Abstract
MicroRNAs (miRNAs) play essential roles in plant development, but the roles in the in vitro plant development are unknown. Leaves of ponkan plantlets derived from mature embryos at in vitro culture conditions were used to sequence small RNA fraction via Solexa sequencing, and the miRNAs expression was analyzed. The results showed that there were 3,065,625 unique sequences in ponkan, of which 0.79% were miRNAs. The RNA sequences with lengths of 18–25 nt derived from the library were analyzed, leading to the identification of 224 known miRNAs, of which the most abundant were miR157, miR156, and miR166. Three hundred and fifty-eight novel miRNA candidates were also identified, and the number of reads of ponkan novel miRNAs varied from 5 to 168,273. The expression of the most known miRNAs obtained was at low levels, which varied from 5 to 4,946,356. To better understand the role of miRNAs during the preservation of ponkan in vitro plantlet, the expression patterns of cre-miR156a/159b/160a/166a/167a/168a/171/398b were validated by quantitative real-time PCR (qPCR). The results showed that not only the development-associated miRNAs, e.g., cre-miR156/159/166/396, expressed highly at the early preservation period in the in vitro ponkan plantlet leaves but also the stress-related miRNAs, e.g., cre-miR171 and cre-miR398b, expressed highly at the same time. The expression levels of most tested miRNAs were found to decrease after 6 months and the amounts of these miRNAs were kept at low levels at 18 months. After analyzing the expression level of their targets during the reservation of the ponkan in vitro plantlet, development-associated cre-ARF6 and stress-related cre-CSD modules exhibited negative correlation with miR167 and miR398, respectively, indicating an involvement of the miRNAs in the in vitro development of ponkan and function in the conservation of ponkan germplasm.
Keywords: in vitro plantlet leaf, miRNA, in vitro conservation, miR398, Citrus reticulate
Introduction
MicroRNAs (miRNAs) are the products of endogenous non-coding RNA genes, which perform regulatory roles in the development of plants and animals. MiRNAs are typically 18–25 nucleotide (nt) in length and exhibit different degrees of sequence complementary to target RNAs at the posttranscriptional level (Bartel, 2004; Jones-Rhoades et al., 2006). More evidences have shown that miRNAs play critical roles in plant cell division, morphogenesis, polarity development, hormone secretion, signal transduction as well as responses to stresses (Chen, 2005; Chuck et al., 2009; Liang et al., 2010; Wu et al., 2010; Zhao et al., 2010; Barciszewska-Pacak et al., 2015). Although deep sequencing to identify miRNAs has been reported in many plants, most of which belonged to model plants or main crops such as Arabidopsis thaliana (Hsieh et al., 2009), Lycopersicon esculentum (Moxon et al., 2008), Oryza sativa (Wei et al., 2011), Zea mays (Wang et al., 2011), Triticum aestivum (Meng et al., 2013), and Sorghum bicolor (Katiyar et al., 2015). There are more than 700 species in woody plants, among which fruits of citrus such as tangerine, citrange, tangelo, orange, pomelo, grapefruit, lemon, lime, ponkan, and citron are of great economic importance in the world. Ponkan (Citrus reticulata, an important citrus cultivar) is the major cultivar of mandarins in China. However, deep sequencing for discovery and identification of miRNAs was only reported in sweet orange flesh color and its rootstock, Poncirus trifoliata (synonym: Citrus trifoliata, but more commonly known as Poncirus trifoliata, which is not edible—a related genus plant of citrus rootstock; Song et al., 2010; Xu et al., 2010). Recently, deep sequencing of sRNA was also reported in embryogenic callus, leaf, flower, and fruit of sweet orange (Liu et al., 2014; Wu et al., 2015).
In the indoor preservation of citrus germplasm resources, the in vitro plantlets are used commonly for a longer subculture period and for relatively stable genetic characteristics compared with cell and tissue culture (Chen, 2011). In our Citrus Germplasm Resources Preservation Center, the in vitro plantlets are initiated from mature embryos and undergo a rapid growth phase and then enter a long and slow-growing period in the preservation. Leaf in vitro morphogenesis is an indicator of plant health in our preservation system. Investigations on in vivo leaves indicate that miRNAs are involved in the plant development (Juarez et al., 2004; Ori et al., 2007; Meng et al., 2013). There is no clue about how miRNAs regulate the development of leaf in in vitro ponkan plantlets. In the present study, the in vitro leaves of ponkan were used as materials for discovery and identification of novel and conserved miRNAs by Solexa deep sequencing.
Solexa sequencing provides solid information of the sequences and whether the length of sequences is in accordance with the length of miRNA (Xie et al., 2011). In order to clarify how miRNAs and their targets influence the process of leaf development in vitro, we established a small RNAs library with mixed leaf samples from the early preservation period covering three stages of leaf development, i.e., 9 days after germination (DAG) corresponding to the initiation stage, 22 DAG of fast-growing stage, and 27 DAG of stable-growing stage, and identified miRNAs and their targets in ponkan in vitro plantlet. To further investigate the role of miRNA in the process of in vitro ponkan preservation, the expression patterns of miRNA and their targets of the leaves sampled, both in 6 months after germination (MAG) corresponding to the early preservation period and in 18 MAG corresponding to the slow-growing stage in the later preservation period, were also detected by means of quantitative real-time PCR (qPCR).
Materials and methods
Plantlets and growth conditions
Fruits of ponkan (C. reticulata “Lugan”) were picked from orchard in Youxi in Youxi Technology Bureau (Fujian Agriculture and Forestry University has approved the study. The land accessed is not privately owned and protected. No protected species were sampled. No specific permissions were required for these locations/activities. The Youxi Technology Bureau is located in Jiefang Road No. 17 in Youxi County, Sanming City). The seeds were immersed in soap water for 3–5 min to get rid of the mucilage on the seeds, then drained and washed with distilled water until they reached neutral pH. Subsequently, the ponkan seeds were sterilized for 30 s in 75% ethanol and washed with sterile water until they reached neutral pH, and then immersed in 0.1% mercuric chloride solution for 8 min, followed by washing five times with sterile water. The episperm and endopleura of seeds were cut and the embryos were placed in culture flask (6 × 9 cm) with solid growth medium [sterilized Murashige-Skoog (MS) salt solution (pH = 5.8) + 0.6% agar + 3% sucrose]. Plants were grown under a photoperiod of 12 h light/12 h dark (1200 lx) in a plant growth chamber at 25°C (Chen, 2011). Finally, 9 DAG, 22 DAG, 27 DAG, 6 MAG, and 18 MAG old leaves of in vitro ponkan plantlets were collected for measurements. Leaf samples were rapidly and gently collected from the surface of the filter paper. Small RNA was isolated from 9 DAG, 22 DAG, and 27 DAG old leaves and used for Solexa sequencing.
