Abstract
Increase in food production viz-a-viz quality of food is important to feed the growing human population to attain food as well as nutritional security. The availability of diverse germplasm of any crop is an important genetic resource to mine the genes that may assist in attaining food as well as nutritional security. Here we used 15 RAPD and 23 SSR markers to elucidate diversity among 51 common bean genotypes mostly landraces collected from the Himalayan region of Jammu and Kashmir, India. We observed that both the markers are highly polymorphic. The discriminatory power of these markers was determined using various parameters like; percent polymorphism, PIC, resolving power and marker index. 15 RAPDs produced 171 polymorphic bands, while 23 SSRs produced 268 polymorphic bands. SSRs showed a higher PIC value (0.300) compared to RAPDs (0.243). Further the resolving power of SSRs was 5.241 compared to 3.86 for RAPDs. However, RAPDs showed a higher marker index (2.69) compared to SSRs (1.279) that may be attributed to their higher multiplex ratio. The dendrograms generated with hierarchical UPGMA cluster analysis grouped genotypes into two main clusters with various degrees of sub clustering within the cluster. Here we observed that both the marker systems showed comparable accuracy in grouping genotypes of common bean according to their area of cultivation. The model based STRUCTURE analysis using 15 RAPD and 23 SSR markers identified a population with 3 sub-populations which corresponds to distance based groupings. High level of genetic diversity was observed within the population. These findings have further implications in common bean breeding as well as conservation programs.
Abbreviations: MI, marker index; PIC, polymorphic information content; PCA, principle component analysis; QTL, quantitative trait loci; RAPD, random amplified polymorphic DNA; Rp, resolving power; SSR, simple sequence repeat; UPGMA, unweighted pair group method with arithmetic averages; RFLP, restriction fragment length polymorphism; PAGE, polyacrylamide gel electrophoresis
Keywords: RAPD, SSR, Population structure, Common bean, Dendrogram, PCA
1. Introduction
For direct human consumption, common bean (Phaseolus vulgaris L.) is the most important legume in the developing world (Broughton et al., 2003). Beans are an inseparable part of the food for millions around the globe, representing a major chunk of dietary protein (Biswas et al., 2010). Beans are also a rich source of essential vitamins and minerals, soluble fiber, starch and phytochemicals, and are also reported to have low fat content (Nyombaire et al., 2007, Svetleva et al., 2006, Beebe et al., 2000). In many regions/countries it provides about 15% of total daily calories and greater than 30% of daily protein intake. Being such an important part of the diet around the world, common bean as a crop is subjected to various improvement programs (Hanai et al., 2010). Edible parts and growing habits of the common bean show a high degree of genetic variation (Biswas et al., 2010). Different molecular markers have been used to study genetic diversity among common bean. RFLP was used in constructing first molecular linkage map of common bean (Adam-Blondon et al., 1994, Nodari et al., 1993; Vallejos et al., 1992). The high density linkage map of common bean was developed using various other markers, mainly RAPD (Freyre et al., 1998), SSR’s or microsatellite (Yu et al., 2000, Blair et al., 2003). SSR markers have also been used to evaluate intra-specific diversity within the genus of Phaseolus (Gaitan-solis ewwwwt al., 2002). Among all the markers, SSR’s have been deployed for population structure studies from time to time in various cereals e.g.: rice (Zhang et al., 2009), maize (Liu et al., 2003), wheat (Liu et al., 2010, Zoric et al., 2012) as well as legume crops. Race structure analysis was done in cultivated Andean and Mesoamerican beans (Díaz and Blair, 2006, Blair et al., 2007). Further, the inferences about population structure of 349 common bean genotypes which includes both cultivated and wild accessions using 26 microsatellite marker was done (Kwak and Gepts, 2009). An effective breeding program essentially requires a good knowledge of the extent and nature of genetic diversity within the crop species. The availability of genetically diverse landraces of a crop is an important genetic resource that can be used for the improvement of that crop. The evaluation of population structure and genetic diversity of germplasm could also provide valuable information for association mapping, allele mining for novel traits and crop breeding.
In the present study we employed two different markers i.e. RAPD and SSR to evaluate the efficiency of these markers in diversity analysis of common bean collected from foot hills of the Himalayan region of Jammu and Kashmir, India. Moreover, we have considered various parameters to elucidate genetic diversity and population structure among these genotypes as detailed in results and discussion.
2. Materials and methods
2.1. Genotypes
Fifty-one genotypes of common bean collected from various unexploited regions of Jammu and Kashmir, India (Zargar et al., 2014), were used in this study.
2.2. DNA extraction
Doyle and Doyle (1987), method with little modifications was followed for extraction of genomic DNA from young leaf tissue of common bean genotypes. The DNA quantity as well as quality was checked by Nanodrop (mySPEC, Wilmington, USA). Isolated high quality DNA was diluted to concentration of 25 ng/μL for further use.
2.3. Molecular analysis
2.3.1. RAPD genotyping
15 RAPD primers synthesized at IDT (Integrated DNA Technologies, Coralville, Iowa, USA) were used for studying polymorphism among 51 common bean genotypes. 25 μL reaction mixture containing 3 μL of template DNA (25 ng/μL), 1X PCR Buffer, 2 mM MgCl2, 0.2 mM of each dNTPs (dTTPs, dGTPs, dCTPs, dATPs), 20 pico molar primer concentration, 1 U Taq DNA polymerase (Taq polymerase from Thermus acquaticus, Sigma Aldrich, USA) was amplified in a 96 well Universal Gradient Thermal Cycler (PEQLAB, Deutschland and Osterrtich, United kingdom). Products were separated on a normal agarose gel along with standard molecular weight marker (100 bp ladder) (Sigma Aldrich, USA). The gel was visually examined under UV and documented using gel documentation system (MiniLumi, Sigma-Svi Bio Solutions Pvt. Ltd. New Delhi, India). The list of RAPD primers used is detailed in Table 1.
Table 1.
