Abstract
Candida albicans is the most common human fungal pathogen and can cause life-threatening infections. Filamentous growth is critical in the pathogenicity of C. albicans, as the transition from yeast to hyphal forms is linked to virulence and is also a pivotal process in fungal biofilm development. Homeodomain-containing transcription factors have been linked to developmental processes in fungi and other eukaryotes. We report here on GRF10, a homeobox transcription factor-encoding gene that plays a role in C. albicans filamentation. Deletion of the GRF10 gene, in both C. albicans SN152 and BWP17 strain backgrounds, results in mutants with strongly decreased hyphal growth. The mutants are defective in chlamydospore and biofilm formation, as well as showing dramatically attenuated virulence in a mouse infection model. Expression of the GRF10 gene is highly induced during stationary phase and filamentation. In summary, our study emphasizes a new role for the homeodomain-containing transcription factor in morphogenesis and pathogenicity of C. albicans.
Keywords: GRF10, filamentation, chlamydospore formation, biofilm formation, virulence, Candida albicans
The homeodomain transcription factor Grf10 is important for formation of hyphae and virulence in the human pathogen Candida albicans.
Graphical Abstract Figure.
The homeodomain transcription factor Grf10 is important for formation of hyphae and virulence in the human pathogen Candida albicans.
INTRODUCTION
Candida albicans is an opportunistic fungal pathogen and the most common cause of human fungal infections (Odds 1987). It causes the superficial infections in healthy individuals and life-threatening infections in immunocompromised people. C. albicans has the ability to grow as three distinctive forms, yeast, pseudohyphae and true hyphae [reviewed in (Sudbery, Gow and Berman 2004)]. The morphological transition of C. albicans from yeast to a filamentous form is closely linked to virulence (Lo et al. 1997; Thompson, Carlisle and Kadosh 2011). The filamentous forms are invasive and cause tissue damage. In addition, filamentation is a pivotal aspect of fungal biofilm development as the hyphal-specific proteins are required for adherence (reviewed in Finkel and Mitchell 2011). Biofilms are inherently resistant to both drugs and host immunity, leading to medically complicated infections (Mathe and Van Dijck 2013). Thus, the healthcare burden of Candida infections is largely associated with the C. albicans in hyphal forms.
C. albicans grows as filamentous forms in response to a wide range of environmental cues such as serum, pH, elevated temperature and loss of nutrients (Sudbery, Gow and Berman 2004; Biswas, Van Dijck and Datta 2007; Whiteway and Bachewich 2007; Sudbery 2011). The Ras/cyclic AMP/protein kinase A pathway is principally involved in transducing extracellular signals to the transcriptional regulators that modulate the cellular response (Inglis and Sherlock 2013). These key transcriptional regulators include activators such as Cph1 (Liu, Köhler and Fink 1994), Efg1 (Stoldt et al. 1997), Tec1 (Schweizer et al. 2000), Ndt80 (Sellam et al. 2010) and Rim101 (Ramon, Porta and Fonzi 1999; Davis et al. 2000). Repressors, such as Tup1 (Braun and Johnson 1997), Rfg1 (Kadosh and Johnson 2001; Khalaf and Zitomer 2001), Nrg1 (Braun, Kadosh and Johnson 2001; Murad, Leng and Straffon 2001) and Rbf1 (Ishii et al. 1997), are also important for negatively regulating hyphal development. The coordinate regulation by these factors is responsible for integrating environmental signals with gene expression to obtain different phenotypic outcomes. While much progress on understanding the signals and factors necessary for hyphal formation has been identified, our understanding of the genetic control of this developmental process is still incomplete.
The homeotic genes are master regulators of development in eukaryotes. The first homeotic genes were identified in Drosophila melanogaster, and numerous genes have been identified in diverse organisms, including animals, plants and fungi since that time (reviewed in Holland 2013). These genes share a common sequence of 180 bp, the homeobox, which encodes a highly conserved DNA binding domain of about 60 amino acids called the homeodomain (Burglin 2011). In animals, homeodomain proteins specify the body plan along the anteroposterior axis, whereas in plants, homeodomain proteins are involved in regulating the patterning and morphogenesis of flowers, vegetative shoots and leaves. The homeobox genes have been linked with developmental pathways such as mating and filamentation in fungi, although they have been less studied in these organisms. The best studied cases are the heterodimers formed between the homeodomain proteins Mtla1 and Mtlα2, which regulates white-opaque switching and mating in C. albicans (Miller and Johnson 2002), and the homologous proteins Mata1 and Matα2 in other fungal species (Herskowitz, Rine and Strathern 1992; Kruzel and Hull 2010).
In C. albicans, GRF10 encodes a homeodomain containing transcription factor. It has been implicated in adenylate biosynthesis, biofilm formation and virulence (Homann et al. 2009; Nobile, Fox and Nett 2012; Romanowski, Zaborin and Valuckaite 2012). In this report, we investigated whether it plays a role in morphogenesis. We found that strains with mutations in the grf10Δ gene exhibit filamentous growth defects in liquid and on solid medium, are unable to form normal chlamydospores, and they exhibit a decrease in biofilm formation. The grf10 null mutants also exhibited attenuated virulence in a mouse model of systemic infections. We also found that expression of GRF10 is induced during stationary phase and under hyphal-inducing conditions. Together, these findings support a critical role for the GRF10 homeobox gene in the regulation of C. albicans morphogenesis.
MATERIALS AND METHODS
Strains
Strains of C. albicans used and generated in this study are listed in Table 1. C. albicans BWP17, a derivative of SC5314 (Fonzi and Irwin 1993), served as the parent strain for constructing the GRF10 mutant strains RAC115, RAC117 and RAC120 using the PCR gene knockout method (Wilson, Davis and Mitchell 1999). Strain DAY185 was obtained from A. Mitchell (Davis et al. 2000), and strains SN152, OHWT and TF021 (Homann et al. 2009) were obtained from the Fungal Genetics Stock Center.
Table 1.
