Abstract
Varietal identification of olives is an intrinsic and empirical exercise owing to the large number of synonyms and homonyms, intensive exchange of genotypes, presence of varietal clones and lack of proper certification in nurseries. A comparative study of morphological characters of eight olive cultivars grown in Saudi Arabia was carried out and analyzed using NTSYSpc (Numerical Taxonomy System for personal computer) system segregated smaller fruits in one clade and the rest in two clades. Koroneiki, a Greek cultivar with a small sized fruit shared arm with Spanish variety Arbosana. Morphologic analysis using NTSYSpc revealed that biometrics of leaves, fruits and seeds are reliable morphologic characters to distinguish between varieties, except for a few morphologically very similar olive cultivars. The proximate analysis showed significant variations in the protein, fiber, crude fat, ash and moisture content of different cultivars. The study also showed that neither the size of fruit nor the fruit pulp thickness is a limiting factor determining crude fat content of olives.
Keywords: Morphology, NTSYSpc, Crude fat, Protein, Fiber
1. Introduction
The olive trees (Olea europaea L.) valued for their esthetics and fruits have been a part of Mediterranean civilization since time immemorial. There are about 45 wild olive species, whose combined range of distribution extends from New Caledonia, New Zealand and Australia through southern Asia and through most of Africa to the Near East and southern Europe. O. europaea L. is conceivably originated as an ancient natural hybrid between two species: Olea ferruginea × Olea laperinii (Sauer, 1993). The wild and domesticated O. europaea intercrosses and results in many different local cultivars, all presumably originated as chance seedling and found to bear superior quality fruits. Hundreds of varieties thus emerged are conserved and cultivated in different agro-climatic regions of the world. They are highly variable in size, shape and form of fruits, their chemical constituents and microclimate requirements. The origin and the geographical distribution of such high variability in the cultivated olive is still under investigation. Most studies agree that after an ancestral spreading of a few olive varieties along the Mediterranean basin, a majority of modern cultivars were derived from the crossing of these ancient cultivars among themselves, or by their breeding with wild plants, followed by local selection (Angiolillo et al., 1999, Besnard and Bervillé, 2000, Besnard et al., 2001).
The olive trees have been cultivated for millennia in the Mediterranean basin and its oil has been an important part of human nutrition in the region. Saudi Arabia is one of the largest consumers of olives and olive oils, but only contributes nominally to the world’s olive oil production. The soil and climatic conditions of northern parts of the Kingdom of Saudi Arabia which resemble that of Mediterranean climate favor olive tree growth and thus production of olive oils with the same high quality of international standards. The production area and volume are on the increase in KSA and over the past 15 years KSA has become one of the world’s new olive oil producing countries. In the last three decades, extensive plantations of olive have been established in the northern parts of the kingdom using the planting materials obtained from the neighboring countries (Al-Khalifah et al., 2012) like Syria, Jordan, where many exotic and indigenous cultivars of olives are extensively cultivated.
The cross-pollinating nature of the species and its secular history contributed to determine a wide germplasm biodiversity with a large number of more than 1200 (Bartolini et al., 2005) cultivars present in the main olive oil producing countries. This genetic diversity could be an important resource for the development of modern olive culture toward typical olive oil and fresh productions (Hegazy et al., 2012). This richness in terms of available biodiversity, however, often has determined some drawbacks in the management and identification of the plant material to distinguish between cultivars, and this has been further complicated by the frequency of homonyms and synonyms.
Morphological and biological characters have been widely used for descriptive purposes and are commonly used to distinguish olive cultivars (Barranco and Rallo, 1985, Cantini et al., 1999, Barranco et al., 2000). Agronomic characterization also allowed the classification of different olive cultivars (Barranco and Rallo, 2000, Del Rio, 1994). According to Bartolini et al. (1998) and Barranco et al. (2000), biometric indexes should always be accompanied by a detailed morphological description of the organs (inflorescence, leaf, fruit, and stone) of olive varieties following the UPOV method. Many researchers observed that different cultivars are morphologically variable based on geographical locations and under various plant growth management practices (Grati et al., 2002, Guerfel et al., 2009, Youssefi et al., 2011).
