Abstract
BsrE/SR5 is a new type I toxin/antitoxin system located on the prophage-like region P6 of the Bacillus subtilis chromosome. The bsrE gene encoding a 30-amino acid hydrophobic toxin and the antitoxin gene sr5 overlap at their 3′ ends by 112 bp. Overexpression of bsrE causes cell lysis on agar plates. Here, we present a detailed in vitro analysis of bsrE/SR5. The secondary structures of SR5, bsrE mRNA, and the SR5/bsrE RNA complex were determined. Apparent binding rate constants (kapp) of wild-type and mutated SR5 species with wild-type bsrE mRNA were calculated, and SR5 regions required for efficient inhibition of bsrE mRNA narrowed down. In vivo studies confirmed the in vitro data but indicated that a so far unknown RNA binding protein might exist in B. subtilis that can promote antitoxin/toxin RNA interaction. Using time course experiments, the binding pathway of SR5 and bsrE RNA was elucidated. A comparison with the previously well characterized type I TA system from the B. subtilis chromosome, bsrG/SR4, reveals similarities but also significant differences.
Keywords: antisense RNA, Bacillus, gene regulation, RNA folding, RNA structure, antisense RNA
Introduction
Small regulatory RNAs (sRNAs)3 are key players in bacterial post-transcriptional gene regulation and have been discovered in a plethora of species (reviewed in Refs. 1–3). They employ either RNA/RNA base pairing or protein binding to inhibit or activate target gene expression. A special case of base pairing sRNAs is type I antitoxins that interact with complementary mRNAs encoding small toxic peptides (reviewed in Refs. 4 and 5).
Originally, type I toxin-antitoxin (TA) systems were discovered on plasmids (e.g. hok/Sok on Escherichia coli plasmid R1 (6, 7) or fst/RNAII on Enterococcus faecalis plasmid pAD1 (reviewed in Refs. 8 and 9)), in which they act as postsegregational killing systems. Subsequently, many chromosome-encoded type I TA systems were found and investigated, e.g. in E. coli tisB/IstR1 (reviewed in Ref. 10); symE/SymR (reviewed in Ref. 11); in ibs/Sib, shoB/OhsC, and zor/Orz (reviewed in Ref. 12); and in B. subtilis txpA/RatA (13) and bsrG/SR4 (14). They are arranged as overlapping, convergently transcribed gene pairs or as divergently transcribed gene pairs located apart. The interaction between RNA antitoxin and toxin mRNA either inhibits translation or facilitates degradation of the toxin mRNA (5). One exception is bsrG/SR4, whose antitoxin SR4 is bifunctional: it promotes degradation of the toxin mRNA and inhibits toxin translation by inducing a structural alteration around the bsrG ribosome binding site (RBS) (15). Another exception is fst/RNAII from E. faecalis, in which antitoxin binding yields a complex that stabilizes both RNAs but prevents toxin translation (16).
Some chromosome-encoded type I TA systems are involved in persister formation (17–20), whereas others are involved in recycling of damaged RNA (21), DNA recombination (22), or antibiotic resistance (23). Of 14 predicted B. subtilis type I systems, 5 are located on prophages and might be required for their maintenance or overcoming metabolic or environmental stress (24). Recently, we published the first temperature-dependent type I TA system bsrG/SR4 and investigated it both in vivo and in vitro (14, 15). The 38-amino acid hydrophobic toxin BsrG causes membrane invaginations that dislocate the cell-wall synthesis machinery, which finally leads to cell death in the absence of the antitoxin SR4 (25). SR4 (180 nt) is complementary to the 3′ end of bsrG mRNA (294 nt) and promotes its degradation by an RNase III-dependent mechanism. In addition, it impedes bsrG translation (Ref. 15 and see above). The amount of bsrG RNA, but not SR4, decreases drastically upon heat shock (48 °C) because of faster degradation at high temperatures (14). Similar to fst/RNAII (8) but unlike hok/Sok (26) or sib/Ibs (27), bsrG RNA/SR4 binding occurs by three progressive interactions between sets of complementary regions (15). bsrE/anti-bsrE in B. subtilis was proposed to be a type I TA system (24, 28), and we renamed it bsrE/SR5. We demonstrated that bsrE/SR5 is a type I TA system in vivo in B. subtilis, i.e. bsrE overexpression causes cell lysis.4 Intracellular amounts, expression profiles, and half-lives of SR5 and bsrE RNA were determined, and their decay by different RNases was investigated. RNase J1 is the major player in degradation of both RNAs, and RNase III cleaves the bsrE/SR5 duplex. Interestingly, SR5 and/or bsrE responds to multiple stresses, e.g. anoxia, Fe2+ limitation, ethanol, and pH alterations. In addition, bsrE RNA is heat shock-sensitive but, unlike bsrG, only in the presence of SR5.4
Here, we provide a detailed in vitro characterization of SR5, bsrE RNA, and the SR5/bsrE complex. Secondary structures of both RNAs, as well as that of the complex, were determined, and the region of initial contact between antitoxin and toxin mRNA was identified. The SR5/bsrE RNA binding pathway was elucidated employing EMSAs and time course experiments. Eventually, in vivo experiments with plasmids expressing bsrE and SR5 derivatives suggest that an unknown RNA binding protein could overcome the deficiency of truncated SR5 species to interact with bsrE RNA. A comparison between bsrE/SR5 and the structurally highly similar bsrG/SR4 type I TA system revealed commonalities but also interesting differences.
Experimental Procedures
Enzymes, Media, and Strains
The chemicals used were of the highest purity available. Taq DNA polymerase from Roche, Firepol polymerase from Solis Biodyne, RNases T1 and V1 from AMBION, RNase T2 from Sigma and nuclease S1 from Fermentas were used. T7 RNA polymerase, calf-intestinal phosphatase, and T4 nucleotide kinase were from New England Biolabs, and ThermoScript reverse transcriptase was from Invitrogen. B. subtilis strains DB104 and DB104(Δsr5ΔbsrE::cat)4 and E. coli strain TG1 were grown in complex TY medium (30). B. subtilis strain DB104(Δhfq::cat) (31) was used for the construction of a Δhfq::cat/ΔbsrEΔsr5::spec strain. B. subtilis strain GP49 (ΔcsrA) was used to construct a ΔcsrA::spec/ΔbsrEΔsr5::cat strain.
In Vitro Transcription and Secondary Structure Analysis
In vitro transcription and partial digestions of in vitro synthesized 5′ end-labeled SR5 and bsrE RNA species with RNases T1 (1 unit/μl), T2 (22 units/μl), and V (0.1 unit/μl) and nuclease S1 (100 units/μl) were carried out as described (15). Additionally, bsrE RNA was 3′ end-labeled with [32P]pCp and RNA ligase for 30 min at 37 °C. For the analysis of SR5/bsrE complexes with T1 and T2, either SR5 or bsrE RNA was 5′ end-labeled, and either an equimolar amount or a 10- or 100-fold excess of unlabeled complementary RNA was added prior to RNase digestion.
Analysis of RNA-RNA Complex Formation and Time Course Experiments
Both bsrE RNA and SR5 were synthesized in vitro either on PCR-generated template fragments or on double strands generated by hybridizing complementary oligodeoxyribonucleotides (supplemental Table S1). SR5/bsrE complex formation studies and time course experiments with in vitro synthesized 5′-labeled SR5 (0.025 μm) and a 2-fold excess of unlabeled bsrE RNA were performed as described previously (15). RNases T1 and T2 were used at 10−2 U and 2.2 × 10−2 units, respectively.
Preparation of Total RNA and Northern Blotting
Preparation of total RNA and Northern blotting were carried out as described (14).