Construction of ponkan In vitro plantlet leaf small RNA library and solexa sequencing
Total RNA was isolated from the samples mentioned above using TRIzol Reagent kit (Invitrogen, Life Technologies, Carlsbad, CA) according to the manufacturer's instructions and pooled in an equal fraction ratio for the construction of ponkan small RNA library. After that, the small RNA fragments of 16–30 nt were isolated from the 15% PAGE gel and purified, and then the small RNAs were ligated to a 5′ RNA adapter (5′-GUUCAGAGUUCUACAGUCCGACGAUC-3′) and a 3′ RNA adapter (5′-pUCGUAUG CCGUCUUCUGCUUGidT-3′; p, phosphate; idT, inverted deoxythymidine) sequentially using T4 RNA ligase. The samples were reversely transcribed to cDNA with the RT primer (5′-CAAGCAGAAGACGGCATACGA-3′) using Superscript II reverse transcriptase (Invitrogen), and amplified by PCR. Finally, the small RNAs from samples were sequenced by using Solexa sequencing technology (Beijing Genomics Institute, China) and submitted to SRA with the accession number SRP066915.
Analysis of small RNA of ponkan In vitro plantlet leaf
In order to predict the potentially conserved and novel miRNAs of ponkan in vitro plantlet leaf, bioinformatics analysis of the Solexa sequencing data was conducted. Firstly, the 35-nt sequence tags from Solexa sequencing gone through the data were cleaned, which eliminated several kinds of contaminants and the low-quality tags. The small RNA tags were mapped to the Citrus clementina genome (http://www.phytozome.net/clementine.php) using Short Oligonucleotide Analysis Package (SOAP) to analyze their expression and distribution in the genome (Li et al., 2008). Secondly, the cleaned tags were compared against non-coding RNAs from Rfam database (http://www.sanger.ac.uk/software/Rfam) and the NCBI GenBank database (http://www.ncbi.nlm.nih.gov/) to classify the degradation fragments of non-coding RNAs. Any small RNAs having exact matches with the degradation fragments were excluded from further analysis. In addition, the unique sRNA sequences were used for searching miRNA sequences using miRBase 21 (http://mirbase.org/) to identify the known miRNAs in ponkan in vitro plantlet leaf. Finally, the novel miRNA of the ponkan in vitro plantlet leaf was predicted from the surplus unannotated small RNAs using MIREAP (http://sourceforge.net/projects/mireap/), and the parameters setting for the identification of novel miRNA are minimal miRNA length: 18, maximal miRNA length: 25, minimal miRNA (reference) length: 20, maximal miRNA (reference) length: 23, uniqueness of miRNA: 20, maximal energy: −18, minimal space: 5, maximal space: 300, minimal mature pair: 16, maximal mature bulge: 4, maximal duplex asymmetry: 4, and flank sequence length: 20.
Prediction of targets of ponkan In vitro plantlet leaf miRNAs
It is necessary to identify and characterize the targets of miRNAs in order to understand the biological functions of them. In the present experiment, the potential targets of ponkan miRNAs were predicted using the psRobot (http://omicslab.genetics.ac.cn/psRobot/) and TargetFinder (http://targetfinder.org/)program (Kiełbasa et al., 2010; Wu et al., 2012). Newly identified ponkan miRNAs sequences were used as custom miRNA sequences and C. clementina genome (http://www.phytozome.net/clementine.php) sequences were used. The rules used for predicting ponkan novel miRNAs targets are (1) no more than four mismatches between the sRNA and the target (G-U bases count as 0.5 mismatches); (2) no more than two adjacent mismatches in the miRNA–target duplex; (3) no adjacent mismatches in positions 2–12 of the miRNA–target duplex (5′ of miRNA); (4) no mismatches in positions 10–11 of miRNA–target duplex; (5) no more than 2.5 mismatches in positions 1–12 of the of the miRNA–target duplex (5′ of miRNA); (6) minimum free energy (MFE) of the miRNA–target duplex should be ≥75% of the MFE of the miRNA bound to its perfect complement.
Real-time quantitative PCR of ponkan miRNAs and their targets
The qPCR was used to validate results obtained from high throughput sequencing of ponkan miRNAs and their targets. RNA samples from the five embryogenic cultures described above were reverse transcribed using an One-Step PrimeScript miRNA cDNA Synthesis Kit (Perfect Real Time) (Takara Code: D350A) and PrimeScript® RT reagent Kit (Takara Code: DRR037A) for miRNA and target genes tested, respectively. Expression profiles of 13 miRNAs and six targets were examined using an SYBR® PrimeScript™ miRNA RT-PCR Kit (Takara Code: RR716) and SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) (Takara Code: RR820A), respectively. All reactions were performed in triplicate in a LightCycler 480 qPCR instrument (Roche Applied Science, Switzerland) with a dissociation curve used to control for primer dimers in the reactions. Mature miRNA abundance was calculated relative to expression of reference genes cre-U6 snRNA; miRNA and target names and primer sequences are provided in Supplementary Table S4.
Statistical analysis
Statistical analysis was performed using the SPSS package program version 11.5 (SPSS, Chicago, IL, USA). The data were analyzed by one-way analysis of variance. The values are reported as means ± standard error (SE) for all results. Differences were considered significant at P < 0.05.
Results
Categories and size distribution of small RNAs in ponkan
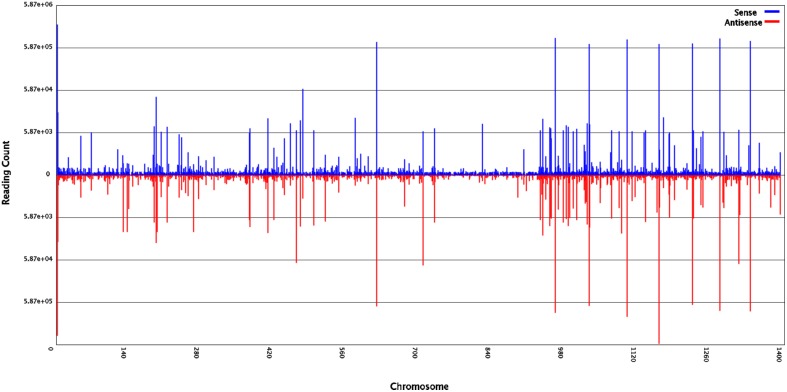
To identify miRNAs involved in the development of ponkan in vitro plantlet leaf, a sRNA library was generated from the leaves of ponkan in vitro plantlet (three stages of pooled samples) and sequenced by a Solexa/Illumina analyzer. In total, 19,145,956 raw reads were generated, including 180,152 low-quality reads and 18,965,804 high-quality reads. After removing low-quality reads and adapter contaminants, 18,033,510 clean reads with lengths ranging from 18 to 30 nt were obtained from the leaves of ponkan in vitro plantlet (Table 1). According to the general principle of miRNA analysis, sRNAs were first compared with C. clementina (http://www.phytozome.net/clementine.php) and then the number of sRNA sequences matching with the genome was identified (Figure 1). After SOAP analysis, 15,865,321 (87.98%) sequences were found to match with the genome perfectly, which contained 2,018,630 (65.85%) unique sequences.
