Abstract
During a study of saprobic fungi from Bagno di Cetica Province, Italy, we collected a pleosporoid ascomycete on stems of Cytisus sp. In morphology, our collection is similar to Cucurbitaria species, but molecular analysis of SSU, LSU and ITS genes reveals it can be referred to Camarosporium. In this study we compare all other Cucurbitaria species from Cytisus sp. and based on both morphology and molecular data, we introduce our collection as a new species in Camarosporium viz. C. arezzoensis.
Keywords: Morphology, Multi-gene analysis, Sexual state
1. Introduction
The genus Camarosporium was introduced by Schulzer (1870) with Camarosporium quaternatum (Hazsl.) Schulz. as the type species. Index Fungorum (2015) lists 508 records as Camarosporium which was formerly recognised as asexual morphs in Botryosphaeriales, Cucurbitariaceae, Phaeosphaeriaceae and related genera (Kirk et al., 2008, Wijayawardene et al., 2012, Doilom et al., 2013, Hyde et al., 2013). However, Wijayawardene et al., 2014a, Wijayawardene et al., 2014b showed that Camarosporium sensu stricto belongs to Pleosporineae, Pleosporales and has cucurbitaria-like sexual morphs.
During our on-going studies, we found a new taxon with bitunicate asci and muriform ascospores which is morphologically similar with members in Cucurbitariaceae, Pleosporales (Doilom et al., 2013, Hyde et al., 2013). The blast results of small subunit rDNA (SSU), large subunit rDNA (LSU) and internal transcribed spacer (ITS) showed this taxon is related to Camarosporium sp. Thus we have carried out molecular analyses viz. maximum-parsimony (MP) and confirmed its placement in Pleosporineae, Pleosporales. As our new collection groups with Camarosporium sensu stricto, we introduce it as a new species of Camarosporium viz. C. arezzoensis.
2. Materials and methods
2.1. Sample collection and morphological study
Fresh fungal specimens were obtained from recent collections made in Italy. Morphological structures were examined under a Carl Zeiss microscopy GmbH (AxioCam ERC 5S) stereo microscope. To observe the fungal structures, ascomata were picked up and put into rehydrated water or lactoglycerol. For hand cross sections 5% KOH was added prior to examination. Microscopic fungal structures were mounted in water for observation under a Nikon ECLIPSE80i compound microscope and photographs were taken with a Cannon 550D digital camera fitted to the microscope. All micro morphologies were measured using Tarosoft® Image Framework program v.0.9.0.7.
2.2. Isolation
Single spore isolation was carried out following the method described in Chomnunti et al. (2014) on potato-dextrose agar (PDA). Germinated spores were transferred to fresh PDA media and incubated at 16 °C. Culture characteristics were observed after four weeks and these cultures were also used for molecular study. The specimens are deposited in the Mae Fah Luang University (MFLU) Herbarium, Chiang Rai, Thailand. Living cultures are deposited at the Mae Fah Luang University Culture Collection (MFLUCC) Chiang Rai, Thailand, Centraalbureau Voor Schimmelcultures, Netherlands (CBS) and International Collection of Microorganisms from Plants, New Zealand (ICMP).
2.3. DNA extraction, PCR amplification and sequencing
Mycelia grown on PDA media at 16 °C for four weeks were used for DNA extraction. Total DNA extraction was established by using a Biospin Fungus Genomic DNA Extraction Kit (Bioer Technology Co., Ltd., Hangzhou, PR China). The concentration of DNA was determined using an ultraviolet spectrophotometer. PCR reactions were carried out according to Telle and Thines (2008) with the primers ITS1-F (Gardes and Bruns, 1993) and ITS4 (White et al., 1990) to amplify the complete internal transcribed spacer (ITS) region. Twenty micro litres (20 μl) of the reaction mixture contained 2 Mix 10 μl, ITS1-F 0.35 μl, ITS4 0.35 μl, 50 ng/μl DNA 0.6 μl, ddH2O 8.7 μl for each sample. The PCR programme was set according to Douanla et al. (2005) with the following modifications: an initial denaturation at 94 °C for 3 min, annealing at 55 °C for 45 s, and extension at 72 °C for 1 min, and a final elongation step of 7 min at 72 °C. To check the PCR products, 1% agarose gel electrophoresis (AGE) for 30 min at 220V was used. All PCR products were sent to Shanghai Majorbio Bio-Pharm Technology Co., Ltd. for purification and sequencing.
