Abstract
Drosophila melanogaster sarcomere length short (SALS) is a recently identified Wiskott-Aldrich syndrome protein homology 2 (WH2) domain protein involved in skeletal muscle thin filament regulation. SALS was shown to be important for the establishment of the proper length and organization of sarcomeric actin filaments. Here, we present the first detailed characterization of the biochemical activities of the tandem WH2 domains of SALS (SALS-WH2). Our results revealed that SALS-WH2 binds both monomeric and filamentous actin and shifts the monomer-filament equilibrium toward the monomeric actin. In addition, SALS-WH2 can bind to but fails to depolymerize phalloidin- or jasplakinolide-bound actin filaments. These interactions endow SALS-WH2 with the following two major activities in the regulation of actin dynamics: SALS-WH2 sequesters actin monomers into non-polymerizable complexes and enhances actin filament disassembly by severing, which is modulated by tropomyosin. We also show that profilin does not influence the activities of the WH2 domains of SALS in actin dynamics. In conclusion, the tandem WH2 domains of SALS are multifunctional regulators of actin dynamics. Our findings suggest that the activities of the WH2 domains do not reconstitute the presumed biological function of the full-length protein. Consequently, the interactions of the WH2 domains of SALS with actin must be tuned in the cellular context by other modules of the protein and/or sarcomeric components for its proper functioning.
Keywords: actin, Drosophila, intrinsically disordered protein, multifunctional protein, severing, skeletal muscle, WH2 domain, sarcomere, sequestration
Introduction
The basic myofibrillar contractile units, sarcomeres, orchestrate the function of striated muscle. Essential components of the sarcomeric protein networks are the thin filaments composed of actin and regulatory proteins. During sarcomerogenesis, the assembly of actin monomers into filaments and the organization of these filaments into higher order complexes result in the formation of thin filaments. Filament barbed ends, anchored to the Z disk by α-actinin, are capped by CapZ, whereas tropomodulin (Tmod)2 caps pointed ends. Therefore, sarcomeric thin filaments with well defined length are arranged in an extremely regular manner. The structural features of sarcomeric actin filaments are well resolved; however, the mechanisms governing the assembly and proper organization of actin filaments are not completely understood.
The Wiskott-Aldrich syndrome protein homology 2 (WH2) domains are found in increasing numbers of proteins that regulate the actin cytoskeleton during morphogenetic and motile processes (1–6). When uncomplexed, the isolated WH2 domain is a short structurally and intrinsically disordered region in solution, which shows partial folding upon binding to actin (7, 8). The central element of the domain is an LKK(T/V) consensus motif (Fig. 1A). This motif is flanked by an N-terminal amphipathic α-helix followed by an FXXXK linker and a C-terminal region with variable length and composition. WH2 can occur as a single module or as tandem regions within the actin-associated proteins. Interesting features of the WH2 domains are their multifunctional character originating from subtle structural differences in their complexes with actin. The single WH2 domain of thymosin β4 sequesters actin monomers into a non-polymerizable complex, whereas the ones from Ciboulot, Actobindin, or Cordon Bleu have profilin-like function by directing monomer assembly to the filament barbed end (8–11). The presence of tandem WH2 domains can lead to a further level of functional complexity by integrating multiple activities in actin dynamics into a single protein. In vitro, tandem WH2 domains, such as found in Spire, Cordon Bleu, N-WASP, and the Vibrio cholerae/parahemeolyticus VopL/F proteins, possess monomer sequestration activity, which can be converted into nucleation and filament depolymerization-promoting functions by other modules surrounding the WH2 domains (10, 12–17). Additionally, the tandem WH2 domains of Junction-mediating and regulatory protein (JMY) seem to be sufficient for nucleation (16).
