Abstract
Viruses have developed distinct strategies to overcome the host defense system. Regulation of apoptosis in response to viral infection is important for virus survival and dissemination. Like other viruses, Crimean-Congo hemorrhagic fever virus (CCHFV) is known to regulate apoptosis. This study, for the first time, suggests that the non-structural protein NSs of CCHFV, a member of the genus Nairovirus, induces apoptosis. In this report, we demonstrated the expression of CCHFV NSs, which contains 150 amino acid residues, in CCHFV-infected cells. CCHFV NSs undergoes active degradation during infection. We further demonstrated that ectopic expression of CCHFV NSs induces apoptosis, as reflected by caspase-3/7 activity and cleaved poly(ADP-ribose) polymerase, in different cell lines that support CCHFV replication. Using specific inhibitors, we showed that CCHFV NSs induces apoptosis via both intrinsic and extrinsic pathways. The minimal active region of the CCHFV NSs protein was determined to be 93–140 amino acid residues. Using alanine scanning, we demonstrated that Leu-127 and Leu-135 are the key residues for NSs-induced apoptosis. Interestingly, CCHFV NSs co-localizes in mitochondria and also disrupts the mitochondrial membrane potential. We also demonstrated that Leu-127 and Leu-135 are important residues for disruption of the mitochondrial membrane potential by NSs. Therefore, these results indicate that the C terminus of CCHFV NSs triggers mitochondrial membrane permeabilization, leading to activation of caspases, which, ultimately, leads to apoptosis. Given that multiple factors contribute to apoptosis during CCHFV infection, further studies are needed to define the involvement of CCHFV NSs in regulating apoptosis in infected cells.
Keywords: apoptosis, caspase, mitochondria, mitochondrial membrane potential, proteasome, Crimean-Congo hemorrhagic fever virus, non-structural protein
Introduction
Crimean-Congo hemorrhagic fever virus (CCHFV),2 an arthropod-borne virus, is a member of the genus Nairovirus in the family Bunyaviridae (1). CCHFV is one of the most widely spread tick-borne viral infection of humans, affecting many countries across Africa, Asia, and Europe (2, 3). Human infection with CCHFV often results in severe hemorrhagic disease that has an approximate fatality rate of 30% (1). Cerebral hemorrhage, severe anemia, myocardial infarction, lung edema, and severe dehydration are the major reasons for the fatal outcome of the disease caused by CCHFV (1, 4). At present, there is no vaccine or specific antiviral therapy against the virus, and progress in the development of new therapeutic agents is slow because of a lack of suitable animal models (2).
The CCHFV envelope is studded with spikes comprising the glycoproteins Gn and Gc, which are responsible for the virus binding to host cell receptors (3). The CCHFV genome contains three negative-sense RNA segments, small (S), medium (M), and large (L), encoding the nucleocapsid protein, Gn and Gc glycoproteins, and the L polymerase, respectively (1). In addition, several Bunyaviridae members encode nonstructural proteins, either on the M (termed NSm) or S segment (termed NSs) of the genome (5). The nonstructural protein NSs of CCHFV is encoded by the positive-sense of the S segment genome (6). The ORF of NSs is conserved in almost all strains of CCHFV, indicating that NSs might have a conserved biological function. Although there are no reports regarding the function of the NSs protein of CCHFV, several studies have been published regarding the role of viral NSs belonging to the same family. La Crosse encephalitis virus, a member of the Orthobunyavirus genus, is an apoptogenic virus causing severe encephalitis in children. Previously, La Crosse encephalitis virus NSs has been shown to induce caspase activation and apoptosis (7, 8). Punta Toro virus, a member of the Phlebovirus genus, induces apoptosis in hepatocytes in vivo and in cultured mammalian cells in vitro (9). Subsequently, the NSs protein of Punta Toro virus has been shown to induce hepatocyte apoptosis by triggering the extrinsic and intrinsic pathways (10). Interestingly, several NSs proteins in the Bunyaviridae family exhibit sequence similarity with a known proapoptotic protein, reaper, from Drosophila (7). Apart from this, NSs proteins in the Bunyaviridae family have also been shown to antagonize interferon α/β production, shut down host cell protein synthesis, and induce the degradation of double-stranded RNA-dependent protein kinase (11–15).
Regulation, namely suppression or induction, of apoptosis during viral infection is crucial for the maintenance of viral latency or dissemination (16, 17). Several viral proteins regulate cell death by altering the mitochondrial membrane potential either directly or indirectly (18). Loss of the mitochondrial membrane potential causes the release of proteins that are usually confined to the intermembrane space of the mitochondria. Most important are the apoptotic-inducing factor and caspase activators, such as cytochrome c and Smac (second mitochondria-derived activator of caspases)/DIABLO (direct IAP-binding protein with low PI). Protease caspases play an important role in apoptosis, and their activation can lead to a series of catabolic reactions resulting in the activation of caspase-3 and -7, which serve as executioners of apoptosis (16, 19). Poly(ADP-ribose) polymerase (PARP), a substrate of activated caspase-3/7, is responsible for the disassembly of cell structures (20, 21). Recently, it has been reported that CCHFV induces caspase-3-dependent apoptosis (22) and modulates both intrinsic as well as extrinsic pathways of apoptosis in hepatocyte cells (23). Here we report the caspase-dependent apoptotic activity of CCHFV NSs. We also determined the minimal active region and the key residues important for its apoptotic activity. In addition, disruption of the mitochondrial membrane potential by the CCHFV NSs protein is dependent on these key residues.
Materials and Methods
Virus Production
SW13 cells (human adrenal cortex adenocarcinoma cells) were maintained in Leibovitz medium (L15) supplemented with 2% fetal bovine serum and antibiotics (10 units/ml penicillin and 10 μg/ml streptomycin). MG132 (C2211) was purchased from Sigma-Aldrich and diluted in PBS to a final concentration of 5 μg/ml. The Nigerian CCHFV Ibar10200 strain, originally isolated in Nigeria, was used in the experiments, and all handling of live virus was performed in a biosafety level 4 facility.
In Vitro Infection
SW13 cells were seeded in 24-well plates and/or in chamber slides and then infected with CCHFV (multiplicity of infection, 1). 1 h post-infection (h.p.i.), mock- and CCHFV-infected cells were treated with MG132 (5 μg/ml) for 48 and 72 h.p.i. Cells were harvested in lysis buffer for Western blot analysis or fixed using acetone for the immunofluorescence assay.
Mouse Polyclonal Serum
BALB/c mice (Biological Resource Center, Agency for Science, Technology, and Research) were immunized with five doses of synthesized KLH-NSs(48–92) and KLH-NSs(120–150) peptides (GL Biochem, Shanghai, China). The primary dose was a mixture of equal volumes of both peptides (200 μg each peptide) with complete Freund adjuvant (Sigma). Subsequent doses were mixtures of equal volumes of the peptides (100 μg each peptide) with incomplete Freund adjuvant (Sigma). The blood was collected from the mice, and serum was obtained by spinning down the cells. All mice were handled according to National Advisory Committee for Laboratory Animal Research guidelines.
Rabbit Polyclonal Serum
The cDNA encoding for residues 48–92 of NSs was cloned into the pGEX-6P1 vector (GE Healthcare). GST-fused NSs(48–92) protein was then expressed in Escherichia coli BL21(DE3) (Novagen, EMD Chemicals Inc.) and purified as described previously (24). The protein was sent to Agrisera AB for the production of rabbit polyclonal serum.