2.4. Molecular phylogenetic analysis
BLAST searches of LSU, SSU and ITS sequence data were carried out to reveal the closest taxa to our strain in GenBank (http://www.ncbi.nlm.nih.gov/). Combined analyses of LSU, SSU and ITS dataset of the closest relatives in Coniothyriaceae, Cucurbitariaceae and Pleosporaceae were used to carry out phylogenetic analyses. Bioedit v.7.2.5 (Hall, 2004), ClustalW v.1.6 (Thompson et al., 1997) and MAFFT v.6 (Katoh et al., 2002, Katoh and Toh, 2008) online sequence alignment editor under the default settings (mafft.cbrc.jp/alignment/server/) were used for aligning the sequences separately for each gene region. The individual datasets were finally combined into one dataset and used PAUP v. 4.0b10 (Swofford, 2002) to perform maximum-parsimony (MP) analysis by bootstrap analysis with 10,000 replicates. All multiple, equally parsimonious trees were saved and descriptive tree statistics for parsimony consistency index (CI), retention index (RI), rescaled consistency index (RC) and homoplasy index (HI) were calculated. The robustness of the best parsimonious tree was estimated by a bootstrap (BT) value with 10,000 replicates, each with 10 replicates of random stepwise addition of taxa (Liu et al., 2011, Phookamsak et al., 2013), and the trees were figured in Treeview v.1.6.6.
3. Results
3.1. Phylogenetic analysis
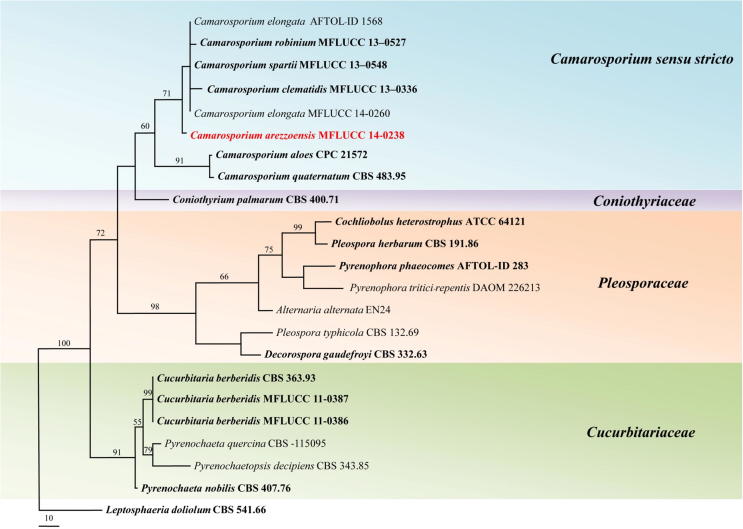
The combined gene data set of SSU, ITS and LSU rDNA consists of 23 taxa including our strain of IT 791 (MFLUCC 14-0238) and the outgroup taxon Leptosphaeria doliolum (CBS 541.66). The dataset consists of 2092 characters including coded alignment gaps; 1835 are constant, and 114 are parsimony informative in the MP analysis. A best scoring tree is shown in Fig. 1. Bootstrap support (BS) values of MP (equal to or above 50% based on 10,000 replicates) are shown above branches (TL = 447, CI = 0.694, RI = 0.700, RC = 0.486, HI = 0.306). Our strain of MFLUCC 14-0238 belongs to the genus Camarosporium sensu stricto and were separated from representative species of the genus with a relatively higher bootstrap values as circumscribed by Wijayawardene et al. (2014b).
Figure 1.
One of the most parsimonious trees generated with SSU, ITS and LSU rDNA combined data analysis. The tree is rooted with Leptosphaeria doliolum (CBS 541.66). Type and ex-type strains are in bold. Newly introduced species in red.
3.2. Taxonomy
Camarosporium arezzoensis Tibpromma, Wijayawardene, Camporesi & K.D. Hyde, sp. nov.