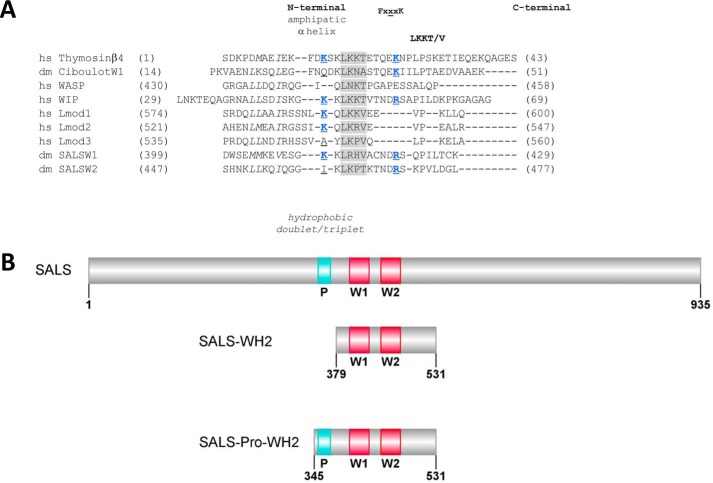
FIGURE 1.
Comparative sequence analysis of the WH2 domains of SALS and domain organization of the constructs investigated in this study. A, comparative sequence alignment of the WH2 domains of SALS. The hydrophobic amino acid doublet/triplet in the N-terminal region is shown by italics; the consensus LKK(T/V) motif is highlighted in gray; amino acids shown previously to form salt bridges with actin are shown in blue; and the central amino acid of the FXXXK region is underlined. hs, Homo sapiens; dm, D. melanogaster. Lmod, leiomodin; WASP, Wiskott-Aldrich syndrome protein. Sequence IDs are as follows: Thymosinβ4 (NP_066932.1); Ciboulot (NP_525065.1); WASP (NP_000368.1); WIP (O43312); Lmod1 (NP_036266); Lmod2 (NP_997046.1); Lmod3 (NP_938012), and SALS (NP_001163588.1). B, SALS constructs used in this study. P, proline-rich sequence region; W1 and W2, WH2 domains. The figure was made by Illustrator for BioSequence software (12).
Recently, members of the WH2 domain protein family emerged as important regulators of muscle actin structures as well. In skeletal muscles two WH2 domain proteins have been implicated in the regulation of assembly and organization of sarcomeric actin filaments as follows: the vertebrate specific leiomodin (Lmod) and the Drosophila melanogaster Sarcomere Length Short (SALS) proteins (18–20). Lmod protein isoforms (Lmod1–3) belong to the Tmod protein family (20). Lmod1 is selectively expressed in smooth muscles, whereas Lmod2 and Lmod3 are present typically in heart and skeletal muscle, respectively. Each Lmod has a single WH2 domain; however, they contain additional actin-binding regions, as well as Tmod-binding domains. Both Lmod2 and Lmod3 were shown to play an important role in the organization of sarcomeric actin filaments (19–21).
SALS exists in two isoforms; the long 935-amino acid isoform (lSALS) is expressed specifically in muscle, and the short one (sSALS) can be found in non-muscle tissues (18). lSALS possesses two tandem WH2 domains and an upstream proline-rich region (Fig. 1B), and the sSALS covers the C-terminal half of the long isoform containing only a single WH2 domain and lacking the proline-rich element. The investigation of the biological activities of the full-length lSALS isoform revealed that it has important roles in the establishment of the proper length and organization of sarcomeric thin filaments. It was proposed that SALS promotes actin filament elongation at the pointed ends by antagonizing the pointed end capping Tmod (18). However, the activities of the WH2 domains of SALS were not analyzed in detail.
In this work, our aim was to better understand how the WH2 domains of SALS contribute to the biological function of the full-length protein. We produced recombinant SALS constructs containing the isolated WH2 domains and analyzed their interactions with actin monomers and filaments by using a combination of protein biochemical, fluorescence spectroscopy, and microscopy approaches. We found that the isolated WH2 domains of SALS possess sequestration and depolymerizing activities. These functions support the accumulation of monomeric actin in vitro, which does not reconstitute the in vivo activity of the full-length protein. Based on these findings, we propose that regions of SALS other than the WH2 domains and/or other sarcomeric proteins (such as tropomyosin) must play essential roles in adjusting and adapting the activity of SALS for its proper biological function. These observations emphasize the role of other modules in tuning the activities of WH2 domains, which is a currently exciting issue to investigate in WH2 domain research.