Proximate analysis used to determine the proximate principles of any substance, as contrasted with an ultimate analysis (Jaafar et al., 2009). Proximate and nutrient analysis of edible fruit and vegetables plays a crucial role in assessing their nutritional significance (Pandey et al., 2006, Adepoju, 2009, Hussain et al., 2009). The macro components are generally analyzed for their proximate amounts (Owusu-Apenten, 2005). Since olive is an oily fruit, extensive studies have been conducted on the fatty acid contents of different varieties (Andrews et al., 2003, Petridis et al., 2012, DeLeonardis et al., 2008) but a proximate analysis showing the other fruit qualities is seldom attempted in olives.
The objectives of this study are to provide a more comprehensive identification and their inter relationships of the most popular cultivars of olives growing in Saudi Arabia using morphological characters and proximate fruit analysis.
2. Materials and methods
Leaf and fruit samples were collected from eight cultivars of olives (Arabic – ‘zeytoon’) grown in the northern region of Saudi Arabia (Al-Jouf) by two leading agriculture companies, viz. National Agriculture Development Company (NADEC) and Al-Jouf Agriculture Company during the harvesting season of 2012. Samples were collected from randomly selected trees of the populations of each cultivar. Cultivars from NADEC Company were named as Arbosana, Arbequina, Picual and Koroneiki and those from Al-Jouf Agriculture Company as Kaissy H-85, Picual H-78, Sorani and K-18. The variety K-18 also belongs to one of the popular named varieties but its proper identification was lost due to mishandling. The leaves and fruits were packed separately in polyethylene bags, immediately placed in ice box and transferred to the laboratory for storage in a refrigerator at −30 °C, processing and further study.
2.1. Morphological analysis
Two hundred leaves and fruits were randomly selected from each cultivar lot. Maximum length and width of leaves were measured using mm scale and maximum length and diameter of the fruits were measured using a screw gauge. Seeds were removed from the measured fruits and their dimensions were also measured using a screw gauge. Length–width ratio of leaves and fruits were calculated. Thickness of fruit-pulp was calculated using the formula: diameter of the fruit-diameter of the seed/2. All measurements were tabulated and LSD among the values of each cultivar corresponding to each character was found out. Data was further analyzed using the software called NTSYSpc (Numerical Taxonomy System for personal computer, Rohlf, 1998) as suggested by Jamshidi and Jamshidi (2011). Under each character, the values of those cultivars that are not significantly different were scored as (1) and the rest as (0). All significantly different values were treated as separate characters and altogether 43 characters pertaining to eight cultivars were entered in the system. Similarity matrix and Cluster analysis, demonstrating relationships of accessions were generated using UPGMA (Unweighted Pair-Group Method using Arithmetic averages) and simple matching coefficient.
2.2. Proximate analysis of olive fruits
Different parameters such as protein, crude fat, ash and moisture content of eight olive fruit samples were analyzed using the protocol given by AOAC (2000) while dietary fiber was determined according to Prosky et al. (1988) using Fibretec™ System (Foss, Denmark). Unblemished fruits were selected, washed and seeds removed. The remaining portion was homogenized in a blender to obtain 100 g fruit pulp from each variety. To determine the dry matter content 2 g samples were dried in an oven (Memmert, Karl Kolb, Dreieich, West Germany) at 105 °C until constant weight is obtained for three consequent weighing. The moisture and dry matter contents were calculated based on weight difference method. Total nitrogen was determined by Kjeldahl (1883) method and the protein content was calculated by multiplying the total nitrogen content by a factor of 5.6. The crude fat content was determined by a Soxtec apparatus (Gerhardt soxtherm, C. Gerhardt GmbH & Co., Bonn, Germany). The ash content was determined by combustion of 2 g samples in silica crucibles in a furnace (Thermolyne, Dubuque, IA, USA) for 24 h at 550 °C.
All analyses were carried out in triplicate and the results were subjected to statistical analysis using Duncan multiple test range and Analysis of variance (ANOVA). The results are presented with their means and standard deviation.