Index Fungorum Number: IF550877; Facesoffungi number: 00382
Etymology: Refers to the name of the province in Italy where the fungus was collected
Saprobic on decaying plant stems of Cytisus sp. Sexual morph: Ascomata 400–500 μm high, 450–550 μm diam. (=449 × 482 μm, n = 10), black, semi-immersed, scattered beneath the host periderm or on decorticated wood, fully or partly erumpent, globose, rough or hairy, with an ostiole. Ostiole central, short, slightly sunken, minute and inconspicuous at the surface, smooth, ostiolar canal filled with hyaline cells. Peridium 30–45 μm wide at the base, 35–70 μm wide in sides, thick, comprising 8–10 layers, outer layer heavily pigmented, thick-walled, comprising blackish to dark brown cells of textura angularis, inner layer composed of hyaline, thin-walled cells of textura angularis. Hamathecium comprising numerous, 5.5 μm (n = 40) wide, filamentous, branched septate, pseudoparaphyses. Asci 180–240 × 10–15 μm (=199 × 13 μm, n = 40), 8-spored, bitunicate, fissitunicate, cylindrical, short-pedicellate, apex rounded with a minute ocular chamber. Ascospores 19–28 × 9–15 μm (=26 × 12 μm, n = 50), partially overlapping, mostly ellipsoidal, muriform, with 5–7 transverse septa, with 4–6 longitudinal septa, constricted at the central septum, initially hyaline, becoming brown at maturity, with slightly paler ends, conical and narrowly rounded at the ends, not surrounded by a mucilaginous sheath.
Culture characteristics: on PDA reaching 2 cm diam. after 4 weeks at 16 °C, later with dense mycelium, circular, rough margin white at first, iron-grey after 6 weeks, reverse cinnamon, flat on the surface, without aerial mycelium. Hyphae septate, branched, hyaline, thin (see Fig. 2).
Figure 2.
Camerosporium arezzoensis (holotype). (a) Ascomata on host substrate. (b) Section of ascoma. (c) Section of peridium. (d) Light brown hyphae around ascomata. (e) Pseudoparaphyses. (f–i) Asci. (j–n) Ascospores. Scale bars: b = 200 μm, c = 50 μm, d–i = 20 μm, j–n = 10 μm.
Material examined: ITALY, Arezzo Province, Bagno di Cetica, on stems of Cytisus sp., 1 October 2012, Erio Camporesi IT791 (MFLU14-0636, holotype), extype living cultures, MFLUCC 14-0238, CBS, ICMP (see Table 1).
Table 1.
Strains used in this study (Type and ex-type strains are in bold, the new taxon is indicated with an asterisk).
| Taxon | Culture collection number | GenBank Accession number |
||
|---|---|---|---|---|
| SSU | ITS | LSU | ||
| Alternaria alternata | EN24 | – | FJ809940 | – |
| Camarosporium aloes | CPC 21572 | – | KF777142 | KF777198 |
| Camarosporium clematidis | MFLUCC 13-0336 | KJ589414 | KJ562213 | KJ562188 |
| Camarosporium elongata | AFTOL-ID 1568 | DQ678009 | – | DQ678061 |
| Camarosporium elongata | MFLUCC 14-0260 | – | – | KJ724249 |
| Camarosporium arezzoensis∗ | MFLUCC 14-0238 | KP120928 | KP120926 | KP120927 |
| Camarosporium quaternatum | CBS 483.95 | GU296141 | – | GU301806 |
| Camarosporium robinium | MFLUCC 13-0527 | KJ589415 | KJ562214 | KJ589412 |
| Camarosporium spartii | MFLUCC 13-0548 | KJ589416 | – | KJ589413 |
| Cochliobolus heterostrophus | ATCC 64121 | – | JX094779 | JX094789 |
| Coniothyrium palmarum | CBS 400.71 | EU754054 | AY720708 | JX681084 |
| Decorospora gaudefroyi | CBS 332.63 | AF394542 | AF394541 | – |
| Leptosphaeria doliolum | CBS 541.66 | – | JF740206 | JF740284 |
| Pleospora herbarum | CBS 191.86 | GU238232 | – | GU238160 |
| Pleospora typhicola | CBS 132.69 | JF740105 | – | JF740325 |
| Pyrenophora phaeocomes | AFTOL-ID283 | – | DQ491507 | DQ499596 |
| Pyrenophora tritici-repentis | DAOM 226213 | – | JN943670 | JN940071 |
| Cucurbitaria berberidis | CBS 363.93 | GQ387545 | JF740191 | GQ387606 |
| Cucurbitaria berberidis | MFLUCC 11-0387 | KC506800 | – | KC506796 |
| Cucurbitaria berberidis | MFLUCC 11-0386 | KC506799 | – | KC506795 |
| Pyrenochaeta nobilis | CBS 407.76 | EU754107 | – | EU754206 |
| Pyrenochaetopsis decipiens | CBS 343.85 | GQ387563 | – | GQ387624 |
