Abstract
Ethylene-responsive factors (ERF) are usually considered to play diverse roles in plant response to biotic and abiotic stresses. In this study, an ERF gene CaPTI1 was isolated from pepper transcriptome database. CaPTI1 contains an open reading frame (ORF) of 543 bp, which encodes a putative polypeptide of 180 amino acids with a theoretical molecular weight of 20.30 kDa. Results of expression profile showed that CaPTI1 had a highest expression level in roots and this gene could not only response to the infection of Phytophthora capsici and the stresses of cold and drought, but also be induced by the signaling molecule (salicylic acid, Methyl Jasmonate, Ethephon, and hydogen peroxide). Furthermore, virus-induce gene silencing (VIGS) of CaPTI1 in pepper weakened the defense response significantly by reducing the expression of defense related genes CaPR1, CaDEF1 and CaSAR82 and also the root activity. These results suggested that CaPTI1 is involved in the regulation of defense response to P. capsici in pepper.
Keywords: pepper, ethylene-responsive factor (ERF), Phytophtora capsici, relative expression, virus-induce gene silencing (VIGS)
Introduction
Pepper (Capsicum annuum L.) is an agriculturally important vegetable crop worldwide. While the production of pepper is severely threatened by a variety of diseases, especially the soil borne disease Phytophthora blight which is caused by Phytophthora capsici (Hausbeck and Lamour, 2004; Granke et al., 2012). P. capsici could not only attack pepper plants, but also other crops such as cucurbits, tomato, eggplant, snap and lima beans (Lamour et al., 2012). P. capsici could affect the plant at any stage of development and cause damping-off, seedling blight, foliar blight and wilting follow by plant death. Infection on mature plants materializes as dark, rapidly expanding, water-soaked lesions (Baysal et al., 2005). Chemical and biological control are the most common strategies used to prevent the spreading of P. capsici (Oelke et al., 2003). Several fungicides have been reported to be effective against P. capsici on vegetable crops (Matheron and Porchas, 2000; Babadoost and Islam, 2003; Kousik et al., 2011). Although many studies have revealed that resistance to P. capsici is polygenic and could be controlled by quantitative trait loci (QTL) (Thabuis et al., 2003; Kim et al., 2008; Rehrig et al., 2014), little is known about pepper–P. capsici interactions at the molecular and genetic levels.
Plants have evolved many sophisticated defense mechanisms by activating multiple defense pathways in order to protect themselves from pathogen infection and cope with the pathogen invasion. salicylic acid (SA), Jasmonate (JA), and Ethylene (ET) are frequently induced in response to infection with various pathogens. The balance of these hormones is dependent on the recognized pathogen and plays a vital role in fine tuning appropriate defense responses (Dong, 1998; Dangl and Jones, 2001). SA typically mediates basal defense to biotrophic pathogens and is essential for the rapid activation of local and systemic resistance (Loake and Grant, 2007; Vlot et al., 2009). While JA controls defense reactions to necrotrophs. Together with ET, JA generally regulates induced systemic resistance (ISR) (Glazebrook, 2005; Alazem and Lin, 2015). ET may regulate pathogenesis-related (PR) genes expression through ERFs, and ERFs were supposed to act as either transcriptional activators or repressors of GCC-box mediated gene expression and be involved in the regulation of plant defense mechanisms (Fujimoto et al., 2000; Chakravarthy et al., 2003; Gutterson and Reuber, 2004; Yang et al., 2005; Alazem and Lin, 2015).
ERFs were firstly isolated in tobacco and found to regulate the expression of PR genes through binding specifically to the GCC-box (Ohmetakagi and Shinshi, 1995). Then the ERFs in other plants were constantly identified, such as AtERFs, StERFs and LeERFs (Zhou et al., 1997; Fujimoto et al., 2000; Wang et al., 2015). Fujimoto et al. (2000) isolated five AtERFs which respond to extracellular signals and modulate GCC box-mediated gene expression. Then Brown et al. (2003) found that AtERF2 possibly play important role in regulating JA-dependent defense response via interaction with the GCC-box. The ERFs in tomato, Pti4, Pti5, and Pti6 which were interacted with the resistance (R) gene Pto kinase were identified by yeast two-hybrid assays (Zhou et al., 1997). Further research found these ERFs could induced by Pseudomonas bacterium and also specifically recognize and bind to GCC-box of PR proteins (Thara et al., 1999). Moreover, Pti4, Pti5, and Pti6 were suggested to indirectly regulate the SA response through interaction with other TFs, and activate the expression of PR genes (Gu et al., 2002; Chakravarthy et al., 2003). The over-expression of Pti5 in tomato demonstrated the positive role of Pti5 in defense genes regulation and disease resistance (He et al., 2001). Recently research revealed that Pti5 contributes to potato aphid resistance in tomato independent of ethylene signaling (Wu et al., 2015). Besides, NtTsi1 and CaERFLP1 were also isolated and demonstrated to bind to the GCC-box (Park et al., 2001; Lee et al., 2004). Over-expression of NtTsi1 in transgenic hot pepper plants induced constitutive expression of several PR genes in the absence of stress or pathogen treatment and enhanced the disease resistance to Xanthomonas campestris pv. vesicatoria and also the P. capsici (Shin et al., 2002).
Most of researches suggest that ERFs were involved in disease resistance responses as activators. There were also some ERFs whose protein contained an ERF-associated amphiphilic repression (EAR) motif in the C-terminal as repressors, such as NtERF3, AtERF3, AtERF4 and StERF3 (Ohta et al., 2001; McGrath et al., 2005; Tian et al., 2015). But recently Dong et al. (2015) found that the GmERF5 which has an EAR motif could not only significantly induced by P. sojae, ETH, ABA and SA, but also bind to the GCC-box. Besides, transgenic soybean of GmERF5 exhibited significant enhanced resistance to P. sojae and positively regulated the expression of the PR genes.
Although much progress has been made in elucidating the functional properties of ERFs, there was less research about ERFs in pepper. Previously, the ERFs CaCBF1A, CaCBF1B and CaDREBLP1 who mainly response to environmental stresses (low temperature and drought) were isolated (Kim et al., 2004; Hong and Kim, 2005). In this study, an ERF gene, CaPTI1 which was induced by pathogen was isolated from the transcriptome of interactions between pepper and P. capsici. The sequence character and structure were analyzed firstly. Then the expression profile of CaPTI1 was analyzed by quantitative Real-Time-PCR (qRT-PCR). Furthermore, in past most of the virus-induced gene silencing (VIGS) concentrated in leaves but here we have performed in leaves as well as in roots of pepper to investigate the function of CaPTI1 gene.
Materials and Methods
Plant Materials and Growth Conditions
Pepper cultivar Y3 was used in this study, which was provided by pepper breeding group in Northwest A&F University, China. First presoaking of the seeds was done and when 80% of seeds germinated, they were transferred to plastic pots having growing media [grass charcoal/perlite (3/1, v/v)]. Pepper plants were grown in a growth chamber with a 16 h light and 8 h dark photoperiod at 25°C.