Yeast strains.
| Strain | Relevant genotype | Source |
|---|---|---|
| SC5314 | URA3/URA3 | Fonzi and Irwin (1993) |
| SN152 | arg4Δ/arg4Δ leu2Δ/leu2Δ his1Δ/his1Δ URA3/ura3Δ | Noble and Johnson (2005) |
| IRO1/iro1Δ | ||
| TF021 | arg4Δ/arg4Δ leu2Δ/leu2Δ his1Δ/his1Δ URA3/ura3Δ | Homann et al. (2009) |
| IRO1/iro1Δ grf10Δ::HIS1/grf10Δ::LEU2 | ||
| OHWT | arg4Δ/arg4Δ::ARG4 leu2Δ/leu2Δ::LEU2 his1Δ/his1Δ::HIS1 | Homann et al. (2009) |
| URA3/ura3Δ IRO1/iro1Δ | ||
| BWP17 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG | Wilson, Davis and Mitchell (1999) |
| arg4::hisG/arg4::hisG | ||
| DAY185 | ura3::λimm434/ura3::λimm434 HIS1::his1::hisG/ his1::hisG | Davis et al. (2000) |
| ARG4::URA3::arg4::hisG/arg4::hisG | ||
| RAC114 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG | This study |
| arg4::hisG/arg4::hisG GRF10/grf10Δ::URA3 | ||
| RAC117 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG | This study |
| arg4::hisG/arg4::hisG grf10Δ::ARG4/grf10Δ::URA3 | ||
| RAC120 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG | This study |
| arg4::hisG/arg4::hisG grf10Δ::ARG4/GRF10::HIS1::grf10Δ::URA3 |
Media and growth conditions
Strains were grown on YPD or SC media (containing 0.17% yeast nitrogen base, 0.5 mM uridine, 0.15 mM adenine and CSM amino acids (Sunrise Science Products, MP Biologicals) (Sherman 1991). All stock strains were maintained at 4°C on YPD plates. Hyphal formation was monitored on the following liquid and solid agar media: Spider medium (Liu, Köhler and Fink 1994), 10% fetal calf serum, Medium-199 (Gibco-BRL) buffered with 155 mM HEPES, pH 7.5, SLAD medium (Gimeno et al. 1992) and Lee's medium (Lee, Buckley and Campbell 1975). The chlamydospore formation was monitored on the corn meal agar (CMA) medium (BD Scientific), supplemented with Tween 80 (1% v/v) and auxotrophic supplements. The final agar concentration for all solid media was 1.5% (w/v).
Hyphal induction
To induce hyphae, cells were grown overnight in YPD medium and washed twice with sterile water. For induction on solid medium, cell densities were adjusted to 2 × 107 cfu/ml, 5 μl of each strain was spotted and plates were incubated for 3 days at 37°C. For induction in liquid medium, cells were inoculated 1:50 into 2 ml of medium in 12-well tissue culture plates and the cultures were incubated at 37°C with mild shaking for 0, 4, 8 and 24 h. Each culture was examined microscopically using an Olympus BH2-RFCA microscope at 40× and photographed.
Quantitative macroscopic colony assay
Strains (DAY185, RAC117, OHWT and TF021) were grown overnight in YPD medium at 30°C in a tube roller. The macroscopic colony spot assay was performed as described (Homann et al. 2009): overnight cultures were diluted to OD600 = 0.08 in sterile water (1X) and a further 1:5 dilution was made in water (5X); 5 μL of these two samples was spotted onto Spider medium and the plates were incubated at 37°C up to 14 days. The macroscopic colony morphology was quantified by measuring the overall colony diameter (mm), central region (mm) and peripheral region (mm) using a regular ruler (Noble et al. 2010). The p-value was determined using the Student's t-Test.
Filamentous growth rate measurement
The macroscopic colony was grown in the same method as described above. The diameter size (mm), central region (mm) and peripheral region (mm) were measured by using a regular ruler daily for 12 days. Graphs and p-values for Student's t-Test were generated by using Excel. The filamentous growth rate was calculated from the slope of the graph, X-axis: time (day) and Y-axis: colony size (mm).
Chlamydospore and biofilm formation
To induce chlamydospore formation, overnight grown cultures or colonies were streaked deep into the surface of CMA plates and a coverslip was placed on top (Joshi, Solanki and Prakash 1993). Plates were incubated at 25°C for 3–5 days and chlamydospore formation was evaluated microscopically and photographed. Biofilm formation was assessed using the protocol as described (Krueger et al. 2004); briefly: overnight cultures were washed in phosphate buffered saline (PBS), diluted to an OD600 of ∼1 in PBS, and 100 μl was added to each well of a 96-well plate that had been coated with 10% fetal bovine serum and allowed to adhere for 90 min at 37°C. Non-adherent cells were removed by two washes with PBS. Biofilms were allowed to form for 48 h in supplemented YNB + glucose medium. After two washes with PBS, adherent cells were measured by absorbance at 595 nm in a spectrophotometric plate reader.
Virulence determination
C. albicans strains were grown in YPD broth at 30°C to stationary phase, washed twice with calcium- and magnesium-free PBS (from Biosource), and resuspended in PBS at a cell density of 5 × 106 cells ml−1 based on hemocytometer counts. Virulence in mice was assessed as described previously (Tsuchimori et al. 2000; Chauhan et al. 2005). Groups of 13 male BALB/c mice (20–22 g from Harlan) were formed and each mouse was injected through the lateral tail vein with a 200 μl inoculum containing 106 cells of wild-type control or mutant yeast. Mice were given food and water ad libitum. Survival of the mice was monitored twice daily and moribund mice were euthanized by asphyxiation with carbon dioxide, as recommended by American Veterinary Medical Association (AVMA 2001). Yeast was recovered from the kidneys of the first three mice from each group to die; PCR analysis showed that the recovered fungi had the same genotype as the inoculum (data not shown). The post-infection morphology of each strain was assessed microscopically from the homogenized kidney treated with 10% KOH (data not shown) (Odds, Van Nuffel and Gow 2000). In addition, three mice from each experimental group were sacrificed by CO2 inhalation at 24, 48 and 72 h post infection; their kidneys were removed, weighed and homogenized in PBS. Serial dilutions of the homogenate were plated on YPD agar supplemented with 50 μg/ml streptomycin (to prevent bacterial growth), and were incubated at 30°C for 48 h. The number of colonies on each plate was determined, and a calculation was made of the number of colonies per gram of tissue. Statistical analyses were performed with the SPSS 15.0 software using the Kaplan–Meier survival analysis.
RNA preparation, cDNA synthesis and quantitative real-time PCR
Strain SC5314 strain was grown in triplicate overnight in YPD medium at 30°C with shaking. The overnight culture was diluted 1:50 into 5 mL of SC + Uri medium, and grown with shaking at 30°C until mid-log phase (6 h, OD600 ∼0.5–1) or stationary phase (24 h, OD600 ∼8–9). To induce filamentation, the overnight culture was diluted 1:50, and cells were grown in 12-well dishes containing Spider medium with mild shaking at 37°C for 6 h; filamentation was confirmed by light microscopy. Cells were quickly cooling on ice and pelleted by centrifugation at 4°C for 5 min; the cell pellet was frozen at −80°C. RNA was extracted from the frozen pellets using the RiboPure Yeast Kit (Ambion) following the manufacturer's instructions, except that less RNA (3 μg) was treated with DNaseI. cDNA was synthesized from 200 ng of RNA using the SensiFAST cDNA synthesis kit (Bioline), following the manufacturer's instructions. The synthesized cDNA was diluted 1:5 in DEPC-treated water.