Table 1.
Distribution of small RNAs among different categories in ponkan in vitro plantlet.
| Category | Unique sRNA | Percent (%) | Total sRNA | Percent (%) |
|---|---|---|---|---|
| Total | 3065625 | 100.00 | 18033510 | 100.00 |
| Exon-antisense | 55429 | 1.81 | 175677 | 0.97 |
| Exon-sense | 91444 | 2.98 | 301569 | 1.67 |
| Intron-antisense | 38444 | 1.25 | 93759 | 0.52 |
| Intron-sense | 59701 | 1.95 | 241971 | 1.34 |
| miRNA | 24179 | 0.79 | 9436789 | 52.33 |
| rRNA | 64266 | 2.10 | 881150 | 4.89 |
| Repeat | 316430 | 10.32 | 721841 | 4.00 |
| snRNA | 1748 | 0.06 | 4499 | 0.02 |
| snoRNA | 1063 | 0.03 | 1894 | 0.01 |
| tRNA | 6487 | 0.21 | 262679 | 1.46 |
| Unann | 2406434 | 78.50 | 5911682 | 32.78 |
Figure 1.
Small RNAs of ponkan in vitro plantlet leaf distribution across different ponkan chromosomes.
Among the unique sequences, 24,179 (0.79%) were found to be similar with the known miRNAs after comparing against all plant miRNA precursors and mature miRNAs in miRBase 21. Other unique sequences, including ribosomal RNA (rRNA, 64,266, 2.10%), small nuclear RNA (snRNA, 1748, 0.06%), small nucleolar RNA (snoRNA, 1063, 0.03%), and transfer RNA (tRNA, 6487, 0.21%), were also identified in ponkan in vitro plantlet leaves by performing a BLASTN search against the Rfam (12.0) database (http://rfam.xfam.org/) (Nawrocki et al., 2014). However, 2,406,434 (78.50%) unique sequences could not be annotated (Table 1).
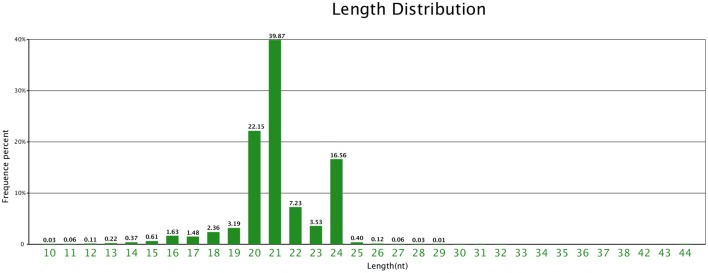
The size distribution patterns of the sRNAs' unique sequences in ponkan were listed in Figure 2. The results showed that the size distribution of sRNAs was uneven. The length of most unique sRNAs varied from 18 to 25 nt, and 39.87% were 21 nt in size, followed by 20 nt (22.15%) and 24 nt (16.56%).
Figure 2.
Length distribution of unique sequences in ponkan in vitro plantlet.
Identification of known miRNAs in ponkan
In order to identify the conserved miRNAs from ponkan sRNA sequences, the unique sRNAs were compared with miRNA precursors or mature miRNAs of all plant data in miRBase 21. A total of 24,179 sequences were found to be orthologs of known miRNAs in other plant species. After Blastn, 224 known miRNAs, belonging to 201 miRNA families, were identified from the leaf of ponkan in vitro plantlet (Supplementary Table S1). There were 178 miRNA families containing only one member, whereas 23 miRNA families contained two members.
The length distribution of known miRNAs in ponkan was listed in Supplementary Table S1, which varied from 18 to 24 nt. The lengths of most miRNAs were 21 nt (40.41%), followed by 24 nt (20.54%) and 22 nt (10.27%).
The expression frequency of known miRNAs in ponkan was found to have large divergences ranging from 5 to 4,946,356. Most of the known ponkan miRNAs (66.52%) were sequenced < 1000 times, and only 2.01% known miRNAs (miR530, miR3623, and miR7757) were sequenced < 10 times. Generally, the 224 known miRNAs were classified into two categories based on their conservation, and conserved miRNAs expressed higher than non-conserved miRNAs. MiR157 was the most abundant one with 4,945,356 reads detected in leaves, followed by miR156 and miR166. High accumulation of miR157 indicated its important roles in the development of ponkan in vitro plantlet leaves. Non-conserved miRNAs such as miR3954 and miR5770 were also highly expressed (177,692 and 110,104, respectively).
Prediction of novel miRNAs in ponkan
The unannotated sequences were mapped with the genome sequences of C. clementina (http://www.phytozome.net/clementine.php), and the perfectly matched sequences were collected to predict the secondary structure of each locus using the software MIREAP. Analyzed sRNAs are considered as candidate miRNA genes only if they fulfill the following criteria: (1) a mature sequence localized in one arm of the stem-loop structure and between 19 and 24 nt; (2) the corresponding miRNA* sequence identified; (3) the pre-miRNA sequence folded into an appropriate stem-loop hairpin secondary structure; (4) the mfe of secondary structures ≤ −20 kcal/mol; and (5) no more than 7-nt mismatches in the miRNA:miRNA* duplex (Meyers et al., 2008). The results revealed that 358 sRNA sequences that perfectly matched citrus genome were able to fold into stem-loop hairpin secondary structures. The number of reads of ponkan novel miRNAs varied from 5 to 168,273, among which cre-miR-m0019 was expressed at the highest level (168,273), followed by cre-miR-m0190 and cre-miR-m0175. Only 5.31% novel miRNAs were expressed over 1000 times (Table 2, Supplementary Table S2).
Table 2.
Ten novel miRNAs identified from ponkan in vitro plantlet.
| Novel miRNA | Location | Mfe | Count | Sequence | Length (nt) | 5′/3′ |
|---|---|---|---|---|---|---|
| Cre-miR-m0019 | scaffold_1:15209053:15209175: − | −55 | 168238 | GTGACAGAAGATAGAGAGCGC | 21 | 5′ |
| Cre-miR-m0190 | scaffold_5:5960222:5960542: − | −75 | 39182 | GCAATGCTCTTGAAGGACTAC | 21 | 3′ |
| Cre-miR-m0175 | scaffold_5:5960245:5960485: + | −59 | 35081 | AAGTCATTAGAAGAACTGCCG | 21 | 5′ |
| Cre-miR-m0053 | scaffold_2:8093618:8093764: − | −78 | 7107 | CCGCAGGGGCGACATGAGATC | 21 | 5′ |
| Cre-miR-m0144 | scaffold_4:3919238:3919484: + | −94 | 5367 | ACTGACAGCGGCTGTACTGTAGT | 23 | 5′ |
| Cre-miR-m0094 | scaffold_3:25587984:25588086: + | −38 | 3689 | TGCTTGTTGATTGTCATCTAA | 21 | 5′ |
| Cre-miR-m0133 | scaffold_3:46661840:46661982: − | −53 | 2946 | CATGTTTCGGGTTTGTGCGTG | 21 | 5′ |
| Cre-miR-m0106 | scaffold_3:50445048:50445335: + | −85 | 2416 | GGTGTCGTGGTGTAGTTGGTT | 21 | 5′ |
| Cre-miR-m0073 | scaffold_2:28637496:28637638: − | −53 | 1574 | GGTCATGGGAGGATTGGCGA | 20 | 5′ |
| Cre-miR-m0238 | scaffold_6:20198376:20198482: + | −66 | 2038 | CTGGATGCAACTGTGGTACGG | 21 | 3′ |
According to Mfold, the precursors of the miRNAs had negative folding free energies ranging from −18.10 to −131.00 to 116.60 kcal/mol. The average Mfold was −45.16 kcal/mol (Table 2).