Pathogen Preparation and Inoculation
The virulent (HX-9) and avirulent strains (PC) of P. capsici, which was compatible and incompatible to cultivar Y3 respectively, were used in this research. Both of the strains were grown on potato dextrose agar (PDA) medium in the dark at 28°C for 7 days as described previously by Zhang et al. (2013). Then zoospores of P. capsici were induced by chilling at 4°C for 30 min and then incubated at room temperature for 30–60 min. The zoospores were collected by filtering through four layers of cheesecloth. After the zoospores were collected, a haemocytometer was used to adjust the concentration to 1 × 105 zoospores mL-1 with sterile water.
For the VIGS of CaPTI1 gene, pepper plants were inoculated with 3 mL zoospore suspension of P. capsici virulent strain HX-9 (1 × 105 zoospores mL-1) using root drenching method. While for inoculation of detached leaves, third to fifth leaf from top of the control (pTRV2:00) and CaPTI1 silenced plants were taken and sterilized with 75% ethanol for 1 min, washed three times with sterile water, then 20 μL zoospore suspension (1 × 105 zoospores mL-1) of HX-9 were injected with a needleless syringe. After inoculation the leaves were kept in petri dishes and sealed with parafilm immediately. All the plants inoculated with P. capsici were put in a growth chamber at 28°C with a photoperiod of 16 h light/8 h dark and the relative humidity was kept 60%.
Cloning and Sequence Analysis of CaPTI1 Gene
Total RNA was extracted from the roots of Y3 plants which were inoculated with virulent strain of P. capsici and reverse transcribed into cDNA using a PrimeScriptTM Kit (Takara, Bio Inc, China) following the manufacturer’s protocols, and then the cDNA was used as a template to isolate the CaPTI1 gene according to the transcriptome database of compatible and incompatible interactions of pepper (Y3) with P. capsici. After that CaPTI1 gene was cloned into pMD19-T vector (Takara) and sequenced by Sangon Biotech (Shanghai) Co. Ltd.
The molecular weight (MW) and isoelectric point (pI) of CaPTI1 were predicted by ExPASy1. Conserved domain of CaPTI1 protein was identified in Conserved Domain of NCBI2. The full length of CaPTI1 and other ERF proteins were used to construct the neighbor joining phylogenetic tree by MEGA 6.05 with 1000 bootstrap replicates. Multiple sequence alignments of the AP2/ERF domain in CaPTI1 and other ERFs were performed by DNAMAN 5.0.
CaPTI1 Gene Expression Profile Analysis
To evaluate the expression levels of CaPTI1 gene in different organs, the roots, stems, leaves, flowers, and immature fruits were collected from Y3 pepper plants, then frozen in liquid nitrogen and kept at -80°C for further RNA extraction and gene expression analysis.
Pepper plants at the six-true leaf stage were used for P. capsici inoculation and treatments of plant signaling molecule (ETH, SA, MeJA, and H2O2) and environmental stresses (drought and cold). For P. capsici inoculation treatment the root-drenching method was used as described previously (Zhang et al., 2013). Then leaves and roots from infected plants were collected at 0, 6, 12, 24, 48, 72, and 96 h after inoculation. All samples were collected and analyzed with three biological replicates.
In case of plant signaling molecule treatments, pepper plants were sprayed with 10 mM ETH, 5 mM SA, 50 μM MeJA or 10 mM H2O2 solutions, while in case of drought stress treatment, the roots of seedlings were watered with 0.4 M mannitol solutions. Pepper plants were placed at 4°C for cold stress treatment. Control plants were sprayed with sterile water. Pepper plants in above treatments were placed in a chamber at 25°C (except cold stress) under a 16 h light/8 h dark photoperiod cycle with 60% relative humidity. Leaves from treated plants were collected at 0, 3, 6, 9, 12, and 24 h after treatment, and frozen in liquid nitrogen immediately and kept at -80°C for RNA extraction. All samples were collected and analyzed with three biological replicates.
RNA Isolation and Quantitative RT-PCR for Gene Expression Analysis
Total RNA was extracted from collected sample after various stress treatments using Trizol (Invitrogen) method. Then the first strand cDNA was synthesized using of prime script TM kit (Takara) following the manufacturer’s protocols. The NanoDrop instrument (Thermo Scientific NanoDrop 2000C, USA) was used to measure the concentration of cDNA. Later on qRT-PCR was carried out using SYBR® Premix Ex TaqTM II (TaKaRa) in an iCycler iQTM Multicolor PCR Detection System (Bio-Rad, USA). The amplification cycling parameters of qRT-PCR were as follow: 95°C for 1 min and followed by 40 cycles at 95°C for 10 s, 56°C for 15 s, and 72°C for 15 s. All the primers used for qRT-PCR were shown in Supplementary Table S1. Relative expression of genes was calculated as described by Livak and Schmittgen (2001) and CaUbi3 gene was used as the reference gene in this study (Wan et al., 2011).
VIGS of CaPTI1 in Pepper
For VIGS assay, the 3′-untranslated region (UTR) of CaPTI1 gene was selected to construct the combined vector pTRV2:CaPTI1 and then this combine vector was used for subsequent VIGS experiment to ensure the specific silencing of CaPTI1 (primers used for vector construction were shown in Supplementary Table S1). Then the pTRV1, pTRV2 (negative control), pTRV2:CaPDS (positive control) (Wang et al., 2013a) and also the combined vector pTRV2:CaPTI1 was transformed into Agrobacterium tumefaciens strain GV3101 by the freeze-thaw method. Agrodrench and leaf infiltration methods were used to silence CaPTI1 in pepper (Ryu et al., 2006; Velasquez et al., 2009). The growth condition used for pepper seedling after inoculation was as described by Wang et al. (2013a). After inoculation for about 5 weeks, leaves and roots from CaPTI1 silenced and control plants were collected for the analysis of silencing efficiency and expression level of relative defense genes. All experiments were performed and analyzed with three biological replicates.
Determination of Root Activity
The root activity was measured by a modified triphenyltetrazolium chloride (TTC) method (Ou et al., 2011). Before the TTC tests, the roots of silenced and control plants at different time points after inoculation with virulent strain of P. capsici were collected and washed with sterile water. Then the root surfaces were dried carefully with absorbent papers. After that root activity was measured by a modified TTC method as described by Wang et al. (2013b). The root samples were placed in small beaker and incubated with 10 ml 1:1 (v/v) mixture of 1% TTC solution and 0.l M phosphate buffer (pH 7.0) at 37°C for 1 h in the dark. After the incubation, 2 ml of 1 M sulphuric acid was to added to inhibit the reaction. The root samples were rinsed twice with distilled water and ground in mortar with 3 ml ethyl acetate, in which the extracts of TTF (a derivative of TTC from the reduced reaction) were obtained and transferred to a 10 ml volumetric flask. The residues were rinsed with ethyl acetate and mixed with the earlier extracts, then the final volume was adjusted to10 ml. The absorption of the extraction was measured with a spectrophotometer at 485 nm. The control was similarly performed except the sulphuric acid was added first, the root sample second, and the mixture last. The reduced TTC amount was obtained from the standard curve and its intensity in the roots was calculated as follows: TTC reduction intensity [mg g-1 h-1] = reduced TTC amount/FW h (FW-fresh root mass; h-the incubation time). Each treatment was conducted in three independent experiments and each measurement was repeated three times.