The RT-qPCR was performed on each cDNA sample following the manufacturer's instructions (SensiFAST SYBR No-ROX Kit, Bioline) using primers to GRF10 (forward 5′-CAAACCGCTCAAATAGTCAAAG-3′ and reverse 5′-GCAAATTGTGGAGAGTTTGTAG-3′) and the reference gene TEF1 (forward 5′-TTCGTCAAATCCGGTGATG-3′ and reverse 5′-CTGACAGCGAATCTACCTAATG-3′). The relative gene expression was calculated by the ΔCT method using a reference gene as described by the Bio-Rad Laboratories qPCR manual. Student's t-test and statistical significance were calculated using Excel.
RESULTS
The GRF10 gene is essential for normal filamentous growth
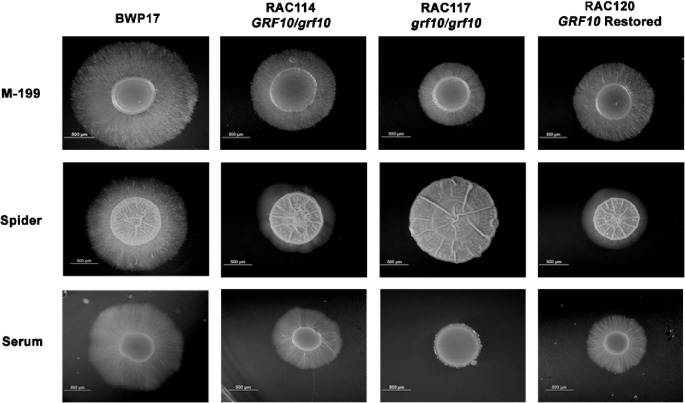
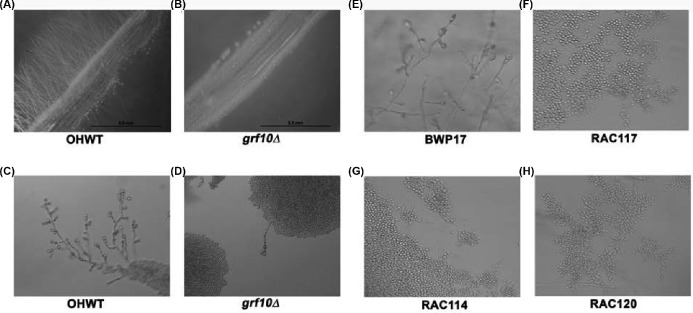
To determine if GRF10 has a role in filamentation, we examined the effects of the GRF10 gene deletion on C. albicans colony morphology on solid media. Homozygous and heterozygous strains were constructed with mutations in GRF10 in the BWP17 background (Table 1). Morphological phenotypes of the grf10Δ mutant were examined on several different solid agar media. We observed striking defects in filamentation in strain RAC117 on 10% serum, M-199 and Spider media (Fig. 1) and found no difference in filamentation on SLAD or Lee's media (data not shown).
Figure 1.
Disruption of GRF10 affects hyphal formation on serum, M-199 and Spider media. Cells (1 × 105) of parental strain BWP17 and mutant strains RAC114 (GRF10/grf10Δ), RAC117 (grf10Δ/grf10Δ) and RAC120 (GRF10 restored) were inoculated on solid 10% serum, Spider medium and M-199, pH 7.5 medium. Plates were incubated at 37°C for 3 days and photographed. The induction of hyphae was performed at least three times, representative examples are shown. Bar, 500 mm.
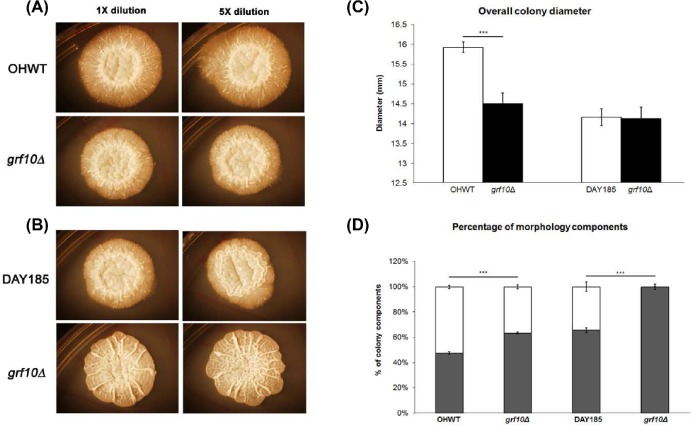
The morphological defects we observed in the BWP17 genetic background were stronger than reported elsewhere for strains in the SN152 background (Homann et al. 2009), so this difference was examined more closely. We grew grf10Δ strains from both genetic backgrounds (RAC117 and TF021) on solid Spider medium, monitored the macroscopic appearance of the C. albicans colony (Fig. 2A and B), and measured the overall colony diameter as well as the central and peripheral regions for two weeks (Fig. 2C and D) (Noble et al. 2010). Regardless of the strain background, the grf10Δ mutant strains had an overall increase in the central region and a significant decrease in the peripheral region; this difference was more evident in strain RAC117 from the BWP17 strain background. The grf10Δ mutant exhibits a leaky adenine auxotrophy (Homann et al. 2009). To see if adenine affected filamentation, we examined morphology on Spider medium with and without adenine supplementation. The addition of adenine did not rescue the filamentation defect (Fig. S1, Supporting Information). Neither of the grf10Δ mutants differed in doubling times from their respective parental strains in SC medium (data not shown), indicating that the defect seen on Spider medium is in filamentation and not in the overall growth.
Figure 2.
The grf10Δ mutant in two genetic backgrounds exhibits filamentation and colony defects. Cells (0.08 OD600) were spotted onto Spider medium; photos of the macroscopic colonies were taken on day 10: (A) strains OHWT and TF021 (SN152 background), (B) strains DAY185 and RAC117 (BWP17 background). (C) Quantification of the overall colony diameter (mm) at day 10. (D) Distribution of the overall colony diameter by percentage into the central region (dark bar) and peripheral region (white bar). All measurements were averaged from six biological samples with two technical replicates (n = 12). Student's t-test was calculated using Excel. *** p-value < 0.0001.