3. Results and discussion
3.1. Morphological analysis
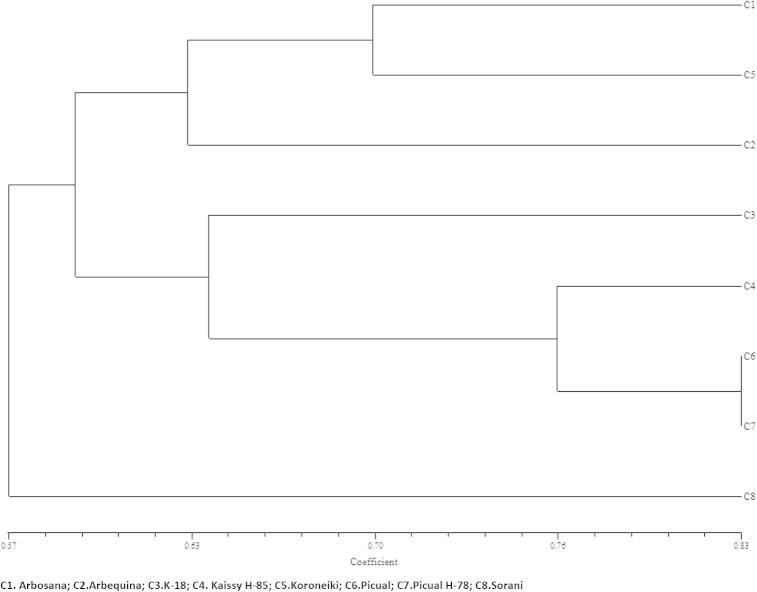
Analysis of the morphological data pertaining to the eight cultivars showed significant variations in many parameters (Table 1). Largest leaf-length was recorded by K-18 followed by Picual H-78 but maximum leaf-width was seen in Kaissy H-85. Leaf length–width ratio was significantly higher in Picual H-78 and lower in Sorani, indicating a higher level of variance in the leaf shape. Five cultivars had a higher fruit-length, measuring nearly 25 mm with Picual H-78 showing the largest fruit-length. Arbosana, Arbequina and Koroneiki have comparatively smaller fruits of size ranging from 15.0 to 17.9 mm long. Length–diameter ratios of different fruit varieties also showed significant variations ranging from 1.2 to 1.55. Longest seed was seen in Sorani and shortest was in Arbequina while their length-diameter ratio was not significantly different in most of the cultivars except Sorani and Arbequina. There were significant variations in the thickness of fruit pulp ranging from 3.1 to 6.0 mm. Kaissy H-85 had the thickest fruit-pulp while Koroneiki had a thin pulp. Similarity matrix (Table 2) and dendrogram based on morphological characters showed maximum similarity between the cultivars Picual and Picual H-78 (82.6). The variety K-18 showed 65.2% of similarity with the two known Picuals. Visual observations also showed close similarity between K-18 and Picual varieties (Fig. 1).
Table 1.
Morphological characters of eight cultivars of Olives (All measurements are in mm scale except the ratios).
| Characters | Arbosana | Arbequina | K-18 | Kaissy H-85 | Koroneiki | Picual | Picual H-78 | Sorani | LSD |
|---|---|---|---|---|---|---|---|---|---|
| Leaf-length | 53.7 ± 5.2b | 42.0 ± 4.4a | 63.5 ± 4.8c | 59.2 ± 6.3b | 42.0 ± 5.1a | 42.4 ± 2.0a | 60.9 ± 5.8c | 54.5 ± 2.9b | 5.4 |
| Leaf-width | 10.8 ± 1.0b | 9.3 ± 0.6a | 12.7 ± 1.1c | 14.5 ± 1.2d | 8.4 ± 1.3a | 8.5 ± 1.4a | 10.6 ± 1.1b | 14.1 ± 1.2c | 1.4 |
| Length/width ratio | 4.97 ± 0.01d | 4.56 ± 0.02c | 5.00 ± 0.15d | 4.08 ± 0.02b | 5.00 ± 0.03d | 4.98 ± 0.02d | 5.74 ± 0.04e | 3.86 ± 0.03a | 0.19 |
| Fruit-length | 17.9 ± 1.1b | 15.0 ± 1.3a | 25.1 ± 1.3c | 24.4 ± 1.4c | 17.1 ± 1.3b | 25.0 ± 1.2c | 26.0 ± 1.3c | 24.3 ± 1.4c | 1.7 |
| Fruit-diameter | 12.5 ± 1.2a | 12.6 ± 1.3a | 17.2 ± 1.6b | 20.0 ± 1.7c | 10.9 ± 1.3a | 19.0 ± 1.4c | 20.0 ± 0.7c | 16.5 ± 1.1b | 1.9 |
| Length/width ratio | 1.43 ± 0.07c | 1.20 ± 0.03a | 1.46 ± 0.06c | 1.22 ± 0.02a | 1.55 ± 0.04d | 1.31 ± 0.02b | 1.30 ± 0.01b | 1.47 ± 0.04c | 0.09 |