Statistical Analysis
All data were expressed as mean ± standard deviation (SD) of three independent replicates. Statistical Analysis System software (SAS 8.2, North Carolina State University, USA) was used for least significant difference (LSD) analysis and values of p < 0.05 was considered statistically significant. All experiments were performed and analyzed separately with three biological replicates.
Results
Cloning and Sequence Analysis of CaPTI1
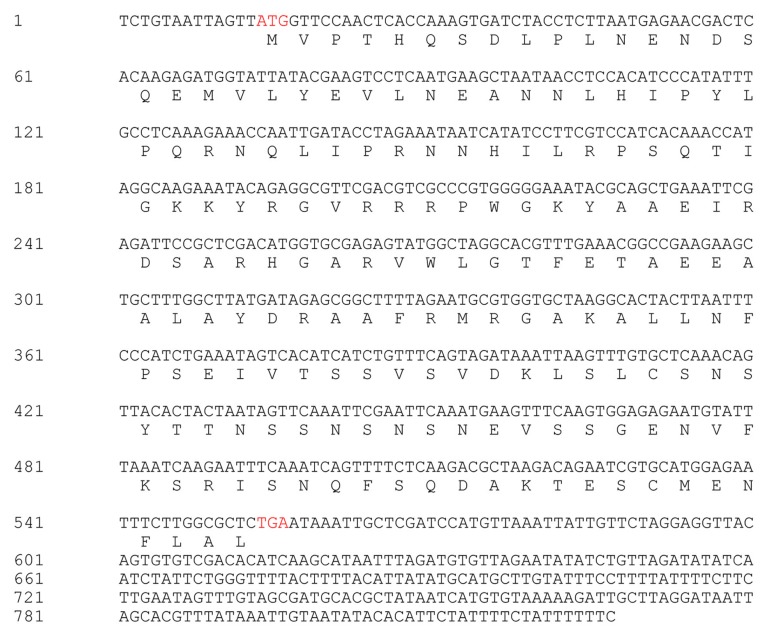
The CaPTI1 gene (Figure 1) (GenBank No. KJ690096), which included an open reading frame (ORF) of 543 base pair (bp), was identified from pepper transcriptome database. The ORF of CaPTI1 was predicted to encode a protein of 180 amino acids. The predicted molecular weight of the encoded protein was 20.30 kDa and pI 8.58. Sequence analysis found a single AP2/ERF domain in the CaPTI1 protein.
FIGURE 1.
The nucleotide sequence and deduced amino acid sequence of CaPTI1.
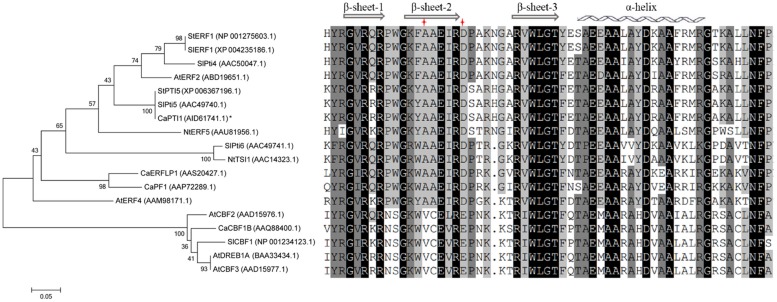
Furthermore the evolutionary relationships and multiple sequence alignment of CaPTI1 in comparison with ERF proteins of other plants were also investigated (Figure 2). The result showed that CaPTI1 had a more closely relationship with SlPti5. Multiple sequence alignment showed the conservation of AP2/ERF domain in ERFs from different plants. There were three β-sheets and an α-helix in the AP2/ERF domain (Allen et al., 1998; Djemal and Khoudi, 2015). The amino acids difference at position 14th and 19th in AP2/ERF domain was also existed in these ERFs (Sakuma et al., 2002).
FIGURE 2.
Phylogenetic tree and multiple alignment of CaPTI1 with other ERFs. Phylogenetic tree of CaPTI1 with other Ethylene-responsive factors (ERF) proteins was generated by the MEGA 6.05 program with 1000 bootstrap replicates. GenBank No. of ERFs is in parentheses after each gene name. While the multiple alignment was conducted by DNAMAN5.0.
Tissue-Specific Expression of CaPTI1 Gene in Pepper Plants
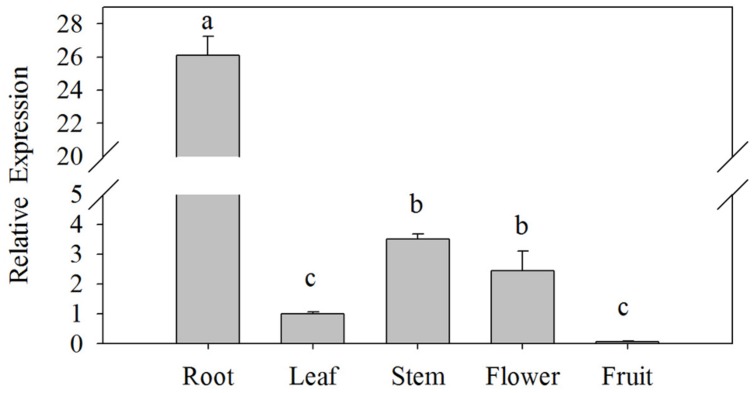
The expression levels of CaPTI1 gene in different tissues of Y3 plants were investigated by qRT-PCR. As shown in Figure 3, significant difference of CaPTI1 gene expression was observed in different tissues. The highest expression was detected in roots, which was almost 26-fold of that in leaves. The expression level of CaPTI1 gene in stems and flowers were also higher than that in leaves, which was 3.50 and 2.45-fold of that in leaves respectively. While lower expression was detected in fruits as compared to leaves which was statistically non-significant.
FIGURE 3.
Tissue specific expression of CaPTI1 in pepper. Error bars represent standard deviation (SD) for three independent replicates. Bars with different lower case letters indicate significant differences using Duncan’s multiple range test (p < 0.05).
Expression Analysis of the CaPTI1 Gene in Response to Phytophthora capsici Infection
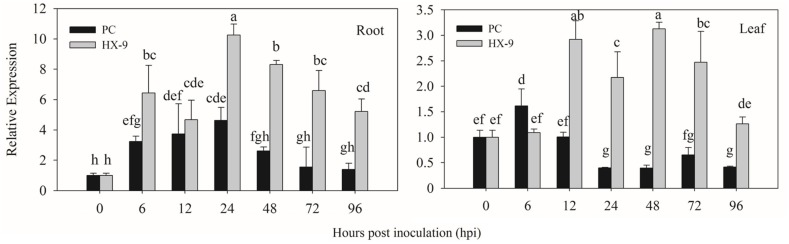
In virulent strain (HX-9) infection (Figure 4), the highest expression of CaPTI1 in roots was detected at 24 h post inoculation (hpi) which was 10.24-fold of control, then decrease gradually and at 96 hpi the expression was 5.22-fold. The same trend of CaPTI1 in roots was observed after inoculation with avirulent strain (PC), in which the expression of CaPTI1 was increasing with the passage of time and reached to peak (4.62-fold) at 24 hpi and then decrease to the lowest (1.39-fold) at 96 hpi. Overall, the expression level of CaPTI1 gene in the whole treatment with virulent strain was significantly higher than that with avirulent strain.