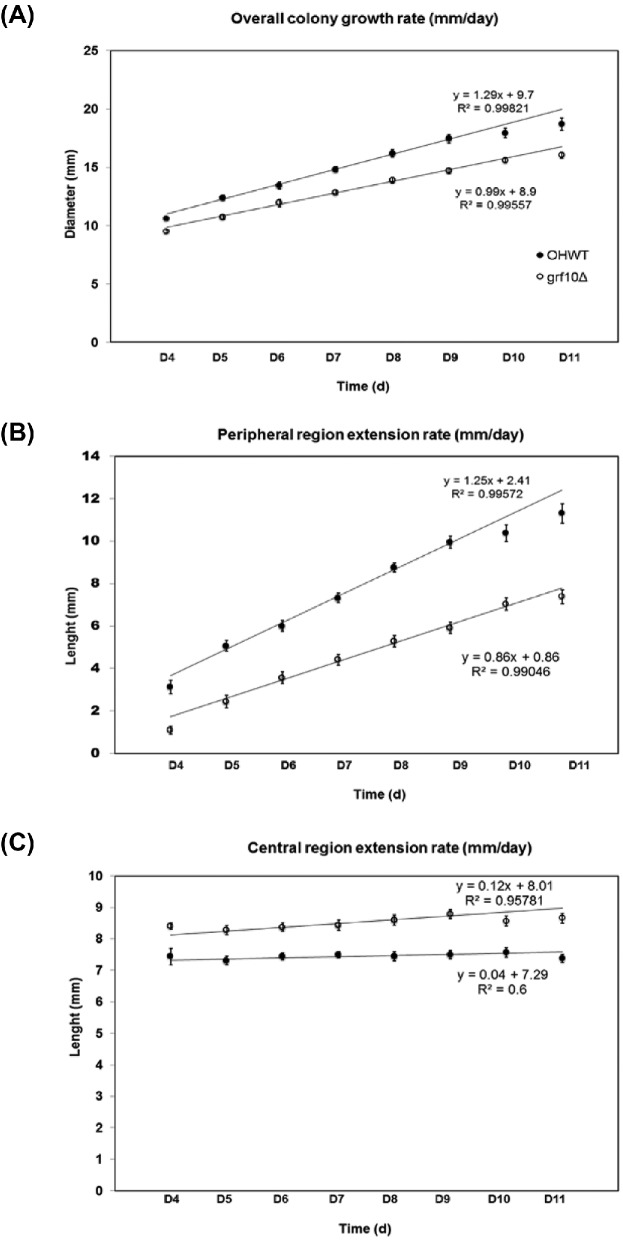
We quantitatively monitored the colony growth rate of the grf10Δ mutant TF021, which had the weaker phenotype, on solid Spider medium daily for up to two weeks (Fig. 3). During the first three days, both the mutant and the wild-type strain have the same appearance and colony size, with no visible hyphae extending from the edges of the spot; hyphae became visible at day 4. The overall rate of growth of the grf10Δ colony was slower by ∼23% (Fig. 3A; 1.0 mm/day for TF021 and 1.3 mm/day for OHWT). This difference in overall colony growth rate comes from differences in the peripheral region; the peripheral region extension rate in the grf10Δ colony was slowed by ∼30% relative to wild type (Fig. 3B; 0.9 mm/day for TF021 and 1.3 mm/day for OHWT), whereas the growth of the central region was constant in the two strains (Fig. 3C). Together these results show a defect in filamentation in the grf10Δ mutant, independent of the strain background.
Figure 3.
The grf10Δ mutant was deficient in the growth of hyphae. Strains (OHWT and TF021) were grown on solid Spider medium and the radial growth was monitored during a time-course of 11 days (D1–D11). Diameter of the overall colony (A), the peripheral region (B) and the central region (C) were measured. The wild-type strain OHWT is represented by the solid circles and the grf10Δ mutant (TF021) is represented by the open circles. The line represents the best fit to the data from D5-D9; the radial rate is the slope of of this line. Data were collected from four biological isolates with two technical replicates (n = 8).
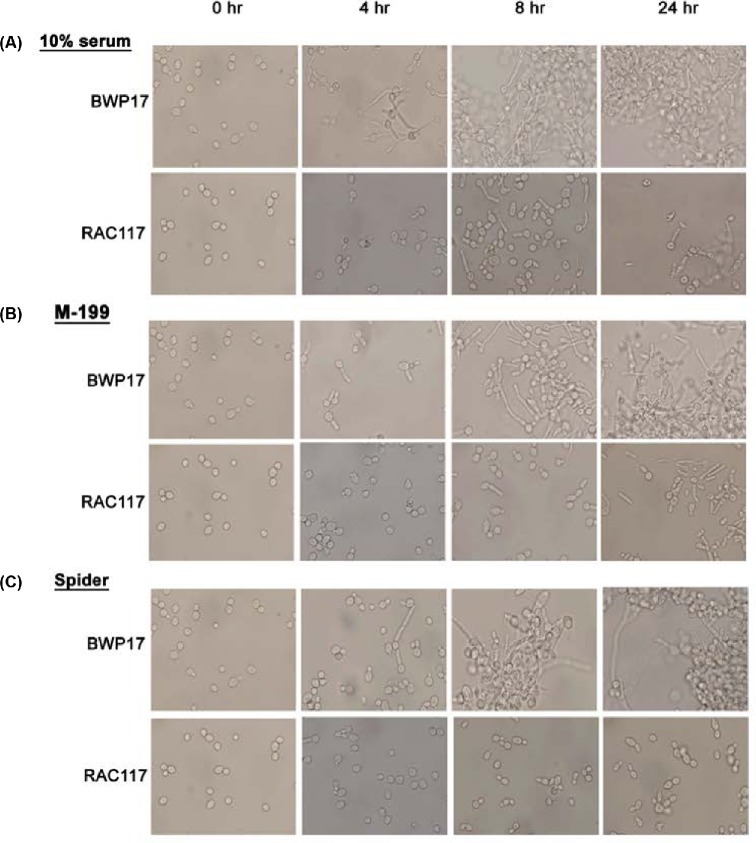
The grf10Δ mutant is delayed in germ-tube formation
The morphological defects of the grf10Δ colony may be due to an inability to initiate germ-tube formation and/or maintain hyphal growth. To examine this, we monitored filamentation of the grf10Δ from the BWP17 strain background, because it had the stronger filamentation phenotype, during a 24-h time course in 10% serum, and in liquid M-199 and Spider media. In serum, the grf10Δ mutant was delayed in inducing filamentation at 4-h and it produced only germ tubes and pseudohyphae at 8 and 24 h (Fig. 4A). In M199 medium at 4 h, the grf10Δ mutant showed only minimal induction of germ tubes, detected in about 10–15% of the cells, and only 20–25% of the grf10Δ cells induced short pseudohyphae after 8- and 24-h incubation time (Fig. 4B). In Spider medium, the grf10Δ mutant exhibited only the formation of germ tubes, few pseudohyphae and no true hyphae (Fig. 4C). Overall, we see delays in initiating filamentation in broth that are consistent with the defects detected at the whole colony level on solid media.
Figure 4.
The grf10Δ mutant is delayed in germ-tube formation. Strains BWP17 (parental wild-type) and RAC117 (grf10Δ) were inoculated in (A) 10% fetal calf serum, (B) M199 medium, and (C) Spider medium, and were grown at 37°C; aliquots were removed microscopy analysis at 0, 4, 8 and 24 h. Cellular morphology was examined at 40× magnification. Note—the photos of the cells at time 0 are the same, and are reproduced here for ease of comparison across the time course.
The grf10Δ mutants are defective in chlamydospore formation
Given the defect that we observed in filamentation, we wondered if chlamydospore formation was also affected. We found that the grf10Δ strains from both strain backgrounds are defective in forming chlamydospores (Fig. 5A–F). We rarely saw hyphae projecting from the streaks in the grf10Δ mutant (Fig. 5B) and the chlamydospores were slow to form or did not form at all at the ends of the hyphal stalks in the null mutants (Fig. 5D and F). The defect in chlamydospore formation was also observed in the heterozygous mutant strains in the BWP17 background (Fig. 5G–H), indicating haploinsufficiency for this trait. In the SN152 background, we observed unusually large, round cells lacking a thick cell wall (Fig. S2, Supporting Information).