| Seed-length | 13.6 ± 0.5b | 11.0 ± 0.8a | 16.6 ± 1.0c | 16.0 ± 1.1c | 12.6 ± 0.7a | 17.3 ± 0.5c | 17.0 ± 0.6c | 18.3 ± 1.3c | 1.6 |
| Seed-diameter | 6.0 ± 0.1a | 6.3 ± 0.5a | 7.3 ± 0.5b | 8.0 ± 0.6b | 5.6 ± 0.5a | 8.3 ± 0.6c | 8.6 ± 0.5c | 7.6 ± 0.4b | 0.8 |
| Length/width ratio | 2.26 ± 0.09b | 1.70 ± 0.22a | 2.26 ± 0.08b | 2.00 ± 0.13b | 2.25 ± 0.11b | 2.08 ± 0.19b | 1.97 ± 0.16b | 2.40 ± 0.24b | 0.5 |
| Fruit-pulp thickness | 3.2 ± 0.1b | 3.1 ± 0.2b | 4.9 ± 0.6d | 6.0 ± 0.5f | 2.6 ± 0.1a | 5.3 ± 0.1e | 5.7 ± 0.2f | 4.4 ± 0.3c | 0.3 |
Means followed by the same letter superscript are not significantly different at 0.05.
Table 2.
Similarity matrix for Nei and Lei’s coefficients of 8 genotypes of Olives based on morphological characters.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|---|---|
| 1. | 1.000 | |||||||
| 2. | 0.652 | 1.000 | ||||||
| 3. | 0.652 | 0.565 | 1.000 | |||||
| 4. | 0.565 | 0.608 | 0.608 | 1.000 | ||||
| 5. | 0.695 | 0.608 | 0.608 | 0.565 | 1.000 | |||
| 6. | 0.565 | 0.652 | 0.652 | 0.739 | 0.652 | 1.000 | ||
| 7. | 0.565 | 0.565 | 0.652 | 0.782 | 0.521 | 0.826 | 1.000 | |
| 8. | 0.652 | 0.521 | 0.622 | 0.565 | 0.521 | 0.565 | 0.521 | 1.000 |
C1. Arbosana; C2. Arbequina; C3. K-18; C4. Kaissy H-85; C5. Koroneiki; C6. Picual; C7. Picual H-78; C8. Sorani.
Figure 1.
Fruits and leaves of eight cultivars of Olives.
Cladistic analysis using morphologic characters with NTSYSpc software recovered three clades. Clade 1 with three cultivars (Arbosana, Koroneiki and Arbequina), Clade 2 with four cultivars (K-18, Kaissy H-85, Picual, Picual H-78) and the third clade with only Sorani. All the cultivars in clade 1 were having smaller fruits while those in clade 2 have bigger fruits. Sorani had an average of 56.67% similarity with the other cultivars and it stood apart in the dendrogram (Fig. 2). Previously, based on morphological traits such as speckles on leaves, color of adaxial leaf surface, shape of leaf blade, leaf apex, leaf margin, leave base, fruit surface type, type of fruit pedicel, presence of wings on fruit surface, shape of fruit, fruit length, fruit diameter and seed size were used to distinguish between closely related Russian olive varieties (Asadiar et al., 2012). The olive genotypes were also evaluated for the morphological traits namely leaf, fruit and endocarp characters (Zaher et al., 2011). Milotic et al. (2005) performed morphologic characterization of 64 olive trees using 23 characters of leaves, inflorescence, fruits and seeds.
Figure 2.
Dendrogram showing relationship of eight cultivars using a similarity matrix obtained from morphological characters.