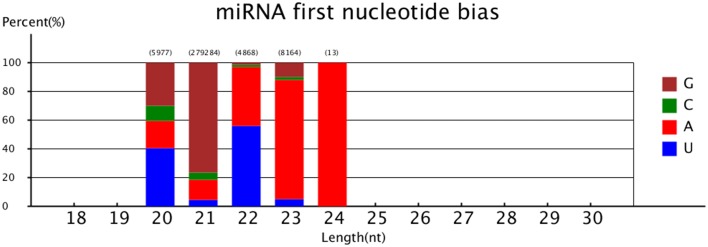
The first nucleotide bias of the different lengths' novel miRNA was also analyzed. The results showed that > 40% miRNAs of 20 and 22 nt began with 5′ uridine, and only <5% miRNAs of 20 and 23 nt began with 5′ uridine (Figure 3).
Figure 3.
MiRNA first nucleotide bias.
Prediction of target genes of miRNA in ponkan
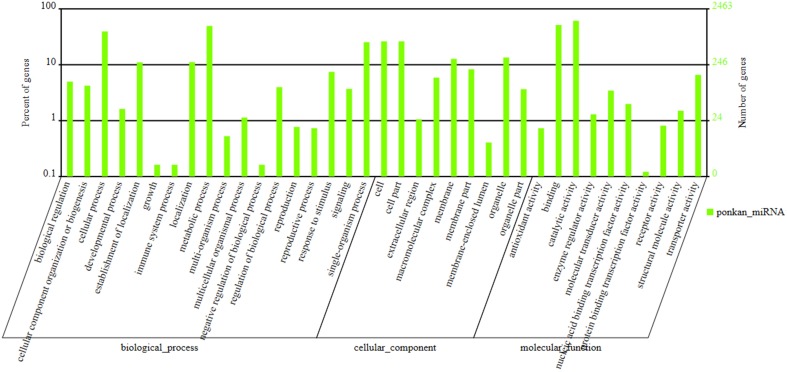
The potential targets of known miRNAs of ponkan were predicted using the psRobot and TargetFinder software. A total of 202 genes and their 7594 corresponding target genes were predicted. Among these genes, 184 known miRNAs and their 2895 corresponding target genes were acquired using psRobot, and 197 known miRNAs and their 5134 corresponding target genes were found after processed with TargetFinder. There are 1398 target genes in common between psRobot and TargetFinder. For novel miRNAs, targets of 358 novel miRNAs were predicted, and targets of 265 novel miRNAs including 4738 corresponding target genes were consistent with the features of target according to psRobot and TargetFinder predictions (Figure 4).
Figure 4.
GO (Gene ontology) annotation of ponkan miRNA.
GO (Gene Ontology) annotation collected from Gene ontology (http://geneontology.org/) and NCBI (http://www.ncbi.nlm.nih.gov/ (Gene ontology)) database information were also conducted in our study. Gene annotation and classification in accordance with the biological pathway (Biology Process), cellular localization (Cellular component), and molecular function (Molecular Function) were listed in Figure 4. Two thousand four hundred and sixty-three genes were classified into 18 biology processes, nine cellular components, and 10 molecular functions. Among these, 1508 genes lied in catalytic activity, 1268 genes in binding section, and 1220 genes in metabolic process.
After enriched analysis in biology process, lignin metabolic process (GO: 0009808) ranked first with 15 differently expressed genes out of 19 genes in this term after comparing its cluster frequency with the genome frequency followed by dicarboxylic acid metabolic process (GO: 0043648) and inorganic anion transport (GO: 0015698). Glutamate receptor activity (GO: 0008066), malate dehydrogenase activity (GO: 0016615), and ion channel activity (GO: 0005216) were the three molecular functions that enriched strongest. For cellular location, AP-type membrane coat (GO: 0030119) and chloroplast outer membrane (GO: 0009707) were the strongest enriched location (Supplementary Table S3).
Development of ponkan In vitro plantlet in the process of conservation
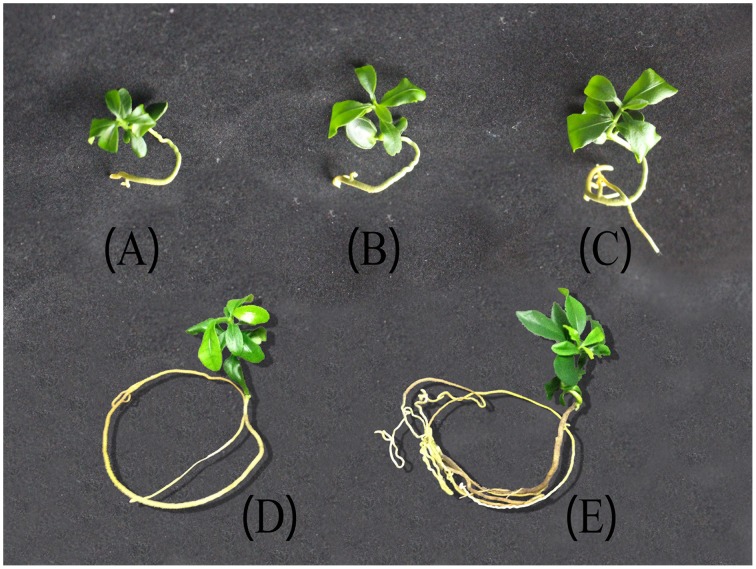
After the mature embryos were incubated on MS and cultured for 4 days, the radicle emerged, and the shoot started to grow after 3 days. After another 9 days, a 4–5 leaf plantlet (9-day-old leaf) was formed. When the leaf was 22 days old, the area was about twice that of a 9-day-old leaf. The area of 27-day-old leaf was as much as that of a 22-day-old leaf and the color became dark green. After 6 months, the growth of plantlet was very slow and the leaf color became darker and darker. In 18 months, the leaf thickness increased and the leaf area enlarged (Figure 5). During the preservation, the ponkan plantlets suffered from nutrient and water stresses. Leaf was the most sensitive organ that reflected the growth status of the in vitro plantlets. Thus, leaves were sampled in the present investigation.
Figure 5.