Details of RAPD primers with various parameters revealing the discriminatory power of each primer.
| S.No. | Primer | Sequence 5′→3′ |
NB | NPB | NMB | NUB | PPB | PIC 2fi (1-fi) |
MI | Rp |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | OPA-02 | TGC CGA GCT G | 12 | 12 | 0 | 02 | 100 | 0.22 | 2.64 | 3.93 |
| 2 | OPA-03 | AGT CAG CCA C | 12 | 12 | 0 | 03 | 100 | 0.24 | 2.88 | 4.08 |
| 3 | OPA-05 | AGG GGT CTT G | 17 | 17 | 0 | 00 | 100 | 0.21 | 3.57 | 4.44 |
| 4 | OPA-07 | GAA ACGGGTG | 11 | 11 | 0 | 02 | 100 | 0.19 | 2.09 | 3.36 |
| 5 | OPA-09 | GGG TAA CGC C | 14 | 14 | 0 | 02 | 100 | 0.24 | 3.36 | 4.92 |
| 6 | OPA-10 | GTG ATC GCA G | 12 | 12 | 0 | 02 | 100 | 0.27 | 3.24 | 4.56 |
| 7 | OPA-11 | CAA TCG CCG T | 14 | 14 | 0 | 04 | 100 | 0.17 | 2.38 | 3.28 |
| 8 | OPB-10 | CTG CTG GGA C | 07 | 07 | 0 | 00 | 100 | 0.32 | 2.24 | 3.24 |
| 9 | OPC-02 | GTG AGG CGT C | 09 | 09 | 0 | 00 | 100 | 0.23 | 2.07 | 2.96 |
| 10 | OPC-08 | TGG ACC GGT G | 10 | 10 | 0 | 00 | 100 | 0.22 | 2.2 | 2.88 |
| 11 | OPD-07 | TTG GCA CGG G | 10 | 10 | 0 | 03 | 100 | 0.24 | 2.24 | 3.30 |
| 12 | OPD-18 | GAG AGC CAA C | 10 | 10 | 0 | 02 | 100 | 0.30 | 3.00 | 4.86 |
| 13 | OPE-01 | CCC AAG GTC C | 15 | 15 | 0 | 05 | 100 | 0.22 | 3.3 | 4.96 |
| 14 | OPE-02 | GGT GCG GGA A | 08 | 08 | 0 | 01 | 100 | 0.28 | 2.24 | 2.80 |
| 15 | OPE-03 | CCA GAT GCA C | 10 | 10 | 0 | 01 | 100 | 0.30 | 3.00 | 4.36 |
| Average | 11.4 | 11.4 | 00 | 1.8 | 100 | 0.243 | 2.69 | 3.86 |
NB: number of bands, NPB: number of polymorphic bands, NMB: number of monomorphic bands, NUB: number of unique bands, PPB: percentage of polymorphic bands, PIC: polymorphism information content, MI: marker index, Rp: resolving power.
2.3.2. SSR genotyping
23 SSR selected from Yu et al., 2000, Gaitan-Solis et al., 2002, Grisi et al., 2007, Hanai et al., 2010, Córdoba et al., 2010, were used for studying polymorphism among common bean genotypes. Details of SSRs are given in Table 2. DNA concentration of primers was adjusted to 25 ng/μL. PCR amplification was carried out in 96 well Universal Gradient Thermal Cycler (PEQLAB, Deutschland and Osterrtich, United Kingdom) in a 25 μL reaction mixture. The reaction mixture contained 5 μM of each forward and reverse primers, 1 U of Taq polymerase (D1806- Sigma Aldrich, USA), 5 μL of 10X PCR buffer with MgCl2, 2.5 mM of each dNTP (dTTPs, dGTPs, dCTPs, dATPs). Amplifications were performed as follows: Initial denaturation of 1 min at 94 °C, followed by 35 cycles of 94 °C 1 min, 50–55 °C 1.30 min, 72 °C 2 min, and a final extension of 10 min at 72 °C. PCR products were mixed with loading dye (3–4 μL). The amplified products of some primers were resolved on 2.5% metaphor agarose gel and those which could not give clear polymorphic pattern on metaphor agarose were further tested on 8% denaturing PAGE. PCR products resolved on metaphor agarose gel were visually examined under UV and documented using gel documentation system (MiniLumi, Sigma-Svi Bio Solutions Pvt. Ltd. New Delhi, India). However, the PCR products that had resolved on PAGE at constant power (120 W) in 1 X TBE running buffer for 3–4 h were visualized by silver-staining method as described by Bassam et al. (1991). Gels were visually scored and scanned for records. The clear and reproducible alleles amplified by each SSR among 51 genotypes were scored according to their fragment size (bp) corresponding to the 50 bp molecular weight marker (Sigma Aldrich, USA).
Table 2.
Details of SSR primers with various parameters revealing the discriminatory power of each primer.