Cell Culture
HeLa (human cervix epithelial cells, ATCC), Vero E6 (African green monkey kidney epithelial cells, ATCC) and 293FT (human embryonic kidney cells transformed with the SV40 large T antigen, Invitrogen) cells were grown in DMEM supplemented with 10% fetal bovine serum (HyClone), nonessential amino acids, and antibiotics (10 units/ml penicillin and 10 μg/ml streptomycin) (Invitrogen). All cells were maintained in a 37 °C incubator with 5% CO2.
Plasmid Preparation
Expression plasmids for the CCHFV NSs proteins were generated by PCR cloning using Q5 high-fidelity DNA polymerase (New England Biolabs Inc.). The RNA extracted from CCHFV (GenBank accession number U88410) was used as template, and DNA encoding NSs was amplified by using NSs-specific primers. The PCR product was digested with the restriction enzymes BamHI and XhoI, followed by ligation into the pXJ40myc vector, which is a myc-tagged plasmid derived from pXJ40 (25). The pXJ40myc vector was used to fuse the human c-Myc tag peptide with the N termini of the proteins for easy detection. Similarly, PCR products for different constructs were cloned into the pXJ40myc vector. The NSs mutants and DN-GRIM19 were generated using the Q5 site-directed mutagenesis kit according to the protocol recommended by the manufacturer (New England Biolabs Inc.). All clones were confirmed by DNA sequencing.
Transient Transfection and Cell Harvesting
Transient transfections of cells were performed using Lipofectamine 2000 reagent (Invitrogen) according to the protocol of the manufacturer. In some experiments, NSs-transfected cells were treated with 10 μm of MG132 (Calbiochem) for 4 h before harvesting the cells. Approximately 24 h after transfection, the cells were harvested by scraping into the media. The cells were washed three times with cold 1× PBS, resuspended in radio-immunoprecipitation assay buffer (50 mm Tris (pH 8.0), 150 mm NaCl, 0.5% Nonidet P-40, 0.5% deoxycholic acid, 0.005% SDS, and 1 mm phenylmethylsulfonyl fluoride) and subjected to freeze-thaw cycles before being spun down at 13,000 rpm to remove cellular debris. The cell lysates were subjected to Western blot analysis and apoptotic assays.
Western Blot Analysis
The cell lysates were separated by SDS-PAGE, and proteins were transferred to membranes. A nitrocellulose membrane was used for the detection of actin and c-Myc-tagged proteins, whereas a PVDF membrane was used for PARP. The membranes were blocked with TBST (20 mm Tris (pH 7.5), 150 mm NaCl, and 0.1% Tween 20) containing 5% skim milk for 1 h and incubated overnight with primary antibody at 4 °C. The blot was washed with TBST, incubated with secondary antibody for 1 h, washed again with TBST, and finally developed through enhanced chemiluminescence. The primary antibodies used in the study included anti-actin monoclonal (Sigma), anti-PARP polyclonal (Cell Signaling Technology Inc.), and anti-myc monoclonal (Santa Cruz Biotechnology) antibodies. Secondary antibodies used were HRP-conjugated goat anti-mouse and goat anti-rabbit IgG (Pierce). For the CCHFV-infected cells, Western blot analysis was performed in a similar manner by using the anti-CCHFV NSs rabbit polyclonal serum described above. A rabbit polyclonal anti-calnexin antibody was used to detect equal amounts of loaded sample.
Caspase-3/7 Assay
The cell lysates were subjected to a caspase-3/7 activity assay using the Apo-ONE homogeneous caspase-3/7 assay kit according to the protocol of the manufacturer (Promega). Briefly, 50 μl of substrate was mixed with 5 ml of buffer to make the caspase-3/7 reagent. Later, 50 μl of reagent was incubated with 50 μl of cell lysates for 3 h with shaking. Subsequently, the fluorescence of each well was measured.
Caspase-8 Assay
Caspase-8 activation was measured using the Caspase-Glo 8 assay system (Promega) according to the instructions of the manufacturer with slight modifications. Cells cultured in a 6-cm dish were transfected with vector or NSs, followed by cell lysis as mentioned above. For the positive control, cells were treated with 50 ng/ml TNF-α (Roche) and 10 μg/ml of cycloheximide (CHX) (Sigma), which was dissolved in dimethyl sulfoxide, for 6 h. For TNF-α + CHX treatment, 0.1% BSA in PBS and dimethyl sulfoxide-treated cells were used as a negative control. Briefly, an equal volume of Caspase-Glo reagent, which contained substrate, buffer, and MG132, was added to an equal amount of cell lysates in an opaque 96-well microplate. The plate was incubated at room temperature for 30 min. Subsequently, luminescence was measured.
Treatment of Cells with Caspase Inhibitors
Cells (293FT) were transfected (as above), followed by treatment with 10 μm of inhibitors of caspase-3 (Z-DEVD-fmk), caspase-8 (Z-IETD-fmk), caspase-9 (Z-LEHD-fmk), or a negative control (Z-FA-fmk) (BD Biosciences). Briefly, the transfection medium was replaced with the fresh medium containing 10 μm of the respective cell-permeable caspase inhibitors 6 h post-transfection. Approximately 24 h after transfection, the cells were harvested, and the cell lysates were subjected to Western blot analysis and apoptotic assays (as above).
Cell Viability Assay
Cell viability was determined using the cell proliferation reagent WST-1 (Roche) according to the instructions of the manufacturer. Briefly, 10 μl of WST-1 was added to cells cultured in a transparent 96-well microplate and incubated for 1 h, followed by measuring absorbance at 440 nm.
Immunofluorescence Assay
Cells (Vero E6 and HeLa) grown on coverslips were transfected as described above. Approximately 24 h after transfection, the medium was aspirated, and the cells were incubated with 500 μl of 200 nm MitoTracker Red CMXRos (Invitrogen) for 40 min at 37 °C. After rinsing with PBS twice, cells were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.1% Triton X-100 for 15 min, followed by blocking with 1% BSA in PBS for 30 min. The cells were incubated with anti-myc (rabbit polyclonal or mouse monoclonal antibodies, Santa Cruz Biotechnology), anti-HA (Roche), or anti-mtTFA (Santa Cruz Biotechnology) primary antibodies for 2 h. After washing, cells were incubated with Alexa Fluor 488-conjugated goat anti-mouse IgG secondary antibodies (Invitrogen) for 1 h. For cells incubated with both anti-myc and anti-mtTFA antibodies, Rhodamine-conjugated goat anti-mouse IgG (Invitrogen) and Alexa Fluor 488-conjugated goat anti-rabbit IgG (Invitrogen) were used as secondary antibodies and incubated for 1 h. After washing, cells were stained with DAPI before mounting. Images were captured with an Olympus FluoView FV1000 laser-scanning confocal microscope. For the CCHFV-infected cells, an immunofluorescence assay was performed in a similar manner by using the anti-CCHFV NSs mouse polyclonal serum described above and a rabbit polyclonal anti-CCHFV nucleocapsid protein antibody (26).
Statistical Analysis
All experiments were repeated at least three times. Statistical analysis was performed using Student's t test. p < 0.05 was considered significant.