Construction of Plasmids for in Vivo Assays
High copy number plasmids were constructed to overexpress wild-type bsrE and truncated sr5 variants in B. subtilis under control of their own promoters. Plasmid pUCBES4 was generated as follows: a PCR on chromosomal DNA from strain DB104 with primer pair SB2045/SB2265 yielded a fragment comprising the sequence for truncated sr51–50 followed by the heterologous bsrF terminator (32). It was digested with BamHI and PstI and inserted into pUC19. After confirmation of the sequence, the fragment was recloned into BamHI/PstI digested pUCBES2 carrying the wild-type bsrE gene. The resulting plasmid pUCBES4 that comprises wild-type bsrE, and truncated sr5 was used for transformation of B. subtilis DB104(ΔbsrE/Δsr5::cat). Plasmids pUCBES7, 8, and 9 containing wild-type bsrE and mutated sr5 variants were constructed likewise using the primer pairs listed in supplemental Table S1. Table 1 summarizes all plasmids. Plasmid pUCBES6 was constructed using three subsequent PCR steps. First, three parallel PCRs were performed on plasmid pUCBES2 as template using primer pairs SB2045/SB2356, SB2417/SB2400 and SB2418/SB2355 yielding fragments F1, F2, and F3, respectively. In the second PCR, fragments F2 and F3 were used as templates with primer pair SB2045/SB2418 resulting in fragment F4. Finally, F1 and F4 were combined and amplified with primer pair SB2045/2400. The resulting fragment, F5, was digested with BamHI and PstI and inserted into pUCBES2 as described above.
TABLE 1.
Plasmids used in this work
| Plasmid | Description | Reference |
|---|---|---|
| pUC19 | E. coli cloning vector, AmpR, MCS | Ref. 29 |
| pUCB2 | pUC19 /pUB110 shuttle vector, NeoR, PhleoR, AmpR | Brantl (unpublished observations) |
| pUCBE1 | pUCB2 with bsrE gene under own promoter and 96 bp upstream of − 35 box of pbsrE | Footnote 4 |
| pUCB5.1 | pUCB2 with sr5 gene under own promoter and 87 bp upstream of − 35 box of psr5 | Footnote 4 |
| pUCBES2 | pUCB5.1 with bsrE gene under own promoter and 96 bp upstream of − 35 box | Footnote 4 |
| pUCBES4 | As pUCBES2, nt 51–163 of SR5 are lacking | This study |
| pUCBES6 | As pUCBES2, SR5 loop sequences of SL2 and SL4 exchanged by heterologous sequences | This study |
| pUCBES7 | As pUCBES2, nt 1–47, 91–104, and 126–163 of SR5 are lacking | This study |
| pUCBES8 | As pUCBES2, nt 49–78 of SR5 are lacking | This study |
| pUCBES9 | As pUCBES2, nt 1–46 and 105–163 of SR5 are lacking | This study |
Results
Secondary Structures of SR5 and bsrE mRNA
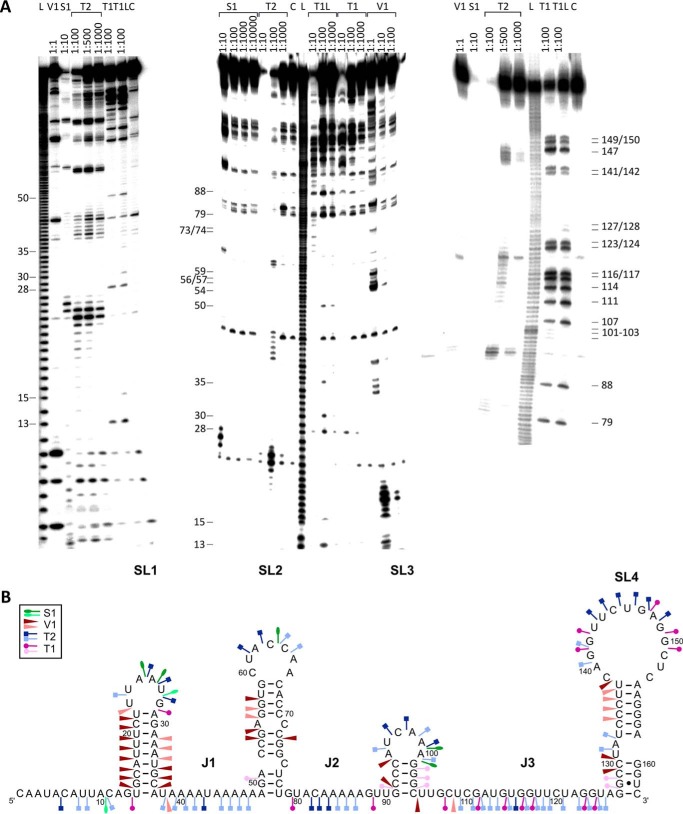
Computer-predicted sRNA structures often differ from experimentally determined ones (15, 33, 34). Therefore, we conducted limited digestions with structure-specific ribonucleases to determine the secondary structures of SR5 and bsrE RNA. Full-length SR5 (163 nt), as well as 5′ truncated SR547–163 (not shown), was 5′ end-labeled, gel-purified, and treated with RNases T1 (cleaves 3′ of unpaired G residues), T2 (unpaired nucleotides with a slight preference for A residues), and V1 (double-stranded and stacked regions) and nuclease S1 (single-stranded nucleotides). Fig. 1A shows the analysis of SR5, and Fig. 1B displays the schematic presentation of the SR5 structure inferred from the cleavage data. SR5 contains four stem loops, SL1, SL2, SL3, and SL4, interrupted by unpaired regions, with SL4 being the terminator stem loop. Structure probing of full-length SR5 (Fig. 1) and a truncated SR547–163 (not shown) revealed that the four stem loops fold independently.
FIGURE 1.
Secondary structure of SR5 (163 nt). A, secondary structure probing of SR5 with RNases. Purified, 5′-labeled wild-type SR5 was subjected to limited cleavage with the RNases indicated. Digested RNAs were separated on 15% (left) or 8% (center and right) denaturing gels. Autoradiograms are shown. C, control without RNase treatment; L, alkaline ladder; T1L, T1 digestion under denaturing conditions. The nucleotide positions are indicated. B, proposed secondary structure of SR5. A structure consistent with the cleavage data in Fig. 1A is depicted. Major (dark symbols) and minor (light symbols in the same color) cuts are indicated (see box). The four stem loops SL1–SL4 and the single-stranded regions J1–J3 are indicated.