| S.No. | Primer | Sequence 5′→3′ |
Chromosome number | NB | NPB | NMB | NUB | PPB | PIC 2fi (1-fi) |
MI | Rp |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Pvm097 | F CAAGAGTGAAGGGGCAGTTT R CGGCCAACCACTACTTTTAG |
1 | 09 | 09 | 00 | 00 | 100 | 0.27 | 0.656 | 3.44 |
| 2 | BM156 | F CTTGTTCCACCTCCCATCATAGC R TGCTTGCATCTCAGCCAGAATC |
2 | 11 | 11 | 00 | 00 | 100 | 0.434 | 2.604 | 7.696 |
| 3 | X59469 | F AAACACACAAAAAGTTGGACGCAC R TTCGTGAGGTAGGAGTTTGGTGG |
2 | 04 | 04 | 00 | 02 | 100 | 0.023 | 0.038 | 0.61 |
| 4 | U77935 | F CGTTAGATCCCGCCCAATAGT R CCGTCCAGGAAGAGCGAGC |
2 | 12 | 12 | 00 | 00 | 100 | 0.339 | 1.374 | 5.885 |
| 5 | BM159 | F GGTGCTGTTGCTGCTGTTAT RGGGAGATGTGGTAAGATAATGAAA |
3 | 21 | 21 | 00 | 00 | 100 | 0.407 | 4.963 | 13.5 |
| 6 | X96999 | F AGTCGCCATAGTTGAAATTTAGGTG R TATTAAAACGTGAGCATATGTATCATTC |
3 | 09 | 09 | 00 | 00 | 100 | 0.34 | 0.752 | 4.253 |
| 7 | X57022 | F AAGGATGGGTTCCGTGCTTG R AAGGATGGGTTCCGTGCTTG |
4 | 27 | 27 | 00 | 00 | 100 | 0.324 | 2.414 | 8.75 |
| 8 | X04660 | F TTGATGACGTGGATGCATTGC R AAAGGGCTAGGGAGAGTAAGTTGG |
4 | 07 | 07 | 00 | 0 | 100 | 0.400 | 1.590 | 4.67 |
| 9 | BM155 | F GTTCATGTTTGTTTGACAGTTCA R CAGAAGTTAGTGTTGGTTTGATACA |
5 | 13 | 12 | 01 | 00 | 92.3 | 0.272 | 1.249 | 4.96 |
| 10 | X74919 | F CCGTTGCCTGTATTTCCCCAT R CGTGTGAAGTCATCTGGAGTGGTC |
5 | 14 | 14 | 00 | 0 | 100 | 0.414 | 2.011 | 9.29 |
| 11 | BM158 | F CCGAGCACCGTAACTGAATGC R CGCTCGCTTACTCACTGTACGC |
6 | 18 | 18 | 00 | 01 | 100 | 0.239 | 0.824 | 5.86 |
| 12 | X61293 | F AATCTGCCGAGAGTGGTCCTGCC R GATTGAAATATCAAAGAGAATTGTTAC |
6 | 16 | 16 | 00 | 2 | 100 | 0.181 | 0.695 | 3.76 |
| 13 | PVBR93 | F TGGGGTGAGAGAGAAAGGTG R TACCATAGCAGGCGTTGTTG |
7 | 10 | 10 | 00 | 00 | 100 | 0.29 | 0.904 | 3.96 |
| 14 | BM150 | F CGAACTATTTGATACTCATGTGC R TTGCAGGACAGATAAGTTAGAAGA |
7 | 8 | 8 | 00 | 0 | 100 | 0.283 | 0.776 | 3.125 |
| 15 | PVBR185 | F TGGTAAAGCAAAAACGATGG R GACAGAAGAGTGAGGGTGTGAA |
8 | 07 | 07 | 00 | 01 | 100 | 0.23 | 0.245 | 2.44 |
| 16 | BM151 | F CACAACAAGAAAGACCTCCT R TTATGTATTAGACCACATTACTTCC |
8 | 8 | 8 | 00 | 0 | 100 | 0.403 | 1.769 | 5.134 |
| 17 | PvBR213 | F ACAATGTAGACAGCGCAGCA R GCTCTTTCTCCTCCCATCCT |
9 | 8 | 8 | 00 | 4 | 100 | 0.152 | 0.252 | 1.577 |
| 18 | X80051 | F GTTAAATTATACGAGGTTAGCCTAAATC R CATTCCCTTCACACATTCACCG |
9 | 9 | 9 | 00 | 0 | 100 | 0.324 | 0.323 | 3.927 |
| 19 | BM154 | F TCTTGCGACCGAGCTTCTCC R CTGAATCTGAGGAACGATGACCAG |
9 | 09 | 09 | 00 | 00 | 100 | 0.447 | 1.937 | 6.501 |
| 20 | BM157 | F ACTTAACAAGGAATAGCCACACA R GTTAATTGTTTCCAATATCAACCTG |
10 | 20 | 20 | 00 | 05 | 100 | 0.217 | 0.442 | 6 |
| 21 | BMb152 | F ACGCAGAGAAATCTCCAATA R CCTTCCATGATTTGTTGTTT |
10 | 13 | 13 | 00 | 0 | 100 | 0.483 | 2.423 | 10.823 |
| 22 | BMb654 | F CGCATCGATCAAAGATAGTC R CTCTTTCCCAACAAATGAAG |
11 | 06 | 06 | 00 | 00 | 100 | 0.279 | 0.689 | 2.65 |
| 23 | M75856 | F GGGAGGGTAGGGAAGCAGTG R GCGAACCACGTTCATGAATGA |
11 | 9 | 9 | 00 | 2 | 100 | 0.163 | 0.501 | 1.732 |
| Average | 11.65 | 11.65 | 0.04 | 0.73 | 99.6 | 0.300 | 1.279 | 5.241 |
NB: number of bands, NPB: number of polymorphic bands, NMB: number of monomorphic bands, NUB: number of unique bands, PPB: percentage of polymorphic bands, PIC: polymorphism information content, MI: marker index, Rp: resolving power.
2.4. Data analysis
The profile developed by each marker was scored (1) for the presence and (0) for the absence of a band for each genotype. In order to compare the efficiency of these two marker systems in genotype identification, differentiation and diversity analysis, we considered the following parameters for each assay unit (U).
-
1.
Number of polymorphic bands (np);
-
2.
Number of monomorphic bands (nnp);
-
3.
Average number of polymorphic bands per unit assay (np/U);
-
4.
Number of loci (L): number of loci in case of RAPD is equal to the total number of bands (np + nnp) obtained;
-
5.
Number of loci per assay unit: nu = L/U;
-
6.
Fraction of polymorphic loci (β) according to Powell et al. (1996): β = np/np + nnp;
-
7.
Effective multiplex ratio (E) according to Powell et al. (1996): E = nuβ;
-
8.
Polymorphic information content (PIC) according to Powell et al. (1996): PIC = 2fi (1-fi);
-
9.
Marker index (MI) according to Powell et al. (1996): MI = PIC × β × α;
-
10.
Resolving power (RP) according to Prevost and Wilkinson (1999): RP = ΣIb
Scored data were used for the estimation of Jaccard’s similarity coefficient using NTSYS-pc version 2.02e (Rohlf, 1998) package to compute pair-wise Jaccard’s similarity coefficient (Jaccard, 1908) and this similarity matrix was used in cluster analysis using the unweighted pair-group method with arithmetic averages (UPGMA) and sequential, agglomerative, hierarchical and nested (SAHN) clustering algorithm to obtain dendrogram.