Experimental Procedures
Protein Purifications
cDNAs of D. melanogaster SALS encoding the WH2 domains (SALS-WH2, 379–531 amino acids) and the proline-rich region upstream to the WH2 domains (SALS-Pro-WH2, 345–531 amino acids) (Fig. 1B) were cloned into pGEX-2T vector using SmaI site. SALS-WH2 and SALS-Pro-WH2 were expressed as glutathione S-transferase (GST) fusion proteins in Escherichia coli BL21(DE3)pLysS strain (Novagen). Transformed bacteria were grown at 37 °C in Luria Broth (Lennox) EZMixTM powder microbial growth medium (Sigma). Protein expression was induced by the addition of 1 mm isopropyl β-d-1-thiogalactopyranoside at A600 = 0.5. After overnight expression at 20 °C, the bacterial extracts were collected by centrifugation (10,000 × g, 15 min, 4 °C) and stored at −80 °C until use. For protein purification, the bacterial pellet was lysed by sonication in phosphate-buffered saline (PBS) supplemented with 0.5% Triton X-100, 0.15 mm PMSF, 1 mm DTT, 5 mm MgCl2, 0.1 mg/ml DNase, 0.5 mg/ml lysozyme, 400 mm NaCl, and Protease Inhibitor Mixture (Sigma, P8465, components: 23 mm 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride, 100 mm EDTA, 2 mm bestatin, 0.3 mm pepstatin A, and 0.3 mm E-64). The cell lysate was ultracentrifuged (Sorvall, T1250 rotor, 100,000 × g, 1 h, 4 °C). The supernatant was incubated with GSH resin (Amersham Biosciences) in batch overnight at 4 °C and washed with PBS and SALS buffer (10 mm Tris-HCl, pH 7.5, 10 mm NaCl, 1 mm DTT, 1% sucrose). The proteins were cleaved with thrombin overnight at 4 °C and eluted by SALS buffer. Thrombin was separated by benzamidine-Sepharose 4 Fast Flow (GE Healthcare). The proteins were concentrated (Amicon Ultra 5-kDa cutoff, Sigma) and clarified by ultracentrifugation (Beckman tabletop ultracentrifuge, MLA80 rotor, 300,000 × g, 30 min, 4 °C). SALS constructs were flash-frozen in liquid N2 and stored at −80 °C until use. Control experiments showed that a freeze/thaw cycle does not affect the functionality of the constructs (data not shown). Typically, a 5–6-g bacterial pellet yielded 1–1.2 mg/ml protein. The protein concentrations were measured spectrophotometrically using the extinction coefficient of ϵ280 nm = 7115 m−1 cm−1 at 280 nm and molecular masses of 17,116 and 21,051 Da for SALS-WH2 and SALS-Pro-WH2, respectively, derived from the amino acid sequence (ExPASy ProtParam tool). The same concentration was obtained from measuring the mass of the lyophilized proteins. In contrast, bicinchoninic acid assay (QuantiProTM BCA assay kit, Sigma) gave underestimated values of concentrations due to the acidic nature of the constructs (22). Actin was purified from rabbit skeletal muscle according to standard procedures (23), gel filtered by Sephadex G-200 (GE Healthcare), and stored in G buffer (4 mm Tris-HCl, pH 7.8, 0.1 mm CaCl2, 0.2 mm ATP, 0.005% NaN3, 0.5 mm β-mercaptoethanol). Human gelsolin (gelsolin), mouse profilin 1 (profilin), and rabbit skeletal muscle tropomyosin were purified as described previously and stored at −80 °C until used (24–26).
Protein Modifications
Actin was labeled at Cys374 with N-(1-pyrene)iodoacetamide (Sigma) and 5-((((2-iodoacetyl)amino)ethyl)amino)naphthalene-1-sulfonic acid (IAEDANS, Life Technologies. Inc.) or with the primary amine-specific Alexa Fluor® 488 carboxylic acid succinimidyl ester (Alexa488NHS, Invitrogen) according to standard protocols (27–29). Alexa488NHS-labeled actin was analyzed by mass spectrometry. The binding site of the probe was found to be the Lys328 residue of actin. Profilin was labeled by Alexa Fluor® C5 568 maleimide (Alexa568C, Invitrogen) as follows. 10-Fold molar excess of dye was added to the protein solution in DTT-free buffer, and the sample was incubated overnight at 4 °C. The reaction was terminated by the addition of 10 mm DTT. The unbound dye was removed by a PD-10 column (GE Healthcare). The protein concentration was determined by correcting for the absorption of the dye at 280 nm.