| Pyrenochaeta quercina | CBS 115095 | GQ387558 | – | GQ387619 |
Notes: Mirza (1968) and Ellis and Ellis (1985) have listed Cucurbitaria cytisi Mirza, Cucurbitaria laburni (Pers.) De Not., Cucurbitaria obducens (Schumach.) Petr. and Camarosporium spartii (Nees ex Fr.) Ces. & De Not. on Cytisus sp. We compared our collection with those species (Table 2). Molecular data analysis confirms our stain groups with C. quaternatum Schulzer (Schulzer, 1870), the type species of Camarosporium and other Camarosporium spp. C. arezzoensis however, differs in having 180–240 × 10–15 μm asci and 19–28 × 9–15 μm brown ascospores. Our new species should be considered as Camarosporium sensu stricto and it is not congeneric with Cucurbitaria sensu stricto (Cucurbitariaceae) (Fig. 1).
Table 2.
Comparison of our strain with the morphologically similar species in Mirza (1968).
| Name | Ascomata | Peridium | Hypostoma | Asci | Ascospore |
|---|---|---|---|---|---|
| Camarosporium arezzoensis (In this study) | Black, semi-immersed, scattered beneath the host periderm or on decorticated wood, fully or partly erumpent, globose, rough or hairy, with an ostiole | Thick, comprising 8–10 layers, outer layer heavily pigmented, thick-walled, comprising blackish to dark brown cells of textura angularis, inner layer composed of hyaline, thin-walled cells of textura angularis | Comprising numerous, filamentous, branched septate, pseudoparaphyses | 8-spored, bitunicate, fissitunicate, cylindrical, short-pedicellate, apex rounded with a minute ocular chamber | Partially overlapped, mostly ellipsoidal, muriform, with 5–7 transverse septa, with 4–6 longitudinal septa, constricted at the central septum, initially hyaline, becoming brown at maturity, with slightly paler ends, conical and narrowly rounded at the ends, not surrounded by a mucilaginous sheath |
| Cucurbitaria ahmadi | Erumpent, globose to subglobose or obovate, papilla bearing a comparatively wide ostiole | Uniform on sides, made up of dark-brown polygonal cells | Well developed, light-brown densely interwoven hyphae | Long stipitate, 4–8 spores, spore overlapped uniseriately or biseriately | Golden-brown, 3–7 transverse septa, one longitudinal septum |
| Cucurbitaria ononidis | Globose to subglobose, forming a slight depression bearing ostiole, papilla lacking | Slightly rough surface sometimes provided with hair-like structures | Poorly developed, a subiculum of dark-brown | Short stipitate, 4–8 spores, spore overlapped uniseriately | Brown, 5–9 transverse septa, 1–3 longitudinal septa |
| Cucurbitaria elaeagni | Erumpent, globose to subglobose | Slightly rough surface, made up of elongated polygonal cells, hyaline | Well developed, brown | Long stipitate, 4–8 spores, spore overlapped uniseriately or biseriately | Golden to dull brown, 5–7 transverse septa, up to 2 longitudinal septa |
4. Discussion
Pleosporales is the largest order of Dothideomycetes (Kirk et al., 2008) and several studies have been carried out using multi-gene phylogeny, providing the groundwork towards a natural classification of the class (Nelsen et al., 2009, Nelsen et al., 2011, Schoch et al., 2009, Boonmee et al., 2011, Boonmee et al., 2012, Boonmee et al., 2014, Chomnunti et al., 2011, Chomnunti et al., 2014, Liu et al., 2011, Liu et al., 2012, Zhang et al., 2011, Zhang et al., 2012, Hyde et al., 2013, Wijayawardene et al., 2014c). Schoch et al. (2009) recognised the suborders Pleosporinae and Massarinae in Pleosporales and Zhang et al. (2012) confirmed it in their molecular data analyses. In their molecular data analyses, Wijayawardene et al., 2014a, Wijayawardene et al., 2014b, Wijayawardene et al., 2014c showed that Camarosporium sensu stricto clusters as a distinct phylogenetic lineage in Pleosporinae. In our molecular data analyses (Fig. 1) we also show Camarosporium sensu stricto is not related to Cucurbitariaceae, Pleosporaceae or/and Leptosphaeriaceae.