Model based cluster analysis was performed to infer genetic structure and to define the number of clusters in the data set using the software STRUCTURE version 2.3.4 (Pritchard et al., 2000). The number of presumed populations (K) was set from 1 to 10, and the analysis was repeated 2 times. For each run the burn-in and MCMC were set to 50,000 each and iterations were set to 5. The run with maximum likelihood was used to assign individual genotypes into groups. Within a group, genotypes with inferred ancestry based on probability values ⩾80% were assigned to a different group, and those with <80% were treated as “admixture”, i.e., these genotypes seem to have a mixed ancestry from parents belonging to different geographical origins or gene pools. The expected heterozygosity (gene diversity) and population differentiation (Fst) between individuals in a sub-population was also worked out using STRUCTURE programme.
3. Results and discussion
3.1. Allele diversity in the common bean using two different marker systems
Both the marker techniques (RAPD and SSR) proved to be highly effective in discriminating the 51 genotypes. Results obtained are summarized in Table 1, Table 2, Table 3. 15 RAPD and 23 SSR primers used in the present study amplified 171 and 268 polymorphic bands for RAPD and SSR respectively. An average number of 11.40 polymorphic bands per assay unit were identified for RAPD, whereas in SSR it was 11.65 (Table 3). The utility of a given marker is a balance between the level of polymorphism it can detect, and its capacity to identify multiple polymorphisms (Powell et al., 1996). Marker index is a feature of a marker which elucidates the discriminatory power of a marker and therefore it was calculated for all the markers. Due to high multiplex ratio component (11.4) for RAPD, higher marker index value was observed for RAPD (2.69) in comparison to SSR (1.279) (Table 3). Maras et al. (2008) observed a higher multiplex ratio for AFLP (11.20) than for SSR (1.00) in 29 common bean accessions. In another study a higher multiplex ratio of 5.19 for RAPD was observed as compared to SSRs (1.00) in 32 olive cultivars (Belaj et al., 2002). For RAPD markers only two alleles per locus are considered, however for SSR an average of 11.65 alleles per locus, ranging from 4 (X59469) to 27 (X57022) was observed. The average number of alleles per locus for SSR’s observed in the present study is higher than earlier studies carried out by Maras et al. (2008), where they found an average of 7.14 alleles per locus for 14 SSR loci scored for 29 common bean accessions. Since a higher number of genotypes as well as SSR primers were used in this study and that can be the reason for higher number of alleles per locus observed in case of SSR’s. PIC is an important feature of a primer which indicates its potential to differentiate various individuals. An average PIC of 0.243 was observed for RAPD where as it was 0.300 for SSR markers (Table 1, Table 2). Highest PIC was observed for primers OPD-18 and OPE-03 (0.300) in RAPD assay (Table 1) while the highest PIC was observed for primer BM154 (0.447) in SSR assay (Table 2). Ahmed et al. (2012) also observed a higher PIC value in SSR (0.39) than in RAPD (0.250) in genetic diversity estimation of 82 walnut cultivars. Further resolving power/discriminatory power of a marker, which indicates the discriminatory potential of the primer to distinguish the genotypes or individuals, was estimated for each primer. An average resolving power of 3.86 was observed for RAPD whereas for SSR it was 5.241. Highest resolving power of 4.96 was observed for primer OPE-01 among RAPD markers while as highest resolving power of 13.5 was observed for primer BM159 among the SSR markers. Higher resolving power for SSR markers can also be attributed to the fact that SSR markers were resolved on low melting agarose (metaphor) and PAGE, both of which have higher resolving capacity than normal agarose.
Table 3.
Levels of polymorphism and comparison of the discriminating power of RAPD and SSR markers.
| Indexes with their abbreviations | Marker systems |
||
|---|---|---|---|
| RAPD | SSR | ||
| Number of assay units | U | 15 | 23 |
| Number of polymorphic bands | np | 171 | 268 |
| Number of monomorphic bands | nnp | 0 | 1 |
| Average number of polymorphic bands/assay unit | np/U | 11.4 | 11.65 |
| Number of loci | L | 171 | 23 |
| Number of loci/assay unit | nu | 11.4 | 1 |
| Average number of alleles per locus | nav | 2 | 11.65 |
| Fraction of polymorphic loci | β | 1 | 0.99 |
| Effective multiplex ratio | E | 11.4 | 0.99 |
| Marker index | MI | 2.69 | 1.279 |
| Expected heterozygosity | He | 0.0878 | 0.147 |
3.2. Genetic relationship among common bean genotypes
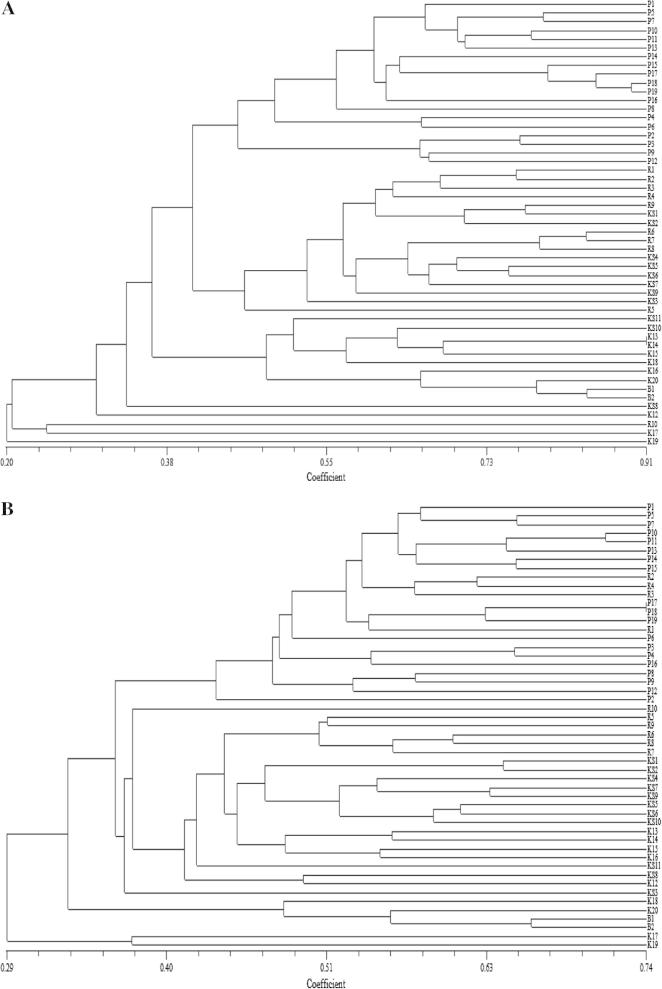
Both the marker systems showed a high degree of similarity in the topology of their respective dendrograms. Although some differences in positioning of some genotypes was observed. However, all the dendrograms reflected similar pattern of relationship among most of the genotypes, depending upon the area of their cultivation (Fig. 1A-C).