Morphological cladistic analyses were successfully employed in many species to detect phylogenetic relationships (Carvalho-Sobrinho and Queiroz, 2011). Mehr et al. (2012) reconstructed phylogenetic relationship among some bifurcate hairy sections of Astragalus L. using 38 vegetative and reproductive morphological characters. Abdali et al. (2014) used 38 morphological characters to distinguish between 10 Iranian Olives and found qualitative properties of fruits and core are powerful tools in separating cultivars than the quantitative characters.
3.2. Proximate analysis of fruits
Table 3 shows the proximate fruit analysis of eight olive cultivars grown in Saudi Arabia. Result shows that Picual has a significantly higher percentage of protein content than the other cultivars. Kaissy recorded the lowest percentage of protein but its value is not significantly different from other cultivars except Picual and K-18. The highest fiber content was seen in Kaissy while Picual exhibited the lowest percentage. There was a negative correlation between the protein and fiber contents of Picual and Kaissy but this was not true in all other cases. There were significant differences between the crude fat contents of eight cultivars with K-18 having the lowest and Sorani having the highest percentage of fat contents. Among the eight cultivars K-18 has the highest ash content and Koroneiki has the lowest percentage of ash content. The mean percentage of moisture content in all the cultivars was 58.3% with the lowest 54 in K-18 and highest 65% in Picual-H78. The higher values for proximate analysis parameter were moisture content and crude fat.
Table 3.
Proximate chemical composition of olive fruits (%).
| No | Sample | Protein | Fiber | Crude Fat | Ash | Moisture |
|---|---|---|---|---|---|---|
| 1 | Koroneiki | 3.2 ± 0.1a | 3.4 ± 0.1b | 21 ± 1.0c | 1.38 ± 0.02a | 59 ± 2b |
| 2 | Picual | 3.9 ± 0.05b | 3.0 ± 0.5a | 19 ± 0.5c | 1.92 ± 0.03b | 56 ± 1a |
| 3 | Arbequina | 3.3 ± 0.02a | 3.5 ± 0.2b | 20 ± 0.7c | 2.07 ± 0.03b | 59 ± 1.5b |
| 4 | Arbosana | 3.5 ± 0.15b | 3.4 ± 0.5b | 22 ± 1.2c | 2.06 ± 0.04b | 58 ± 0.5a |
| 5 | Kaissy | 3.1 ± 0.05a | 3.6 ± 0.3c | 15 ± 1.0b | 1.95 ± 0.03b | 59 ± 1.7b |
| 6 | Sorani | 3.3 ± 0.1a | 3.1 ± 0.3a | 24 ± 1.0d | 3.50 ± 0.02c | 57 ± 0.7a |
| 7 | K-18 | 3.6 ± 0.07b | 3.2 ± 0.06a | 11.1 ± 0.4a | 5.22 ± 0.4d | 54 ± 1.2a |
| 8 | Picual H-78 | 3.2 ± 0.5a | 3.3 ± 0.2b | 22 ± 1.4c | 1.96 ± 0.6b | 65 ± 3.9c |
| Mean | 3.38 | 3.31 | 19.26 | 2.50 | 58.3 | |
| LSD | 0.25 | 0.23 | 2.24 | 0.38 | 4.87 |
Means followed by the same letter superscript are not significantly different at 0.05.
Oleic acid is the main monounsaturated fatty acid present in olive oil and its concentration varies from variety to variety (Zarrouk et al., 2009). Fatty acid composition of the olive oil samples depended mainly on the variety rather than place of collection, maturity index and other edaphic factors. Rondanini et al. (2011) observed lowest oleic acid values with the Spanish variety Arbequina (51.8%) and highest values in Picual (71.9%) grown in Northwestern Argentina. The present study of eight olive cultivars of exotic origin, cultivated in Saudi Arabia showed a lower percentage of crude fatty acid content than in their original locations (>70). This is mainly attributed to the prevailing higher temperature and lower humidity. Percentage of saturated and unsaturated fatty acid contents are influenced by environmental conditions such as temperature, rainfall, and genotypes (Esmaeili et al., 2012). There was a negative correlation between oleic acid values in Arbequina and other varieties with the mean temperature during oil accumulation, indicating that oleic acid content decreased at 2% per °C increase in mean temperatures (Esmaeili et al., 2012). The lower values for proximate analysis parameter were crude protein, fiber and ash content. Since Olive is an oily fruit the crude fat content was higher than that of non-oily fruit such as Dragon fruit (Jaafar et al., 2009). Water activity plays the main role where it controls the microbial activity and thus moisture content decreases the keeping quality and shelf life of any fruits.