Results
NSs Expression in CCHFV-infected Cells
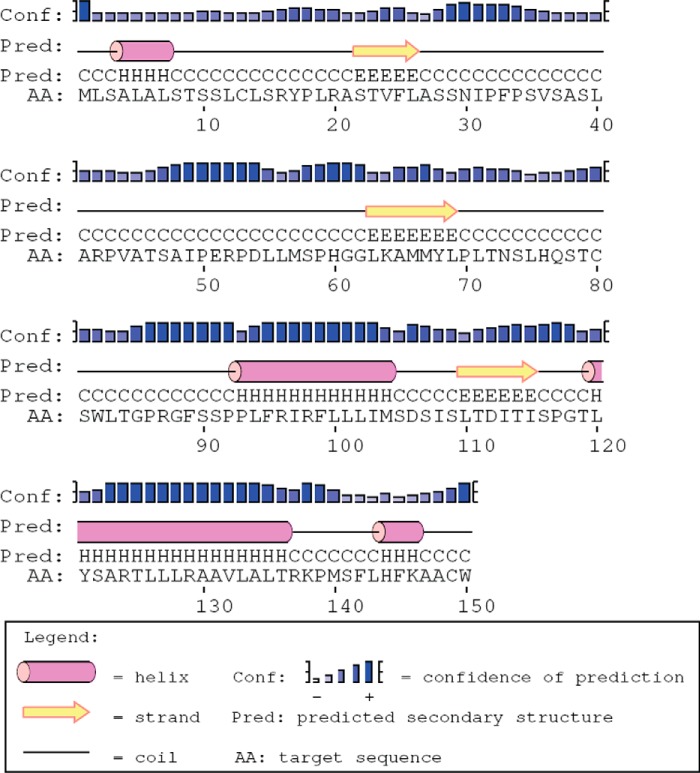
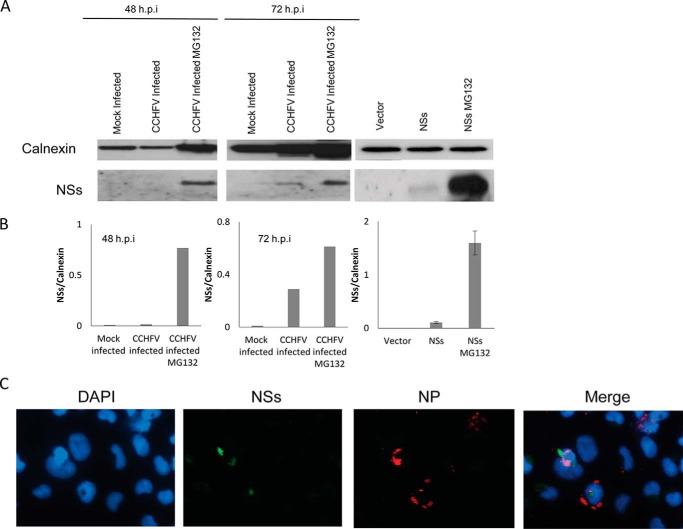
The CCHFV NSs protein is composed of 150 amino acid residues, with its secondary structure predicted by PSIPRED as shown in Fig. 1 (27). To examine the expression levels of NSs during infection, cells were infected with CCHFV and subjected to Western blot analysis. NSs was detected in the infected cells at 72 h.p.i., whereas it was below the detection limits at 48 h.p.i. (Fig. 2A). Interestingly, treating the CCHFV-infected cells with the proteasome inhibitor MG132 seemed to result in accumulation of NSs (Fig. 2A). However, the Western blot analysis showed that there was unequal loading of samples, as reflected by the level of calnexin, and this occurred because our biosafety level 4 facility does not have suitable equipment to accurately quantify the amount of protein in the lysates. Nevertheless, normalization of NSs band intensities to those of calnexin by densitometric analysis revealed that there was more NSs protein in cells treated with MG132, especially at 48 h.p.i. (Fig. 2B). In transfected cells, a significant increase in NSs expression was also observed upon MG132 treatment. This indicates that NSs is subjected to proteasomal protein degradation. Similarly, van Knippenberg et al. (28) have reported the active degradation of NSs during Bunyamwera virus infection.
FIGURE 1.
Predicted secondary structure of CCHFV NSs. The secondary structure of NSs containing 150 amino acid residues was predicted by PSIPRED.
FIGURE 2.
NSs expression in CCHFV-infected or NSs-transfected cells. A, SW13 cells were mock-infected or infected with CCHFV and either left untreated or treated with MG132, followed by cell harvesting at 48 and 72 h.p.i. Alternatively, SW13 cells were transfected with empty vector or myc-NSs and either left untreated or treated with MG132, after which cells were harvested 24 h post-transfection. Western blot analysis was performed to determine the expression levels of CCHFV NSs (bottom row) and calnexin as a loading control (top row). The Western blots shown represent one of three independent experiments. B, densitometric analysis of NSs band intensities normalized to calnexin. C, immunofluorescence analysis was performed on CCHFV-infected cells. NSs (green) and nucleocapsid protein (NP, red) were analyzed by fluorescence microscopy. Nuclei were counterstained with DAPI (blue). Three independent experiments were performed, with a representative set of images shown here.
To confirm the expression of CCHFV NSs, infected cells were also subjected to an immunofluorescence assay. Few of the infected cells expressing nucleocapsid protein showed NSs protein expression (Fig. 2C). This could be attributed to the active degradation of NSs in the infected cells.
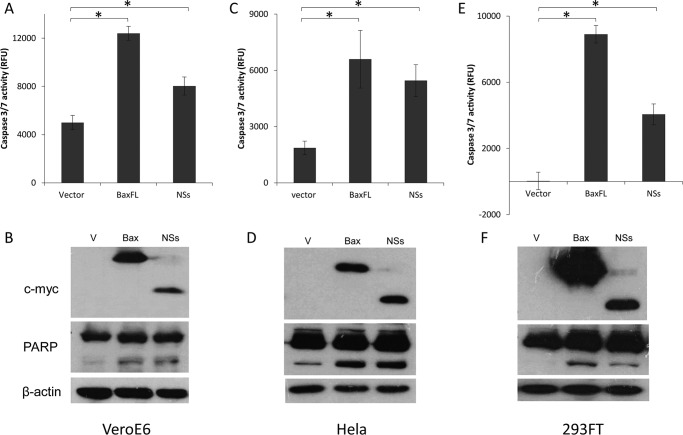
CCHFV NSs Protein Induces Apoptosis in Vero E6, HeLa, and 293FT Cells
To determine the apoptotic activity of NSs protein, Vero E6, HeLa, and 293FT cells, which can be infected by CCHFV, were transiently transfected with the pXJmyc-NSs expression plasmid, empty vector as a negative control, and pXJmyc-Bax as a positive control. 24 h post-transfection, the harvested cells were lysed, and the cell lysates were used for Western blot analysis and measurement of the activation of caspase-3/7 enzyme. Cells overexpressing NSs protein or Bax had significantly higher level of caspase-3/7 activity than the vector control cells. Similarly, the cleavage of endogenous PARP, a substrate of activated caspase-3/7, was clearly observed in cells overexpressing Bax and NSs, whereas cells transfected with empty vector showed a significantly lower cleavage of PARP (Fig. 3). Among the three cell lines, 293FT cells exhibited higher levels of protein expression, and cells transfected with the vector control showed negligible caspase-3/7 activity. Therefore, 293FT cells were used for subsequent experiments involving caspase-3/7 activity and Western blot analysis.
FIGURE 3.