Figure 1.
(A) Cluster tree derived by SHAHN method based on 15 RAPD markers among 51 genotypes of common bean, (B) Cluster tree derived by SHAHN method based on 23 SSR markers among 51 genotypes of common bean, (C) Cluster tree derived by SHAHN method based on 15 RAPD and 23 SSR markers among 51 genotypes of common bean.
In order to find out the genetic relationship among the common bean genotypes, analysis was done separately as well as in combination for RAPD and SSR data sets. The Jaccard’s similarity coefficient for RAPD based diversity analysis ranged from 0.20 to 0.91 (Fig. 1A), whereas for SSR it ranged from 0.29 to 0.74 (Fig. 1B). Further, the Jaccard’s similarity coefficient ranged from 0.25 to 0.79 for the combined RAPD and SSR based data sets (Fig. 1C). The dendrogram generated from RAPD data grouped genotypes in two main clusters as represented in Fig. 1A, in which K-19 was totally distinguished from remaining other genotypes that had grouped together. The similarity coefficients of the common bean genotypes based on 15 RAPD ranged from to 0.185 to 0.905. Among the 51 pair-wise combinations of genotypes, K-13 and K-14 showed the highest similarity index (0.905), while the genotypes P1 and K19 showed the lowest (0.185). As such RAPD markers generated a mean similarity index of 0.545 among 51 diverse common bean genotypes.
The dendrogram obtained with SSR markers as represented in Fig. 1B, also divided the genotypes into two main clusters. Cluster-I represented only two genotypes (K-17 and K-19) whereas rest of the genotypes were grouped in the Cluster-II. Cluster-II further divided the genotypes into two sub clusters. Most of the genotypes from Poonch, along with a few genotypes collected from Rajouri were grouped together. Whereas most of the genotypes collected from Kashmir along with some from Rajouri gathered together. Certainly the genotypes from Rajouri and Bhaderwah formed separate sub-clusters within these groups. SSR based similarity coefficient of the common bean genotypes ranged from 0.260 to 0.738. Of the 51 pair wise combinations of genotypes, P-17 and P-18 showed the highest similarity index (0.738), while the genotypes P1 and K19 showed the lowest similarity index (0.260).
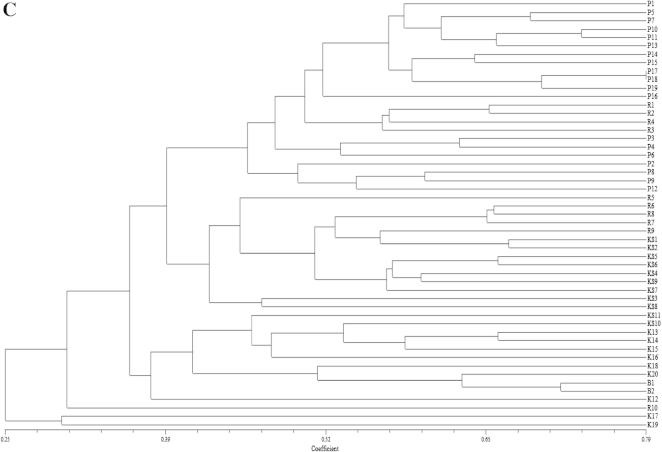
The dendrogram generated from the combined RAPD and SSR based data sets exhibited a pattern almost similar to that obtained from the SSR data as represented in Fig. 1C. In this dendrogram K-17 and K-19 clustered together as observed in case of SSR based dendrogram. Here the similarity coefficients among 51 genotypes ranged from 0.232 to 0.788. Among all the pair-wise combinations, P-17 and P-18 showed the highest similarity index (0.788), while the genotypes P1 and K19 showed the lowest similarity index (0.232). In all the three data sets P-1 and K-19 were farthest from one another and as such they showed lowest similarity coefficient values as revealed in Fig. 1A-C. Principal component analysis (PCA) of 51 common bean genotypes using 15 RAPD and 23 SSR markers revealed similar results as observed by UPGMA based clustering (Fig. 2). Belaj et al. (2002) got similar results regarding the dendrogram topologies in diversity studies of 32 olive cultivars using RAPD, AFLP and SSR markers. Combination of the data sets of RAPD, AFLP and SSR revealed a better representation of the relationship for most of the olive cultivars as represented in the dendrogram, according to the geographic area of diffusion.
Figure 2.
PCA analysis based results of 51 common bean genotypes using 15 RAPD and 23 SSR primers.