Our combined LSU, SSU and ITS analyses show that our stain clusters with C. quaternatum, the type species of Camarosporium, with high bootstrap support 71% (Fig. 1). Recently introduced species of Camarosporium have been treated as host-specific (Wijayawardene et al. 2014b), but it is essential to re-collect and carry out generic revision. There are about 500 species epithets of Camarosporium and Cucurbitaria in Index Fungorum (2015) but most of the species lack good illustrations and descriptions, thus it is difficult to compare all the species with our collection. However, Mirza (1968) has accepted only 28 species based on morphological characteristics. We have compared our collection with accepted species in Mirza (1968) which have closer morphologies with our collection i.e. Cucurbitaria ahmadi Mirza, Cucurbitaria ononidis Massenot and Cucurbitaria elaeagni Mirza. (Table. 2). Furthermore, we compared the morphology of C. cytisi, C. laburni, C. obducens and C. spartii on Cytisus sp. (Mirza, 1968, Ellis and Ellis, 1985) with our strain (Table 3). Our collection has narrowly fusiform didymosporous ascospores, with mostly ellipsoidal, 5–7 transversely septate, with 4–6 vertical septa, constricted at the central septum, with 1–2 longitudinal septa, with acute ends constricted at the septum.
Table 3.
Comparison of Cucurbitaria species on Cytisus sp.
| Characters | Cucurbitaria cytisi (Mirza, 1968) | Cucurbitaria laburni (Pers.) De Not. 1862 | Cucurbitaria obducens (Schumach.) Petr. 1927 | Cucurbitaria spartii (Nees ex Fr.) Ces. & De Not. 1863 | Camarosporium arezzoensis MFLUCC 14-0238 |
|---|---|---|---|---|---|
| Fruiting bodies (Ascomata) | Pseudothecia 300–700 μm, gregarious in groups of 2–8, erumpent, papilla | Pseudothecia 500–700 μm, black, papillate, usually in large groups seated on a black hyphalsubiculum | Pseudothecia 300–500 μm, black, papillate, usually in large groups seated on a black hyphalsubiculum | Pseudothecia300–700 × 350–610 μm diam., black or blackish brown, erumpent in clusters seated on a scanty brown subiculum | Pseudothecia 450 × 480 μm, black, semi-immersed, scattered beneath the host |
| Peridium | Prominently rough 55–100 μm | Prominently rough 60–100 μm | Prominently rough up to 130 μm | Prominently rough 75–160 μm | Prominently rough 30–70 μm |
| Asci | 140–200 × 13–15 μm | 156–260 × 11–16 μm | 100–160 × 17–22 μm | 140–200 × 13–15.5 μm | 180–240 × 10–15 μm |
| Spore | Dark- to light-golden brown, 18–26 × 7,5–10 μm, muriform, 3 to 7 transverse septa, constricted at the central septum, longitudinal septa 1 or continuos or dis-continuos | Golden brown, 25–35 × 9–15 μm, muriform, 5 to 7 transverse septa, constricted at the central septum, 1 to 2 longitudinal septa | Olive brown, 21–30 × 8.5–13 μm, muriform, 3 to 7 transverse septa, usually 5–7 transverse septa, constricted at the central septum, 1 to 2 longitudinal septa | Golden brown, 25–30 × 11–12 μm, muriform, 5 to 7 transverse septa, constricted at the central septum, with 1 longitudinal septa | Brown 19–28 × 9–15 μm, muriform, mostly ellipsoidal, 5–7 transversely septate, with 4–6 vertical septa, constricted at the central septum, with 1–2 longitudinal septa |
| Host species (Cytisus sp.) |
C. pendulinus, C. scoparius, C. sessilifolius |
C. alpinss, C. laburnum, C. radiatus |
C. scoparius |
C. capitatus, C. scoparius, Cytisus sp. |
Cytisus sp. |
| Country | Portugal, Spain, France, Italy, Sweden | Germany, England, Italy, Switzerland | Spain | Germany, Portugal, Spain, Sweden | Italy |
| References | Mirza, 1968, Ellis and Ellis, 1985 | Mirza, 1968, Ellis and Ellis, 1985 | Mirza, 1968, Ellis and Ellis, 1985 | Mirza, 1968, Ellis and Ellis, 1985 | This study |
In this study we used morphology and phylogenetic analyses for the identification of our collection. Thus it is important to carry out molecular analyses to confirm the taxonomic and phylogenetic placement. According to the morphological and phylogenetic analysis results, we introduce our taxon (MFLUCC 14-0238) as a new species of Camarosporium sensu stricto. Other Cucurbitaria spp. should be recollected and subjected to morphological and molecular analyses as Camarosporium sensu stricto has cucurbitaria-like sexual states (Wijayawardene et al., 2014a, Wijayawardene et al., 2014b).