Ponkan plantlets under different storage time. (A) 9 days after germination (DAG) corresponding to the initiation stage, (B) 22 DAG of fast-growing stage, (C) 27 DAG of stable-growing stage, (D) 6 months after germination (MAG), (E) 18 MAG. Both (D,E) were corresponding to slow-growing stage.
Expression patterns of miRNAs and targets in the preservation of ponkan
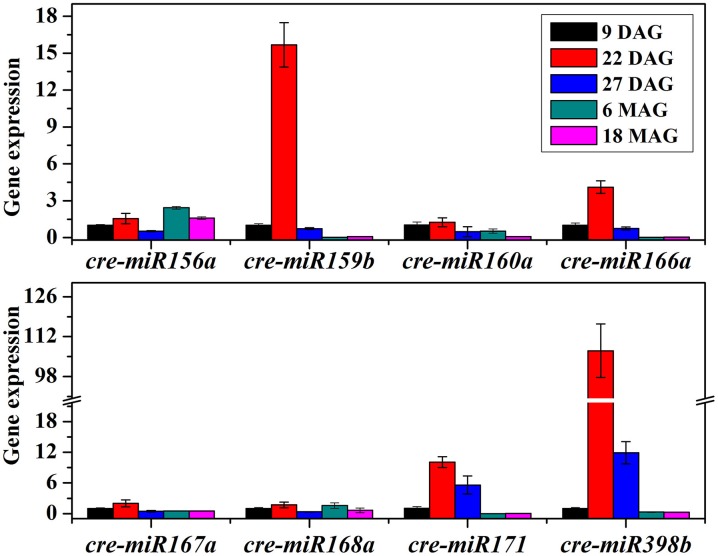
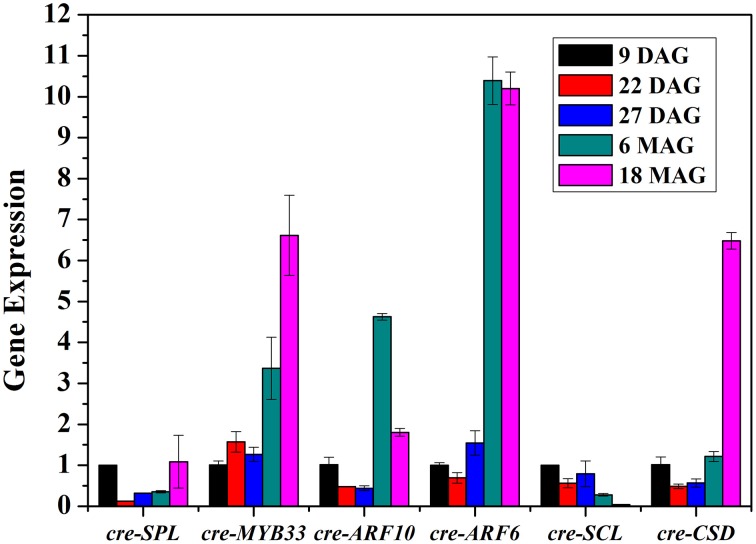
To better understand the role of miRNAs during the preservation of ponkan in vitro plantlet, the expressions of miRNA and some target genes in 9 DAG, 22 DAG, 27 DAG, 6 MAG, and 18 MAG in ponkan in vitro plantlet leaf were analyzed using qPCR (Figures 6, 7).
Figure 6.
QPCR analysis of relative expressions of miRNAs in ponkan in vitro plantlet leaf.
Figure 7.
QPCR analysis of relative expressions of the targets of miRNAs in ponkan in vitro plantlet leaf.
The results showed that the tested miRNAs were expressed at various levels in the early preservation period, including the initial phase and fast-growing stage of the in vitro ponkan plantlets, i.e., 9 & 22 DAG, and the stable-growing stage, i.e., 27 DAG. The expression pattern can be divided into three categories (Figure 6): (i) the expression levels of cre-miR159b, cre-miR166a, cre-miR171, and cre-miR398b first reached the highest level in 22 DAG. Then, during the stable-growing stage, i.e., 27 DAG, the expression levels of cre-miR171, cre-miR398b, cre-miR159b, and cre-miR166a decreased. (ii) The expression levels of cre-miR156a, cre-miR167a, and cre-miR168a did not change significantly from 9 to 22 DAG, and then decreased in 27 DAG. (iii) The expression levels of cre-miR160a and cre-miR167a did not change significantly during 9–27 DAG. After six MAG, the in vitro plantlets arrived at the slow-growing stage, which was corresponding to the later preservation period, in which most of the tested miRNAs expressed at lower levels. The decreased expression level of cre-miR156a seemed postponed.
The expressions of cre-SPL, cre-MYB33, cre-ARF10, cre-ARF6, cre-SCL, and cre-CSD, target genes of cre-miR156, cre-miR159, cre-miR160, cre-miR167, and cre-miR171, respectively, were at varied levels (Figure 7). The amounts of cre-SPL and cre-SCL were kept at low level during the conservation of ponkan in vitro plantlet. Other target genes such as cre-MYB33, cre-ARF10, cre-ARF6, and cre-CSD exhibited similar expression pattern, decreased at the early stage, and increased at the later stage except unchanging cre-MYB33 level at the early stage.
Discussion
A complex miRNA population existed in ponkan In vitro plantlet leaf
It is well known that miRNA play key roles in the development of leaf. In the present study, a total of 3,065,625 unique sequences containing 224 conserved miRNAs and 358 novel miRNAs were detected. It was obvious that the percentage of sRNAs with 21 nt in length was much higher than that with 24 nt in ponkan. Similar results were found in wheat (Yao et al., 2007), populus (Barakat et al., 2007), and tomato (Moxon et al., 2008), and the length distribution of sRNAs was mainly at 21 nt. The sRNAs from leaf, flower, and fruit in sweet orange showed that 24 nt sRNAs were the most abundant (Liu et al., 2014), which was also found in the majority of angiosperms (Morin et al., 2008; Szittya et al., 2008; Chen et al., 2009; Donaire et al., 2011; Lin and Lai, 2013). The size distribution of known miRNAs was mainly at 21 nt, thereby accounting for 42.41%, and 20-nt miRNA take the proportion of 8.48%. Although 20-nt miRNA does not represent a major class of ponkan miRNAs, highly conserved 20-nt miRNAs appear in most plant species. miR156 and miR394 are predominantly 20-nt long in many land plants, which were also found in ponkan in vitro plantlet leaves.
There were many conserved miRNAs in angiosperm plants. In the present study, most of the identified miRNA families in ponkan also existed in other plant species. For example, cre-miR156 family had a perfect match with miR156 in other plants, such as Arabidopsis thaliana, Oryza, Sorghum, Medicago, Populus, Gossypium, and Vitis. The cre-miR160/166/396 families were also found to be highly homologous to those forms in other plants. Furthermore, there were some specific miRNAs belonging to monocotyledon or dicotyledon. For example, miR444 was regarded as monocot-specific miRNAs (Yao et al., 2007; Sunkar and Jagadeeswaran, 2008; Zhang et al., 2009), whereas we detected the expression of miR444 in ponkan. On the other hand, miR158/163/173/403/472/479 was considered to be dicot-specific miRNAs (Jones-Rhoades and Bartel, 2004; Yin and Shen, 2010), which were also found in our experiment except for miR163. The inexistence of miR163 may be because of severely repressed leaves (Ha et al., 2009).