Until recently, identification of cultivars of O. europaea was mainly based on morphological, biochemical and agronomic traits which are known to be deeply influenced by environmental factors (Contento et al., 2002). Neither chemical analysis of different clusters of compounds, nor analysis of bio-morphological traits have led to cultivar identification, due to environmental effects on the chemical composition and phenotype (Alessandri et al., 1997). Genetic identity seems to be the only possibility for identifying the cultivar and the products deriving from it (Busconia et al., 2003). However, the probability of finding the same molecular profiles for more than one morphologically different variety cannot be ruled out and was attributed to the small differences at the DNA level (Besnard et al., 2001). In the present study also morphological data of some of the cultivars showed similar distance among cultivars.
4. Conclusion
Out of the eight exotic olive cultivars grown in Saudi Arabia, one was Greek (Koroneiki), five were Spanish (Arbequina, Arbosana, Picual, Picual H-78, K-18) and two were Syrian (Kaissy, Sorani) in origin. In the new desert environment of Saudi Arabia they showed significant variations in the morphology and fatty acid contents. Morphologic analysis using NTSYSpc revealed that dimensions of leaves, fruits and seeds are reliable morphologic characters to distinguish between varieties provided a large number of replicates are measured per sample. However, biometric values alone were not able to detect differences among some morphologically similar varieties characterized by different agronomical traits. Proximate analysis of fruits clearly indicates that neither the size of fruits nor the fruit pulp thickness is a limiting factor controlling fatty acid contents. Among the eight studied cultivars, Arbosana, Arbequina and Koroneiki with comparatively smaller fruits and less fruit-pulp thickness showed above 20% fatty acid contents while two bigger fruit yielding cultivars (K-18 and Kaissy H-85) showed less than 15% crude fat content. These two cultivars with higher fiber content and lower crude fat content can be used as table olives for edible purpose than for oil production.
Acknowledgements
Authors express thanks to the NADEC and Al-Jouf Agricultural Company of Saudi Arabia for providing olive samples and the King Abdulaziz City for Science and Technology, Riyadh for infrastructure facilities.
Footnotes
Peer review under responsibility of King Saud University.
References
- Abdali N., Ataei S., Keihan A., Motaghi L., Naghavi M., Kazemitabar S.K., Rezazade A., Hosseini-Mazinani M. Comparative study of the morphological and molecular characteristics of 10 Iranian olive (Olea europaea L.) cultivars. Ann. Biol. Res. 2014;5(3):76–83. [Google Scholar]
- Adepoju O.T. Proximate composition and micronutrient potentials of three locally available wild fruits in Nigeria. Afr. J. Agric. Res. 2009;4(9):887–892. [Google Scholar]
- Alessandri S., Cimato A., Modi G., Mattei A., Crescenzi A., Caselli S., Tracchi S. Univariate models to classify Tuscan virgin olive oils by zone. Riv. Ital. Sostanze Gr. 1997;74:155–164. [Google Scholar]
- Al-Khalifah N.S., Askari E., El-Kholy M. Following olive footprints in Saudi Arabia. In: El-Kholy M., editor. Aarinena, IOC, and ISHS; 2012. (Following Olive Footprints (Olea europaea L.) Cultivation and Culture, Folklore and History, Traditions and Uses). (Scripta Horticulturae N. 13) [Google Scholar]
- Andrews P., Busch J.L.H.C., Joode T.D., Groenewegen A., Alexandre H. Sensory properties of virgin olive oil polyphenols: identification of deacetoxyligstroside aglycon as a key contributor to pungency. J. Agric. Food Chem. 2003;51:1415–1420. doi: 10.1021/jf026042j. [DOI] [PubMed] [Google Scholar]
- Angiolillo A., Mencuccini M., Baldoni L. Olive genetic diversity assessed using amplified fragment length polymorphisms. Theor. Appl. Genet. 1999;98:411–414. [Google Scholar]
- AOAC . Association of Official Analytical Chemists; Virginia, USA: 2000. Official Methods of Analysis of AOAC International. [Google Scholar]
- Asadiar L.S., Rahmani F., Siami A. Assessment of genetic diversity in the Russian olive (Elaeagnus angustifolia) based on ISSR genetic markers. Rev. Ciênc. Agron. 2012;44(2):310–316. [Google Scholar]
- Barranco D., Rallo L. Las variedades de olivo cultivades en Espana. Olivae. 1985;9:16–22. [Google Scholar]
- Barranco D., Rallo L. Olive cultivars in Spain. Hortechnology. 2000;10(1):107–110. [Google Scholar]
- Barranco D., Cimato A., Fiorino P., Rallo L., Touzani A., Castañeda C., Serafín F., Trujillo I. International Olive Oil Council; Madrid, Spain: 2000. World Catalogue of Olive Varieties. 360 pp. [Google Scholar]
- Bartolini, G., Prevost, G., Messeri, C., Carignani, G., 2005. Olive germplasm: cultivars and world-wide collections. Available via DIALOG <http://www.apps3.fao.org/wiews/olive/oliv.jsp> (accessed November 2014).