General Experimental Considerations
Samples at each concentration were prepared individually for experiments. All measurements were performed at 20 °C. The sum of the volume of the proteins and the volume of their storing buffer were constant in the samples and represented 50% of the total volume of the sample. The concentrations given in the text are final concentrations, unless stated otherwise. The measurements were performed using Mg2+-ATP-actin. The actin monomer bound calcium was replaced with magnesium by adding 200 μm EGTA and 50 μm MgCl2 and incubating the samples for 5–10 min at room temperature. Actin-polymerizing conditions were 1 mm MgCl2 and 50 mm KCl. The data presented were derived from at least two independent experiments.
Fluorescence Spectroscopy Experiments
Steady-state Fluorescence Emission and Anisotropy Measurements
The fluorescence emission and the steady-state anisotropy (anisotropy) of IAEDANS or Alexa488NHS-labeled G-actin was measured to study the interaction of SALS constructs with monomeric actin. First, IAEDANS- or Alexa488NHS-Mg2+-G-actin (1 μm or 20 μm) was incubated with latrunculin A (4 or 20 μm, respectively) for 30 min to avoid polymerization. Then SALS-WH2 or SALS-Pro-WH2 at different concentrations was added, and the samples were incubated overnight at 4 °C. The measurements were performed using a Horiba Jobin Yvon spectrofluorometer (IAEDANS-G-actin, λex = 350 nm and λem = 460 nm; Alexa488NHS-G-actin, λex = 488 nm and λem = 516 nm). To derive the dissociation equilibrium constant (KD) of the SALS·G-actin complex, the SALS-WH2 concentration dependence of the fluorescence emission (I) and wavelength (λ) values were fit using Equations 1 and 2 (30, 31),
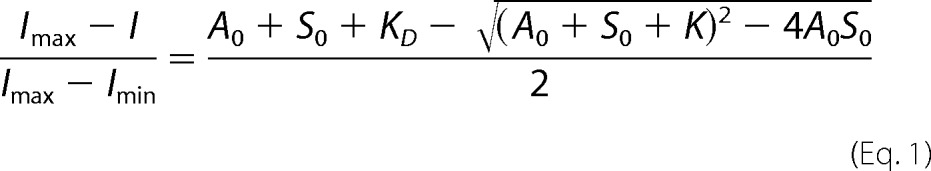 |
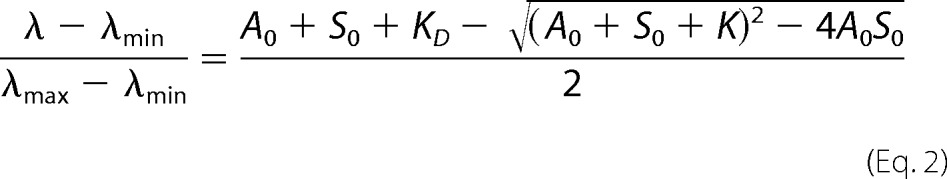 |
where A0 and S0 are the total G-actin and SALS-WH2 concentration, respectively; Imax and λmin are the relative fluorescence emission; wavelength values were measured in the absence of SALS-WH2; Imin and λmax are the relative fluorescence emission; and wavelength values are measured in the presence of saturating amount of SALS-WH2. The relative values of the fluorescence parameters were derived by dividing the values measured in the presence of SALS-WH2 by the value measured in the absence of SALS-WH2.