Acknowledgements
Mae Fah Luang University Grant “Taxonomy and Phylogeny of selected families of Dothideomycetes (Grant number: 56101020032)” is thanked for supporting this study and Plant Germplasm and Genomics Center in Germplasm Bank of Wild Species, Kunming Institute of Botany is thanked for the help with the molecular work. Erio Camporesi thanks Gigi Stagioni for his invaluable field assistance. Saowaluck Tibpromma would like to thank the Mushroom Research Foundation (MRF), Chiang Rai Province, Thailand for its continuous support. Dhanushka Udayanga, Samantha C. Karunarathna, Belle Damodara Shenoy, Jian-Kui Liu, and Hiran A. Ariyawansa are thanked for their valuable suggestions and help.
Footnotes
Peer review under responsibility of King Saud University.
References
- Boonmee S., Zhang Y., Chomnunti P., Chukeatirote E., Tsui C.K.M., Bahkali A.H., Hyde K.D. Revision of lignicolous Tubeufiaceae based on morphological re-examination and phylogenetic analysis. Fungal Divers. 2011;51:63–102. [Google Scholar]
- Boonmee S., Ko-Ko T.W., Chukeatirote E., Hyde K.D., Chen H., Cai L., Mckenzie E.H.C., Jones E.B.G., Kodsueb R., Hassan B.A. Two new Kirschsteiniothelia species with Dendryphiopsis anamorphs cluster in Kirschsteiniotheliaceae fam. nov. Mycologia. 2012;104:698–714. doi: 10.3852/11-089. [DOI] [PubMed] [Google Scholar]
- Boonmee S., Bhat J.D., Maharachchikumbura S.S.N., Hyde K.D. Clavatispora thailandica gen. et sp. nov., a novel taxon of Venturiales (Dothideomycetes) from Thailand. Phytotaxa. 2014;176:092–101. [Google Scholar]
- Chomnunti P., Hongsanan S., Hudson B.A., Tian Q., Peršoh D., Dhami M.K., Alias A.S., Xu J., Liu X., Stadler M., Hyde K.D. The sooty moulds. Fungal Divers. 2014;66:1–36. [Google Scholar]
- Chomnunti P., Schoch C.L., Aguirre-Hudson B., Ko-Ko T.W., Hongsanan S., Jones E.B.G., Kodsub R., Chukeatirote E., Bahkali A.H., Hyde K.D. Capnodiaceae. Fungal Divers. 2011;51:103–134. doi: 10.1007/s13225-011-0145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doilom M., Liuk K., Jaklitsch W.M., Ariyawansa H., Wijayawardene N.N., Chukeatirote E., Zhang M., Mckenzie E.H.C., Geml J., Voglmayr H., Hyde K.D. An outline of the family Cucurbitariaceae. Sydowia. 2013;65:167–192. [Google Scholar]
- Douanla M.C., Langer E., Calonge F.D. Geastrumpleosporus sp. nov., a new species of Geastraceae identified by morphological and molecular phylogenetic data. Mycol. Prog. 2005;4(3):239–250. [Google Scholar]
- Ellis M.B., Ellis J.P. Richmond Publishing; 1985. Microfungi on Land Plants: An Identification Handbook. [Google Scholar]
- Gardes M., Bruns T.D. ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- Hall, T., 2004. Bioedit version 6.0.7. Available from: <http://www.mbio.-ncsu.edu/bioedit/bioedit.html>.