Figure 5.
The grf10Δ mutant is defective in forming chlamydospores. Chlamydospore formation was induced on corn meal agar medium. View of the edge of a streak showing filamentation from (A) OHWT and (B) the grf10Δ mutant at low magnification (1.25× magnification). Representative chlamydospores or individual cells observed from (C) strain OHWT, (D) the grf10Δ mutant TF021, (E) BWP17, (F) the grf10Δ mutant RAC117, (G) the GRF10/grf10Δ heterozygous strain RAC114, (H) and the GRF10 restored strain RAC120. Panels (C) and (D), 20× magnification; panels (E–H), 40× magnification.
The grf10Δ mutants are deficient in biofilm formation
Filamentation is required for the formation of the Candida biofilm (Blankenship and Mitchell 2006). Since the grf10Δ mutant exhibited a filamentation defect, we investigated whether the ability to form a biofilm was also affected in the mutants. We compared biofilm formation in the null and heterozygous mutants with the parental strain BWP17 using a spectrophotometric assay (Krueger et al. 2004). As shown in Fig. 6, the parental strain BWP17 produced a dense biofilm (OD595 of 0.58 ± 0.02). We observed a significantly reduced biofilm formation in the grf10Δ mutant (OD595 of 0.2 ± 0.016, p = <0.001) as well as in the heterozygote strains RAC114 (OD595 of 0.39 ± 0.01, p = <0.001) and RAC120 (OD595 of 0.4 ± 0.02, p = 0.009), indicating a gene dosage effect for Grf10.
Figure 6.
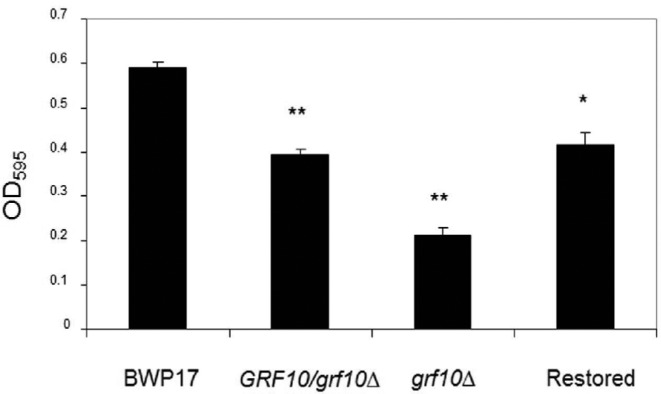
grf10Δ mutants were reduced in biofilm formation. Homozygous and heterozygous mutant and wild-type strains were allowed to form a biofilm, and the extent of biofilm formation was assessed by measuring the OD595. Assays were performed in triplicate, the mean and standard deviation of the OD595 measurements are plotted. *, p-value = 0.009; **, p-value < 0.001 relative to BWP17.
The grf10Δ mutation attenuates mortality in a mouse model of infection
To determine whether GRF10 is required for virulence, we used a mouse model of disseminated candidiasis (Tsuchimori et al. 2000), comparing the survival of mice infected with the wild-type, grf10Δ null and grf10 heterozygous mutant strains. Mice infected with either of the two heterozygous GRF10 mutants (RAC114 and RAC120) survived to 12 days, whereas the wild-type strain (DAY185) caused mortality in 8 days (Fig. 7A). Remarkably, 80% of mice infected with the grf10Δ mutant (RAC117) survived for 4 weeks post infection, at which point the experiment was terminated. We repeated this result by infecting 10 additional mice with the grf10Δ mutant; again, we found that 70% of the mice survived for 4 weeks post infection. We note that these strains differ in their auxotrophies for arginine and histidine (see Table 1). We think that these differences are unlikely to have affected virulence because the two heterozygotes RAC114 and RAC120 had similar virulence profiles in spite of their auxotrophic differences, and secondly, Noble and Johnson found that neither the arg4Δ or his1Δ mutations had an effect on virulence in the mouse systemic infection model (Noble and Johnson 2005). The survival of the grf10Δ null infected mice is statistically different from the survival of the DAY185-infected mice, with a p-value of <0.0001.
Figure 7.
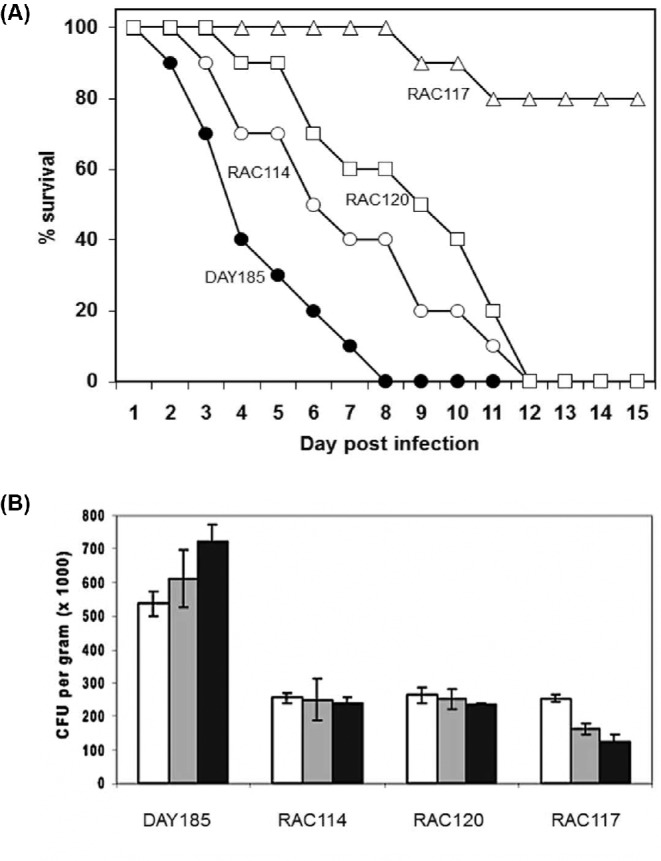
The grf10Δ mutants are less virulent in a mouse model of infections. (A) Ten Balb/c mice were injected with 1 × 106 cells through the lateral tail vein for each strain and survival was monitored for 15 d post-infection. •, wild type (DAY185); º, heterozygote (RAC114); Δ, homozygous null (RAC117); □, restored strain (RAC120). (B) Mice were sacrificed during the first three days after infection, and kidney homogenates were prepared and plated to determine the extent of colonization (colony forming units per gram; CFU/g). Histogram color: white, 24 h after infection; grey, 48 h after infection; black, 72 h after infection. The GRF10 genotype of the strains used in both panels are wild-type DAY185 (GRF10/GRF10), RAC114 (GRF10/grf10Δ), RAC120 (grf10Δ/grf10Δ::GRF10) and RAC117 (grf10Δ/ grf10Δ).