The regulatory role of miRNAs during the development of ponkan In vitro plantlet leaf
Leaf was the first lateral organ produced by the activity of shoot apical meristem. Leaf development is a multifaceted process during which a small group of undifferentiated cells get recruited in the meristem (Tsukaya, 2006). A complicated regulatory network involved in transcription factors and hormones is necessary to ensure the process of leaf development on the rails. To ensure the normal operation of various physiological processes of organisms, a tight regulatory network must operate precisely, which is constituted by the interaction of miRNAs and their target genes (Alvarez-Garcia and Miska, 2005). Some miRNAs have been found to play critical roles during the development of leaf in recent years, such as miR156, miR159, miR160, miR166, and miR396. In the conditions of present in vitro preservation, cre-miR156, cre-miR159, cre-miR160, cre-miR166, and cre-miR396 were all detected. The most abundant miRNAs belong to the families miR157, followed by miR156, miR166, and mir167, which are described as the most widespread miRNA families in plants (Sun, 2012). However, in leaf of cork oak (Quercus suber), the most abundant is miR167 and not miR157, and miR167 is one of the miRNA families involved in the regulation of auxin signaling.
MiR157 was predicted to target squamosa promoter-binding protein (SBP) or squamosal promoter binding-like protein (SPL) to function in multiple stress responses in tomato (Luan et al., 2014). In our study, the abundant reads of miR157 were also detected in ponkan in vitro plantlet leaves, indicating its key role in preservation of citrus germplasm resources.
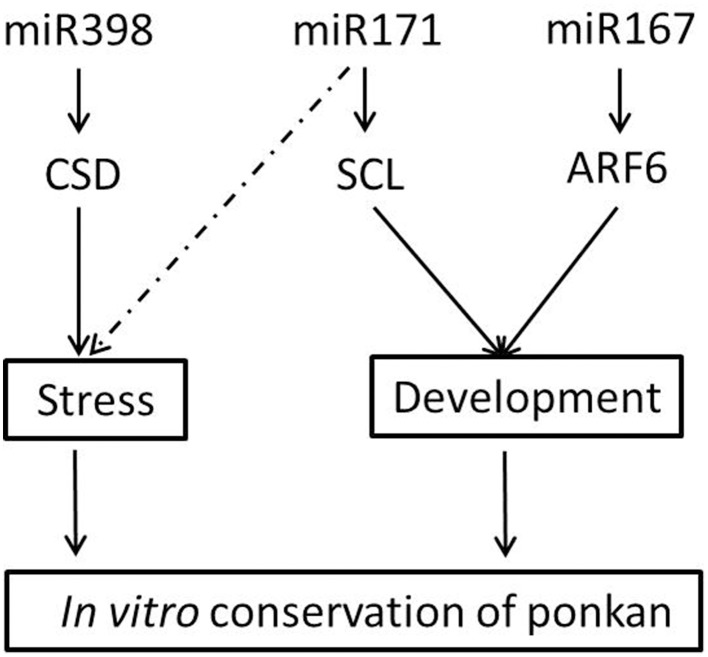
MiR167 targets the members of AUXIN RESPONSE FACTOR (ARF) family of transcription factors, namely ARF6/8, involved in early auxin response through activation of auxin-responsive genes (Wu et al., 2006), as well as IAA-Ala Resistant3 (IAR3), which hydrolyzes an inactive form of auxin (Kinoshita et al., 2012). The regulation of auxin signaling pathways by miR167 and its targets has been related to male and female flower development (Wu et al., 2006) and osmotic stress-induced root architecture changes (Kinoshita et al., 2012). In our study, the expression of cre-ARF6 was down-regulated at nine DAG, and afterwards, it was up-regulated until 18 MAG, which is consistent with the first high and then low expression level of cre-miR167, indicating that miR167–ARF6 was involved in the regulation of leaf development of ponkan in vitro plantlet leaf (Figure 8).
Figure 8.
Possible roles of miRNA in the in vitro conservation of ponkan.
Beside miR167 in ponkan, miR156 is one of the most abundant and evolutionarily conserved miRNAs in plants, which regulated plastochron length by quantitatively modulating the levels of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factors (Schwarz et al., 2008; Wang et al., 2008). Overexpression of miR156 accelerated the rate of leaf initiation. A similar effect was observed in the spl9 spl15 double mutant, indicating that SPL genes are targeted by miR156 in Arabidopsis. SPL proteins could modulate auxin accumulation or response (Bencivenga et al., 2012). In our study, the expression of cre-SPL was down-regulated from 22 DAG to 6 MAG. This could be due to the different function of SPL. It was reported that the miR156-SPL module could be responsible for the lateral root development (Yu et al., 2015). The root development in ponkan in vitro plantlet is different from that grown in outdoor environment. The same conflicting expression trends were also observed in the expression of cre-miR159-MYB33 and cre-miR160-ARF10.
Stress-responding miRNAs were involved in the preservation of ponkan In vitro plantlet
MiRNAs not only function as plant developmental regulators but also play important roles in adaption to stress conditions. MiR171 can regulate shoot branching through targeting GRAS gene family members SCARECROW-LIKE6 (SCL6) in Arabidopsis (Wang et al., 2010). Transgenic plants overexpressing Arabidopsis miR171a have been reported to exhibit multiple developmental defects, including reduced cauline leaf and rosette leaf formations and reduced shoot branching (Wang et al., 2010). In our study, the high expression level of cre-miR171 combined with the low expression level of cre-SCL during the early stage indicated that miR171-SCL might mainly act as a leaf formation regulator at the early stage as the leaf area is expanding and the number of blades is increasing (Figure 8). After reserved for 6 months up to 18 months, the growth of in vitro ponkan plantlet decreased, and cre-SCL exhibited a very low expression level. At the same time, the expression level of cre-miR171 was very low possibly due to stress from the limited nutrition and environment. It was reported that the expression of miR171 was involved in drought response in Solanum tuberosum and can be regulated by light, temperature, and salt stress (Hwang et al., 2011).
MiR398 is required for embryo morphogenesis in citrus and longan (Wu et al., 2011; Lin and Lai, 2013) and is well known for its role in resistance (Sunkar et al., 2006; Jagadeeswaran et al., 2009). MiR398 was proposed to be directly linked to the plant stress regulatory network and regulates plant responses to oxidative stress, water deficit, salt stress, abscisic acid stress, ultraviolet stress, copper and phosphate deficiency, high sucrose, as well as bacterial infection. MiR398 targets Copper/zinc Superoxide Dismutase (CSD) and is down-regulated under stress conditions to permit up-regulation of its target genes (Sunkar et al., 2006; Jagadeeswaran et al., 2009). The negative correlation of the expression pattern of cre-miR398b and cre-CSD was also found in our experiment, indicating that the regulation of CSD by miR398b plays important roles in the in vitro conservation of ponkan (Figure 8). The change of expression levels of cre-miR398b and its target cre-CSD in in vitro plantlet during the conversation of C. reticulata manifested that stress did exist in the preservation of in vitro plantlet and miRNAs acted as a stress-regulator.