- Bartolini G., Prevost G., Messeri C., Carignani G., Menini U.G. FAO; Rome: 1998. Olive Germplasm. Cultivars and World-Wide Collections. [Google Scholar]
- Besnard G., Baradat P., Chevalier D., Tagmount A., Bervillé A. Genetic differentiation in the olive complex (Olea europaea) revealed by RAPDs and RFLPs in the rRNA genes. Genet. Resour. Crop. Ev. 2001;48:165–182. [Google Scholar]
- Besnard G., Bervillé A. Multiple origins for Mediterranean olive (Olea europaea L. ssp. europaea) based upon mitochondrial DNA polymorphisms. Life Sci. 2000;323:173–181. doi: 10.1016/s0764-4469(00)00118-9. [DOI] [PubMed] [Google Scholar]
- Busconia M., Foronib C., Corradib M., Bongiornia C., Cattapanc F., Foghera C. Analytical, Nutritional and Clinical Methods: DNA extraction from olive oil and its use in the identification of the production cultivar. Food Chem. 2003;83:127–134. [Google Scholar]
- Cantini C., Cimato A., Sani G. Morphological evaluation of olive germplasm present in Tuscany region. Euphytica. 1999;109:173–181. [Google Scholar]
- Carvalho-Sobrinho J.G.D., Queiroz L.P.D. Morphological cladistic analysis of Pseudobombax Dugand (Malvaceae, Bombacoideae) and allied genera. Revista Brasil. Bot. 2011;34(2):197–209. [Google Scholar]
- Contento A., Ceccarelli M., Gelati M.T., Maggini F., Baldoni L., Cionini P.G. Diversity of Olea genotypes and the origin of cultivated olives. Theor. Appl. Genet. 2002;104:1229–1238. doi: 10.1007/s00122-001-0799-7. [DOI] [PubMed] [Google Scholar]
- Del Rio C. Preliminary agronomical characterization of 131cultivars introduced in the olive germplasm bank of Cordoba in March 1987. Acta Hortic. 1994;356:110–115. [Google Scholar]
- DeLeonardis A., Acetini A., Alfano G., Macciola V., Ranalli G. Isolation of hydroxytyrosol rich extract from Olive leaves (Olea europaea L.) and evaluation of its antioxidant properties and bioactivity. Eur. Food Res. Technol. 2008;226:653–659. [Google Scholar]
- Esmaeili A., Shaykhmoradi F., Naseri R. Comparison of oil content and fatty acid composition of native olive genotypes in different region of Liam. Iran. Int. J. Agri. Crop Sci. 2012;4(8):434–438. [Google Scholar]
- Grati K.N., Ouazzani N., Trigui A. Characterizing isozymes of some Tunisian olive (Olea europaea L.) cultivars. Acta Hortic. 2002;586:137–140. [Google Scholar]
- Guerfel M., Ouni Y., Taamalli A., Boujnah D., Stefanoudaki E., Zarrouk M. Effect of location on virgin olive oils of the two main Tunisian olive cultivars. Eur. J. Lipid Sci. Tech. 2009;111:926–932. [Google Scholar]
- Hegazi E.S., Hegazi A.A., Tawfik A.A., Sayed H.A. Molecular characterization of local and imported olive cultivars grown in Egypt using ISSR technique. J. Hortic. Sci. Orn. Plants. 2012;4(2):148–154. [Google Scholar]
- Hussain J., Khan A., Najeeb ur Rehman L., Hamayun M., Shah T., Nisar M., Bano T., Shinwari Z.K., Lee I.J. Proximate and nutrient analysis of selected vegetable species: a case study of Karak region. Pakistan. Afr. J. Biotechnol. 2009;8(12):2725–2729. [Google Scholar]