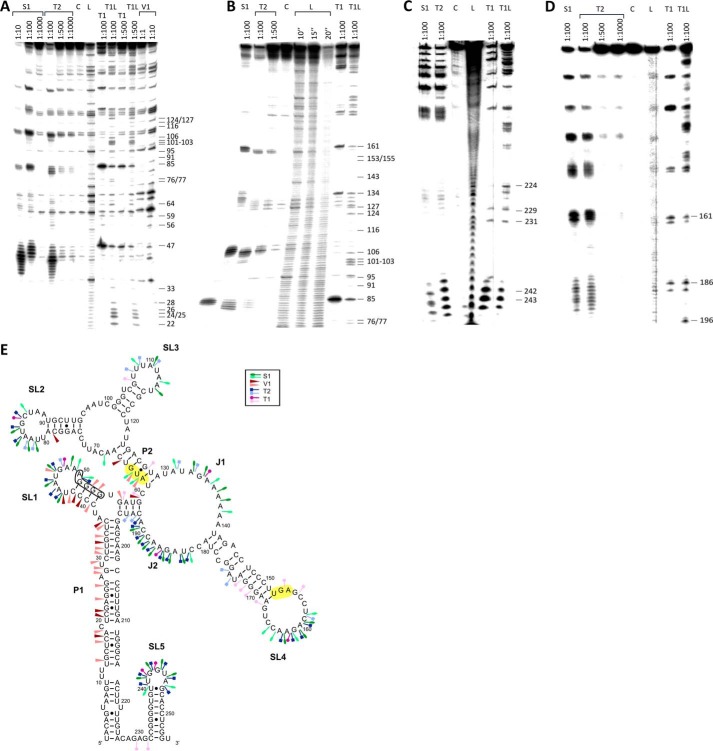
The secondary structure of full-length bsrE mRNA (255 nt) was determined by enzymatic probing of 5′ and 3′ end-labeled RNA species using nucleases T1, T2, V1, and S1 as described under “Experimental Procedures.” Representative gels are shown in Fig. 2,A–D, the schematic representation of the bsrE structure in Fig. 2E. Similar to bsrG RNA (15), bsrE RNA is highly structured and barely contains single-stranded regions: the 5′ nt 1–36 and the 3′ nt 196–226 base pair in the long helix P1, which is divided by bulged out nt into five subhelices. P1 branches on top into stem loop structure SL1 and a short paired region that itself branches into helix P2 and a large internal loop J1/J2 that is topped by stem loop SL4. On top of P2, an internal loop branches into SL2 and SL3. Alternatively, T2 signals at positions 192/193 could indicate that nt 57–59 might interact with nt 131–133 in J1 instead of with nt 192–194; however, this would result in 3 bp—among them one GU bp—instead of 4 bp, which would decrease the stability of this structure. The start codon is situated in the short paired region upstream of P2 and the stop codon is located in L4 (Fig. 2E). The detection of weak T1 cuts additionally to V1 cuts indicates certain breathing around the AUG start codon. The terminator stem loop SL4 comprises nt 232–254 and is separated from P1 by 5 unpaired nt. The sizes of loops L2, L3, and L4, as well as the length of single-stranded regions J1 and J2, were confirmed by using single-strand specific RNases and DMS-modified bsrE RNA (not shown).
FIGURE 2.
Secondary structure of bsrE mRNA (255 nt). A–D, secondary structure probing of bsrE RNA with RNases as in Fig. 1. Digested RNAs were separated on 8% (A, B, and D) or 15% (C) denaturing gels. A and B, 5′-labeled bsrE; C and D, 3′-labeled bsrE. Autoradiograms are shown. C, L, and T1L are as defined in the legend to Fig. 1. E, proposed secondary structure of bsrE mRNA. A structure inferred from the cleavage data in A–D is depicted. Symbols are as defined in the legend to Fig. 1. The long double-stranded helix P1, helix P2, the single-stranded regions J1 and J2, and the four main stem loops SL1–SL5 are indicated. The bsrE SD sequence is boxed. Start and stop codon are shaded in yellow.
Secondary Structure of the SR5/bsrE RNA Complex
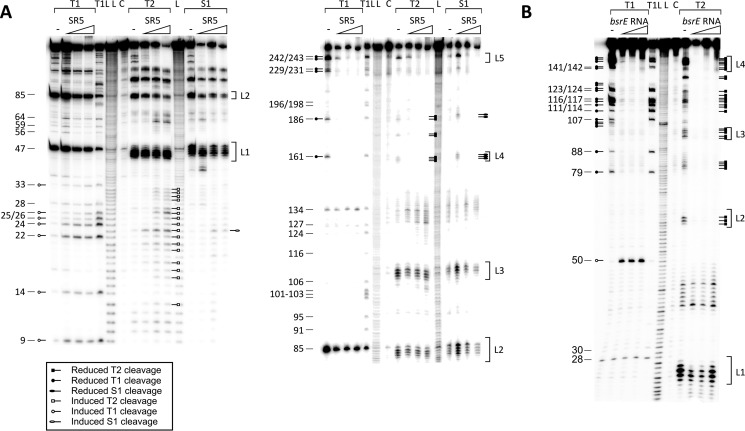
SR5 and bsrE RNA are complementary over 112 nt and therefore expected to form a stable complex. To investigate conformational alterations in the secondary structures of both RNAs upon pairing, the secondary structure of the SR5/bsrE RNA complex was determined. To ascertain alterations in the bsrE RNA structure, 5′-labeled bsrE RNA was incubated with an excess of unlabeled SR4, and the complex was allowed to form for 5 min at 37 °C and subsequently partially digested with T1, T2 and S1 (Fig. 3A). On the other hand, 5′-labeled SR5 was incubated with an excess of unlabeled bsrE RNA and treated likewise (Fig. 3B). In Fig. 4, the schematic representation of the complex is displayed. As expected, no alterations in SL1 of SR5 were observed, because this stem loop is not part of the region complementary to bsrE RNA. By contrast, the entire region of SR5 complementary to bsrE (nt 51–163) was found to be double-stranded in the complex, visible by a significant reduction of T1 and T2 specific signals already upon addition of equimolar amounts of the toxin mRNA.
FIGURE 3.
Secondary structure probing of the SR5/bsrE complex. 80 nm of purified 5′-labeled bsrE RNA (A) or 5′-labeled SR5 (B) were incubated with increasing amounts of the complementary unlabeled RNA (equimolar, 10- and 100-fold excess), and the complex allowed to form for 10 min at 37 °C and subjected to limited cleavage with T1 (10−2 units/μl), T2 (2.2 × 10−2 units/μl), or nuclease S1 (10 units/μl). Digested RNAs were separated on 8% denaturing gels. Autoradiograms are shown. C, L, and T1L are as defined in the legend to Fig. 1. Nucleotide positions are included. Altered T1 and T2 cleavages are indicated by the symbols in the box.
FIGURE 4.
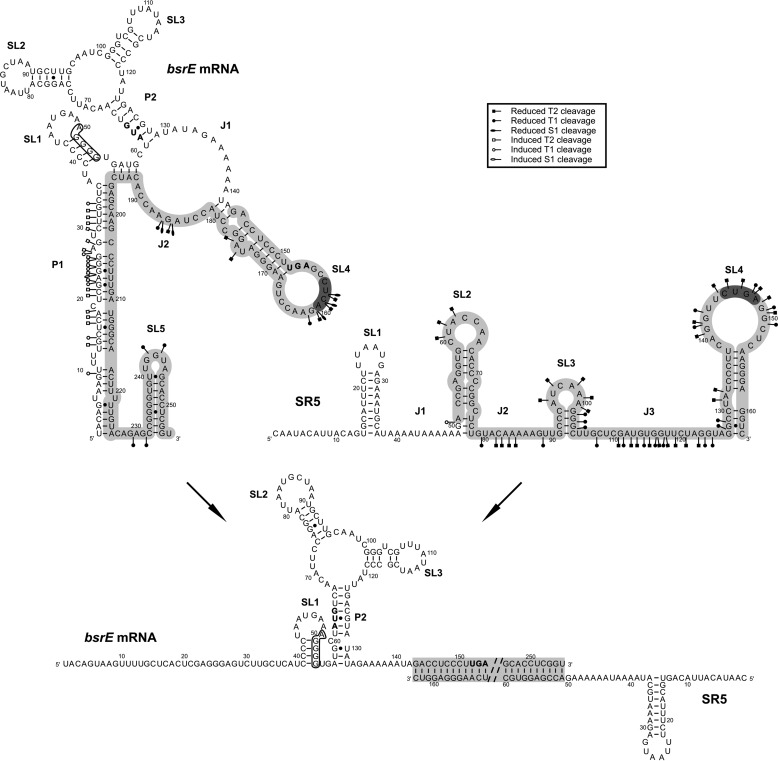
Alterations in the structures of bsrE RNA and SR5 upon complex formation. Altered cleavages are shown in bsrE RNA and SR5 separately (see box). The bsrE SD sequence is boxed, and complementary regions are shown in gray. 5′ YUNR motifs are highlighted in dark gray. The presentation of individual RNAs and the complex are inferred from the cleavage data in Figs. 1–3. The complementary region was completely double-stranded; therefore, only part of it is shown, and instead of the rest, two dashed lines are included.