To examine the ability of the mice to clear the infection, we prepared kidney homogenates from the mice sacrificed during the first three days after infection, and plated serial dilutions of the homogenate to determine the extent of colonization. For mice infected with DAY185, we found a high colony count (5.6 × 105 cfu/gm tissue) with a trend that suggested an increase in colony number (Fig. 7B). Interestingly, the two grf10-heterozygous strains (RAC114 and RAC120) displayed a cell count that was about half of DAY185 (2.5 × 105 cfu/gm tissue) and that was the same finding in the animals sacrificed at 48 and 72 h. Infection with the grf10Δ strain (RAC117) displayed the same lower cfu/gm tissue at 24 h as found in the heterozygous mutant strains, but the cell counts decreased from the mice sacrificed during the next two days. These findings are consistent with the attenuation of virulence that we observed.
Expression of GRF10 gene increases during stationary phase and filamentation
Given the important role for Grf10 during filamentation, we wondered whether its gene expression changed during filamentation. To investigate this, we examined expression of GRF10 gene during planktonic yeast growth in log phase and during stationary phase in SC medium, and after hyphal induction on Spider medium. In this analysis, strain SC5314 strain was used as it was parental to both parental strains SN152 and BWP17, and the derived grf10 mutants. We found that expression of GRF10 was induced approximately 4-fold during stationary phase and filamentous growth (Fig. 8).
Figure 8.
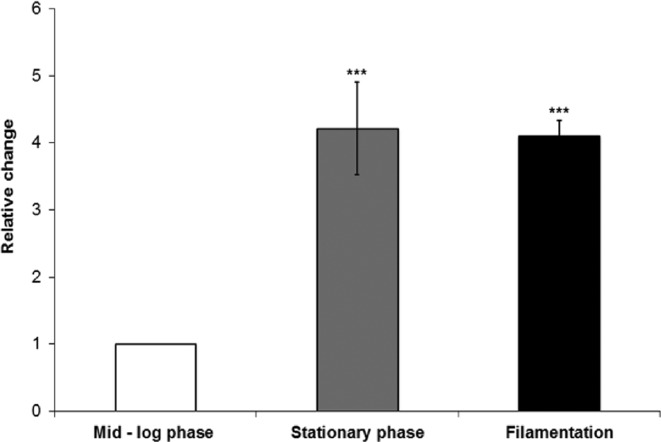
The GRF10 gene is induced during stationary phase and filamentation. Strain SC5314 was grown in SC + Uri medium to mid-log phase, stationary phase, or induced for filamentation in Spider medium. GRF10 expression was quantified by qRT-PCR and normalized to TEF1 (EFT3). Three biological samples and two technical replicates (n = 6) were performed. Student's t-test was performed using Excel. ***p-value < 0.0001.
DISCUSSION
In eukaryotes, the evolutionarily conserved homeobox genes play critical roles in pattern formation and morphology in animals, plants and fungi. Filamentation and spore formation are crucial developmental steps in the life cycle of most fungi. Strikingly, these developmental processes are controlled by homeobox genes in many diverse fungal species (Berben, Legrain and Hilger 1988; Torres-Guzmán and Domínguez 1997; Arnaise et al. 2001; Colot, Park and Turner 2006; Kim, Park and Kim 2009; Liu, Xie and Zhao 2010; Antal et al. 2012; Kim, Kwon and Roe 2012; Lee et al. 2014). The absence of the homeobox GRF10 gene leads to defects in filamentous growth in C. albicans. Although it was reported that the grf10Δ mutant did not impair filamentation (Homann et al. 2009), we found morphology defects in both the SN152 background as well as in the BWP17 background. The involvement of GRF10 in C. albicans morphology is supported by the finding that overexpression of GRF10 triggers filamentous growth both on solid and in liquid media under conditions that normally support yeast growth (Chauvel, Nesseir and Cabral 2012).
Consistent with a defect in filamentation, we found that processes dependent upon filamentation were also affected in the grf10Δ mutant. These included chlamydospore development (Fig. 5), biofilm formation (Fig. 6), and virulence in two different mouse infection models (Fig. 7 and Romanowski, Zaborin and Valuckaite 2012). The chlamydospore defect, which had not been reported previously in either strain background, was a very robust phenotype and exhibited haploinsufficiency. The role for chlamydospores is currently unknown, but they may represent a specialized growth form (Citiulo et al. 2009). Chlamydospore formation is generally induced when the cells are grown on nutrient-poor and under oxygen-limited conditions, suggesting that the Grf10 transcription factor may control hyphal development under specific environmental cues.
Expression of the GRF10 gene is upregulated during filamentation and stationary phase. It was previously shown to be upregulated during C. albicans biofilm formation (Yeater, Chandra and Cheng 2007; Nobile, Fox and Nett 2012; Desai, Bruno and Ganguly 2013) and it was identified as one of eight core target genes whose expression was regulated by the biofilm master regulators (Nobile, Fox and Nett 2012). Five of the six biofilm master regulators—Efg1, Bcr1, Brg1, Ndt80 and Tec1—bind to the promoter of GRF10. As the overexpression of GRF10 can rescue the biofilm defects in only the tec1Δ and bcr1Δ strains, Nobile and colleagues suggest that it acts downstream of Tec1p and Bcr1p (Nobile, Fox and Nett 2012). Tec1p is required for hyphal formation whereas Bcr1p regulates the expression of genes related to adherence (Nobile and Mitchell 2005). The upregulation of GRF10 gene during stationary and filamentous phases may be required to support the additional roles for the Grf10 transcription factor during filamentation.
In many fungal species, entry into stationary phase by environmental stress or nutrient starvation can trigger mating, meiotic division and spore formation, and/or invasive growth by pseudohyphae and true hyphae (Gimeno et al. 1992; Chu et al. 1998; Mata et al. 2002). Our results reveal that the GRF10 gene is induced by at least 4-fold during stationary phase and in response to hyphal-inducing conditions. Interestingly, Phx1 gene, the GRF10 homolog in the fission yeast S. pombe, is upregulated during stationary phase; Phx1 induces expression of pyruvate decarboxylase and genes for the biosynthesis of its co-factor thiamine (Kim, Kwon and Roe 2012; Kim et al. 2014). These gene products are proposed to shift carbohydrate metabolism from respiration to fermentation, which the authors suggest decreases the reactive oxygen species (ROS) and promotes long-term survival. It still remains unclear if Grf10 transcription factor plays a role in metabolic control in addition to filamentation during C. albicans stationary phase.