- Hyde K.D., Jones E.B.G., Liu J.K., Ariyawansa H., Boehm E., Boonmee S., Braun U., Chomnunti P., Crous P.W., Dai D.Q., Diederich P., Dissanayake A., Doilom M., Doveri F., Hongsanan S., Jayawardena R., Lawrey J.D., Li Y.M., Liu Y.X., Lücking R., Monkai J., Muggia L., Nelsen M.P., Pang K.L., Phookamsak R., Senanayake I.C., Shearer C.A., Suetrong S., Tanaka K., Thambugala K.M., Wijayawardene N.N., Wikee S., Wu H.X., Zhang Y., Hudson B.A., Alias S.A., Aptroot A., Bahkali A., Bezerra J.L., Bhat D.J., Camporesi E., Chukeatirote E., Gueidan C., Hawksworth D.L., Hirayama K., Hoog S.D., Kang J.C., Knudsen K., Li W.J., Li X.H., Liu Z.Y., Mapook A., Mckenzie E.H.C., Miller A.N., Mortimer P.E., Phillips A.J.L., Raja H.A., Scheuer C., Schumm F., Taylor J.E., Tian Q., Tibpromma S., Wanasinghe D.N., Wang Y., Xu J.C., Yacharoen S., Yan J.Y., Zhang M. Families of Dothideomycetes. Fungal Divers. 2013;63:1–313. [Google Scholar]
- Index Fungorum., 2015 http://www.indexfungorum.org/names/NamesRecord.asp?-RecordID=489566.
- Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008;9:276–285. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Kirk P.M., Cannon P.F., Minter D.W., Stalpers J.A. 10th ed. CABI; Wallingford: 2008. Dictionary of the Fungi. [Google Scholar]
- Liu J.K., Phookamsak R., Jones E.B.G., Zhang Y., Ko-Ko T.W., Hu H.L., Boonmee S., Doilom M., Chukeatirote E., Bahkali A.H., Wang Y., Hyde K.D. Astrosphaeriella is polyphyletic, with species in Fissuroma gen. nov., and Neoastrosphaeriella gen. nov. Fungal Divers. 2011;51:135–154. [Google Scholar]
- Liu J.K., Phookamsak R., Doilom M., Wiki S., Mei L.Y., Ariyawansa H.A., Boonmee S., Chomnunti P., Dai D.Q., Bhat D.J., Romero A.I., Xhuang W.Y., Monkai J., Jones E.B.G., Chukeatirote E., Ko-Ko T.W., Zhoa Y.C., Wang Y., Hyde K.D. Towards a natural classification of Botryosphaeriales. Fungal Divers. 2012;57:149–210. [Google Scholar]
- Mirza F. Taxonomic investigations on the ascomycetous genus Cucurbitaria. Nova Hedwigia. 1968;16:161–213. [Google Scholar]
- Nelsen M.P., Lücking R., Grube M., Mbatchou J.S., Muggia L., Rivas-Plata E., Lumbsch H.T. Unravelling the phylogenetic relationships of lichenised fungi in Dothideomyceta. Stud. Mycol. 2009;64:135–144. doi: 10.3114/sim.2009.64.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelsen M.P., Lücking R., Mbatchou J.S., Andrew C.J., Spielmann A.A., Lumbsch H.T. New insights into relationships of lichen-forming Dothideomycetes. Fungal Divers.. 2011;51:155–162. [Google Scholar]
- Phookamsak R., Liu J.K., Chukeatirote E., McKenzie E.H.C., Hyde K.D. Phylogeny and morphology of Leptosphaerulina saccharicola sp. nov. and Pleosphaerulina oryzae and relationships with Pithomyces. Cryptogam Mycol. 2013;34(4):303–319. [Google Scholar]