In the bsrE/SR5 complex, the 5′ end of bsrE RNA until nt 36—in the absence of SR5 paired with the 3′ end forming helix P1—is single-stranded, as demonstrated by a multitude of binding-induced S1, T1, and T2 cuts. In the complex, helix P2 is extended at its bottom by 3 bp, because during complex formation nt 192–195 are no longer available for interaction with nt 56–59, and instead, nt 57–59 can base pair with nt 130–132. Further structural changes in the region complementary to SR5 were observed in SL4, SL5, and J2 in bsrE RNA and in the short single-stranded region that connects P1 and SL5. Interestingly, bsrE SL1 comprising the SD sequence was not subject to structural changes. Taken together, experimental probing of the bsrE/SR5 complex demonstrated that the complementary regions of both interaction partners form a perfect duplex.
Binding Assays of Wild-type and Truncated SR5/bsrE mRNA Pairs
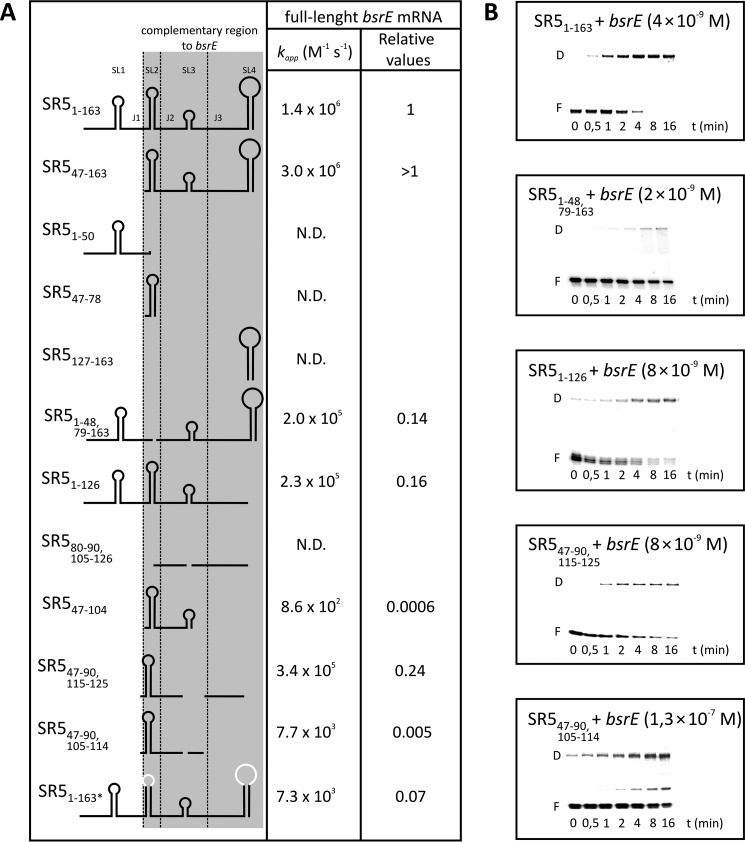
To analyze the kinetics of stable complex formation in vitro, we first studied binding between labeled full-length SR5 and unlabeled full-length bsrE RNA and vice versa by gel shift assays (EMSAs) as described under “Experimental Procedures.” Fig. 5A summarizes the calculated second order binding rate constants (kapp), and Fig. 5B shows representative EMSAs. For the full-length SR5/bsrE pair, a kapp value of ≈1.4 × 106 m−1 s−1 was determined, independent of which of the interacting RNAs was labeled and which was provided unlabeled in excess. To investigate which structural elements are required for stable complex formation, different truncations were introduced into SR5. bsrE RNA could not be truncated, because this altered its secondary structure (not shown). SR547–163 lacking only SL1 and J1 that are not complementary to bsrE RNA showed an even higher kapp value than full-length SR5, whereas SR51–50 comprising only SL1 and J1 was not able to bind bsrE RNA. Likewise, very short species comprising only SL2 (SR547–78) or the terminator stem loop SL4 (SR5127–163) proved to be unable to form a stable complex with bsrE RNA. However, individual deletions of either SL2 (SR51–48, 79–163) or SL4 (SR51–126) in an otherwise wild-type context decreased kapp to 14 or 16%, respectively, revealing that both loops contribute equally to stable pairing. That at least one stem loop of SR5 is required for complex formation was corroborated by an SR5 species composed of only single-stranded regions J2 and J3 (SR580–90, 102–126), which was unable to form a duplex.
FIGURE 5.
Binding assays of wild-type and truncated SR5/bsrE RNA pairs. Binding experiments were performed as described under “Experimental Procedures.” A, summary of pairing rate constants (kapp). The conformation of the used SR5 species is shown schematically in the left column, and sequence alterations are indicated. The kapp values were calculated as described previously (15). B, representative binding assays with wild-type and mutated SR5 derivatives. SR5 species were 5′-labeled with [γ-32P]ATP and used in at least 10-fold lower equimolar amounts compared with full-length bsrE RNA. The concentrations of unlabeled wild-type bsrE RNA species are indicated. F, free labeled RNA; D, SR5/bsrE RNA duplex. Replaced loop sequences are shown in white.
Because SL2 alone was unable to form a stable complex, a species containing additionally J2 and SL3 was tested (SR547–104). For this species, very weak binding was detected with a kapp value 3 orders of magnitude lower than that of full-length SR5. This could not be explained simply by the lack of SL4. Therefore, we added the long single-stranded region J3 (SR547–126) (not shown), and binding increased to 20% of the wild-type value, demonstrating that J3 might contribute significantly to stable pairing. Apparently, J3 of SR5 is important for efficient pairing. To find out whether SL3 is needed, we first deleted this stem loop (SR547–90,115–125). Indeed, there was no difference between the kapp value of this species and SR547–126. Subsequently, we further shortened the SR5-J3 region to only the 5′ part (SR547–90,105–114), which decreased the kapp value ≈50-fold. Therefore, the entire region single-stranded region J3 is crucial, because it interacts with a single-stranded bulge region in the complementary bsrE RNA. A full-length SR5163 with simultaneous nt exchanges in both loops L2 and L4 was significantly (≈14-fold) impaired in binding (Fig. 5).
In conclusion, SL2 and SL4 are fundamental for the formation of a stable duplex with bsrE RNA, because the lack of both SLs hardly allowed binding, and deletion of either of them or of both loop sequences decreased binding 6–7-fold. Furthermore, J3 of SR5 is essential for stable complex formation.
Binding Pathway of SR5 and bsrE mRNA
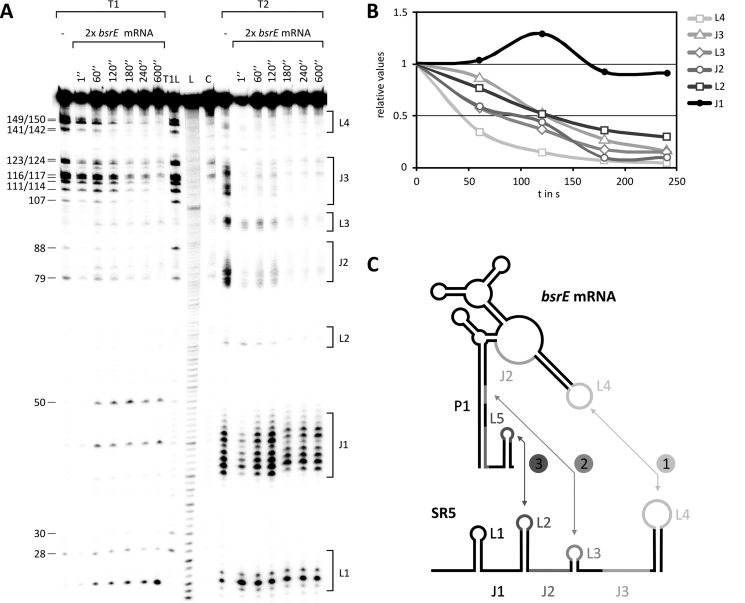
In vitro binding assays do not allow conclusions regarding whether binding between both RNAs initiates with one or two simultaneous loop-loop contact(s). Therefore, a time course experiment (see “Experimental Procedures”) using 5′-labeled SR5 and unlabeled bsrE RNA was performed to assay the sequential pairing between antisense and sense (toxin) RNA. As shown in Fig. 6 (A and B), T1 and T2 cleavage signals specific for SR5 loop L4 decreased 2-fold already 40 s after addition of bsrE RNA, indicating that the initial interaction occurs between this loop and bsrE L4.