Induction of apoptosis by NSs in different cell lines that support CCHFV replication. The cell lines used were Vero E6 (A and B), HeLa (C and D), and 293FT (E and F). A, C, and E, the apo-ONE homogeneous caspase-3/7 assay was used to measure the activation of caspase-3/7 in three different cell lines that were transiently transfected with empty vector, myc-NSs, or myc-Bax. All experiments were performed in triplicate, and the results are expressed as mean ± S.D. Three independent experiments were performed, and a representative dataset is shown. *, p < 0.05. B, D, and F, Western blot analysis was performed to determine the expression levels of the myc-tagged proteins (top panels), the cleavage of endogenous PARP (center panels), and the levels of endogenous actin as a loading control (bottom panels). The Western blots shown represent one of three independent experiments. RFU, relative fluorescence units; V, vector.
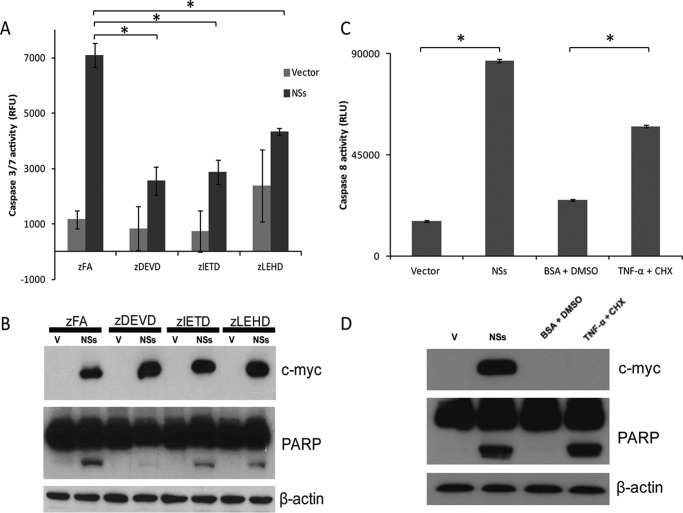
NSs Induces Apoptosis via the Intrinsic and Extrinsic Pathways
Apoptosis may be triggered by two primarily different signaling pathways, commonly termed the extrinsic and intrinsic signaling cascades. To determine which of the two pathways is activated in NSs-induced apoptosis, non-toxic, cell-permeable peptide caspase inhibitors that bind irreversibly and specifically to activated caspases (29, 30), namely caspase-3 (Z-DEVD-fmk), caspase-8 (Z-IETD-fmk), and caspase-9 (Z-LEHD-fmk), were added to the cells 6 h post-transfection. The extent of NSs-induced apoptosis, as reflected by the caspase-3/7 activity and cleaved PARP, was inhibited significantly by all three caspase inhibitors but not by the irrelevant peptide (Z-FA-fmk) (Fig. 4, A and B). Caspase-3 inhibition has a greater effect on NSs-induced cleavage of PARP. Caspase-8 and caspase-9 are upstream of caspase-3 in the cascade, and our results suggest that caspase-dependent, NSs-induced apoptosis involves the activation of both caspase-8 and caspase-9, which are initiator caspases of the extrinsic and intrinsic signaling cascades, respectively. To confirm the activation of caspase-8 during NSs-induced apoptosis, a caspase-8 activity assay was performed using cells treated with TNF-α + CHX as a positive control. In agreement with our inhibitor study results, cells expressing NSs showed a significantly higher level of caspase-8 activity in comparison with that of vector control cells (Fig. 4C). TNF-α + CHX treatment also induced PARP cleavage, as shown in Fig. 4D.
FIGURE 4.
NSs induces apoptosis via both the intrinsic and extrinsic pathways. A, the apo-ONE homogeneous caspase-3/7 assay was used to measure the activation of caspase-3/7 in 293FT cells that were transiently transfected with empty vector or myc-NSs. The cells were treated with an irrelevant peptide (Z-FA-fmk) or caspase-3, caspase-8, and caspase-9 inhibitors (Z-DEVD-fmk, Z-IETD-fmk, and Z-LEHD-fmk, respectively). All experiments were performed in triplicate, and the results are expressed as mean ± S.D. Three independent experiments were performed, and a representative dataset is shown. *, p < 0.05. B, Western blot analysis was performed to determine the expression levels of myc-NSs (top panel), the cleavage of endogenous PARP (center panel), and the levels of endogenous actin as a loading control (bottom panel). The Western blots shown represent one of three independent experiments. C, the caspase-Glo 8 assay was used to measure the activation of caspase-8 in 293FT cells that were transiently transfected with empty vector or NSs. Cells treated with TNF-α + CHX were used as a positive control, and those treated with 0.1% BSA in PBS + dimethyl sulfoxide (DMSO) were used as a negative control. All experiments were performed in triplicate, and the results are expressed as mean ± S.D. Four independent experiments were performed, and a representative dataset is shown. *, p < 0.05. D, Western blot analysis was performed to determine the expression levels of myc-NSs (top panel), the cleavage of endogenous PARP (center panel), and the levels of endogenous actin as a loading control (bottom panel). The Western blots shown represent one of three independent experiments. RFU, relative fluorescence units; V, vector.
The Minimal Active Region of the NSs Protein
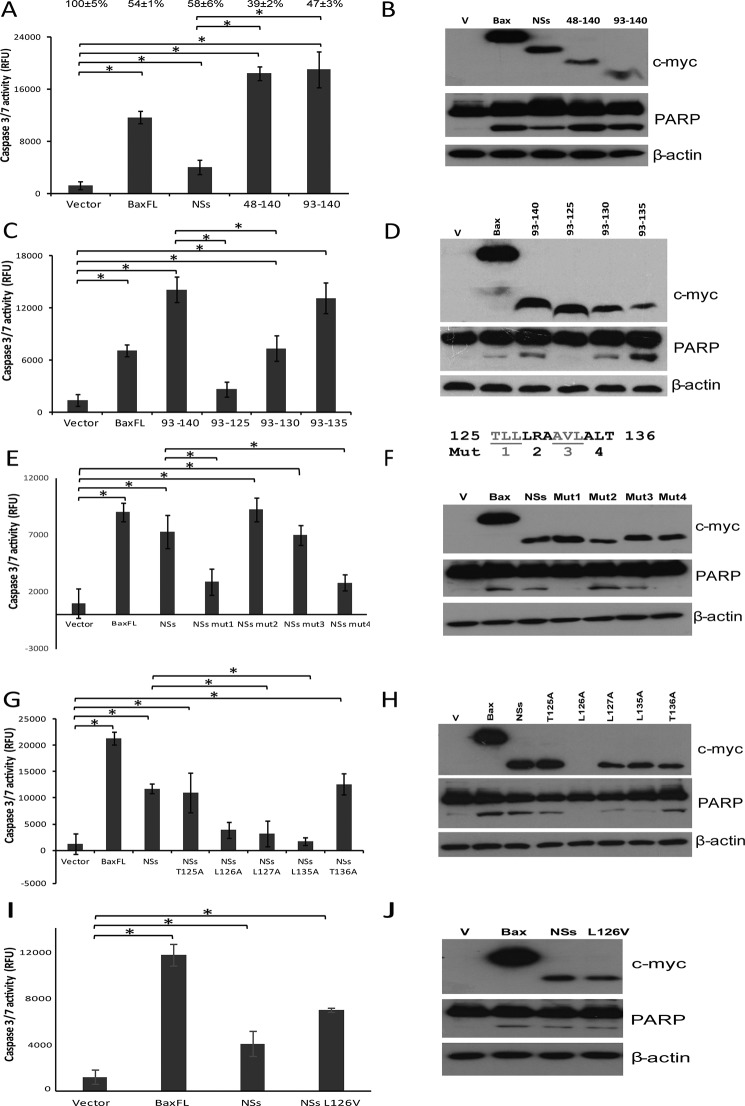
To determine the minimal active fragment of NSs, we made various constructs of NSs in the pXJmyc vector and transiently transfected them into 293FT cells. NSs(48–140) and NSs(93–140) were found to be functionally active in inducing apoptosis (Fig. 5, A and B), and the caspase-3/7 activity for cells expressing both proteins was around five times higher than that of cells expressing full-length NSs, indicating autoinhibitory effects by the N terminus of NSs (Fig. 5A). Similarly, the cleavage of PARP was significantly higher in cells expressing NSs(48–140) and NSs(93–140) compared with cells expressing NSs. To determine the degree of cell death induced by these proteins, cell viability was determined using WST-1. Overexpression of full-length NSs, NSs(48–140), and NSs(93–140) led to cell death, as evidenced by the decrease in cell viability to 58%, 39%, and 47%, respectively (Fig. 5A).