FIGURE 4.
Relative expression of CaPTI1 in pepper plants exposed to P. capsici infection. Relative expression of CaPTI1 in the roots and leaves of pepper cultivar Y3 after inoculation with virulent (HX-9) or avirulent (PC) strains of P. capsici. Error bars presented means ± SD from three replicates. Bars with different lower case letters indicate significant differences using Duncan’s multiple range test (p < 0.05).
While in leaves, CaPTI1 was up-regulated by virulent strain inoculation and the expression level was increased to 2.92-fold of control at 12 hpi, followed by a bit decrease, then reached to the peak (3.13-fold) at 48 hpi and at last decreased to 1.26-fold of control at 96 hpi. Less up regulation of CaPTI1 was detected in the infection of avirulent strain except for a little increase at 6 hpi. Finally, the results showed that CaPTI1 could be induced by P. capsici, especially in roots.
Expression Analysis of CaPTI1 Gene Under Abiotic Stresses
The signaling molecules (ETH, SA, MeJA and H2O2) were used to investigate whether CaPTI1 could be induced or not. As shown in Figure 5, CaPTI1 in the leaves were strongly induced by foliar spraying of ETH, SA, and MeJA. As an ERF, CaPTI1 in leaves showed a huge increase by ETH induction. The highest expression of CaPTI1 was detected at 3 h post treatment (hpt), which was 102.57-fold of control. In the treatment of SA and MeJA, both the highest expression of CaPTI1 was detected at 6 hpt, which were 114.53-fold and 108.80-fold of control respectively. While in the treatment with H2O2, the significant up regulation of CaPTI1 was observed at 3 and 6 hpt, which was 6.26-fold and 5.80-fold of control respectively.
FIGURE 5.
Relative expression of CaPTI1 responsed to various abiotic stresses. Relative expression of CaPTI1 gene in leaves of pepper plants after different treatments [Ethephon (ETH), salicylic acid (SA), Methyl Jasmonate (MeJA), hydogen peroxide (H2O2), cold stress and drought stress]. Error bars represent the mean ± SD of three independent biological replicates. Bars with different lower case letters indicate significant differences using Duncan’s multiple range test (p < 0.05).
Moreover, in the stresses of cold and drought, the expression of CaPTI1 in leaves also showed a significant increase and rise to the peak at 12 hpt, which were 11.32-fold and 6.82-fold of control respectively.
VIGS of CaPTI1 Gene in Pepper Plants
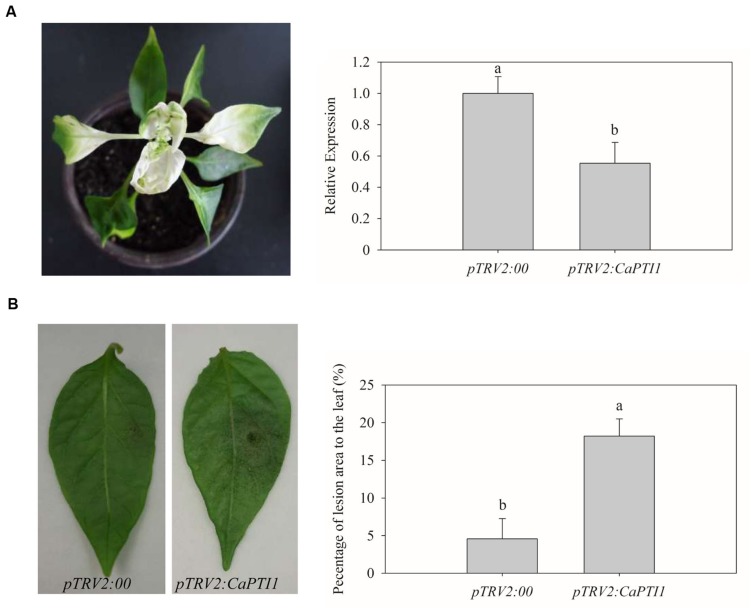
To examine the effect of loss-of-function of the CaPTI1 gene in pepper plants, VIGS was performed in pepper cultivar Y3. Empty vector (pTRV2:00) was used as a negative control. The pTRV2:CaPDS vector was used as a positive control, which will cause the silencing of CaPDS and induces a photo-bleaching phenotype in leaves.
Five weeks post inoculation, the phenotype of photo-bleaching was observed in leaves of positive control plants (Figure 6A). Then qRT-PCR analyses were performed to investigate the silencing efficiency of CaPTI1 in leaves. CaPTI1 in CaPTI1 gene silenced plant exhibited half expression level of that in control (pTRV2:00) (Figure 6B). Then the third to fifth leaves from the top of control plants and CaPTI1 silenced pepper plants (pTRV2:CaPTI1) were detached and inoculated by 20 μL zoospores suspension (105 zoospores mL-1) of virulent strain of P. capsici. Disease symptoms were observed at 2 days post inoculation (dpi). The leaves of control plants exhibited less disease symptoms. While larger and more numerous lesions were observed in the leaves of CaPTI1 silenced plants (Figure 6C). The quantitative analysis of the lesion area in the detached leaves showed a significant increase in CaPTI1 silenced plants (Figure 6D).
FIGURE 6.
Loss of function analysis of CaPTI1 in leaves of pepper. (A) The photo-bleaching phenotype in leaves of positive control pepper plants (pTRV2:CaPDS); (B) The relative expression of CaPTI1 in leaves of CaPTI1 silenced and negative control plants; (C) Disease symptoms developed on the detached leaves of silenced and control plants at 2 day post inoculation (dpi); (D) Percentage of the lesion area of the leaves inoculated with P. capsici. Error bars represent the mean ± SD of three independent biological replicates. Bars with different lower case letters indicate significant differences using Duncan’s multiple range test (p < 0.05).
Besides, we also checked the expression of CaPTI1 in roots of CaPTI1 silenced plants and found it was only about 40% of the control (Figure 7A). After that we investigated whether silencing of CaPTI1 altered the expression of defense related genes in roots during P. capsici infection. Significant decrease of CaPR1 (a SA dependent gene) (Kim and Hwang, 2000), CaDEF1 (a JA dependent gene) (Do et al., 2004) and CaSAR82 (systemic acquired resistance gene) (Lee and Hwang, 2003) were detected in CaPTI1 silenced plants. The time was expands to 4 dpi. Especially the CaPR1 gene showed a really low expression (0.87-fold) in silenced plant, but a highest expression (29.23-fold) in control at 4 dpi. While at the same time, the expression of CaPTI1 in silenced plant (2.02-fold) was still lower than that in control (6.88-fold).
FIGURE 7.