FIGURE 6.
Time course of interaction between SR5 and bsrE RNA. A, autoradiogram of the time course experiment performed as described under “Experimental Procedures.” C, L, and T1L are as defined in the legend to Fig. 1. Loop regions L1–L4 and single-stranded regions J1 and J2 of SR5 are indicated at the right. B, graphic representation of progressive pairing between different regions of SR5 and bsrE RNA as derived from A. C, schematic representation of the binding pathway. Numbers indicate sequences of interaction.
This result is consistent with the data from the binding assays (Fig. 5) and not unexpected, because both loops contain 5′ YUNR motifs known to provide a scaffold for rapid RNA/RNA interactions (35, 36). The next contact occurs between loop L3 and the 3′ part of helix P1 of the bsrE RNA, because a 2-fold reduction of the L3 signals was observed 75 s after addition of bsrE RNA, indicating a conversion to a double-stranded region. Finally, intermolecular helix progression reached SR5 loop L2 and the 3′ part of J3 that binds to the terminator loop and J2 of bsrE RNA, respectively (50% bound after 120 s). After addition of bsrE RNA, duplex formation in the J3 region of SR5 was slowest. In direct comparison, the interaction with L2 was faster in the beginning, but went down to 50% also after 120 s. As expected, no alterations in the cleavage pattern were observed over 600 s for SR5-L1 that is not complementary to bsrE RNA. Fig. 6B shows the quantification of the gel in Fig. 6A, and Fig. 6C represents the binding pathway schematically.
In Vivo Studies
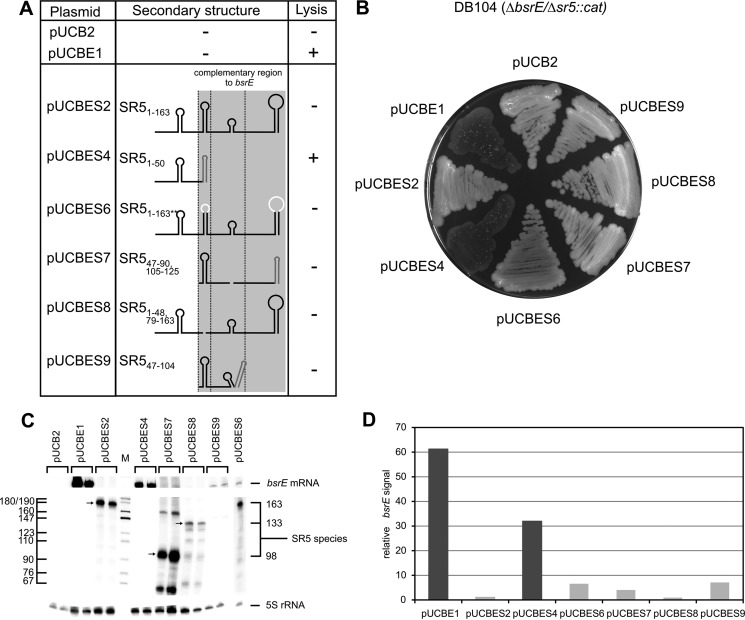
To corroborate the in vitro data in vivo, both bsrE and wild-type or mutated sr5 were expressed under their own promoters from plasmid pUCB2 (≈50 copies/cell) in B. subtilis DB104(Δsr5/ΔbsrE). Transformants were analyzed for lysis on agar plates after overnight incubation at 37 °C followed by 24 h at 24 °C (summarized in Fig. 6A). As expected, DB104(Δsr5/ΔbsrE) containing pUCBE1 lysed on agar plates. Likewise, DB104(Δsr5/ΔbsrE::cat) with pUCBES4 encoding bsrE and the noncomplementary 5′ part of sr5 (SL1 and J1) displayed a lysis phenotype. By contrast, the additional expression of full-length sr5 (plasmid pUCBES2) could perfectly compensate the lysis effect (Fig. 7B). Surprisingly, sr5 variants lacking SL2 (pUCBES8) or composed of only SL2 and single-stranded regions J2 and J3 (pUCBES7), which were still able to form a duplex with bsrE RNA in vitro, albeit with 7- or 4-fold reduced kapp values, respectively, could compensate lysis. Most surprisingly, however, an sr5 mutant comprising only SL2, J2, and SL3 (pUCBES9) that displayed a 1000-fold reduced kapp value in vitro (see SR547–104 in Fig. 5) did not lyse either. Similarly, both behaved like pUCBES6 carrying altered L2 and L4 loop sequences, which were >100-fold impaired in complex formation in vitro. This unexpected result could be either due to reduced expression of bsrE RNA or altered expression or stability of mutated SR5 species in the Δsr5/ΔbsrE strain. Alternatively, an RNA chaperone might be present in vivo that facilitates the interaction between bsrE RNA and the severely truncated SR5 species.
FIGURE 7.
Summary of in vivo data. A, plasmids listed were used to express wild-type bsrE RNA and wild-type or mutated SR5 species in trans from plasmid pUCB2 in a ΔbsrE/Δsr5 background. Test was for lysis at 37 °C followed by 1 day at room temperature. Lysis is indicated by +. The heterologous bsrF terminator is drawn in dark gray. B, agar plate illustrating lysis or wild-type phenotype of the different mutants. C, Northern blot analysis of ΔbsrE/Δsr5 strains grown until A560 = 3.0 expressing wild-type bsrE and wild-type or mutated sr5. [α-32P]UTP-labeled riboprobes were used for SR5 and bsrE RNA. Two independently grown cultures were used for RNA preparation, and samples from these cultures were loaded in parallel. Reprobing was performed with a [γ-32P]ATP-labeled oligonucleotide complementary to 5S rRNA. D, quantification of bsrE signals from the Northern blot in C. Large columns reflect high amounts of bsrE RNA indicative for an impaired ability of mutant SR5 to bind to and promote degradation of toxin mRNA. Dark columns, lysis; light columns, wild type.
First, we analyzed expression of bsrE and sr5 in the corresponding strains by Northern blotting (Fig. 7C) and quantified the relative amount of bsrE (Fig. 7D). In strains containing pUCBES2 and pUCBES8, the amount of bsrE RNA was ≈30-fold lower than in the strain with pUCBES4, which showed lysis. This suggests that a small stretch of complementarity on an SR5 variant is sufficient to direct bsrE RNA into the degradation pathway in vivo. For pUCBES7 and pUCBES9, a 5–8-fold higher amount of bsrE RNA was calculated compared with pUCBES2 and 8, although it was still 4–6-fold lower than in the case of pUCBES4, again indicative for the ability of the severely truncated SR5 variants to promote bsrE degradation. Despite these differences, for all strains (with either pUCBES2, 6, 7, 8, or 9) no lysis was observed. The amount of bsrE seems to be a more sensitive measure for the inhibitory effect of SR5 than cell lysis. Apparently, as soon as a small region of SR5 complementary to bsrE is expressed, it can promote its degradation in vivo.
Truncated SR5 species might display altered stabilities in vivo. However, SR5 species of the expected size were expressed from pUCBES2, 7, and 8 (Fig. 7C). The only exception was pUCBES9, in which the expected 60-nt-long species was not visible. However, this species was able to compensate lysis.