Homeodomain proteins commonly interact with another protein partner to regulate target genes (Mann, Lelli and Joshi 2009). Pho2, the GRF10 orthologue from S. cerevisiae, interacts with at least three transcription factor partners, Bas1, Pho4, and Swi5 (Bhoite, Allen and Garcia 2002). Orthologues of these protein partners are conserved in C. albicans (Bas1, Pho4 and Ace2). C. albicans BAS1 mutants have an adenine auxotrophy (Homann et al. 2009), and we have data showing that Bas1 regulates purine biosynthetic genes with Grf10 (Wangsanut et al. manuscript in preparation). In C. albicans, the Pho4 transcription factor plays a role in the phosphate response and its role in filamentation has been reported (Romanowski, Zaborin and Valuckaite 2012); however, it does not require Grf10 to upregulate phosphate metabolic genes (unpublished data and (Homann et al. 2009; Kerwin and Wykoff 2009)). In S. cerevisiae, SWI5 and ACE2 are paralogs, arising from the whole genome duplication; Swi5 cooperatively binds with Pho2 to activate HO gene to initiate mating type switching while Ace2 activates its target genes independently from Pho2 cooperation (Dohrmann, Voth and Stillman 1996). C. albicans possesses a single orthologue Ace2 (Butler et al. 2004; Kelly et al. 2004) that plays a direct role in morphogenesis through the RAM pathway (Regulation of Ace2 Morphogenesis). Ace2 is required for cell separation, adherence and virulence in animal studies and is a positive regulator of biofilm formation (Kelly et al. 2004; Finkel, Xu and Huang 2012). We do not know if Grf10 interacts with Ace2 or another transcription factor to regulate its target genes. Further experiments on protein–protein interaction and identification of specific target genes during morphogenesis are needed to enlighten the functions of the homeobox GRF10 gene in C. albicans.
Acknowledgments
Thank you to Jessica Quick and Lilli Seabol for technical assistance.
SUPPLEMENTARY DATA
FUNDING
This work was supported by NIH grant no. 1R03AI075226 and Georgetown University Pilot Project funds to RJR.
Conflict of interest. None declared.
REFERENCES
- Antal Z, Rascle C, Cimerman A, et al. The homeobox BcHOX8 gene in Botrytis cinerea regulates vegetative growth and morphology. PLoS One. 2012;7:e48134. doi: 10.1371/journal.pone.0048134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaise S, Zickler D, Poisier C, et al. pah1: a homeobox gene involved in hyphal morphology and microconidiogenesis in the filamentous ascomycete Podospora anserina. Mol Microbiol. 2001;39:54–64. doi: 10.1046/j.1365-2958.2001.02163.x. [DOI] [PubMed] [Google Scholar]
- AVMA. Report of the AVMA panel on euthanasia. J Am Vet Med Assoc. 2000;2001;218:669–96. doi: 10.2460/javma.2001.218.669. [DOI] [PubMed] [Google Scholar]
- Berben G, Legrain M, Hilger F. Studies on the structure, expression and function of the yeast regulatory gene PHO2. Gene. 1988;66:307–12. doi: 10.1016/0378-1119(88)90367-8. [DOI] [PubMed] [Google Scholar]
- Bhoite LT, Allen J, Garcia E, et al. Mutations in the Pho2 (Bas2) transcription factor that differentially affect activation with its partner proteins Bas1, Pho4, and Swi5. J Biol Chem. 2002;277:37612–8. doi: 10.1074/jbc.M206125200. [DOI] [PubMed] [Google Scholar]
- Biswas S, Van Dijck P, Datta A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol Mol Biol R. 2007;71:348–76. doi: 10.1128/MMBR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship JR, Mitchell AP. How to build a biofilm: a fungal perspective. Curr Opin Microbiol. 2006;9:588–94. doi: 10.1016/j.mib.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Braun BR, Johnson AD. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277:105–9. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- Braun BR, Kadosh D, Johnson AD. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 2001;20:4753–61. doi: 10.1093/emboj/20.17.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burglin TR. Homeodomain subtypes and functional diversity. Sub-cell Biochem. 2011;52:95–122. doi: 10.1007/978-90-481-9069-0_5. [DOI] [PubMed] [Google Scholar]
- Butler G, Kenny C, Fagan A, et al. Evolution of the MAT locus and its Ho endonuclease in yeast species. P Natl Acad Sci U S A. 2004;101:1632–7. doi: 10.1073/pnas.0304170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan N, Ciudad T, Rodriquez-Alejandre A, et al. Virulence and karyotype analyses of rad52 mutants of Candida albicans: regeneration of a truncated chromosome of a reintegrant strain (rad52/RAD52) in the host. Infect Immun. 2005;73:8069–78. doi: 10.1128/IAI.73.12.8069-8078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvel M, Nesseir A, Cabral V, et al. A versatile overexpression strategy in the pathogenic yeast Candida albicans: identification of regulators of morphogenesis and fitness. PLoS One. 2012;7:e45912. doi: 10.1371/journal.pone.0045912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, et al. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Citiulo F, Moran GP, Coleman DC, et al. Purification and germination of Candida albicans and Candida dubliniensis chlamydospores cultured in liquid media. FEMS Yeast Res. 2009;9:1051–60. doi: 10.1111/j.1567-1364.2009.00533.x. [DOI] [PubMed] [Google Scholar]
- Colot HV, Park G, Turner GE, et al. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. P Natl Acad Sci U S A. 2006;103:10352–7. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D, Edwards JE, Mitchell AP, et al. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect Immun. 2000;68:5953–9. doi: 10.1128/iai.68.10.5953-5959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai JV, Bruno VM, Ganguly S, et al. Regulatory role of glycerol in Candida albicans biofilm formation. MBio. 2013;4:ej00637–00612. doi: 10.1128/mBio.00637-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrmann PR, Voth WP, Stillman DJ. Role of negative regulation in promoter specificity of the homologous transcriptional activators Ace2p and Swi5p. Mol Cell Biol. 1996;16:1746–58. doi: 10.1128/mcb.16.4.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel JS, Mitchell AP. Genetic control of Candida albicans biofilm development. Nat Rev Microbiol. 2011;9:109–18. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel JS, Xu W, Huang D, et al. Portrait of Candida albicans adherence regulators. PLoS Pathog. 2012;8:e1002525. doi: 10.1371/journal.ppat.1002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–28. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, et al. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–90. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Herskowitz I, Rine J, Strathern J. Mating-type determination and mating-type interconversion in Saccharomyces cerevisiae. In: Jones EW, Pringle JR, Broach JR, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1992. pp. 583–656. [Google Scholar]
- Holland PWH. Evolution of homeobox genes. WIREs Dev Biol. 2013;2:31–45. doi: 10.1002/wdev.78. [DOI] [PubMed] [Google Scholar]
- Homann OR, Dea J, Noble SM, et al. A phenotypic profile of the Candida albicans regulatory network. PLos Genet. 2009;5:e1000783. doi: 10.1371/journal.pgen.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis DO, Sherlock G. Ras signaling gets fine-tuned: Regulation of multiple pathogenic traits of Candida albicans. Euk Cell. 2013;12:1316–25. doi: 10.1128/EC.00094-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N, Yamamoto M, Yoshihara F, et al. Biochemical and genetic characterization of Rbf1p, a putative transcription factor of Candida albicans. Microbiology. 1997;143:429–35. doi: 10.1099/00221287-143-2-429. [DOI] [PubMed] [Google Scholar]