The dissociation equilibrium constant (KD) of the SALS·G-actin complex was derived from the SALS concentration dependence of the steady-state anisotropy of G-actin (r), in conjunction with Equation 3 (30, 31),
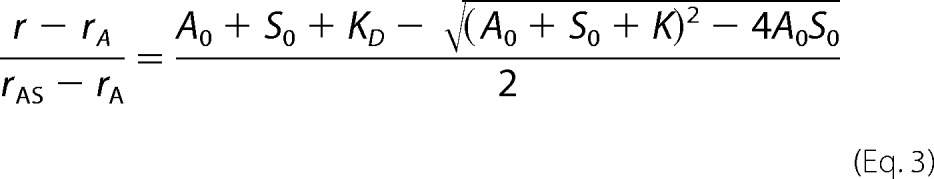 |
where A0 and S0 are the total G-actin and SALS-WH2/SALS-Pro-WH2 concentration, respectively; rA is the steady-state anisotropy of free G-actin; and rAS is the steady-state anisotropy of G-actin in the presence of saturating amount of SALS-WH2/SALS-Pro-WH2.
Actin Polymerization Measurements
Polymerization of Mg2+-G-actin (2.5 μm, containing 5% pyrenyl-actin) was initiated by the addition of 1 mm MgCl2 and 50 mm KCl in the absence and presence of different concentrations of SALS constructs. Actin assembly was measured by monitoring the change in pyrenyl fluorescence emission using a Safas Xenius FLX spectrofluorimeter (λex = 365 nm and λem = 407 nm). The initial rate of actin assembly (v) was derived from the initial slope (0–500 s) of the pyrenyl traces and plotted as the function of SALS-WH2 concentration ([S]). Data were fitted using Equation 4 (30),
 |
where v0 is the initial rate of actin assembly in the absence of SALS-WH2; A0 is the total actin concentration, and [SA] is the SALS-WH2·G-actin complex concentration, which was derived using the quadratic binding Equations 1–3.
To study actin assembly at the pointed ends, gelsolin-actin seeds (GA2) were prepared by incubating 0.5 μm gelsolin and 1.1 μm G-actin in G buffer in the presence of excess CaCl2 (2.5 mm) (27). The assembly of G-actin (1.2 μm, containing 5% pyrenyl-actin) was monitored from 33 nm GA2 in the absence or presence of different amounts of SALS-WH2 using a Safas Xenius FLX spectrofluorimeter (λex = 365 nm and λem = 407 nm).
Dilution-induced Depolymerization Measurements
Pre-polymerized actin filaments (1 μm, containing 50% pyrenyl-actin) were diluted with F buffer (G buffer supplemented with 1 mm MgCl2 and 50 mm KCl) to 20 nm in the absence or presence of different amounts of SALS constructs, gelsolin (0.1 or 0.5 μm), or latrunculin A (20 μm). When tropomyosin was used first, the actin filaments were incubated with excess tropomyosin (4.5 μm) overnight at 4 °C and then the filaments were diluted with F buffer containing the same amount of tropomyosin (4.5 μm) to avoid dissociation of tropomyosin from the filaments due to dilution. Kinetics of actin filament disassembly was monitored by the decrease in pyrenyl fluorescence emission using a Horiba Jobin Yvon spectrofluorometer or a Safas Xenius FLX spectrofluorimeter (λex = 365 nm and λem = 407 nm). The fluorescence emission values were normalized by the value measured at t = 0 s (relative pyrenyl fluorescence).
Critical Concentration Measurements
Mg2+-G-actin (containing 5% pyrenyl-actin) at different concentrations was polymerized and then either SALS-WH2 (0.5 μm or 1.5 μm) or gelsolin (0.01 μm) was added, and the samples were incubated overnight at 4 °C. The fluorescence emission of the samples was measured using a Horiba Jobin Yvon spectrofluorometer (λex = 365 nm and λem = 407 nm) and plotted as the function of the actin concentration ([actin]). The amount of unassembled actin (AU) is reflected by the breaking point of the [actin]-fluorescence intensity curve (J(c) plot). The steady-state pool of unassembled actin is composed of free G-actin (critical concentration, cC) and SALS-WH2-bound G-actin (SA). The concentration of SA ([SA]) can be given by Equation 5 (30),
 |
where S0 is the total SALS-WH2 concentration, and K is the dissociation equilibrium constant of the SALS-WH2·G-actin complex.