As shown in a recent paper,4 the RNA chaperone Hfq stabilized SR5 but not bsrE RNA. Furthermore, Dambach et al. (37) published an Hfq peak in their co-immunoprecipitation experiments to bsrE/SR5. Although cis-encoded sRNAs with a long stretch of complementarity do usually not need Hfq to promote the efficient interaction with their target RNA, it could not be excluded that SR5/bsrE is an exception. Therefore, we constructed a Δhfq::cat, Δsr5/ΔbsrE::spec strain, transformed it with all pUCBES derivatives and analyzed lysis on agar plates. Furthermore, as csrA is the only other protein known in B. subtilis to bind mRNAs and thus might be able to affect antisense/target RNA interaction in vivo, a ΔcsrA::spec Δsr5/ΔbsrE::cat strain was also generated and assayed with all pUCBES derivatives. However, both knock-out strains behaved like the isogenic wild-type strains, i.e. no lysis was observed when SR5 species even with a very short region complementary to bsrE were expressed, whereas the control without SR5 revealed lysis (not shown). Based on these results, we conclude that a so far unidentified RNA binding protein exists in B. subtilis that can overcome the inefficient interaction of severely truncated SR5 species with wild-type bsrE RNA.
Discussion
So far, 14 putative type I TA systems were predicted in B. subtilis (24), and three of them were confirmed in vivo and analyzed in vitro: txpA/RatA (13, 38), bsrG/SR4 (14, 15), and bsrE/SR5 (this report).4 These three systems show commonalities but also interesting differences. In all cases, the toxin is a small hydrophobic peptide that causes cell lysis on agar plates. Toxin action is neutralized by an RNA antitoxin that binds to the 3′ end of the toxin mRNA via a long (>100 nt) stretch of complementarity, thereby promoting its degradation. Degradation is initiated by RNase III cleavage. However, whereas RatA and SR5 seem to act exclusively by facilitating toxin RNA degradation, SR4 additionally affects toxin translation by inducing a structural change in the RBS that further obstructs ribosome binding. Binding between cis-encoded sRNAs and their target RNAs can either initiate with one loop-loop contact (e.g. CopA/CopT of plasmid R1 (39)), two simultaneous loop/loop contacts (e.g. RNAII/RNAIII of plasmid pIP501 (31)), or an interaction between a loop and a single-stranded region (e.g. hok/Sok (40)). The first applies to both bsrE/SR5 (Fig. 5) and bsrG/SR4 (15). Binding starts with one loop/loop interaction between toxin L3 and antitoxin terminator loop L4, the so-called recognition loops. Subsequently, it progresses toward the 3′ end of both complementary molecules via L3 and later the single-stranded region J3 and L3 of the antitoxin. However, both binding pathways reveal a few differences. Although antitoxin loop L2 is bound last by both bsrG and bsrE RNA, both L4 and L2 of SR5 are important for efficient interaction with bsrE RNA (Fig. 5), whereas in bsrG/SR4, a shorter antitoxin comprising only SL4 and the 3′ half of SL3, sufficed for inhibition. In txpA/RatA, binding seems to initiate via a loop/loop contact, too, but the binding pathway has not been analyzed in detail (38).
In type I TA systems with trans-encoded antitoxins such as E. faecalis fst/RNAII, E. coli ibs/Sib, or zorO/OrzO, only partial duplexes between antitoxin and toxin mRNA can be formed. fst/RNAII is similar to bsrG/SR4 or bsrE/SR5 as binding starts with one loop/loop contact at the two terminator loops, afterward involves the direct-repeat-a (Dra) region, reaches the direct-repeat-b (DRb) region, and from there progresses into the final complex (41). By contrast, in ibs/Sib, two simultaneous contacts between TRD1 and TRD2 of Sib antitoxin and their complementary domains in ibs RNA were observed (27). For E. coli zorO/OrzO and zorP/OrzP, the immediate 5′ end of the RNA antitoxin is required for specific pairing, but no binding pathway has been elucidated so far (42).
Apparent binding constants of cis-encoded antisense/sense RNA pairs are usually in the range of 1 × 105 m−1 s−1 to 1–3 × 106 m−1 s−1 (43). This was also confirmed for several type I TA systems (reviewed in Ref. 5). Accordingly, binding constants of ≈106 m−1 s−1 have been determined for wild-type bsrG/SR4 (15) and bsrE/SR5 (Fig. 5). For txpA/RatA, such calculations have not been performed.
U-turn motifs (5′ YUNR) have been predicted and sometimes experimentally verified in a variety of sense/antisense RNA systems in either antisense or target RNA (35). Whereas in some instances, they are important to provide a scaffold for the rapid interaction between the two complementary RNAs and consequently the biological function of the corresponding systems (e.g. hok/Sok of plasmid R1 (44) and RNAII/RNAIII of plasmid pIP501 (36)), they could be mutated in other systems without consequences for pairing rates and inhibitory function of the antisense RNA (45, 46). In a number of type I TA systems, loops displaying 5′ YUNR motifs are involved in the initial contact between toxin mRNA and RNA antitoxin. This was asserted for the fst terminator loop of E. faecalis fst/RNAII (reviewed in Refs. 8 and 9). Likewise, in the E. coli ibs/Sib TA system, the loop within the ibs mRNA ORF (27), decisive for discrimination between different ibs/Sib systems carries a 5′ YUNR motif. In txpA/RatA, neither of the interacting loops displays a 5′ YUNR motif (38). By contrast, in bsrE/SR5, two 5′ YUNR motifs, one in L4 of bsrE RNA and the other in L4 of SR5, are involved in the initial interaction between antitoxin and toxin mRNA (Fig. 6), and SR5 species without or with mutated L4 sequences were impaired in complex formation (Fig. 5) and pairing in vivo (Fig. 7). Because bsrE and bsrG mRNA have a highly similar secondary structure (Fig. 2 and Ref. 15), it is not surprising that loop L3 of bsrG RNA carrying a 5′ YUNR motif is also required for recognition by SR4. However, in bsrG/SR4, only the toxin RNA but not the complementary antitoxin loop L4 displays such a motif. Another potential U-turn motif in L2 of SR4 was neither required for efficient pairing with bsrG RNA in vitro nor for the neutralizing activity of SR4 in vivo (15). Taken together, in all but one (txpA/RatA) type I TA system, U-turn motifs play an important role in rapid and efficient primary contacts between RNA antitoxin and toxin mRNA. However, only in hok mRNA has the predicted U-turn been corroborated experimentally (44).
Different strategies exist that prevent premature toxin expression in type I TA systems. Although in some cases, processing events are required to generate shorter, translatable toxin mRNA (reviewed in Ref. 5), in others, the RBS is sequestered by complementary base pairing. The latter applies to E. faecalis fst/RNAII (41) and also to the three B. subtilis systems; the SD sequences of txpA (38), bsrG (15), and bsrE (Fig. 2) are located in a 4- or 5-bp GC-rich stems making them inaccessible to 30 S ribosomal subunits, reflected by our failure to detect a toeprint or in vitro translation product from bsrG mRNA unless mutations unclosed the double-stranded region (15). However, whereas antitoxin SR4 induces a conformational change around the bsrG SD that extends the double-stranded region to 8 bp and further impedes translation, this was the case neither for bsrE (Fig. 3) nor for txpA (38). Therefore, the corresponding antitoxins RatA and SR5 act—as stated above—solely by promoting toxin RNA degradation. In addition, txpA has a perfect RBS (≥11 bp complementarity to anti-SD in 16S rRNA), which is proposed to efficiently recruit but slowly release ribosomes (47).
