as mentioned in the Table 2. Amplification was carried out with the following thermal cycling profile: initial denaturation for 4 min at 94 °C, 35 cycles of amplification consisting of 30 s at 94 °C, annealing for 1 min at 45 °C, and extension for 1 min at 72 °C, with 7 min at 72 °C for the final extension. Post amplification analysis by gel electrophoresis revealed band at 83bp size (blaNDM) (Fig. 1). As the amplicon size is less than 100bp, it could not be sequenced.
Table 1.
DNA sequences of forward and reverse primers used for PCR directed against blaNDM gene.5
| Primer | Sequence (5′→ 3′) | Size of PCR product |
|---|---|---|
| NDM-F | CATTAGCCGCTGCATTGATG | 83 |
| NDM-R | GTCGCCAGTTTCCATTTGCT |
Table 2.
Various reagents used in PCR mixture (50 μl).
| Reagent | Volume (μl) | Final concentration |
|---|---|---|
| Autoclaved distilled water | 31.85 | – |
| PCR buffer (10X) with MgCl2 (15 mM) | 5 | 1X, 1.5 mM |
| dNTP (10 mM) | 0.25 | 50 μM |
| Primer blaNDM (forward) | 1.25 | 0.5 μM |
| Primer blaNDM (reverse) | 1.25 | 0.5 μM |
| Taq DNA polymerase (250U/μl) | 0.4 | 2U/μl |
| DNA template | 10 | – |
| Total | 50 | - |
Fig. 1.
Gel electrophoresis showing band at 83bp denoting blaNDM in clinical isolate of Raoultella ornithinolytica. A, L – 100 bp molecular marker. B – Positive control for blaNDM gene. C – Negative control for blaNDM gene. I – blaNDM gene (83bp) in clinical isolate of Raoultella ornithinolytica. D, E, F, H, J – Other isolates showing presence of blaNDM gene.
Discussion
Raoultella species are aerobic, Gram-negative, capsulated, non-motile bacilli belonging to the family Enterobacteriaceae that are closely related to Klebsiella spp.1, 2 They were largely regarded as environmental bacteria found in soil and water, and were only recently distinguished phylogenetically from Klebsiella spp.6 Many studies have revealed that 0.2–19.0% of isolates initially identified as Klebsiella spp. were Raoultella spp. by 16S rRNA analysis and that the prevalence of these organisms in clinical settings varies geographically.2, 7 Raoultella spp are a rare cause of human infections and only a few case reports of causing soft tissue and bloodstream infections have been reported.1, 4 The most common species implicated in human infections are R. ornithinolytica, R. planticola and R. terrigena.2
R. ornithinolytica has been isolated from the gut of the fish, termites and aquatic environments. It has an ability to convert histidine to histamine, leading to fish poisoning, cases of which have been reported previously.8 This bacterium was first described in 1989 by Kosako et al9 and since then there have been very few case reports of human infections by R. ornithinolytica.8
In this case report, we report a case of post-operative sub-hepatic space infection caused by R. ornithinilytica in a 75 year old diabetic male patient, who underwent Whipple's pancreaticoduodenectomy for mass head of pancreas causing obstructive jaundice. The fluid from the drain was collected aseptically on two consecutive days (13th and 14th post-op days) and was processed in microbiology laboratory according to the standard protocol. Both the specimens grew similar colonies on MacConkey agar which were further processed and identified as R. ornithinolytica with the help of biochemical reactions. It was differentiated from Klebsiella spp as it was produced indole and was methyl red test positive. Differentiation from other species in the genus Raoultella was made by the ornithine decarboxylase test which is positive in case of R. ornithinolytica and negative in R. planticola and R. terrigena. Antimicrobial susceptibility testing by standard Kirby–Bauer disc diffusion method showed it to be resistant to all the antibiotics tested including aminoglycosides, fluoroquinolones, cephalosporins and carbapenems. The isolate was also tested by Vitek-2 automated identification and antimicrobial susceptibility testing system, which confirmed it as R. ornithinolytica and the antimicrobial resistance profile was identical to the Kirby–Bauer method. In addition, Vitek-2 showed that the isolate was susceptible only to colistin and tigecycline. PCR showed the presence of blaNDM gene in this isolate.