FIGURE 5.
Leu-127 and Leu-135 are key residues for the apoptotic activity of NSs protein. A, C, E, G, and I, the apo-one homogeneous caspase-3/7 assay was used to measure the activation of caspase-3/7 in 293FT cells that were transiently transfected with empty vector, Bax, NSs, NSs(48–140), or NSs(93–140) (A); empty vector, Bax, NSs(93–140), NSs(93–135), NSs(93–130), or NSs(93–125) (C); empty vector, Bax, NSs, or triple alanine substitution mutants such as NSs mut1 (TLL(125–127)AAA), NSs mut2 (LRA(128–130)AAA), NSs mut3 (AVL(131–133)AAA), or NSs mut4 (ALT(134–136)AAA) (E); empty vector, Bax, NSs, or single alanine substitution mutants like NSs T125A, L126A, L127A, L135A, or T136A (G); and empty vector, Bax, NSs, or NSs L126V (I). The percentages of live cells compared with that of the empty vector, which was normalized to 100%, are shown above each column. All experiments were performed in triplicate, and the results are expressed as mean ± S.D. At least three independent experiments were performed, and a representative dataset is shown.*, p < 0.05. B, D, F, H, and J, Western blot analysis was performed for the samples to determine the expression levels of myc-tagged proteins (top panels), the cleavage of endogenous PARP (center panels), and the levels of endogenous actin as a loading control (bottom panels). The Western blots shown represent one of three independent experiments. RFU, relative fluorescence units; Mut, NSs triple alanine substitution mutant; V, vector.
To further narrow down the active region, three more proteins, NSs(93–125), NSs(93–130), and NSs(93–135), were examined for their abilities to induce apoptosis (Fig. 5, C and D). NSs(93–135)-induced caspase-3/7 activity was comparable with that of NSs(93–140), indicating that residues 136–140 do not have a role in NSs-induced apoptosis. On the other hand, cells overexpressing NSs(93–130) had a significantly lower level of caspase-3/7 activity and cleaved PARP in comparison with that of cells expressing NSs(93–140). In addition, NSs(93–125) was found to be completely inactive, as reflected by the abrogated caspase-3/7 activity and cleaved PARP. These results indicate that residues 126–135 are important for the apoptotic activity of NSs.
Leu-127 and Leu-135 Are the Key Residues for the Apoptotic Activity of NSs Protein
Next, alanine scanning was used to determine the key residues important for the apoptotic activity of NSs. Triple alanine substitution mutants between 125–136 amino acids residues were prepared. Of four NSs mutants, two mutants, mut2 and mut3, were still functional in inducing apoptosis, but the cells overexpressing the other two mutants, mut1 and mut4, had a significantly lower level of caspase-3/7 activity and cleaved PARP in comparison with that of cells expressing wild-type NSs (Fig. 5, E and F). On the basis of these results, single alanine substitution mutants for the Thr-125, Leu-126, Leu-127, Leu-135, and Thr-136 residues were prepared to determine the residues important for the apoptotic activity of NSs. Two mutations, T125A and T136A, had no effect on the apoptotic activity of the NSs protein, which indicates that Thr-125 and Thr-136 are not involved in NSs-induced apoptosis (Fig. 5, G and H). However, cells overexpressing NSs L127A and L135A had a significantly lower level of caspase-3/7 activity and cleaved PARP in comparison with that of cells expressing wild-type NSs, indicating that these two residues are very important for the activation of caspases leading to the cleavage of PARP, resulting in apoptosis. NSs L126A protein expression could not be detected in transfected cells, making it difficult to determine the potential role of Leu-126 in NSs-induced apoptotic activity. However, successful expression was achieved with the NSs L126V mutant, the expression of which was almost equivalent to that of wild-type NSs (Fig. 5, I and J). NSs L126V was found to be as active as NSs wild-type, as reflected by the comparable level of caspase-3/7 activity and cleaved PARP, suggesting that residue Leu-126 is not essential in inducing apoptosis. Therefore, Leu-127 and Leu-135 are the key residues responsible for apoptosis induced by NSs.
NSs Disrupts the Mitochondrial Membrane Potential
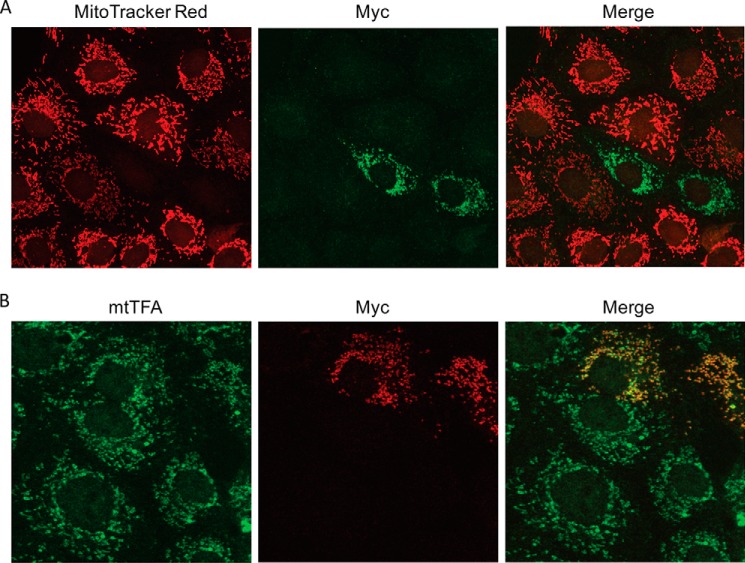
NSs induces apoptosis by triggering both the extrinsic as well as the intrinsic pathway, also termed the mitochondrial pathway. To determine the ability of NSs to disrupt the mitochondrial membrane potential, we transfected Vero E6 cells with pXJmyc-NSs and examined the cellular localization of Myc-NSs by immunofluorescence assay. MitoTracker Red was used to stain mitochondria in live cells because its accumulation is dependent on the integrity of the mitochondrial membrane potential. Cells overexpressing the NSs protein did not show any MitoTracker staining, which indicates that NSs disrupted the mitochondrial membrane potential (Fig. 6A). To further substantiate this result and locate the mitochondria in NSs-transfected cells, another set of cells was transfected with pXJmyc-NSs and stained with an antibody against mitochondrial transcription factor A (mtTFA), which is an endogenous transcription factor. Mitochondria in NSs-transfected cells could be stained with anti-mtTFA antibody, and NSs was found to co-localize with mtTFA (Fig. 6B). These results indicate that NSs disrupts the mitochondrial membrane potential and localizes within mitochondria.
FIGURE 6.