In many trans-encoded sense/antisense RNA systems from Gram-negative bacteria, Hfq plays an important role in either stabilizing the antisense RNA or promoting its interaction with the target RNA (reviewed in Refs. 1, 48, and 49). The only type I TA system in which a role for Hfq has been discovered is E. coli ralA/RalR (23). This might be due to the rather short (16 nt) sequence complementarity between ralR and RalA. RalA binds Hfq at high concentrations, but it is unclear whether Hfq promotes ralR/RalA complex formation and prevents degradation of either RNA or ralR translation (23). By contrast, in Gram-positives, only one case—LhrA from Listeria monocytogenes (50)—has been discovered so far in which Hfq was needed for sense/antisense RNA interaction. cis-encoded antisense RNAs display a long stretch of complementarity to their targets and generally do not depend on RNA chaperones to stabilize complementary base pairing. This does also apply for txpA/RatA, bsrG/SR4, and bsrE/SR5. Whereas Δhfq strains did not lyse on agar plates, indicating that all three antitoxins could neutralize toxin activity in the absence of Hfq in vivo (13, 14),4 Hfq stabilized SR5 but not SR4 and neither of the toxin RNAs bsrE or bsrG (14).4 Whereas in bsrG/SR4, we found a good correlation between in vitro and in vivo data for the functionality of truncated antitoxin species (15), this was, unexpectedly, not always the case for bsrE/SR5 (Figs. 5 and 7): SR5 species barely able to base pair with bsrE RNA could complement BsrE-induced lysis on agar plates, indicating that an RNA binding protein might help overcome this pairing deficiency in vivo. Since we could exclude Hfq (see above), we tested CsrA, which is known to bind sequence specifically to mRNAs and to be sequestered by sRNAs in Gram-negative bacteria (reviewed in Ref. 51). E. coli CsrA was recently found to be titrated by an sRNA, McaS, which could also bind a complementary target RNA and Hfq (52), indicating that both Hfq and CsrA might affect regulation by base pairing sRNAs. B. subtilis CsrA was shown to bind hag mRNA, but also FLiW protein, thereby establishing a checkpoint in flagellum synthesis (53). However, a B. subtilis ΔcsrA strain could still compensate lysis induced by truncated SR5. Therefore, we hypothesize that a so far unknown RNA binding protein exists in B. subtilis that is responsible for the observed effects. Currently, experiments are underway to identify this protein.
In summary, all three B. subtilis type I TA systems, bsrE/SR5, bsrG/SR4, and txpA/RatA, are regulated by RNA antitoxins that recruit the double-strand specific RNase III for toxin mRNA degradation. A peculiarity is bsrG/SR4, whose antitoxin SR4 has a second function; it additionally induces a structural change at the toxin SD that further impedes translation. In all cases, antitoxin/toxin RNA pairing occurs progressively, starting with one loop/loop contact followed by helix progression in the 3′ direction. Binding pathways have been elucidated for bsrG/SR4 and bsrE/SR5. The kapp values for complex formation are with ≈106 m−1 s−1 in the range of other antisense/sense RNA systems. Premature toxin translation is not prevented by toxin RNA processing, but by sequestration of the RBS in a GC-rich 4–5-bp double-stranded region. In addition, txpA has an almost perfect RBS that might efficiently bind but slowly release ribosomes, which further obstructs translation. Similar to most other type I TA systems (e.g. hok/Sok, ibs/Sib, and fst/RNAII), the initial antitoxin/toxin RNA interaction in bsrG/SR4 and bsrE/SR5 involves a potential U-turn in the toxin-RNA loop, with the latter system even using a U-turn/U-turn interaction. This makes txpA/RatA, in which no U-turn was involved, an exception. Hfq is not required in either system for the inhibitory action of the antitoxin. Interestingly, in contrast to bsrG/SR4, in bsrE/SR5, a presently unknown RNA chaperone allows for efficient antitoxin/toxin-RNA base pairing when the antitoxin is severely corrupted.
Author Contributions
S. B. and N. J. planned the experiments; C. M. performed the experiments; N. J. taught C. M. the methods, supervised the experiments, and prepared all figures; and S. B. wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Matthias Gimpel (AG Bakteriengenetik Jena) for helpful and critical suggestions, Sophie Dennhardt (Jena) for construction of plasmid pUCBES6, and Jörg Stülke, (Göttingen) for the csrA knock-out strain.
This work was supported by Grant BR1552/10-1 from the Deutsche Forschungsgemeinschaft (to S. B.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1.
P. Müller, N. Jahn, C. Maiwald, C. Ring, R. Neubert, C. Meißner, and S. Brantl, submitted for publication.
- sRNA
- small regulatory RNA
- TA
- toxin-antitoxin
- nt
- nucletide(s)
- RBS
- ribosome binding site.
References
- 1.Brantl S. (2009) Bacterial chromosome-encoded regulatory RNAs. Future Microbiol. 4, 85–103 [DOI] [PubMed] [Google Scholar]
- 2.Brantl S. (2012) Acting antisense: plasmid- and chromosome-encoded sRNAs from Gram-positive bacteria. Future Microbiol. 7, 853–871 [DOI] [PubMed] [Google Scholar]
- 3.Brantl S., and Brückner R. (2014) Small regulatory RNAs from low-GC Gram-positive bacteria. RNA Biol. 11, 443–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brantl S. (2012) Bacterial type I toxin-antitoxin systems. RNA Biol. 9, 1488–1490 [DOI] [PubMed] [Google Scholar]
- 5.Brantl S., and Jahn N. (2015) sRNAs in bacterial type I and type III toxin/antitoxin systems. FEMS Microbiol. Rev. 39, 413–427 [DOI] [PubMed] [Google Scholar]
- 6.Gerdes K., Rasmussen P. B., and Molin S. (1986) Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. U.S.A. 83, 3116–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerdes K., and Wagner E. G. (2007) RNA antitoxins. Curr. Opin. Microbiol. 10, 117–124 [DOI] [PubMed] [Google Scholar]
- 8.Weaver K. E. (2012) The par toxin-antitoxin system from Enterococcus faecalis plasmid pAD1 and its chromosomal homologs. RNA Biol. 9, 1498–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weaver K. E. (2015) The type I toxin-antitoxin par locus from Enterococcus faecalis plasmid pAD1: RNA regulation by both cis- and trans-acting elements. Plasmid 78, 65–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagner E. G., and Unoson C. (2012) The toxin-antitoxin system tisB-istR1: expression, regulation and biological role in persister phenotypes. RNA Biol. 9, 1513–1519 [DOI] [PubMed] [Google Scholar]
- 11.Kawano M. (2012) Divergently overlapping cis-encoded antisense RNA regulating toxin-antitoxin systems from E. coli: hok/sok, ldr/rdl, symE/symR. RNA Biol. 9, 1520–1527 [DOI] [PubMed] [Google Scholar]
- 12.Fozo E. M. (2012) New type I toxin-antitoxin families from “wild” and laboratory strains of E. coli: Ibs-Sib, ShoB-OhsC and Zor-Orz. RNA Biol. 9, 1504–1512 [DOI] [PubMed] [Google Scholar]
- 13.Silvaggi J. M., Perkins J. B., and Losick R. (2005) Small untranslated RNA antitoxin in Bacillus subtilis. J. Bacteriol. 187, 6641–6650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jahn N., Preis H., Wiedemann C., and Brantl S. (2012) BsrG/SR4 from Bacillus subtilis: the first temperature-dependent type I toxin-antitoxin system. Mol. Microbiol. 83, 579–598 [DOI] [PubMed] [Google Scholar]
- 15.Jahn N., and Brantl S. (2013) One antitoxin–two functions: SR4 controls toxin mRNA decay and translation. Nucleic Acids Res. 41, 9870–9880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weaver K. E., Ehli E. A., Nelson J. S., and Patel S. (2004) Antisense RNA regulation by stable complex formation in the Enterococcus faecalis plasmid pAD1 par addiction system. J. Bacteriol. 186, 6400–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dörr T., Vulić M., and Lewis K. (2010) Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLos Biol. 8, e1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurnev P. A., Ortenberg R., Dörr T., Lewis K., and Bezrukov S. M. (2012) Persister-promoting bacterial toxin TisB produces anion-selective pores in planar lipid bilayers. FEBS Lett. 586, 2529–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verstraeten N., Knapen W. J., Kint C. I., Liebens V., Van den Bergh B., Dewachter L., Michiels J. E., Fu Q., David C. C., Fierro A. C., Marchal K., Beirlant J., Versées W., Hofkens J., Jansen M., Fauvart M., and Michiels J. (2015) Obg and membrane polarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol. Cell 59, 9–21 [DOI] [PubMed] [Google Scholar]