- Joshi KR, Solanki A, Prakash P. Morphological identification of Candida species on glucose agar, rice extract agar and corm meal agar with and without Tween-80. Indian J Pathol Micr. 1993;36:48–52. [PubMed] [Google Scholar]
- Kadosh D, Johnson AD. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, control filamentous growth and virulence in Candida albicans. Mol Cell Biol. 2001;21:2496–505. doi: 10.1128/MCB.21.7.2496-2505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly MT, MacCallum DM, Clancy SD, et al. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol Microbiol. 2004;53:969–83. doi: 10.1111/j.1365-2958.2004.04185.x. [DOI] [PubMed] [Google Scholar]
- Kerwin CL, Wykoff DD. Candida glabrata PHO4 is necessary and sufficient for Pho2-independent transcription of phosphate starvation genes. Genetics. 2009;182:471–9. doi: 10.1534/genetics.109.101063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalaf RA, Zitomer RS. The DNA binding protein Rfg1 is a repressor of filamentation in Candida albicans. Genetics. 2001;157:1503–12. doi: 10.1093/genetics/157.4.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Kwon ES, Roe JH. A homeobox protein Phx1 regulates long-term survival and meiotic sporulation in Schizosaccharomyces pombe. BMC Microbiol. 2012;12:86. doi: 10.1186/1471-2180-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Kim EJ, Lopez-Maury L, et al. A metabolic strategy to enhance long-term survival by Phx1 through stationary phase-specific pyruvate decarboxylases in fission yeast. Aging. 2014;6:587–601. doi: 10.18632/aging.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Park SY, Kim KS, et al. Homeobox transcription factors are required for conidiation and appressorium development in the rice blast fungus Magnaporthe oryzae. PLoS Genet. 2009;5:e1000757. doi: 10.1371/journal.pgen.1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger KE, Ghosh AK, Krom BP, et al. Deletion of the NOT4 gene impairs hyphal development and pathogenicity in Candida albicans. Microbiology+ 2004;150:229–40. doi: 10.1099/mic.0.26792-0. [DOI] [PubMed] [Google Scholar]
- Kruzel EK, Hull CM. Establishing an unusual cell type: how to make a dikaryon. Curr Opin Microbiol. 2010;13:706–11. doi: 10.1016/j.mib.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KL, Buckley HR, Campbell CC. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–53. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- Lee M, Kwon N, Choi JM, et al. NsdD is a key repressor of asexual development in Aspergillus nidulas. Genetics. 2014;197:159–73. doi: 10.1534/genetics.114.161430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Köhler JR, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–26. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- Liu W, Xie S, Zhao X, et al. A homeobox gene is essential for conidogenesis of the rice blast fungus Magnaporthe oryzae. Mol Plant Microbe In. 2010;23:366–75. doi: 10.1094/MPMI-23-4-0366. [DOI] [PubMed] [Google Scholar]
- Lo H-J, Köhler JR, Didomenico B, et al. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–49. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Mann RS, Lelli KM, Joshi R. Hox specificity: Unique roles for cofactors and collaborators. Curr Top Dev Biol. 2009;88:63–101. doi: 10.1016/S0070-2153(09)88003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Lyne R, Burns G, et al. The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet. 2002;32:143–7. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- Mathe L, Van Dijck P. Recent insights into Candida albicans biofilm resistance mechanisms. Curr Genet. 2013;59:251–64. doi: 10.1007/s00294-013-0400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allow efficient mating. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- Murad AM, Leng P, Straffon M, et al. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001;20:4742–52. doi: 10.1093/emboj/20.17.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Fox EP, Nett JE, et al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–38. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Mitchell AP. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol. 2005;15:1150–5. doi: 10.1016/j.cub.2005.05.047. [DOI] [PubMed] [Google Scholar]
- Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell. 2005;4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, French SW, Kohn LA, et al. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 2010;42:590–8. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds FC. Candida infections: an overview. Crit Rev Microbiol. 1987;15:1–5. doi: 10.3109/10408418709104444. [DOI] [PubMed] [Google Scholar]
- Odds FC, Van Nuffel L, Gow NA. Survival in experimental Candida albicans infections depends on inoculum growth conditions as well as animal host. Microbiology. 2000;146:1881–9. doi: 10.1099/00221287-146-8-1881. [DOI] [PubMed] [Google Scholar]
- Ramon AM, Porta A, Fonzi WA. Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J Bacteriol. 1999;181:7524–30. doi: 10.1128/jb.181.24.7524-7530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski K, Zaborin A, Valuckaite V, et al. Candida albicans isolates from the gut of critically ill patients respond to phosphate limitation by expressing filaments and a lethal phenotype. PLoS One. 2012;7:e30119. doi: 10.1371/journal.pone.0030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer A, Rupp S, Taylor BN, et al. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol Microbiol. 2000;38:435–45. doi: 10.1046/j.1365-2958.2000.02132.x. [DOI] [PubMed] [Google Scholar]
- Sellam A, Askew C, Epp E, et al. Role of transcription factor CaNdt80p in cell separation, hyphal growth and virulence in Candida albicans. Eukaryot Cell. 2010;9:634–44. doi: 10.1128/EC.00325-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting Started with Yeast. In: Gutherie C, Fink GR, editors. Methods of Enzymology. Vol. 194. San Diego, CA: Academic Press; 1991. pp. 3–21. [DOI] [PubMed] [Google Scholar]
- Stoldt VR, Sonneborn A, Leuker C, et al. Efg1, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–91. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery P, Gow NA, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–24. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9:737–48. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- Thompson DS, Carlisle PL, Kadosh D. Coevolution of morphology and virulence in Candida species. Eukaryot Cell. 2011;10:1173–82. doi: 10.1128/EC.05085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Guzmán JC, Domínguez A. HOY1, a homeo gene required for hyphal formation in Yarrowia lipolytica. Mol Cell Biol. 1997;17:6283–93. doi: 10.1128/mcb.17.11.6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimori N, Sharkey LL, Fonzi WA, et al. Reduced virulence of HWP1-deficient mutants of Candida albicans and their interactions with host cells. Infect Immun. 2000;68:1997–2002. doi: 10.1128/iai.68.4.1997-2002.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M, Bachewich C. Morphogenesis in Candida albicans. Annu Rev Microbiol. 2007;61:529–53. doi: 10.1146/annurev.micro.61.080706.093341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–74. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeater KM, Chandra J, Cheng G, et al. Temporal analysis of Candida albicans gene expression during biofilm development. Microbiology+ 2007;153:2373–85. doi: 10.1099/mic.0.2007/006163-0. [DOI] [PubMed] [Google Scholar]