3.3. Population structure and relationship among 51 genotypes
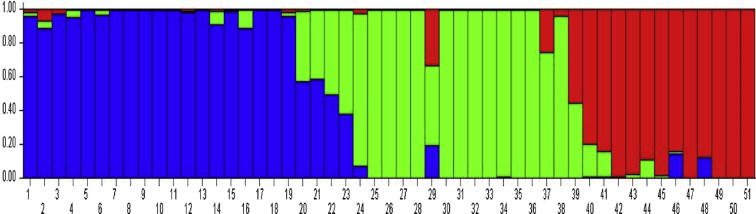
Further STRUCTURE analysis was carried out to observe the number of populations that may be generated from 51 genotypes using 15 RAPD and 23 SSR markers. Here we acquired three populations with slight mixing of genotypes as represented in Fig. 3 and Table 4. Since the locations of collection (Rajouri, Poonch and Kashmir) are connected to each other, as such this may be a reason of having admixture among 3 distinguished populations. Moreover, population structure analysis confirmed the grouping of the genotypes, as observed by PCA and UPGMA clustering analyses. The STRUCTURE simulations were carried out by varying K from 1 to 10 with 10 run for each K using all 51 genotypes. In this analysis, the two populations initially separated at K = 2 and then further subgroups were formed at K = 3. The admixtures obtained in three distinguished populations are an indication of sub grouping of genotypes as evident from UPGMA based analysis. The sub grouping can be owed to geographic structuring or adaptation in different seasons. This confirms the classification of 51 common bean genotypes into three distinct population groups with high resolution population structure. Using this approach, 51 accessions were assigned to the corresponding A-C sub-populations, representing 37.25% (19), 25.49% (13) and 21.56% (11) of the total germplasm analyzed. Of the 51 genotypes, only 13.7% (7) showed admixtures (membership probability <0.8, Table 4). Similar results were observed by Sharma and coworkers while analyzing genetic diversity of two Indian common bean germplasm collections based on morphological and microsatellite markers (Sharma et al., 2013). A total of 149 genotypes were evaluated using 24 microsatellites and initial separation of the gene pools was observed at K = 2 and the further sub-groups were formed at K = 3, which indicated some level of sub-grouping in each gene pool and they also found it to be in tone with UPGMA analysis. The expected heterozygosity which measures the probability that two randomly chosen individual will be different (heterozygous) at a given locus ranged from 0.219 in the first sub-population to 0.282 in the third sub-population with an average of 0.2379 (Table 5). Similarly population differentiation measurements (Fst) which is the summary of genetic differentiation among groups, and on the basis of which two different clusters or populations corresponding closely are assigned to different populations ranged from 0.2059 (in the 3rd sub-population) to 0.4047 (in the 1st sub-population) with an average of 0.3301 (Table 5, Table 6), which is relatively high confirming the separation of all the sub-populations and their diversity in RAPD and SSR alleles. Blair et al. (2012), analyzed 108 common bean genotypes using 36 fluorescently labeled SSRs and they also observed a high Fst value (0.203) for genetic differentiation between all the five populations. Yet in our study we have obtained a higher Fst value and it may be due to the use of different types of markers in our study.
Figure 3.
Graphical representation of population structure. Each common bean genotype is shown by a vertical line representing membership of subgroup 1 (blue), subgroup 2 (green), and subgroup 3 (red). Genotypes are arranged as per estimated membership coefficients (q) in K = 3 clusters.
Table 4.
Assignment of individuals to the sub populations (K) based on probability.
| Code | Genotype | K-1 | K-2 | K-3 | Assignment to sub-populations |
|---|---|---|---|---|---|
| 1 | P1 | 0.967 | 0.016 | 0.017 | 1 |
| 2 | P2 | 0.896 | 0.034 | 0.070 | 1 |
| 3 | P3 | 0.975 | 0.003 | 0.022 | 1 |
| 4 | P4 | 0.950 | 0.046 | 0.005 | 1 |
| 5 | P5 | 0.999 | 0.001 | 0.000 | 1 |
| 6 | P6 | 0.971 | 0.024 | 0.005 | 1 |
| 7 | P7 | 0.999 | 0.000 | 0.001 | 1 |
| 8 | P8 | 0.998 | 0.001 | 0.001 | 1 |
| 9 | P9 | 0.997 | 0.001 | 0.002 | 1 |
| 10 | P10 | 0.999 | 0.001 | 0.000 | 1 |
| 11 | P11 | 0.998 | 0.001 | 0.001 | 1 |
| 12 | P12 | 0.985 | 0.006 | 0.009 | 1 |
| 13 | P13 | 0.998 | 0.001 | 0.001 | 1 |
| 14 | P14 | 0.909 | 0.083 | 0.008 | 1 |
| 15 | P15 | 0.990 | 0.009 | 0.001 | 1 |
| 16 | P16 | 0.893 | 0.104 | 0.003 | 1 |
| 17 | P17 | 0.997 | 0.002 | 0.001 | 1 |
| 18 | P18 | 0.998 | 0.002 | 0.001 | 1 |
| 19 | P19 | 0.970 | 0.015 | 0.015 | 1 |
| 20 | R1 | 0.577 | 0.413 | 0.010 | ADMIXTURE |
| 21 | R2 | 0.587 | 0.412 | 0.001 | ADMIXTURE |
| 22 | R3 | 0.497 | 0.501 | 0.002 | ADMIXTURE |
| 23 | R4 | 0.389 | 0.608 | 0.003 | ADMIXTURE |
| 24 | R5 | 0.091 | 0.891 | 0.018 | 2 |
| 25 | R6 | 0.004 | 0.995 | 0.001 | 2 |
| 26 | R7 | 0.001 | 0.998 | 0.001 | 2 |
| 27 | R8 | 0.001 | 0.998 | 0.001 | 2 |
| 28 | R9 | 0.002 | 0.998 | 0.001 | 2 |
| 29 | R10 | 0.198 | 0.472 | 0.330 | ADMIXTURE |
| 30 | KS1 | 0.003 | 0.997 | 0.001 | 2 |
| 31 | KS2 | 0.001 | 0.996 | 0.003 | 2 |
| 32 | KS3 | 0.007 | 0.990 | 0.003 | 2 |
| 33 | KS4 | 0.002 | 0.997 | 0.001 | 2 |
| 34 | KS5 | 0.018 | 0.981 | 0.001 | 2 |
| 35 | KS6 | 0.002 | 0.996 | 0.001 | 2 |
| 36 | KS7 | 0.003 | 0.995 | 0.002 | 2 |
| 37 | KS8 | 0.007 | 0.731 | 0.262 | ADMIXTURE |
| 38 | KS9 | 0.003 | 0.965 | 0.032 | 2 |
| 39 | KS10 | 0.002 | 0.447 | 0.551 | ADMIXTURE |
| 40 | KS11 | 0.010 | 0.190 | 0.800 | 3 |
| 41 | K12 | 0.013 | 0.149 | 0.838 | 3 |
| 42 | K13 | 0.001 | 0.010 | 0.989 | 3 |
| 43 | K14 | 0.001 | 0.031 | 0.968 | 3 |
| 44 | K15 | 0.002 | 0.114 | 0.884 | 3 |
| 45 | K16 | 0.003 | 0.013 | 0.984 | 3 |
| 46 | K17 | 0.148 | 0.008 | 0.844 | 3 |
| 47 | K18 | 0.002 | 0.003 | 0.995 | 3 |
| 48 | K19 | 0.122 | 0.003 | 0.875 | 3 |
| 49 | K20 | 0.003 | 0.001 | 0.996 | 3 |
| 50 | B1 | 0.003 | 0.001 | 0.996 | 3 |
| 51 | B2 | 0.001 | 0.001 | 0.998 | 3 |
Table 5.