The CaPTI1 silenced pepper plants exhibit reduced resistance to P. capsici. (A) The expression of CaPTI1, CaPR1, CaDEF1, and CaSAR82 in the root of CaPTI1 silenced and control plants were analyzed; (B) Root activity in CaPTI1 silenced and control plants after inoculation with P. capsici. Values are the means ± SD from three independent experiments. Small letters represent significant differences (p < 0.05); (C) Disease symptoms of control and silenced plants were observed at 7 day post inoculation (dpi).
In addition, the roots activity of silenced and control plants after inoculation with P. capsici were measured by TTC method. The CaPTI1 silenced plants showed a lower root activity as compared to control, and a significant difference between silenced and control plants were detected at 4 dpi and enlarged to 7 dpi (Figure 7B). At 7 dpi disease symptoms of Phytophthora blight in pepper plants were observed. The CaPTI1 silenced plants showed a serious disease symptom, such as the blacking and constricting of stem basement, the wilting and abscission of leaves and wilting of growing point of plant. While less and slightly disease symptoms were observed in the control plants (Figure 7C).
Discussion
The AP2/ERF family is one of the largest transcription factor family and demonstrated to be involved in the regulation of plant development and response to biotic and abiotic stresses (Gutterson and Reuber, 2004; Wessler, 2005; Mizoi et al., 2012). In this study, we identified a new ERF in pepper, which named CaPTI1 and belonged to the ERF family.
Sakuma et al. (2002) divided the ERF family into ERF subfamily and DREB subfamily according to the amino acids difference at position 14th and 19th in AP2/ERF domain. In addition, several ERFs which contain alanine and aspartic acid at 14th and 19th position of the AP2/ERF domains could bind to both the DRE and GCC boxes (Park et al., 2001; Wang et al., 2004). The 14th alanine and the 19th aspartic acid in the AP2/ERF domain of CaPTI1 suggested a member of ERF subfamily. And sequence character analysis showed that CaPTI1 does not contain an EAR motif, which maybe indicate the activation role of CaPTI1 in plant defense response.
Phytophthora capsici mainly invades the pepper plant by roots and CaPTI1 has a highest expression in roots before inoculation. After infection of P. capsici the expression level of CaPTI1 in roots got a significant increase. These possibly indicated that CaPTI1 is involved in the interaction of pepper and P. capsici. CaPTI1 could not only be induced by P. capsici, but also strongly induced by ETH, SA and MeJA. These hormones as signaling molecules had been showed to play crucial roles in defense response pathway (Dong, 1998; Reymond and Farmer, 1998; Pieterse and van Loon, 1999). The SA signaling transduction pathway usually acts as antagonism with ET/JA pathway. ERF1 of Arabidopsis was found to be a key element in the integration of both ET and JA pathways for the regulation of defense response genes (Lorenzo et al., 2003). Besides, AtERFs may be involved in the cross-talk between SA and JA signaling pathways and important for plant defense to pathogen attack (Onate-Sanchez and Singh, 2002). ERF transcription factor ORA59 was supposed to be the integrator of JA and ET signaling pathways (Zarei et al., 2011). The ERF transcription factor in tomato, Pti4 could be induced by SA, JA, and ETH. While Pti5 and Pti6 could not be induced by SA in tomato (Gu et al., 2000). Further studies found that Pti4, Pti5, and Pti6 may be implicated indirectly in regulating the SA response through interaction with other TFs, and all of them could activate the expression of PR genes and play important roles in plant defense (Gu et al., 2002; Chakravarthy et al., 2003). All of these indicated that CaPTI1 may be involved in the pathways which were mediated by the signaling molecules SA, MeJA and ETH.
We also detected the up regulation of CaPTI1 by the spraying of H2O2. Reactive oxygen species (ROS) are produced constantly during normal plant growth and development and also perform essential roles as highly specific signaling molecules under stress conditions (Apel and Hirt, 2004). Plant defense hormones SA and JA could also modulate the plants ROS status (Mittler et al., 2011). ERF6 was turned out to be a regulator of ROS in Arabidopsis and response to biotic and abiotic stresses (Sewelam et al., 2013). Moreover, CaPTI1 could be induced by the cold and drought stresses which indicated CaPTI1 response to abiotic stresses. Over-expression of the tobacco Tsi1 gene also enhances resistance against pathogen attack and osmotic stress in tobacco (Park et al., 2001). StERFs also were shown to be regulated by drought stress (Wang et al., 2015). All these suggested the multiple role of CaPTI1 in pepper plants response to diverse stresses.
We silenced the CaPTI1 gene in leaves and roots of pepper plants successfully by VIGS. Detached leaves assay were frequently used to investigate the plant disease resistance to the invasion of pathogens (Wang et al., 2013c; Dong et al., 2015; Tian et al., 2015). After inoculation of P. infestans larger disease lesions were observed on the leaves of StERF3 over-expressed potato lines as compared to the wild type, which indicated the repressor role of StERF3 in defense response to P. infestans in potato. Less disease lesion were observed on the leaves of GmERF5 transgenic soybean lines after infection with P. sojae (Dong et al., 2015). Detached leaves assay with the inoculation of P. capsici were also performed in this study after the silencing of CaPTI1 in pepper, and then larger lesion area was observed in leaves of CaPTI1 silenced plants as compared to control. This indicated that silencing of CaPTI1 in pepper leaves make it more susceptible to P. capsici infection.
In addition, the roots of pepper were focused on because Phytophthora blight is a soil borne disease. Significant decrease of defense related genes CaPR1, CaDEF1, and CaSAR82 was detected in roots of CaPTI1 silenced plants when compared to the control plants, especially for the CaPR1, which is a SA dependent gene (Kim and Hwang, 2000). As mentioned before, ERFs could specially recognize and bind to the GCC-box. Pti4 could regulate defense related genes expression via both GCC-box and non-GCC-box (Chakravarthy et al., 2003). Two GCC-box were found in the upstream promoter region (about 1500 bp) of CaPR1 according the database of pepper genome (Kim et al., 2014). The silencing of CaPTI1 maybe decrease the expression of CaPR1 by affecting the binding of GCC-box which is in the promoter region of CaPR1. Moreover, Kim and Hwang (2000) found that treatment with SA in combination with ETH, caused a lower expression of the CaPR1 in pepper leaves than the treatment with ETH alone, and pepper stems will synthesize ethylene at an exponential rate soon after infection with either virulent or avirulent strains of P. capsici. As Pti4, Pti5, and Pti6 were supposed to be involved in the regulation of ethylene production in the plant singly or in combination (Gu et al., 2000). The silencing of CaPTI1 may reduce the ethylene production of pepper plants. As a result, the silencing of CaPTI1 may enhance the antagonism between SA and ET pathways by changing the ratio of SA to ET in CaPTI1 silenced pepper plants and then decrease the expression of CaPR1. The enhanced antagonism may also affect the expression of CaPTI1 in turn.