NSs co-localizes in mitochondria and disrupts the mitochondrial membrane potential. A, myc-NSs was transiently transfected into Vero E6 cells. Subcellular localization of NSs was detected by anti-myc antibody and Alexa Fluor 488-conjugated secondary antibody (green). Mitochondria were labeled with MitoTracker Red CMXRos (red). B, the subcellular localization of NSs in Vero E6 cells was detected by anti-myc antibody and Rhodamine-conjugated secondary antibody (red). Endogenous mtTFA was detected by anti-mtTFA antibody and Alexa Fluor 488-conjugated secondary antibody (green). Three independent experiments were performed, with a representative set of images shown here.
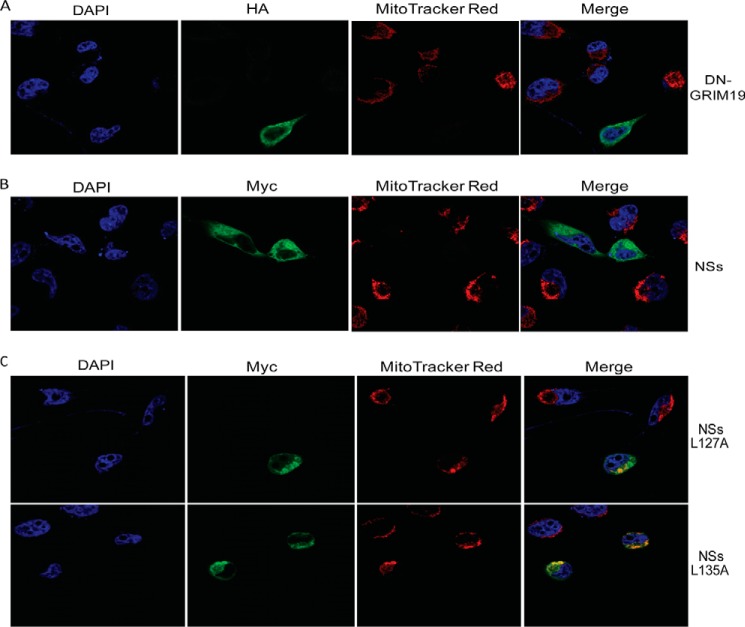
To corroborate our finding that NSs disrupts the mitochondrial membrane potential, we transfected HeLa cells with pXJmyc-NSs and pXJ40-DN-GRIM19-HA and subjected them to immunofluorescence assay. DN-GRIM19, a potent dominant-negative mutant of GRIM19, was used as a positive control because its overexpression has been shown to disrupt the mitochondrial membrane potential (31). Most of the HeLa cells overexpressing DN-GRIM19 showed no MitoTracker staining (Fig. 7A). Similarly, HeLa cells overexpressing NSs protein did not show MitoTracker staining, which further confirms that NSs disrupts the mitochondrial membrane potential (Fig. 7B).
FIGURE 7.
NSs disrupts the mitochondrial membrane potential in HeLa cells. A, the subcellular localization of transiently transfected DN-GRIM19-HA was detected by anti-HA antibody and Alexa Fluor 488-conjugated secondary antibody (green). The subcellular localization of transiently transfected myc-NSs (B) and myc-NSs L127A and L135A (C) was detected by anti-myc antibody and Alexa Fluor 488-conjugated secondary antibody (green). Mitochondria were labeled with MitoTracker Red CMXRos (red), and nuclei were counterstained with DAPI (blue). At least three independent experiments were performed, with a representative set of images shown here.
NSs L127A and L135A Mutants Do Not Disrupt the Mitochondrial Membrane Potential
Both substitution mutations, L127A and L135A, disrupt the apoptotic activity of the NSs. To understand the role of these two key residues in the disruption of the mitochondrial membrane potential, HeLa cells were transfected with pXJmyc-NSs L127A and pXJmyc-NSs L135A. Cells overexpressing NSs L127A and L135A showed MitoTracker staining, which indicates that these two mutants are not able to disrupt the mitochondrial membrane potential (Fig. 7C). Therefore, Leu-127 and Leu-135 are important residues for disruption of the mitochondrial membrane potential by NSs.
Discussion
In the process of host-pathogen co-evolution, viruses have developed distinct strategies to overcome the immunological and biochemical defenses of the host. Viruses have acquired an ability to inhibit host cell apoptosis, a defense mechanism against infection, for their survival. On the contrary, viruses may stimulate apoptosis to kill uninfected immune cells or favor viral dissemination (16). CCHFV suppresses caspase activation at early stages of the infection and induces host cellular pro-apoptotic molecules at late stages post-infection (32). In the past, numerous viral proteins have been shown to modulate, either positively or negatively, the apoptotic response of the hosts (33–37).
The NSs protein was not detected, or detected much less, in La Crosse encephalitis virus-infected cells compared with that of snowshoe hare virus-infected cells (38). Our results indicate that CCHFV NSs is expressed but that it undergoes active proteasomal degradation during infection. Recently, Bunyamwera virus NSs has been shown to actively degrade during infection, and mutation of all the four lysine residues to arginine stabilizes the Bunyamwera virus NSs protein (28). The degradation of NSs protein during infection might be a host defense mechanism against viral infection.
In this report, we have studied the apoptotic properties of CCHFV NSs. Although NSs proteins of other Bunyaviridae family members have been reported to induce apoptosis (7, 8, 10), this is the first time that the NSs protein of CCHFV has been shown to induce apoptosis. Using three different cell lines, we demonstrated that CCHFV NSs protein induces caspase-3/7 activation and PARP cleavage, which are hallmarks of apoptosis (Fig. 3). Earlier, it has been reported that CCHFV induces apoptosis, which may be dependent on caspase-3 activation (22). Recently, NSs protein of Schmallenberg virus (SBV), a member of the Orthobunyavirus genus, has been reported to induce caspase-3/7 activity, leading to apoptosis (39). Similarly, La Crosse encephalitis virus NSs induces caspase-3 activation and nuclear fragmentation, which are the hallmark of apoptosis (7). On the contrary, NSs protein of Bunyamwera virus, a member of the same genus, can delay cell death by counteracting IRF-3-mediated apoptosis (40).
Further characterization of the apoptosis induced by CCHFV NSs reveals that NSs-induced apoptosis is blocked by the specific inhibitors of caspase-3, -8, and -9 (Fig. 4). Apoptosis can be induced via two major pathways: the mitochondrial death pathway (intrinsic pathway), triggered by the mitochondria in response to oxidative stress, DNA damage, and viral proteins, with caspase-9 as the initiator caspase, and the death receptor pathway (extrinsic pathway), triggered by the activation of the death receptors of the TNF family, with caspase-8 as the initiator caspase (41). Therefore, our results suggest that caspase-dependent NSs-induced apoptosis involves both intrinsic and extrinsic pathways. Similarly, Punta Toro virus NSs also induces apoptosis by triggering both intrinsic and extrinsic pathways (10).