Steady-state Measurements of F-actin
Actin filaments (2.5 μm, containing 5% of pyrenyl-actin) were polymerized in the absence and presence of phalloidin (2.5 μm), jasplakinolide (2.5 μm), or tropomyosin (6 μm) overnight at 4 °C. Then different amounts of SALS-WH2 were added, and the samples were further incubated overnight at 4 °C. The fluorescence emission of the samples were measured using a Horiba Jobin Yvon spectrofluorometer (λex = 365 nm and λem = 407 nm). The value measured in the absence of SALS-WH2 was used to normalize emission values (relative pyrenyl fluorescence), and these were plotted against the SALS-WH2 concentration.
High Speed Sedimentation Experiments
Actin filaments (1, 1.5, or 2.5 μm) or phalloidin-stabilized actin filaments were incubated with different amounts of SALS-WH2 overnight at 4 °C. Phalloidin was added to actin filaments in an equimolar amount and incubated overnight at 4 °C prior to the addition of SALS-WH2. The samples were ultracentrifuged (Beckman tabletop ultracentrifuge, TLA 100 rotor, 300,000 × g, 30 min, 20 °C). The pellets (P) and supernatants (SN) were separated and processed for SDS-PAGE analysis. The protein content of the pellets and the supernatants was derived from Coomassie Blue-stained gels (Syngene Bioimaging System). Quantification of Coomassie Blue intensities was performed within the linear range of exposure identified by a calibration curve. The intensity values were corrected for the molecular weight of each protein. Then the values were converted to actin concentration ([G-actin] or [F-actin]) using a calibration curve. The slope of the linear fit to the [SALS-WH2]-[G-actin] or [SALS-WH2]-[F-actin] curves (i.e. [SA]/[S0]) can be related to the critical concentration of actin assembly (cC) and the dissociation equilibrium constant (KD) of the SALS-WH2·G-actin complex ([SA]) by Equation 5.
Total Internal Reflection Fluorescence Microscopy (TIRFM)
TIRFM experiments were carried out as described previously (27). Briefly, glass flow cells (∼50 μl) were incubated with 1 volume of N-ethylmaleimide myosin, washed extensively with 2 volumes of myosin buffer (F buffer supplemented with 0.5 m KCl) and 2 volumes of 0.1% (w/v) BSA (dissolved in F buffer), and equilibrated with 2 volumes of TIRFM buffer (0.5% (w/v) methylcellulose, 10% (w/v) BSA, 1 mm 1,4-diazabicyclo-[2,2,2]octane, 100 mm DTT dissolved in F buffer). Polymerization of 0.5 μm Mg2+-G-actin (containing 10% Alexa488NHS-G-actin) was performed in the absence and presence of different amounts of SALS-WH2. Images were captured in every 10 s with an Olympus IX81 microscope equipped with a laser-based (491 nm) TIRFM system, an apoTIRFM ×60 NA 1.45 oil objective, the Cell∧R acquisition software, and a Hamamatsu CCD camera. Time-lapse images were analyzed by Fiji. The growth rates of actin filaments in microns/s were determined using the multiple kymograph plugin. Filament length was converted to subunits using 370 subunits/μm filament (32). The spontaneous elongation rate of actin filaments (v0) can be related to the critical concentration (cC) of actin assembly, to the association rate constant of actin monomers to filament barbed ends (k+), and to the total actin concentration ([G0]) as shown by Equation 6 (30),
For further analysis, the elongation rates of actin filaments (v) were plotted as the function of SALS-WH2 concentration [S0]. Data were fitted using Equation 4.