Heterozygosity and Fst value calculated for 3 common bean sub-populations.
| Sub-population (K) | Expected heterozygosity | Fst value |
|---|---|---|
| 1 | 0.2192 | 0.4047 |
| 2 | 0.2124 | 0.3799 |
| 3 | 0.2821 | 0.2059 |
| Average | 0.2379 | 0.3301 |
Table 6.
Genetic differentiation based on Fst values between three common bean sub-populations identified by population structure analysis.
| Pop A | Pop B | Pop C | |
|---|---|---|---|
| Pop A | – | 0.0969 | 0.1028 |
| Pop B | 0.0969 | – | 0.0737 |
| Pop C | 0.1028 | 0.0737 | – |
4. Conclusion
Both RAPD and SSR marker techniques have provided useful information regarding the level of polymorphism in common bean. Thus they have a higher utility in characterizing the common bean genotypes. RAPD based analysis have the limitation of reliability and transferability (Jones et al., 1997). But if a standard protocol is followed the reliability of RAPD data can become high. Both the marker systems have comparable accuracy in grouping genotypes according to their origin of cultivation. And this has a high significance with respect to the management of germplasm from different geographic locations (Singh et al., 1991). However, it is worth to note here that SSRs proved to be better by showing higher values for most of the parameters that determine the potential of markers in diversity analysis. Further, the information obtained from the population structure analysis will be useful in carrying out association mapping in common bean for various traits. All the observations made in this study will provide valuable evidence for decision making in choosing of markers for future work, characterization of germplasm, breeding and common bean germplasm management.
Conflict of interest
Authors have no conflict of interest.
Submission declaration
The work described has neither been published nor is under consideration for publication in any other journal, and if accepted, it will not be published anywhere in this form or any other form.
Contribution of authors
SF, RM, AB and AS have contributed equally in conducting the experiment. SMZ has designed the experiment and prepared the manuscript along with SF. SMZ and SF did data analysis.
Acknowledgements
SMZ is grateful to SERB, DST New Delhi for financial support of this work (Project sanction order No. SR/FT/LS-27/2011). SF is thankful to Mr. VK Yadav for helping in data analysis.
Footnotes
Peer review under responsibility of King Saud University.
References
- Adam-Blondon A.F., Sevignac M., Bannnerot H., Dron M. SCAR, RAPD and RFLP markers linked to a dominant gene (are) conferring resistance to anthracnose in common bean. Theor. Appl. Genet. 1994;88:865–870. doi: 10.1007/BF01253998. [DOI] [PubMed] [Google Scholar]
- Ahmed N., Mir J.I., Mir R.R., Rather N.A., Rashid R., Wani S.H., Shafi W., Mir H., Sheikh M.A. SSR and RAPD analysis of genetic diversity in walnut (Juglansregia L.) genotypes from Jammu and Kashmir, India. Physiol. Mol. Biol. Plants. 2012;18(2):149–160. doi: 10.1007/s12298-012-0104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassam B.J., Caetano-Anolles G., Gresshoffer P.M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Annal Biochem. 1991;196(1):80–83. doi: 10.1016/0003-2697(91)90120-i. [DOI] [PubMed] [Google Scholar]
- Beebe S., Gonzalez A.V., Rengifo J. Research on trace minerals in the common bean. Food Nutrition Bulletin. 2000;21:387–391. [Google Scholar]
- Belaj A., Satovic Z., Rallo L., Trujillo Genetic diversity and relationship in olive (Olea europaea L.) germplasm collections as determined by randomly amplified polymorphic DNA. Theor. Appl. Genet. 2002;105(4):638–644. doi: 10.1007/s00122-002-0981-6. [DOI] [PubMed] [Google Scholar]
- Biswas M.S., Hassan J., Hossain M.M. Assessment of genetic diversity in French bean (Phaseolus vulgaris L.) based on RAPD marker. Afr. J. Biotechnol. 2010;9(32):5073–5077. [Google Scholar]
- Blair M.W., Diaz J.M., Hidalgo R., Diaz L.M., Duque M.C. Microsatellite characterization of Andean races of common bean (Phaseolus vulgaris L.) Theor. Appl. Genet. 2007;116:29–43. doi: 10.1007/s00122-007-0644-8. [DOI] [PubMed] [Google Scholar]
- Blair M.W., Perzada F., Buendia H.F., Gaitan–Solis E., Beebe S.E., Gepts P., Tohme J. Development of a genome wide anchored microsatellite map for common bean (Phaseolus vulgaris L.) Theor. Appl. Genet. 2003;107:1362–1374. doi: 10.1007/s00122-003-1398-6. [DOI] [PubMed] [Google Scholar]
- Blair M.W., Soler A., Corté s A.J. Diversification and population structure in common beans (Phaseolus vulgaris L.) PLoS ONE. 2012;7(11):e49488. doi: 10.1371/journal.pone.0049488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton W.J., Hernandez G., Blair M., Beebe S., Gepts P., Vanderleyden J. Beans (Phaesolus spp.) – model food legumes. Plant Soil. 2003;252:55–128. [Google Scholar]
- Córdoba J.M., Chavarro C., Schlueter J.A., Jackson S.A., Blair M.W. Integration of physical and genetic maps of common bean through BAC-derived microsatellite markers. BMC Genomics. 2010;11:436. doi: 10.1186/1471-2164-11-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz L.M., Blair M.W. Race structure within the Mesoamerican gene pool of common bean (Phaseolus vulgaris L.) as determined by microsatellite markers. Theor. Appl. Genet. 2006;114:143–154. doi: 10.1007/s00122-006-0417-9. [DOI] [PubMed] [Google Scholar]