This case report is only one of few case reports of hospital acquired infections caused by R. ornithinolytica. A report of blaNDM-1 in R. ornithinlolytica causing post-operative soft tissue infections has been reported by Khajuria et al1 However, to the best of our knowledge and worldwide literature search, this is only the second report of NDM producing R. ornithinolytica in the world, which was isolated from post-operative sub-hepatic space infection following Whipple's procedure.
As it has already been discussed, it has many similarities with Klebsiella spp. in the form of capsule and colony morphology, so it can be frequently misidentified as Klebsiella spp. if biochemical reactions are not routinely carried out in the laboratory, which can lead to under-estimation of the real importance of R. ornithinolytica as a human pathogen.
R. ornithinolytica is an emerging pathogen and a cause of hospital acquired infection, which can be resistant to many antimicrobial agents. Antimicrobial resistance to most of the antibiotics tested including aminoglycosides, fluoroquinolones, cephalosporins and carbapenems is a cause of concern. Similar extensively drug-resistant isolate of R. ornithinolytica carrying the blaNDM-1 gene has been reported earlier by Khajuria et al,1 which were also sensitive only to colistin and tigecycline.
The present case illustrates for the first time, isolation of extensively drug resistant R. ornithinolytica from drain fluid in a case of post-operative sub-hepatic space infection following Whipple's pancreaticoduodenectomy. In addition, this is only the second case report of New Delhi Metallo-β-lactamase producing R. ornithinolytica. The presence of blaNDM in R. ornithinolytica poses a serious threat for the spread of hospital-acquired infections. These findings warrant proper identification of isolates by biochemical reactions or confirmation by Vitek-2 and detection of genes responsible for carbapenem resistance. Early recognition of these isolates and proper infection control measures are mandatory for controlling their spread.
Conflicts of interest
All authors have none to declare.
References
- 1.Khajuria A., Praharaj A.K., Grover N., Kumar M. First Report of blaNDM-1 in Raoultella ornithinolytica. Antimicrob Agents Chemother. 2013;57:1092. doi: 10.1128/AAC.02147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castanheira M., Deshpande L.M., DiPersio J.R., Kang J., Weinstein M.P., Jones R.N. First Descriptions of blaKPC in Raoultella spp. (R. planticola and R. ornithinolytica): report from the SENTRY Antimicrobial Surveillance Program. J Clin Microbiol. 2009;47:4129. doi: 10.1128/JCM.01502-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nada B., Areej M. Raoultella planticola, a central venous line exit site infection. J Talibah Univ Med Sci. 2013 http://dx.doi.org/10.1016/j.jtumed.2013.11.008 [Google Scholar]
- 4.Al-Hulu S.M., Al-Charrakh A.H., Al-Saadi M.A.K. Isolation and characterization of Raoultella ornithinolytica from Clinical specimens in Hilla city. Iraq Med J Babylon. 2009;7:42–47. http://www.biomedcentral.com/1471-2334/14/27 [Google Scholar]
- 5.Van der Zee A., Roorda L., Bosman G. Multi-centre evaluation of real-time multiplex PCR for detection of carbapenemase genes OXA-48, VIM, IMP, NDM and KPC. BMC Infect Dis. 2014;14:27. doi: 10.1186/1471-2334-14-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drancourt M., Bollet C., Carta A., Rousselier P. Phylogenetic analyses of Klebsiella species delineate Klebsiella and Raoultella genus with description of Raoultella ornithinolytica, Raoultella terrigena and Raoultella planticola. Int J Syst Evol Microbiol. 2001;51:925–932. doi: 10.1099/00207713-51-3-925. [DOI] [PubMed] [Google Scholar]
- 7.Murray P.R., Baron E.J., Jorgensen J.H., Landry M.L., Pfaller M.A. 9th ed. vol. 1. ASM Press; Washington, DC: 2007. (Manual of Clinical Microbiology). [Google Scholar]
- 8.Sandal G., Ozen M. Fatal Raoultella ornithinolytica sepsis and purpura fulminans in preterm newborn. Indian J Pediatr Dermatology. 2014;15:24–26. [Google Scholar]
- 9.Kosako Y., Tamura K., Sakazaki R., Miki K. Klebsiella ornithinolytica formerly known as ornithine-positive Klebsiella oxytoca. Curr Microbiol. 1989;18:201–206. [Google Scholar]