Results
WH2 Domains of SALS Bind Actin Monomers
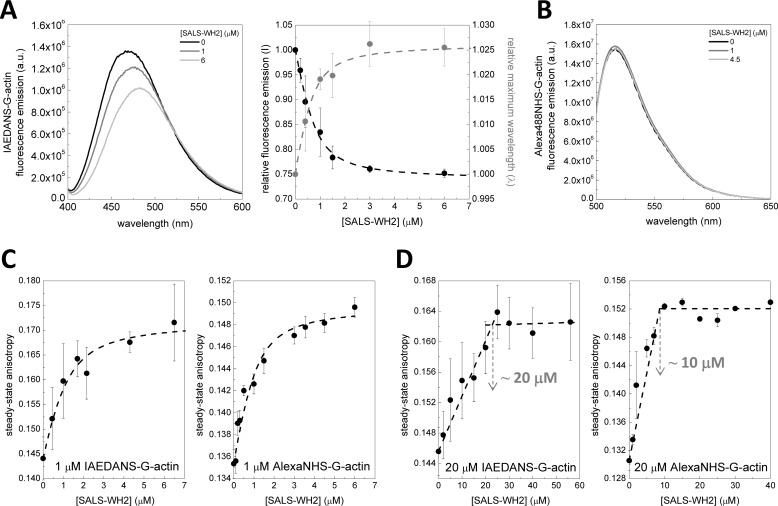
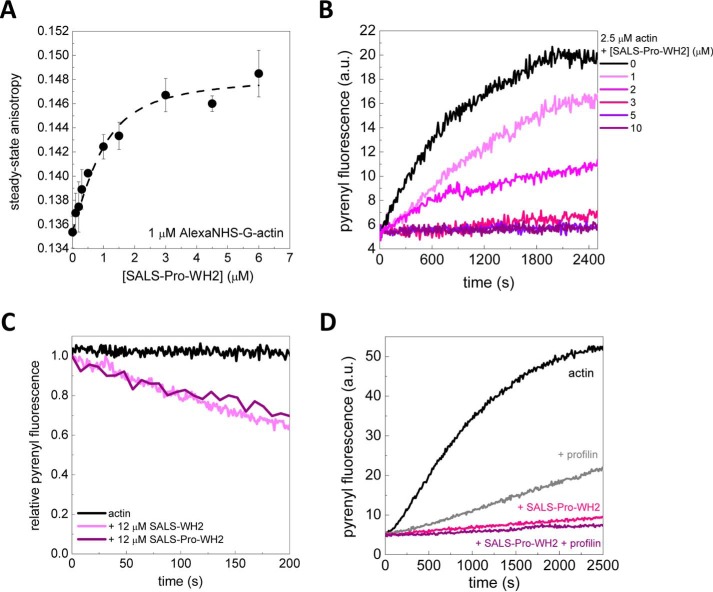
To begin the biochemical characterization of the tandem WH2 domains of SALS (SALS-WH2), we addressed the question whether, similar to other WH2 domains, SALS-WH2 is able to interact with actin monomers. First, the fluorescence emission of IAEDANS-labeled Mg2+-ATP-G-actin (1 μm) was measured in the presence of different concentrations of SALS-WH2. The data revealed that the fluorescence emission of IAEDANS was reduced by ∼25% and the wavelength corresponding to the emission maximum (maximum wavelength) was red-shifted by ∼10 nm in the presence of saturating amounts of SALS-WH2 (Fig. 2A). These changes are consistent with complex formation between actin and SALS-WH2. For quantitative analysis, the fluorescence emission and the maximum wavelength values were derived from the spectra and plotted as the function of SALS-WH2 concentration (Fig. 2A, right panel). The analysis revealed a dissociation equilibrium constant (KD) of the SALS-WH2·G-actin complex of ∼0.23 μm (Fig. 2A, right panel, Equations 2 and 3).
FIGURE 2.