The minimal active region of the CCHFV NSs protein responsible for its apoptotic activity was determined to be 93–140 amino acid residues. Moreover, the caspase-3/7 activity for cells expressing NSs(48–140) and NSs(93–140) were five times higher than that of cells expressing NSs (Fig. 5A), indicating autoinhibitory effects by the N terminus of NSs. The role of the N terminus of NSs in apoptosis is currently unknown, but it is possible that it is involved in the interaction with some host apoptosis regulatory proteins or that it may fold back and interfere with the function of the minimal active region found at the C terminus (see below). Further truncation of C-terminal residues led us to narrow down the segment responsible for inducing apoptosis to 126–135 amino acid residues (Fig. 5, C and D). In the quest to pinpoint the key residues involved, Leu-127 and Leu-135 were found to be critical for NSs-induced apoptosis. Several NSs of Orthobunyavirus induce apoptosis and have a reaper-like region at the C-terminal, but CCHFV NSs does not exhibit any sequence similarity with reaper (7). However, CCHFV NSs has been predicted to possess a transmembrane α helix domain (118–135 amino acids) that contains the Leu-127 and Leu-135 residues important for CCHFV NSs-induced apoptosis (transmembrane helices predicted by PHDhtm (42)). Interestingly, these two residues fall on the same face of the putative transmembrane helix in a predicted three-dimensional structure (data not shown).
Recently, NSs of Rift Valley fever virus has been reported to associate with the mitochondria of infected cells, contributing to an increase in reactive oxygen species, leading to apoptosis (43). On the other hand, NSm of the same virus integrates into the mitochondrial outer membrane and exerts an antiapoptotic function. The C-terminal domain of NSm contains a putative transmembrane domain that targets the protein to the mitochondrial outer membrane (44). Our study shows that CCHFV NSs co-localizes with mtTFA, indicating the localization of NSs in the mitochondria (Fig. 6B). Vero E6 cells expressing the NSs protein did not show any MitoTracker staining, revealing that NSs disrupts the mitochondrial membrane potential (Fig. 6A), and this was confirmed further using HeLa cells (Fig. 7B). Interestingly, we found that residues Leu-127 and Leu-135 are also important for NSs-induced disruption of the mitochondrial membrane potential (Fig. 7C). Therefore, we speculate that the NSs protein localizes in mitochondria and triggers mitochondrial membrane permeabilization by an unknown mechanism. This leads to the release of intermembrane space proteins into the cytosol, causing a mitochondrial membrane potential reduction that results in the activation of caspases and apoptosis. Loss of the mitochondrial membrane potential induced by different viral proteins has been reported previously (16). In one such report, non-structural protein 4A (NS4A) of the hepatitis C virus has been shown to accumulate in mitochondria, causing a mitochondrial transmembrane potential reduction, release of cytochrome c, and induction of apoptosis through activation of caspase-3 but not caspase-8 (45). Unlike NS4A of the hepatitis C virus, NSs of CCHFV seems to activate both caspase-8 and -9, which, in turn, activates caspase-3. Similarly, p7 protein of the hepatitis C virus activates both caspase-8 and -9 to induce apoptosis (46).
In summary, this study demonstrated the expression of NSs in infected cells for the first time and used ectopic expression of NSs wild-type and mutant proteins to yield insights into the ability of NSs to induce apoptosis. Our results indicate that ectopic expression of CCHFV NSs induces apoptosis through both intrinsic and extrinsic pathways, but the exact mechanisms for the activation of these pathways have to be investigated further. The putative transmembrane α helix domain (118–135 amino acids), containing the Leu-127 and Leu-135 residues, is important for disruption of the mitochondrial membrane potential, leading to apoptosis. However, further studies are needed to define the exact role played by NSs in regulating apoptosis during infection and determine whether this has implications for viral replication/pathogenesis.
Author Contributions
B. B., A. M., and Y. J. T. designed the experiments. B. B., H. K., A. M., and Y. J. T. performed the experiments. B. B. and Y. J. T. wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgment
We thank X. Cao for the GRIM19 construct.
This work was supported by NUS/NUHS Startup Grants R-182-000-156-720 and R-182-000-156-133 (to Y. J. T.) and intramural funds from the Agency for Science, Technology, and Research. The authors declare that they have no conflicts of interest with the contents of this article.
- CCHFV
- Crimean-Congo hemorrhagic fever virus
- PARP
- poly(ADP-ribose) polymerase
- h.p.i.
- hours post-infection
- CHX
- cycloheximide
- Z-DEVD-fmk
- benzyloxycarbonyl-DEVD-fluoromethyl ketone
- mtTFA
- mitochondrial transcription factor A.
References
- 1.Whitehouse C. A. (2004) Crimean-Congo hemorrhagic fever. Antiviral Res. 64, 145–160 [DOI] [PubMed] [Google Scholar]
- 2.Keshtkar-Jahromi M., Kuhn J. H., Christova I., Bradfute S. B., Jahrling P. B., and Bavari S. (2011) Crimean-Congo hemorrhagic fever: current and future prospects of vaccines and therapies. Antiviral Res. 90, 85–92 [DOI] [PubMed] [Google Scholar]
- 3.Bente D. A., Forrester N. L., Watts D. M., McAuley A. J., Whitehouse C. A., and Bray M. (2013) Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antiviral Res. 100, 159–189 [DOI] [PubMed] [Google Scholar]
- 4.Ergönül O. (2006) Crimean-Congo haemorrhagic fever. Lancet Infect. Dis. 6, 203–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elliott R. M. (1990) Molecular biology of the Bunyaviridae. J. Gen. Virol. 71, 501–522 [DOI] [PubMed] [Google Scholar]
- 6.Hewson R., Chamberlain J., Mioulet V., Lloyd G., Jamil B., Hasan R., Gmyl A., Gmyl L., Smirnova S. E., Lukashev A., Karganova G., and Clegg C. (2004) Crimean-Congo haemorrhagic fever virus: sequence analysis of the small RNA segments from a collection of viruses world wide. Virus Res. 102, 185–189 [DOI] [PubMed] [Google Scholar]
- 7.Colón-Ramos D. A., Irusta P. M., Gan E. C., Olson M. R., Song J., Morimoto R. I., Elliott R. M., Lombard M., Hollingsworth R., Hardwick J. M., Smith G. K., and Kornbluth S. (2003) Inhibition of translation and induction of apoptosis by Bunyaviral nonstructural proteins bearing sequence similarity to reaper. Mol. Biol. Cell 14, 4162–4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blakqori G., and Weber F. (2005) Efficient cDNA-based rescue of La Crosse bunyaviruses expressing or lacking the nonstructural protein NSs. J. Virol. 79, 10420–10428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu F., Liang X., Tesh R. B., and Xiao S. Y. (2008) Characterization of cell-death pathways in Punta Toro virus-induced hepatocyte injury. J. Gen. Virol. 89, 2175–2181 [DOI] [PubMed] [Google Scholar]
- 10.Li G., Ren J., Xu F., and Ferguson M. R. (2010) Non-structural and nucleocapsid proteins of Punta Toro virus induce apoptosis of hepatocytes through both intrinsic and extrinsic pathways. Microbiol. Immunol. 54, 20–30 [DOI] [PubMed] [Google Scholar]