- 20.Koyanagi S., and Lévesque C. M. (2013) Characterization of a Streptococcus mutans intergenic region containing a small toxin peptide and its cis-encoded antisense small RNA antitoxin. PLoS One 8, e54291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawano M., Aravind L., and Storz G. (2007) An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 64, 738–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weel-Sneve R., Kristiansen K. I., Odsbu I., Dalhus B., Booth J., Rognes T., Skarstad K., and Bjørås M. (2013) Single transmembrane peptide DinQ modulates membrane-dependent activities. PLoS Genet. 9, e1003260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y., Quiroga C., Chen Q., McAnulty M. J., Benedik M. J., Wood T. K., and Wang X. (2014) RalR (a DNase) and RalA (a small RNA) form a type I toxin-antitoxin system in Escherichia coli. Nucleic Acids Res. 42, 6448–6462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durand S., Jahn N., Condon C., and Brantl S. (2012a) Type I toxin-antitoxin systems in Bacillus subtilis. RNA Biol. 9, 1491–1497 [DOI] [PubMed] [Google Scholar]
- 25.Jahn N., Brantl S., and Strahl H. (2015) Against the mainstream: the membrane associated type I toxin BsrG from Bacillus subtilis interferes with cell envelope biosynthesis without increasing membrane permeability. Mol. Microbiol. 98, 651–666 [DOI] [PubMed] [Google Scholar]
- 26.Thisted T., and Gerdes K. (1992) Mechanism of post-segregational killing by the hok/sok system of plasmid R1. Sok antisense RNA regulates hok gene expression indirectly through the overlapping mok gene. J. Mol. Biol. 223, 41–54 [DOI] [PubMed] [Google Scholar]
- 27.Han K., Kim K.-S., Bak G., Park H., and Lee Y. (2010) Recognition and discrimination of target mRNAs by Sib RNAs, a cis-encoded sRNA family. Nucleic Acids Res. 38, 5851–5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irnov I., Sharma C. M., Vogel J., and Winkler W. C. (2010) Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res. 38, 6637–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J., Fritsch E. F., and Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 30.Heidrich N., Chinali A., Gerth U., and Brantl S. (2006) The small untranslated RNA SR1 from the B. subtilis genome is involved in the regulation of arginine catabolism. Mol. Microbiol. 62, 520–536 [DOI] [PubMed] [Google Scholar]
- 31.Heidrich N., and Brantl S. (2007) Antisense RNA-mediated transcriptional attenuation in plasmid pIP501: the simultaneous interaction between two complementary loop pairs is required for efficient inhibition by the antisense RNA. Microbiology 153, 420–427 [DOI] [PubMed] [Google Scholar]
- 32.Preis H., Eckart R. A., Gudipati R. K., Heidrich N., and Brantl S. (2009) CodY activates transcription of a small RNA in Bacillus subtilis. J. Bacteriol. 191, 5446–5457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brantl S., and Wagner E. G. (2000) Antisense RNA-mediated transcriptional attenuation: an in vitro study of plasmid pT181. Mol. Microbiol. 35, 1469–1482 [DOI] [PubMed] [Google Scholar]
- 34.Heidrich N., Moll I., and Brantl S. (2007) In vitro analysis of the interaction between the small RNA SR1 and its primary target ahrC mRNA. Nucleic Acids Res. 35, 4331–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Franch T., and Gerdes K. (2000) U-turns and regulatory RNAs. Curr. Opin. Microbiol. 3, 159–164 [DOI] [PubMed] [Google Scholar]
- 36.Heidrich N., and Brantl S. (2003) Antisense-RNA mediated transcriptional attenuation: importance of a U-turn loop structure in the target RNA of plasmid pIP501 for efficient inhibition by the antisense RNA. J. Mol. Biol. 333, 917–929 [DOI] [PubMed] [Google Scholar]
- 37.Dambach M., Irnov I., and Winkler W. C. (2013) Association of RNAs with Bacillus subtilis Hfq. PLoS One 8, e455156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durand S., Gilet L., and Condon C. (2012b) The essential function of B. subtilis RNase III is to silence foreign toxic genes. PLoS Genet. 8, e1003181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolb F. A., Engdahl H. M., Slagter-Jäger J. G., Ehresmann B., Ehresmann C., Westhof E., Wagner E. G., and Romby P. (2000) Progression of a loop-loop complex to a four-way junction is crucial for the activity of a regulatory antisense RNA. EMBO J. 19, 5905–5915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thisted T., Sørensen N. S., Wagner E. G., and Gerdes K. (1994) Mechanism of post-segregational killing: Sok antisense RNA interacts with Hok mRNA via its 5′-end single stranded leader and competes with the 3′-end of Hok mRNA for binding to the mok translational initiation region. EMBO J. 13, 1960–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greenfield T. J., Franch T., Gerdes K., and Weaver K. E. (2001) Antisense RNA regulation of the par post-segregational killing system: structural analysis and mechanism of binding of the antisense RNA, RNAII and its target, RNAI. Mol. Microbiol. 42, 527–537 [DOI] [PubMed] [Google Scholar]
- 42.Wen J., Won D., and Fozo E. M. (2014) The ZorO-OrzO type I toxin-antitoxin locus: repression by the OrzO antitoxin. Nucleic Acids Res. 42, 1930–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brantl S. (2007) Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 10, 102–109 [DOI] [PubMed] [Google Scholar]
- 44.Franch T., Petersen M., Wagner E. G., Jacobsen J. P., and Gerdes K. (1999) Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J. Mol. Biol. 294, 1115–1125 [DOI] [PubMed] [Google Scholar]
- 45.Shokeen S., Johnson C. M., Greenfield T. J., Manias D. A., Dunny G. M., and Weaver K. E. (2010) Structural analysis of AntiQ-Qs interaction: RNA-mediated regulation of E. faecalis plasmid pCF10 conjugation. Plasmid 64, 26–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwong S. M., and Firth N. (2015) Structural and sequence requirements for the antisense RNA regulating replication of staphylococcal multiresistance plasmid pSK41. Plasmid 78, 17–25 [DOI] [PubMed] [Google Scholar]
- 47.Li G. W., Oh E., and Weissman J. S. (2012) The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484, 538–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gottesman S., and Storz G. (2011) Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 3, a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papenfort K., and Vanderpool C. K. (2015) Target activation by regulatory RNAs in bacteria. FEMS Microbiol. Rev. 39, 362–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nielsen J. S., Lei L. K., Ebersbach T., Olsen A. S., Klitgaard J. K., Valentin-Hansen P., and Kallipolitis B. H. (2010) Defining a role for Hfq in Gram-positive bacteria: evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic Acids Res. 38, 907–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romeo T., Vakulskas C. A., and Babitzke P. (2013) Posttranscriptional regulation on a global scale: form and function of Csr/Rsm systems. Environ. Microbiol. 15, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jørgensen M. G., Thomason M. K., Havelund J., Valentin-Hansen P., and Storz G. (2013) Dual function of the McaS small RNA in controlling biofilm formation. Genes Dev. 27, 1132–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mukherjee S., Yakhnin H., Kysela D., Sokoloski J., Babitzke P., and Kearns D. B. (2011) CsrA-FliW interaction governs flagellin homeostasis and a checkpoint on flagellar morphogenesis in Bacillus subtilis. Mol. Microbiol. 82, 447–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.