CaPTI1 silenced pepper plants showed a significant decrease in the expression of CaDEF1 (a JA-dependent gene) compare to the control. JA and ETH usually appear to function synergistically to induce PDF1.2 (a JA-dependent gene) in Arabidopsis, and osmotin and PR1b in tobacco (Xu et al., 1994; Penninckx et al., 1998; Norman-Setterblad et al., 2000). CaDEF1 may act as the downstream of JA/ET pathway and be regulated by CaPTI1 indirectly. The silencing of CaPTI1 maybe also decreases the synergism between JA and ET by decreasing the ethylene production in pepper plants. CaSAR82 serves as a molecular marker for the onset of ISR in pepper plants and could not only be induced by pathogens (Xanthomonas campestris pv. vesicatoria, Colletotrichum coccodes, and Phytophthora capsici), but also the signaling molecule (SA, JA, ETH and H2O2) and various stresses (Lee and Hwang, 2003). ISR was generally regulated by JA and ET. The significant decrease of CaSAR82 in CaPTI1 silenced pepper plants maybe indicated the systemic resistance mediated by JA/ET was weakened by the silencing of CaPTI1.
Besides a significant decrease of root activity was observed at 4 dpi in CaPTI1 silenced plants as compared to control. The root activity which was measured by the TTC method was used as a cell vitality indicator (Chen et al., 2006). The cell vitality of root indicated the injury degree of pepper roots after the infection of P. capsici. Yeom et al. (2011) used this assay to measure TTC reductase activity in the roots of pepper cultivars CM334 (resistant to P. capsici) and Chilsungcho (susceptible to P. capsici) after inoculation with P. capsici. They found that significant differences were observed from 24 to 48 hpi, showing two or three times more activity in CM334 than in Chilsungcho. Moreover Wang et al. (2013b) also found that there is a significant difference in pepper plants after inoculation with the incompatible and compatible strains of P. capsici, and the root activity was much higher in the infection of incompatible strain than that of compatible strain. In this study, a significant decrease of root activity was observed at 4 dpi in CaPTI1 silenced plants when compared to the control plants. This may indicate the serious injury to the roots of CaPTI1 silenced pepper plants by P. capsici. Both of the significant decreased root activity and the reduced expression of defense related genes indicated the weakness of defense response in CaPTI1 silenced pepper plants and suggested the significant role of CaPTI1 in defense response to the infection of P. capsici.
Taken together, CaPTI1 contains an AP2/ERF domain and belongs to the ERF family. CaPTI1 could not only responses to the infection of P. capsici and the stresses of cold and drought, but also be induced by SA, MeJA, ETH and H2O2. VIGS of CaPTI1 in pepper could weaken the defense response significantly by reducing the expression of defense related genes CaPR1, CaDEF1 and CaSAR82 and also the root activity. However, more detailed researches are still needed to reveal the exact role of CaPTI1 in the regulation of defense response to P. capsici in pepper.
Author Contributions
J-HJ, D-WL, and Z-HG conceived the research. J-HJ, H-XZ, J-YT, and M-JY performed the research. J-HJ and H-XZ performed statistical analyses. J-HJ wrote the paper. J-HJ, AK and Z-HG revised the paper. D-WL and Z-HG provided the materials and resources for the research. J-HJ, H-XZ, and Z-HG performed the integrity of the work. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31272163, 31401879), the Shaanxi Provincial Science and Technology Coordinating Innovative Engineering Project (2012KTCL02-09) and Northwest A&F University Basic Fund for Scientific Research (QN2013016).
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.01217
References
- Alazem M., Lin N. S. (2015). Roles of plant hormones in the regulation of host-virus interactions. Mol. Plant Pathol. 16 529–540. 10.1111/mpp.12204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M. D., Yamasaki K., Ohme-Takagi M., Tateno M., Suzuki M. (1998). A novel mode of DNA recognition by a β-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 17 5484–5496. 10.1093/emboj/17.18.5484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55 373–399. 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- Babadoost M., Islam S. Z. (2003). Fungicide seed treatment effects on seedling damping-off of pumpkin caused by Phytophthora capsici. Plant Dis. 87 63–68. 10.1094/Pdis.2003.87.1.63 [DOI] [PubMed] [Google Scholar]
- Baysal O., Turgut C., Mao G. (2005). Acibenzolar-S-methyl induced resistance to Phytophthora capsici in pepper leaves. Biol. Plant. 49 599–604. 10.1007/s10535-005-0055-0 [DOI] [Google Scholar]
- Brown R. L., Kazan K., Mcgrath K. C., Maclean D. J., Manners J. M. (2003). A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 132 1020–1032. 10.1104/pp.102.017814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy S., Tuori R. P., D’Ascenzo M. D., Fobert P. R., Despres C., Martin G. B. (2003). The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell 15 3033–3050. 10.1105/Tpc.017574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. W., Yang Y. W., Lur H. S., Tsai Y. G., Chang M. C. (2006). A novel function of abscisic acid in the regulation of rice (Oryza sativa L.) root growth and development. Plant Cell Physiol. 47 1–13. 10.1093/pcp/pci216 [DOI] [PubMed] [Google Scholar]
- Dangl J. L., Jones J. D. G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411 826–833. 10.1038/35081161 [DOI] [PubMed] [Google Scholar]
- Djemal R., Khoudi H. (2015). Isolation and molecular characterization of a novel WIN1/SHN1 ethylene-responsive transcription factor TdSHN1 from durum wheat (Triticum turgidum. L. subsp. durum). Protoplasma 252 1461–1473. 10.1007/s00709-015-0775-8 [DOI] [PubMed] [Google Scholar]
- Do H. M., Lee S. C., Jung H. W., Sohn K. H., Hwang B. K. (2004). Differential expression and in situ localization of a pepper defensin (CADEF1) gene in response to pathogen infection, abiotic elicitors and environmental stresses in Capsicum annuum. Plant Sci. 166 1297–1305. 10.1016/j.plantsci.2004.01.008 [DOI] [Google Scholar]
- Dong L., Cheng Y., Wu J., Cheng Q., Li W., Fan S., et al. (2015). Overexpression of GmERF5 a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. J. Exp. Bot. 66 2635–2647. 10.1093/jxb/erv078 [DOI] [PubMed] [Google Scholar]
- Dong X. N. (1998). SA, JA, ethylene, and disease resistance in plants. Curr. Opin. Plant Biol. 1 316–323. 10.1016/1369-5266(88)80053-0 [DOI] [PubMed] [Google Scholar]