Overall, we reported the data on miRNAs in in vitro plant materials and found that both stress-associated and development-associated miRNAs show similar expression patterns during the plant in vitro morphogenesis and preservation. The miRNAs are considered to be involved in the development via regulation of the target genes and function in the conservation of ponkan germplasm.
Author contributions
ZL, RG, and XC designed research; RG and XC performed research and wrote the paper, and they contributed equally to this study; ZL, RG, XC, YL, XX, and MT analyzed data. ZL, XX, YL, and MT participated in the sequence analysis and helped to modify the manuscript. All authors have read and approved the manuscript for publication.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer Jeng-Shange Lin and handling Editor Keqiang Wu declared their shared affiliation, and the handling Editor states that, nevertheless, the process met the standards of a fair and objective review.
Acknowledgments
We acknowledge the support from the National Natural Science Foundation of China (31401859), Major Science and Technology Project in Fujian Province (2015NZ0002-1), Horticulture Postdoctoral Funding from Fujian Agriculture and Forestry University (132300155), National Science and Technology Supporting Project (2007BAD07B01), Fujian Provincial Science and Technology Platform Construction Project (2008N2001), and Fujian Provincial Special Program for Key Science and Technology (2004NZ02-2).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.01212
Known miRNA in ponkan sRNA library.
Novel miRNA in ponkan sRNA library.
Enriched analysis of gene ontology annotation of ponkan miRNA.
Primers for qPCR.
References
- Alvarez-Garcia I., Miska E. A. (2005). MicroRNA functions in animal development and human disease. Development 132, 4653–4662. 10.1242/dev.02073 [DOI] [PubMed] [Google Scholar]
- Barakat A., Wall P. K., Diloreto S., dePamphilis C. W., Carlson J. E. (2007). Conservation and divergence of microRNAs in Populus. BMC Genomics 8:481. 10.1186/1471-2164-8-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barciszewska-Pacak M., Milanowska K., Knop K., Bielewicz D., Nuc P., Plewka P., et al. (2015). Arabidopsis microRNA expression regulation in a wide range of abiotic stress responses. Front. Plant Sci. 6:410. 10.3389/fpls.2015.00410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bencivenga S., Simonini S., Benková E., Colombo L. (2012). The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in Arabidopsis. Plant Cell 24, 2886–2897. 10.1105/tpc.112.100164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Zheng H., Zhang H. (2009). Identification of microRNAs in wild soybean (Glycine soja). J. Integr. Plant Biol. 51, 1071–1079. 10.1111/j.1744-7909.2009.00887.x [DOI] [PubMed] [Google Scholar]
- Chen X. (2005). MicroRNA biogenesis and function in plants. FEBS Lett. 579, 5923–5931. 10.1016/j.febslet.2005.07.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. D. (2011). Germplasm Conservation and microRNA Identification of in Vitro Plantlets in Citrus Trees. Thesis, Fujian Agriculture and Forestry University. Available online at: http://d.g.wanfangdata.com.cn/Thesis_Y1878643.aspx (Accessed August 24, 2011).
- Chuck G., Candela H., Hake S. (2009). Big impacts by small RNAs in plant development. Curr. Opin. Plant Biol. 12, 81–86. 10.1016/j.pbi.2008.09.008 [DOI] [PubMed] [Google Scholar]
- Donaire L., Pedrola L., de la Rosa R., Llave C. (2011). High-throughput sequencing of RNA silencing-associated small RNAs in olive (Olea europaea L.). PLoS ONE 6:e27916. 10.1371/journal.pone.0027916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M., Lu J., Tian L., Ramachandrand V., Kasschaue K. D., Chapmane E. J., et al. (2009). Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc. Natl. Acad. Sci. U.S.A. 106, 17835–17840. (Ha et al., 2009) 10.1073/pnas.0907003106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh L. C., Lin S. I., Shih A. C. C., Chen J. W., Lin W. Y., Tseng C. Y., et al. (2009). Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant physiol. 151, 2120–2132. 10.1104/pp.109.147280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang E., Shin S., Yu B., Byun M. O., Kwon H. B. (2011). MiR171 family members are involved in drought response in Solanum tuberosum. J. Plant Biol. 54, 43–48. 10.1007/s12374-010-9141-8 [DOI] [Google Scholar]
- Jagadeeswaran G., Saini A., Sunkar R. (2009). Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta 229, 1009–1014. 10.1007/s00425-009-0889-3 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M. W., Bartel D. P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14, 787–799. 10.1016/j.molcel.2004.05.027 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M. W., Bartel D. P., Bartel B. (2006). MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 57, 19–53. 10.1146/annurev.arplant.57.032905.105218 [DOI] [PubMed] [Google Scholar]
- Juarez M. T., Kui J. S., Thomas J., Heller B. A., Timmermans M. C. (2004). MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428, 84–88. 10.1038/nature02363 [DOI] [PubMed] [Google Scholar]
- Katiyar A., Smita S., Muthusamy S. K., Chinnusamy V., Pandey D. M., Bansal K. C. (2015). Identification of novel drought-responsive microRNAs and trans-acting siRNAs from Sorghum bicolor (L.) Moench by high-throughput sequencing analysis. Front. Plant Sci. 6:506. 10.3389/fpls.2015.00506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiełbasa S. M., Blüthgen N., Fähling M., Mrowka R. (2010). Targetfinder. org: a resource for systematic discovery of transcription factor target genes. Nucleic Acids Res. 38(Suppl. 2), W233–W238. 10.1093/nar/gkq374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita N., Wanga H., Kasaharac H., Liu J., MacPherson C., Machid Y., et al. (2012). IAA-Ala Resistant3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress. Plant Cell 24, 3590–3602. 10.1105/tpc.112.097006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Li Y., Kristiansen K., Wang J. (2008). SOAP: short oligonucleotide alignment program. Bioinformatics 24, 713–714. 10.1093/bioinformatics/btn025 [DOI] [PubMed] [Google Scholar]
- Liang C., Zhang X., Zou J., Xu D., Su F., Ye N. (2010). Identification of miRNA from Porphyra yezoensis by high-throughput sequencing and bioinformatics analysis. PLoS ONE 5:e10698. 10.1371/journal.pone.0010698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. L., Lai Z. X. (2013). Comparative analysis reveals dynamic changes in miRNAs and their targets and expression during somatic embryogenesis in longan (Dimocarpus longan Lour.) PloS ONE 8:e60337. 10.1371/journal.pone.0060337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wang L., Chen D., Wu X., Huang D., Chen L., et al. (2014). Genome-wide comparison of microRNAs and their targeted transcripts among leaf, flower and fruit of sweet orange. BMC Genomics 15:695. 10.1186/1471-2164-15-695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan Y., Wang W., Liu P. (2014). Identification and functional analysis of novel and conserved microRNAs in tomato. Mol. Biol. Rep. 41, 5385–5394. 10.1007/s11033-014-3410-4 [DOI] [PubMed] [Google Scholar]