- Schoch C.L., Sung G.H., López-Giráldez F., Townsend J.P., Miadlikowska J., Hofstetter V., Robbertse B., Matheny P.B., Kauff F., Wang Z., Gueidan C., Andrie R.M., Trippe K., Ciufetti L.M., Wynns A., Fraker E., Hodkinson B.P., Bonito G., Groenewald J.Z., Arzanlou M., De-Hoog G.S., Crous P.W., Hewitt D., Pfister D.H., Peterson K., Gryzenhout M., Wingfield M.J., Aptroot A., Suh S.O., Blackwell M., Hillisdm, Griffith G.W., Castlebury L.A., Rossman A.Y., Lumbsch H.T., Lücking R., Büdel B., Rauhut A., Diederich P., Ertz D., Geiser D.M., Hosaka K., Inderbitzin P., Kohlmeyer J., Volkmann-Kohlmeyer B., Mostert L., O’Donnell K., Sipman H., Rogers J.D., Shoemaker R.A., Sugiyama J., Summerbell R.C., Untereiner W., Johnston P.R., Stenroos S., Zuccaro A., Dyer P.S., Crittenden P.D., Cole M.S., Hansen K., Trappe J.M., Yahr R., Lutzoni F., Spatafora J.W. The Ascomycota tree of life: a phylum–wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst. Biol. 2009;58:224–239. doi: 10.1093/sysbio/syp020. [DOI] [PubMed] [Google Scholar]
- Schulzer S. Mykologische Beiträge. Verh. Zoologisch-Botanischen Ges. Wien. 1870;20:635–658. [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, MA: 2002. PAUP: Phylogenetic Analysis using Parsimony, Version 4.0 b10. [Google Scholar]
- Telle S., Thines M. Amplification of cox2 (∼620 bp) from 2 mg of up to 129 years old herbarium specimens, comparing 19 extraction methods and 15 polymerases. PLoS ONE. 2008;3:3584. doi: 10.1371/journal.pone.0003584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T.J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; New York, USA: 1990. pp. 315–322. [Google Scholar]
- Wijayawardene N.N., Mckenzie E.H.C., Hyde K.D. Towards incorporating anamorphic fungi in a natural classification – checklist and notes for 2011. Mycosphere. 2012;3(2):157–228. [Google Scholar]
- Wijayawardene N.N., Hyde K.D., Bhat D.J., Camporesi E., Schumacher R.K., Chethana K.W.T., Wikee S., Bahkali A.H., Wang Y. Camarosporium-like species are polyphyletic in Pleosporales; introducing Paracamarosporium and Pseudocamarosporium gen. nov. in Montagnulaceae. Cryptogamie Mycol. 2014;35(2):177–198. [Google Scholar]
- Wijayawardene N.N., Bhat D.J., Hyde K.D., Camporesi E., Wikee S., Chethana K.W.T., Tangthirasunun N., Wang Y. Camarosporium sensu stricto in Pleosporinae, Pleosporales with two new species. Phytotaxa. 2014;183(1):16–26. [Google Scholar]
- Wijayawardene N.N., Crous P.W., Kirk P.M., Hawksworth D.L., Boonmee S., Braun U., Chomnunti P., Dai D.Q., D’Souza M.J., Diederich P., Dissanayake A., Doilom M., Hongsanan S., Jones E.B.G., Groenewald J.Z., Jayawardena R., Lawrey J.D., Liu J.K., Lücking R., Madrid H., Manamgoda D.S., Muggia L., Nelsen M.P., Phookamsak R., Suetrong S., Tanaka K., Thambugala K.M., Wikee S., Zhang Y., Aptroot A., Ariyawansa H.A., Bahkali A.H., Bhat J.D., Gueidan C., De-Hoog G.S., Knudsen K., Mckenzie E.H.C., Miller A.N., Mortimer P.E., Wanasinghe D.N., Phillips A.J.L., Raja H.A., Slippers B., Shivas R.S., Taylor J.E., Wang Y., Woudenberg J.H.C., Piątek M., Cai L., Jaklitsch W.M., Hyde K.D. Naming and outline of Dothideomycetes – 2014c. Fungal Divers. 2014;68 doi: 10.1007/s13225-014-0309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Crous P.W., Schoch C.L., Bahkali A.H., Guo L.D., Hyde K.D. A molecular, morphological and ecological re-appraisal of Venturiales a new order of Dothideomycetes. Fungal Divers. 2011;51:249–277. doi: 10.1007/s13225-011-0141-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Crous P.W., Schoch C.L., Hyde K.D. Pleosporales. Fungal Divers. 2012;52:1–225. doi: 10.1007/s13225-011-0117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]