- Fujimoto S. Y., Ohta M., Usui A., Shinshi H., Ohme-Takagi M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12 393–404. 10.1105/tpc.12.3.393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43 205–227. 10.1146/annurev.phyto.43.040204.135923 [DOI] [PubMed] [Google Scholar]
- Granke L. L., Quesada-Ocampo L., Lamour K., Hausbeck M. K. (2012). Advances in research on Phytophthora capsici on vegetable crops in the United States. Plant Dis. 96 1588–1600. 10.1094/Pdis-02-12-0211-Fe [DOI] [PubMed] [Google Scholar]
- Gu Y. Q., Wildermuth M. C., Chakravarthy S., Loh Y. T., Yang C. M., He X. H., et al. (2002). Tomato transcription factors Pti4 Pti5 and Pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 14 817–831. 10.1105/Tpc.000794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y. Q., Yang C., Thara V. K., Zhou J., Martin G. B. (2000). Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 12 771–785. 10.1105/Tpc.12.5.771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterson N., Reuber T. L. (2004). Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opin. Plant Biol. 7 465–471. 10.1016/j.pbi.2004.04.007 [DOI] [PubMed] [Google Scholar]
- Hausbeck M. K., Lamour K. H. (2004). Phytophthora capsici on vegetable crops: research progress and management challenges. Plant Dis. 88 1292–1303. 10.1094/Pdis.2004.88.12.1292 [DOI] [PubMed] [Google Scholar]
- He P., Warren R. F., Zhao T., Shan L., Zhu L., Tang X., et al. (2001). Overexpression of Pti5 in tomato potentiates pathogen-induced defense gene expression and enhances disease resistance to Pseudomonas syringae pv. tomato. Mol. Plant Microbe. Interact. 14 1453–1457. 10.1094/MPMI.2001.14.12.1453 [DOI] [PubMed] [Google Scholar]
- Hong J. P., Kim W. T. (2005). Isolation and functional characterization of the Ca-DREBLP1 gene encoding a dehydration-responsive element binding-factor-like protein 1 in hot pepper (Capsicum annuum L. cv. Pukang). Planta 220 875–888. 10.1007/s00425-004-1412-5 [DOI] [PubMed] [Google Scholar]
- Kim H. J., Nahm S. H., Lee H. R., Yoon G. B., Kim K. T., Kang B. C., et al. (2008). BAC-derived markers converted from RFLP linked to Phytophthora capsici resistance in pepper (Capsicum annuum L.). Theor. Appl. Gen. 118 15–27. 10.1007/s00122-008-0873-5 [DOI] [PubMed] [Google Scholar]
- Kim S., An C. S., Hong Y. N., Lee K. W. (2004). Cold-inducible transcription factor, CaCBF, is associated with a homeodomain leucine zipper protein in hot pepper (Capsicum annuum L.). Mol. Cells 18 300–308. [PubMed] [Google Scholar]
- Kim S., Park M., Yeom S. I., Kim Y. M., Lee J. M., Lee H. A., et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46 270–278. 10.1038/Ng.2877 [DOI] [PubMed] [Google Scholar]
- Kim Y. J., Hwang B. K. (2000). Pepper gene encoding a basic pathogenesis-related 1 protein is pathogen and ethylene inducible. Physiol. Plant. 108 51–60. 10.1034/j.1399-3054.2000.108001051x./ [DOI] [Google Scholar]
- Kousik C. S., Adams M. L., Jester W. R., Hassell R., Harrison H. F., Holmes G. J. (2011). Effect of cultural practices and fungicides on Phytophthora fruit rot of watermelon in the Carolinas. Crop Prot. 30 888–894. 10.1016/j.cropro.2011.03.012 [DOI] [Google Scholar]
- Lamour K. H., Stam R., Jupe J., Huitema E. (2012). The oomycete broad-host-range pathogen Phytophthora capsici. Mol. Plant Pathol. 13 329–337. 10.1111/j.1364-3703.2011.00754.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Hong J. P., Oh S. K., Lee S., Choi D., Kim W. T. (2004). The ethylene-responsive factor like protein 1 (CaERFLP1) of hot pepper (Capsicum annuum L.) interacts in vitro with both GCC and DRE/CRT sequences with different binding affinities: possible biological roles of CaERFLP1 in response to pathogen infection and high salinity conditions in transgenic tobacco plants. Plant Mol. Biol. 55 61–81. 10.1007/s11103-004-0417-6 [DOI] [PubMed] [Google Scholar]
- Lee S. C., Hwang B. K. (2003). Identification of the pepper SAR8.2 gene as a molecular marker for pathogen infection, abiotic elicitors and environmental stresses in Capsicum annuum. Planta 216 387–396. [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Loake G., Grant M. (2007). Salicylic acid in plant defence-the players and protagonists. Curr. Opin. Plant Biol. 10 466–472. 10.1016/j.pbi.2007.08.008 [DOI] [PubMed] [Google Scholar]
- Lorenzo O., Piqueras R., Sanchez-Serrano J. J., Solano R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15 165–178. 10.1105/Tpc.007468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheron M. E., Porchas H. (2000). Comparison of five fungicides on development of root, crown, and fruit rot of chile pepper and recovery of Phytophthora capsici from soil. Plant Dis. 84 1038–1043. 10.1094/Pdis.2000.84.9.1038 [DOI] [PubMed] [Google Scholar]
- McGrath K. C., Dombrecht B., Manners J. M., Schenk P. M., Edgar C. I., Maclean D. J., et al. (2005). Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 139 949–959. 10.1104/pp.105.068544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Suzuki N., Miller G., Tognetti V. B., Vandepoele K., et al. (2011). ROS signaling: the new wave? Trends Plant Sci. 16 300–309. 10.1016/j.tplants.2011.03.007 [DOI] [PubMed] [Google Scholar]
- Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Et Biophys. Acta-Gene Regul. Mech. 1819 86–96. 10.1016/j.bbagrm.2011.08.004 [DOI] [PubMed] [Google Scholar]
- Norman-Setterblad C., Vidal S., Palva E. T. (2000). Interacting signal pathways control defense gene expression in Arabidopsis in response to cell wall-degrading enzymes from Erwinia carotovora. Mol. Plant Microbe Interact. 13 430–438. 10.1094/Mpmi.2000.13.4.430 [DOI] [PubMed] [Google Scholar]
- Oelke L. M., Bosland P. W., Steiner R. (2003). Differentiation of race specific resistance to Phytophthora root rot and foliar blight in Capsicum annuum. J. Am. Soc. Hortic. Sci. 128 213–218. [Google Scholar]
- Ohmetakagi M., Shinshi H. (1995). Ethylene-inducible DNA-binding proteins that interact with an ethylene-responsive element. Plant Cell 7 173–182. 10.2307/3869993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Matsui K., Hiratsu K., Shinshi H., Ohme-Takagi M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13 1959–1968. 10.1105/tpc.13.8.1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onate-Sanchez L., Singh K. B. (2002). Identification of Arabidopsis ethylene-responsive element binding factors with distinct induction kinetics after pathogen infection. Plant Physiol. 128 1313–1322. 10.1104/Pp.010862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou L. J., Dai X. Z., Zhang Z. Q., Zou X. X. (2011). Responses of pepper to waterlogging stress. Photosynthetica 49 339–345. 10.1007/s11099-011-0043-x [DOI] [Google Scholar]