- Jaafar R.A., Ridhwan A.A.R., Mahmod N.Z.C., Vasudevan R. Proximate analysis of Dragon Fruit (Hylecereus polyhizus) Am. J. Appl. Sci. 2009;6(7):1341–1346. [Google Scholar]
- Jamshidi S., Jamshidi S. NTSYSpc implementation in molecular biodata analysis (clustering, screening and individual selection). International conference on Environmental and Computer Science. IPCBEE. 2011;19:165–169. [Google Scholar]
- Kjeldahl Johan Z. A new method for the determination of nitrogen in organic bodies. Anal. Chem. 1883;22:366. [Google Scholar]
- Mehr R.S.A., Maassoumi A.A., Saidi A., Osaloo S.K., Nohooji M.G. Morphological cladistic analysis of some bifurcate hairy sections of Astragalus (Fabaceae) in Iran. Turk. J. Bot. 2012;36:434–442. [Google Scholar]
- Milotic A., Setic E., Persuric D., Poljuha D., Sladonja B., Brscic K. Identification and characterization of autochthonous olive varieties in Istria (Croatia) Ann. Ser. His. Nat. 2005;14(2):251–256. [Google Scholar]
- Owusu-Apenten, R.K., 2005. Introduction to Food Chemistry. Hardback Publisher, CRC PR INC., ISBN: 13: 9780849317248. ISBN: 084931724X.
- Pandey M., Abidi A.B., Singh S., Singh R.P. Nutritional evaluation of leafy vegetable paratha. J. Hum. Ecol. 2006;19(2):155–156. [Google Scholar]
- Petridis A., Therios I., Samouris G. Genotypic variation of total phenol and oleuropein concentration and antioxidant activity of 11 Greek olive cultivars (Olea europaea L.) Hortscience. 2012;47(3):339–342. [Google Scholar]
- Prosky P., Asp N.G., Schweizer T.F., Devries J.W., Furuda I. Determination of insoluble, soluble and total dietary fiber in foods, food products: inter laboratory study. J. Assoc. Off. Anal. Chem. 1988;71:1017–1023. [PubMed] [Google Scholar]
- Rohlf F.J. Applied Biotatistics Inc.; Setauket, New York: 1998. (NTSYSpc Numerical Taxonomy and Multivariate Analysis System Version 2.0 User Guide). pp. 37. [Google Scholar]
- Rondanini D.P., Castro D.N., Searles P.S., Rousseaux M.C. Fatty acid profiles of varietal virgin olive oils (Olea europaea L.) from mature orchards in warm arid valleys of Northwestern Argentina (La Rioja) Grasas Aceites. 2011;62(4):399–409. [Google Scholar]
- Sauer J.D. CRC Press; Washington DC: 1993. (Historical Geography of Crop plants – A select roster). pp 106–107. [Google Scholar]
- Youssefi O., Guido F., Daoud D., Mokhtar Z. Effect of cultivar on minor components in Tunisia olive fruits cultivated in microclimate. J. Hortic. For. 2011;3(1):13–20. [Google Scholar]
- Zaher H., Boulouha B., Baaziz M., Sikaoui L., Gaboun F., Udupa S.M. Morphological and genetic diversity in olive (Olea europaea subsp. europaea L.) clones and varieties. Plant Omics J. 2011;4(7):370–376. [Google Scholar]
- Zarrouk W., Baccouri B., Taamalli W., Trigui A., Daoud D., Zarrouk M. Oil fatty acid composition of eighteen Mediterranean olive varieties cultivated under the arid conditions of Boughrara (southern Tunisia) Grasas Aceites. 2009;60(5):498–506. [Google Scholar]