- Meng F., Liu H., Wang K., Liu L., Wang S., Zhao Y., et al. (2013). Development-associated microRNAs in grains of wheat (Triticum aestivum L.). BMC Plant Biol. 13:140. 10.1186/1471-2229-13-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B. C., Axtell M. J., Bartel B., Bartel D. P., Baulcombe D., Bowman J. L., et al. (2008). Criteria for annotation of plant MicroRNAs. Plant Cell 20, 3186–3190. 10.1105/tpc.108.064311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin R. D., Aksay G., Dolgosheina E., Ebhardt H. A., Magrini V., Mardis E. R., et al. (2008). Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res. 18, 571–584. 10.1101/gr.6897308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon S., Jing R., Szittya G., Schwach F., Pilcher R. L. R., Moulton V., et al. (2008). Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 18, 1602–1609. 10.1101/gr.080127.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki E. P., Burge S. W., Bateman A., Daub J., Eberhardt R. Y., Eddy S. R., et al. (2014). Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 43, D130–D137. 10.1093/nar/gku1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N., Cohen A. R., Etzioni A., Brand A., Yanai O., Shleizer S., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 39, 787–791. 10.1038/ng2036 [DOI] [PubMed] [Google Scholar]
- Schwarz S., Grande A. V., Bujdoso N., Saedler H., Huijser P. (2008). The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 67, 183–195. 10.1007/s11103-008-9310-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Wang C., Zhang C., Korir N., Yu H., Ma Z., et al. (2010). Deep sequencing discovery of novel and conserved microRNAs in trifoliate orange (Citrus trifoliata). BMC Genomics 11:431. 10.1186/1471-2164-11-431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G. (2012). MicroRNAs and their diverse functions in plants. Plant Mol. Biol. 80, 17–36. 10.1007/s11103-011-9817-6 [DOI] [PubMed] [Google Scholar]
- Sunkar R., Jagadeeswaran G. (2008). In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biol. 8:37. 10.1186/1471-2229-8-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Kapoor A., Zhu J. K. (2006). Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18, 2051–2065. 10.1105/tpc.106.041673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya G., Moxon S., Santos D. M., Jing R., Fevereiro M., Moulton V., et al. (2008). High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genomics 9:593. 10.1186/1471-2164-9-593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H. (2006). Mechanism of leaf shape determination. Annu. Rev. Plant Biol. 57, 477–496. 10.1146/annurev.arplant.57.032905.105320 [DOI] [PubMed] [Google Scholar]
- Wang J. W., Schwab R., Czech B., Mica E., Weigel D. (2008). Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20, 1231–1243. 10.1105/tpc.108.058180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Liu H., Li D., Chen H. (2011). Identification and characterization of maize microRNAs involved in the very early stage of seed germination. BMC Genomics 12:154. 10.1186/1471-2164-12-154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Mai Y., Zhang Y., Luo Q., Yang H. Q. (2010). MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis. Mol. Plant. 3, 794–806. 10.1093/mp/ssq042 [DOI] [PubMed] [Google Scholar]
- Wei L. Q., Yan L. F., Wang T. (2011). Deep sequencing on genome-wide scale reveals the unique composition and expression patterns of microRNAs in developing pollen of Oryza sativa. Genome Biol. 12:R53. 10.1186/gb-2011-12-6-r53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. J., Ma Y.-K., Chen T., Wang M., Wang X. J. (2012). PsRobot: a web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res, 40, W22–W28. 10.1093/nar/gks554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Zhou H. Y., Zhang Q. Q., Zhang J. G., Ni F. R., Liu C., et al. (2010). DNA methylation mediated by a microRNA pathway. Mol. Cell 38, 465–475. 10.1016/j.molcel.2010.03.008 [DOI] [PubMed] [Google Scholar]
- Wu M. F., Tian Q., Reed J. W. (2006). Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133, 4211–4218. 10.1242/dev.02602 [DOI] [PubMed] [Google Scholar]
- Wu X. M., Kou S. J., Liu Y. L., Fang Y. N., Xu Q., Guo W. W. (2015). Genomewide analysis of small RNAs in nonembryogenic and embryogenic tissues of citrus: microRNA−and siRNA−mediated transcript cleavage involved in somatic embryogenesis. Plant Biotechnol. J. 13, 383–394. 10.1111/pbi.12317 [DOI] [PubMed] [Google Scholar]
- Wu X. M., Liu M. Y., Ge X. X., Xu Q., Guo W. W. (2011). Stage and tissue-specific modulation of ten conserved miRNAs and their targets during somatic embryogenesis of Valencia sweet orange. Planta 233, 495–505. 10.1007/s00425-010-1312-9 [DOI] [PubMed] [Google Scholar]
- Xie S., Li X. Y., Liu T., Cao J. H., Zhong Q., Zhao S. H. (2011). Discovery of porcine microRNAs in multiple tissues by a Solexa deep sequencing approach. PLoS ONE 6:e16235. 10.1371/journal.pone.0016235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Liu Y., Zhu A., Wu X., Ye J., Yu K., et al. (2010). Discovery and comparative profiling of microRNAs in a sweet orange red-flesh mutant and its wild type. BMC Genomics 11:246. 10.1186/1471-2164-11-246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Guo G., Ni Z., Sunkar R., Du J., Zhu J. K., et al. (2007). Cloning and characterization of microRNAs from wheat (Triticum aestivum L.). Genome Biol. 8:R96. 10.1186/gb-2007-8-6-r96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z. J., Shen F. F. (2010). Identification and characterization of conserved microRNAs and their target genes in wheat (Triticum aestivum). Genet. Mol. Res. 9, 1186–1196. 10.4238/vol9-2gmr805 [DOI] [PubMed] [Google Scholar]
- Yu N., Niu Q., Ng K., Chua N. (2015). The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J. 83, 673–685. 10.1111/tpj.12919 [DOI] [PubMed] [Google Scholar]
- Zhang J., Xu Y., Huan Q., Chong K. (2009). Deep sequencing of Brachypodium small RNAs at the global genome level identifies microRNAs involved in cold stress response. BMC Genomics 10:449. 10.1186/1471-2164-10-449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C. Z., Xia H., Frazier T. P., Yao Y. Y., Bi Y. P., Li A. Q., et al. (2010). Deep sequencing identifies novel and conserved microRNAs in peanuts (Arachis hypogaea L.). BMC Plant Biol. 10:3. 10.1186/1471-2229-10-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Known miRNA in ponkan sRNA library.
Novel miRNA in ponkan sRNA library.
Enriched analysis of gene ontology annotation of ponkan miRNA.
Primers for qPCR.