- Park J. M., Park C. J., Lee S. B., Ham B. K., Shin R., Paek K. H. (2001). Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-Type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13 1035–1046. 10.1105/tpc.13.5.1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx I. A. M. A., Thomma B. P. H. J., Buchala A., Metraux J. P., Broekaert W. F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10 2103–2113. 10.1105/tpc.10.12.2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C. M. J., van Loon L. C. (1999). Salicylic acid-independent plant defence pathways. Trends Plant Sci. 4 52–58. 10.1016/S1360-1385(98)01364-8 [DOI] [PubMed] [Google Scholar]
- Rehrig W. Z., Ashrafi H., Hill T., Prince J., Van Deynze A. (2014). CaDMR1 Cosegregates with QTL Pc5.1 for resistance to Phytophthora capsici in pepper (Capsicum annuum). Plant Genome 7 1–12. 10.3835/plantgenome2014.03.0011 [DOI] [Google Scholar]
- Reymond P., Farmer E. E. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1 404–411. 10.1016/S1369-5266(98)80264-1 [DOI] [PubMed] [Google Scholar]
- Ryu C. M., Anand A., Kang L., Mysore K. S. (2006). Agrodrench: a novel and effective agroinoculation method for virus-induced silencing in roots and diverse Solanaceous species. (vol 40 pg 322 2004). Plant J. 45 869–869. [DOI] [PubMed] [Google Scholar]
- Sakuma Y., Liu Q., Dubouzet J. G., Abe H., Shinozaki K., Yamaguchi-Shinozaki K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290 998–1009. 10.1006/bbrc.2001.6299 [DOI] [PubMed] [Google Scholar]
- Sewelam N., Kazan K., Thomas-Hall S. R., Kidd B. N., Manners J. M., Schenk P. M. (2013). Ethylene response factor 6 is a regulator of reactive oxygen species signaling in Arabidopsis. PLoS ONE 8:e70289 10.1371/journal.pone.0070289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R., Park J. M., An J. M., Paek K. H. (2002). Ectopic expression of Tsi1 in transgenic hot pepper plants enhances host resistance to viral, bacterial, and oomycete pathogens. Mol. Plant Microbe Interact. 15 983–989. 10.1094/Mpmi.2002.15.10.983 [DOI] [PubMed] [Google Scholar]
- Thabuis A., Palloix A., Pflieger S., Daubeze A. M., Caranta C., Lefebvre V. (2003). Comparative mapping of Phytophthora resistance loci in pepper germplasm: evidence for conserved resistance loci across Solanaceae and for a large genetic diversity. Theor. Appl. Gen. 106 1473–1485. [DOI] [PubMed] [Google Scholar]
- Thara V. K., Tang X. Y., Gu Y. Q., Martin G. B., Zhou J. M. (1999). Pseudomonas syringae pv tomato induces the expression of tomato EREBP-like genes Pti4 and Pti5 independent of ethylene, salicylate and jasmonate. Plant J. 20 475–483. 10.1046/j.1365-313x.1999.00619.x [DOI] [PubMed] [Google Scholar]
- Tian Z., He Q., Wang H., Liu Y., Zhang Y., Shao F., et al. (2015). The potato ERF transcription factor StERF3 negatively regulates resistance to Phytophthora infestans and salt tolerance in potato. Plant Cell Physiol. 56 992–1005. 10.1093/pcp/pcv025 [DOI] [PubMed] [Google Scholar]
- Velasquez A. C., Chakravarthy S., Martin G. B. (2009). Virus-induced gene silencing (VIGS) in Nicotiana benthamiana and tomato. J. Vis. Exp. 2009:1292 10.3791/1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot A. C., Dempsey D. A., Klessig D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47 177–206. 10.1146/annurev.phyto.050908.135202 [DOI] [PubMed] [Google Scholar]
- Wan H. J., Yuan W., Ruan M. Y., Ye Q. J., Wang R. Q., Li Z. M., et al. (2011). Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.). Biochem. Biophys. Res. Commun. 416 24–30. 10.1016/j.bbrc.2011.10.105 [DOI] [PubMed] [Google Scholar]
- Wang H., Huang Z., Chen Q., Zhang Z., Zhang H., Wu Y., et al. (2004). Ectopic overexpression of tomato JERF3 in tobacco activates downstream gene expression and enhances salt tolerance. Plant Mol. Biol. 55 183–192. 10.1007/s11103-004-0113-6 [DOI] [PubMed] [Google Scholar]
- Wang J. E., Li D. W., Gong Z. H., Zhang Y. L. (2013a). Optimization of virus-induced gene silencing in pepper (Capsicum annuum L.). Genet. Mol. Res. 12 2492–2506. 10.4238/2013.July.24.4 [DOI] [PubMed] [Google Scholar]
- Wang J. E., Li D. W., Zhang Y. L., Zhao Q., He Y. M., Gong Z. H. (2013b). Defence responses of pepper (Capsicum annuum L.) infected with incompatible and compatible strains of Phytophthora capsici. Eur. J. Plant Pathol. 136 625–638. 10.1007/s10658-013-0193-8 [DOI] [Google Scholar]
- Wang J. E., Liu K. K., Li D. W., Zhang Y. L., Zhao Q., He Y. M., et al. (2013c). A novel peroxidase CanPOD gene of pepper is involved in defense responses to phytophtora capsici infection as well as abiotic stress tolerance. Int. J. Mol. Sci. 14 3158–3177. 10.3390/ijms14023158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. M., Zhang N., Zhou X. Y., Fan Q., Si H. J., Wang D. (2015). Isolation and characterization of StERF transcription factor genes from potato (Solanum tuberosum L.). C. R. Biol. 338 219–226. 10.1016/j.crvi.2015.01.006 [DOI] [PubMed] [Google Scholar]
- Wessler S. R. (2005). Homing into the origin of the AP2 DNA binding domain. Trends Plant Sci. 10 54–56. 10.1016/j.tplants.2004.12.007 [DOI] [PubMed] [Google Scholar]
- Wu C. J., Avila C. A., Goggin F. L. (2015). The ethylene response factor Pti5 contributes to potato aphid resistance in tomato independent of ethylene signalling. J. Exp. Bot. 66 559–570. 10.1093/Jxb/Eru472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Chang P. F. L., Liu D., Narasimhan M. L., Raghothama K. G., Hasegawa P. M., et al. (1994). Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell 6 1077–1085. 10.1105/Tpc.6.8.1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Tian L. N., Latoszek-Green M., Brown D., Wu K. Q. (2005). Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol. Biol. 58 585–596. 10.1007/s11103-005-7294-5 [DOI] [PubMed] [Google Scholar]
- Yeom S. I., Baek H. K., Oh S. K., Kang W. H., Lee S. J., Lee J. M., et al. (2011). Use of a secretion trap screen in pepper following Phytophthora capsici infection reveals novel functions of secreted plant proteins in modulating cell death. Mol. Plant-Microbe Interact. 24 671–684. 10.1094/MPMI-08-10-0183 [DOI] [PubMed] [Google Scholar]
- Zarei A., Korbes A. P., Younessi P., Montiel G., Champion A., Memelink J. (2011). Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol. Biol. 75 321–331. 10.1007/s11103-010-9728-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. L., Jia Q. L., Li D. W., Wang J. E., Yin Y. X., Gong Z. H. (2013). Characteristic of the pepper CaRGA2 gene in defense responses against Phytophthora capsici leonian. Int. J. Mol. Sci. 14 8985–9004. 10.3390/ijms14058985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. M., Tang X. Y., Martin G. B. (1997). The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. Embo J. 16 3207–3218. 10.1093/emboj/16.11.3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.