- Doyle J.J., Doyle J.L. A rapid DNA isolation procedure to small amounts of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- Freyre R., Skroch W.P., Geffroy V., Adam-Blondon A.F., Shirmohamadali A., Johnson W.C., Llaca V., Nodari R.O., Pereira P.A., Tsai S.M., Tohme J., Dron M., Nienhuis J., Vallejos C.E., Gepts P. Towards an integrated linkage map of common bean. 4. Development of a core linkage map and alignment of RFLP maps. Theor. Appl. Genet. 1998;97:847–856. [Google Scholar]
- Gaitan-Solis E., Duque M.C., Edwards K.J., Tohme J. Microsatellite repeats in common bean (Phaseolus vulgaris): isolation, characterization, and cross-species amplification in Phaseolus ssp. Crop Sci. 2002;42:2128–2136. [Google Scholar]
- Grisi M.C.M., Blair M.W., Gepts P., Brondani C., Pereira P.A.A., Brondadi R.P.V. Genetic mapping of a new set of microsatellite markers in a reference common bean (Phaseolus vulgaris) population BAT93 x Jalo EEP558. Genet. Mol. Res. 2007;3:691–706. [PubMed] [Google Scholar]
- Hanai L.R., Santini L., Camargo L.E.A., Fungaro M.H.P., Gepts P., Tsai S.M., Vieira M.L.C. Extension of the core map of common bean with EST-SSR, RGA, AFLP and putative functional markers. Mol. Breed. 2010;25(1):25–45. doi: 10.1007/s11032-009-9306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaccard P. Nouvelles researches sur la distribution aflorale. Bulletin Societe Vaudoise des Sciences Naturelle. 1908;44:223–270. [Google Scholar]
- Jones C.J., Edwards K.J., Castaglione S., Winfield M.O., Sala F., van deWiel C., Bredemeijer G., Vosman B., Matthes M., Daly A., Brettschneider R., Bettini P., Buiatti M., Maestri E., Malcevschi A., Marmiroli N., Aert R., Volckaert G., Rueda J., Linacero R., Vazquez A., Karp A. Reproducibility testing of RAPD, AFLP and SSR markers in plants by a network of European laboratories. Mol. Breed. 1997;3:381–390. [Google Scholar]
- Kwak M., Gepts P. Structure of genetic diversity in the two major gene pools of common bean (Phaseolus vulgaris L., Fabaceae) Theor. Appl. Genet. 2009;118:979–992. doi: 10.1007/s00122-008-0955-4. [DOI] [PubMed] [Google Scholar]
- Liu K., Goodman M., Muse S., Smith J.S., Buckler E., Doebley J. Genetic structure and diversity among maize inbred lines as inferred from DNA microsatellites. Genetics. 2003;165:2117–2128. doi: 10.1093/genetics/165.4.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang L., Yao J., Zheng J., Zhao C. Association mapping of six agronomic traits on chromosome 4A of wheat (Triticum aestivum L.) Mol. Plant Breeding. 2010;1(5):12. [Google Scholar]
- Maras M., Suštar-Vozlič J., Javornik B., Meglič V. The efficiency of AFLP and SSR markers in genetic diversity estimation and gene pool classification of common bean (Phaseolus vulgaris L.) Acta Agriculturae Slovenica. 2008;91:87–96. [Google Scholar]
- Nodari R.O., Tsai S.M., Guzmán P., Gilbertson R.L., Gepts P. Towards an integrated linkage map of common bean mapping genetic factors controlling host-bacteria interactions. Genetics. 1993;134:341–350. doi: 10.1093/genetics/134.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyombaire G., Siddiq M., Dolan K. Effect of soaking and cooking on the oligosaccharides and lectins of red kidney beans (Phaseolus vulgaris L.) Bean Improv. Coop. Ann. Rep. 2007;50:31–32. [Google Scholar]
- Powell W., Morganate M., Andre C., Hanafey M., Vogel J., Tingey S., Rafalski A. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol. Breed. 1996;2:225–238. [Google Scholar]
- Prevost A., Wilkinson M.J. A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor. Appl. Genet. 1999;98(1):107–112. [Google Scholar]
- Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multi locus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlf, F.J., 1998. NTSYS- pc. Numerical taxonomy and multivariate analysis system. Applied Biostatistics. NewYork.
- Sharma P.N., Díaz L.M., Blair M.W. Genetic diversity of two Indian common bean germplasm collections based on morphological and microsatellite markers. Plant Genetic Resour. 2013;11(2):121–130. [Google Scholar]
- Singh S.P., Gepts P., Debouck D.G. Races of common bean (Phaseolus vulgaris, Fabaceae) Econ. Bot. 1991;45(3):379–396. [Google Scholar]
- Svetleva D., Pereira G., Carlier J., Cabrita L., Leitao J., Genchev D. Molecular characterization of Phaseolus vulgaris L. genotypes included in Bulgarian collection by ISSR and AFLP analyses. Sci. Hortic. 2006;109:198–206. [Google Scholar]
- Vallejos E.C., Sakiyama N.S., Chase C.D. A molecular marker based linkage map of Phaseolus vulgaris L. Genetics. 1992;131:737–740. doi: 10.1093/genetics/131.3.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Park S.J., Poysa V., Gepts P. Integration of simple sequence repeat (SSR) markers into a molecular linkage map of common bean (Phaseolus vulgaris L.) Heredity. 2000;91:429–434. doi: 10.1093/jhered/91.6.429. [DOI] [PubMed] [Google Scholar]
- Zargar S.M., Sharma A., Sadhu A., Agrawal G.K., Rakwal R. Exploring genetic diversity in common bean from unexploited regions of Jammu & Kashmir-India. Mol. Plant Breed. 2014;5(2):5–9. [Google Scholar]
- Zhang D., Zhang H., Wang M., Sun J., Qi Y., Wang F., Wei X., Han L., Wang X., Li Z. Genetic structure and differentiation of Oryza sativa L. in China revealed by microsatellites. Theor. Appl. Genet. 2009;119:1105–1117. doi: 10.1007/s00122-009-1112-4. [DOI] [PubMed] [Google Scholar]
- Zoric M., Dejan D., Kobiljski B., Quarrie S., Barnes J. Population structure in a wheat core collection and genomic loci associated with yield under contrasting environments. Genetica. 2012;140:259–275. doi: 10.1007/s10709-012-9677-2. [DOI] [PubMed] [Google Scholar]