- 11.Bridgen A., Weber F., Fazakerley J. K., and Elliott R. M. (2001) Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 98, 664–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouloy M., Janzen C., Vialat P., Khun H., Pavlovic J., Huerre M., and Haller O. (2001) Genetic evidence for an interferon-antagonistic function of Rift Valley fever virus nonstructural protein NSs. J. Virol. 75, 1371–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber F., Bridgen A., Fazakerley J. K., Streitenfeld H., Kessler N., Randall R. E., and Elliott R. M. (2002) Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of α/β interferon. J. Virol. 76, 7949–7955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Billecocq A., Spiegel M., Vialat P., Kohl A., Weber F., Bouloy M., and Haller O. (2004) NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J. Virol. 78, 9798–9806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Habjan M., Pichlmair A., Elliott R. M., Overby A. K., Glatter T., Gstaiger M., Superti-Furga G., Unger H., and Weber F. (2009) NSs protein of Rift Valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J. Virol. 83, 4365–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galluzzi L., Brenner C., Morselli E., Touat Z., and Kroemer G. (2008) Viral control of mitochondrial apoptosis. PLoS Pathog. 4, e1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teodoro J. G., and Branton P. E. (1997) Regulation of apoptosis by viral gene products. J. Virol. 71, 1739–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irusta P. M., Chen Y. B., and Hardwick J. M. (2003) Viral modulators of cell death provide new links to old pathways. Curr. Opin. Cell Biol. 15, 700–705 [DOI] [PubMed] [Google Scholar]
- 19.Kroemer G., and Reed J. C. (2000) Mitochondrial control of cell death. Nat. Med. 6, 513–519 [DOI] [PubMed] [Google Scholar]
- 20.Thornberry N. A. (1998) Caspases: key mediators of apoptosis. Chem. Biol. 5, R97–103 [DOI] [PubMed] [Google Scholar]
- 21.Thornberry N. A., and Lazebnik Y. (1998) Caspases: enemies within. Science 281, 1312–1316 [DOI] [PubMed] [Google Scholar]
- 22.Karlberg H., Tan Y. J., and Mirazimi A. (2011) Induction of caspase activation and cleavage of the viral nucleocapsid protein in different cell types during Crimean-Congo hemorrhagic fever virus infection. J. Biol. Chem. 286, 3227–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodrigues R., Paranhos-Baccalà G., Vernet G., and Peyrefitte C. N. (2012) Crimean-Congo hemorrhagic fever virus-infected hepatocytes induce ER-stress and apoptosis crosstalk. PLoS ONE 7, e29712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan Z., Akerstrom S., Wee B. Y., Lal S. K., Mirazimi A., and Tan Y. J. (2010) A new panel of NS1 antibodies for easy detection and titration of influenza A virus. J. Med. Virol. 82, 467–475 [DOI] [PubMed] [Google Scholar]
- 25.Xiao J. H., Davidson I., Matthes H., Garnier J. M., and Chambon P. (1991) Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell 65, 551–568 [DOI] [PubMed] [Google Scholar]
- 26.Andersson I., Simon M., Lundkvist A., Nilsson M., Holmström A., Elgh F., and Mirazimi A. (2004) Role of actin filaments in targeting of Crimean Congo hemorrhagic fever virus nucleocapsid protein to perinuclear regions of mammalian cells. J. Med. Virol. 72, 83–93 [DOI] [PubMed] [Google Scholar]
- 27.Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]
- 28.van Knippenberg I., Fragkoudis R., and Elliott R. M. (2013) The transient nature of Bunyamwera orthobunyavirus NSs protein expression: effects of increased stability of NSs protein on virus replication. PLoS ONE 8, e64137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ekert P. G., Silke J., and Vaux D. L. (1999) Caspase inhibitors. Cell Death Differ. 6, 1081–1086 [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Calvo M., Peterson E. P., Leiting B., Ruel R., Nicholson D. W., and Thornberry N. A. (1998) Inhibition of human caspases by peptide-based and macromolecular inhibitors. J. Biol. Chem. 273, 32608–32613 [DOI] [PubMed] [Google Scholar]
- 31.Lu H., and Cao X. (2008) GRIM-19 is essential for maintenance of mitochondrial membrane potential. Mol. Biol. Cell 19, 1893–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karlberg H., Tan Y. J., and Mirazimi A. (2015) Crimean-Congo haemorrhagic fever replication interplays with regulation mechanisms of apoptosis. J. Gen. Virol. 96, 538–546 [DOI] [PubMed] [Google Scholar]
- 33.Tarodi B., Subramanian T., and Chinnadurai G. (1994) Epstein-Barr virus BHRF1 protein protects against cell death induced by DNA-damaging agents and heterologous viral infection. Virology 201, 404–407 [DOI] [PubMed] [Google Scholar]
- 34.Skaletskaya A., Bartle L. M., Chittenden T., McCormick A. L., Mocarski E. S., and Goldmacher V. S. (2001) A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. U.S.A. 98, 7829–7834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacotot E., Ferri K. F., El Hamel C., Brenner C., Druillennec S., Hoebeke J., Rustin P., Métivier D., Lenoir C., Geuskens M., Vieira H. L., Loeffler M., Belzacq A. S., Briand J. P., Zamzami N., Edelman L., Xie Z. H., Reed J. C., Roques B. P., and Kroemer G. (2001) Control of mitochondrial membrane permeabilization by adenine nucleotide translocator interacting with HIV-1 viral protein rR and Bcl-2. J. Exp. Med. 193, 509–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaddy D. F., and Lyles D. S. (2005) Vesicular stomatitis viruses expressing wild-type or mutant M proteins activate apoptosis through distinct pathways. J. Virol. 79, 4170–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan Y. X., Tan T. H., Lee M. J., Tham P. Y., Gunalan V., Druce J., Birch C., Catton M., Fu N. Y., Yu V. C., and Tan Y. J. (2007) Induction of apoptosis by the severe acute respiratory syndrome coronavirus 7a protein is dependent on its interaction with the Bcl-XL protein. J. Virol. 81, 6346–6355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuller F., and Bishop D. H. (1982) Identification of virus-coded nonstructural polypeptides in bunyavirus-infected cells. J. Virol. 41, 643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barry G., Varela M., Ratinier M., Blomström A. L., Caporale M., Seehusen F., Hahn K., Schnettler E., Baumgärtner W., Kohl A., and Palmarini M. (2014) NSs protein of Schmallenberg virus counteracts the antiviral response of the cell by inhibiting its transcriptional machinery. J. Gen. Virol. 95, 1640–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kohl A., Clayton R. F., Weber F., Bridgen A., Randall R. E., and Elliott R. M. (2003) Bunyamwera virus nonstructural protein NSs counteracts interferon regulatory factor 3-mediated induction of early cell death. J. Virol. 77, 7999–8008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar S. (2007) Caspase function in programmed cell death. Cell Death Differ. 14, 32–43 [DOI] [PubMed] [Google Scholar]
- 42.Rost B., Fariselli P., and Casadio R. (1996) Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 5, 1704–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narayanan A., Amaya M., Voss K., Chung M., Benedict A., Sampey G., Kehn-Hall K., Luchini A., Liotta L., Bailey C., Kumar A., Bavari S., Hakami R. M., and Kashanchi F. (2014) Reactive oxygen species activate NFκB (p65) and p53 and induce apoptosis in RVFV infected liver cells. Virology 449, 270–286 [DOI] [PubMed] [Google Scholar]
- 44.Terasaki K., Won S., and Makino S. (2013) The C-terminal region of Rift Valley fever virus NSm protein targets the protein to the mitochondrial outer membrane and exerts antiapoptotic function. J. Virol. 87, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nomura-Takigawa Y., Nagano-Fujii M., Deng L., Kitazawa S., Ishido S., Sada K., and Hotta H. (2006) Non-structural protein 4A of Hepatitis C virus accumulates on mitochondria and renders the cells prone to undergoing mitochondria-mediated apoptosis. J. Gen. Virol. 87, 1935–1945 [DOI] [PubMed] [Google Scholar]
- 46.Aweya J. J., Mak T. M., Lim S. G., and Tan Y. J. (2013) The p7 protein of the hepatitis C virus induces cell death differently from the influenza A virus viroporin M2. Virus Res. 172, 24–34 [DOI] [PMC free article] [PubMed] [Google Scholar]