SALS-WH2 binds actin monomers. A, left panel, representative fluorescence emission spectra of IAEDANS-Mg2+-ATP-G-actin (1 μm, containing 4 μm latrunculin A) recorded in the absence and presence of different amounts of SALS-WH2, as indicated. Note the SALS-WH2-induced decrease in the fluorescence emission and the red shift in the wavelength corresponding to the maximum intensity. a.u., arbitrary units. Right panel, SALS-WH2 concentration dependence of the relative fluorescence emission (black circles, I) and the relative wavelength corresponding to the maximum intensity (gray circles, λ). Dashed lines in the corresponding colors show the fit to the data according to Equations 1 and 2. The fit to the intensity and wavelength data set gave dissociation equilibrium constants of K = 0.24 ± 0.1 μm and 0.22 ± 0.2 μm, respectively. Error bars, standard deviation. B, representative fluorescence emission spectra of Alexa488NHS-Mg2+-ATP-G-actin (1 μm, containing 4 μm latrunculin A) recorded in the absence and presence of different amounts of SALS-WH2, as indicated. C, steady-state anisotropy of IAEDANS- and Alexa488NHS-Mg2+-ATP-G-actin (1 μm, containing 4 μm latrunculin A) in the absence and presence of increasing amounts of SALS-WH2, as indicated. Dashed lines show the fits to the data, according to Equation 3. Dissociation equilibrium constants of K = 0.48 ± 0.2 μm (IAEDANS-G-actin) and K = 0.34 ± 0.2 μm (Alexa488NHS-G-actin) were obtained. Error bars, standard deviation. D, steady-state anisotropy of IAEDANS- and Alexa488NHS-Mg2+-ATP-G-actin (20 μm, containing 20 μm latrunculin A) in the absence and presence of increasing amount of SALS-WH2, as indicated. Note the non-hyperbolic sharply linear tendency of the curves. Importantly the breaking points (highlighted by gray arrows) are observed at different SALS-WH2/G-actin molar ratios, depending on the fluorophore. Dashed lines show the fits to the two linear parts of the curves. Error bars, standard deviation.
In contrast, the fluorescence emission parameters of Alexa488NHS-labeled Mg2+-ATP-G-actin (1 μm) were not affected significantly by SALS-WH2 binding (Fig. 2B). These results show that the local environment around IAEDANS (attached to Cys374), but not around Alexa488NHS, is affected by SALS-WH2 binding. Our MS analysis identified Lys328 in subdomain 3 as the target site of the NHS labeling (data not shown). We concluded that SALS-WH2 binds near the Cys374 residue to actin, whereas the protein region around the Lys328 is not directly involved in the interaction.
To further support the binding of SALS-WH2 to G-actin, steady-state anisotropy measurements were performed. The steady-state anisotropy of both IAEDANS and Alexa488NHS-labeled Mg2+-ATP-G-actin (1 μm) increased upon the addition of increasing concentrations of SALS-WH2 (Fig. 2C). Because the measurements were performed in the presence of saturating amount of latrunculin A, which interferes with filament formation (12, 33), the SALS-WH2 concentration-dependent increase in the steady-state anisotropy could not be attributed to filament formation. It is more consistent with the binding of SALS-WH2 to monomeric actin. In agreement with the analysis of the fluorescence emission data (Fig. 2A), the experiments with either of the two fluorescent probes indicated that the affinity (KD) of SALS-WH2 to monomeric actin is ∼0.4 μm (Equation 3).
Because SALS has two WH2 domains, we tested whether both WH2 domains can interact with actin monomers. The anisotropy measurements were performed at relatively high actin monomer concentrations (20 μm) compared with the KD ∼0.4 μm (Fig. 2D). Under these conditions, most of the WH2 binds to actin, and the experiments are expected to result in break-point titrations, where the breaking point indicates the binding stoichiometry of actin and SALS-WH2 in their complexes (12, 34, 35). For both IAEDANS and Alexa488NHS-labeled Mg2+-ATP-G-actin, the data followed non-hyperbolic saturation curves (Fig. 2D). We noted that the breaking point was reached at different SALS-WH2 concentrations depending on the applied fluorophore; IAEDANS and Alexa488NHS-labeled Mg2+-ATP-G-actin were saturated by ∼20 and ∼10 μm SALS-WH2, respectively (Fig. 2D, gray arrows). The concentration dependence of the fluorescence emission and maximum wavelength of IAEDANS-G-actin revealed saturation at 20 μm SALS-WH2 (data not shown). Considering these observations and those we made in the fluorescence emission measurements, we suggest that the location of the fluorophore can affect the binding of the WH2 domains to G-actin, which can lead to artificial stoichiometry. In conclusion, it is more likely that both of the WH2 domains of SALS can bind to monomeric actin, forming a one SALS-WH2·two G-actin complex.
WH2 Domains of SALS Inhibit Actin Assembly
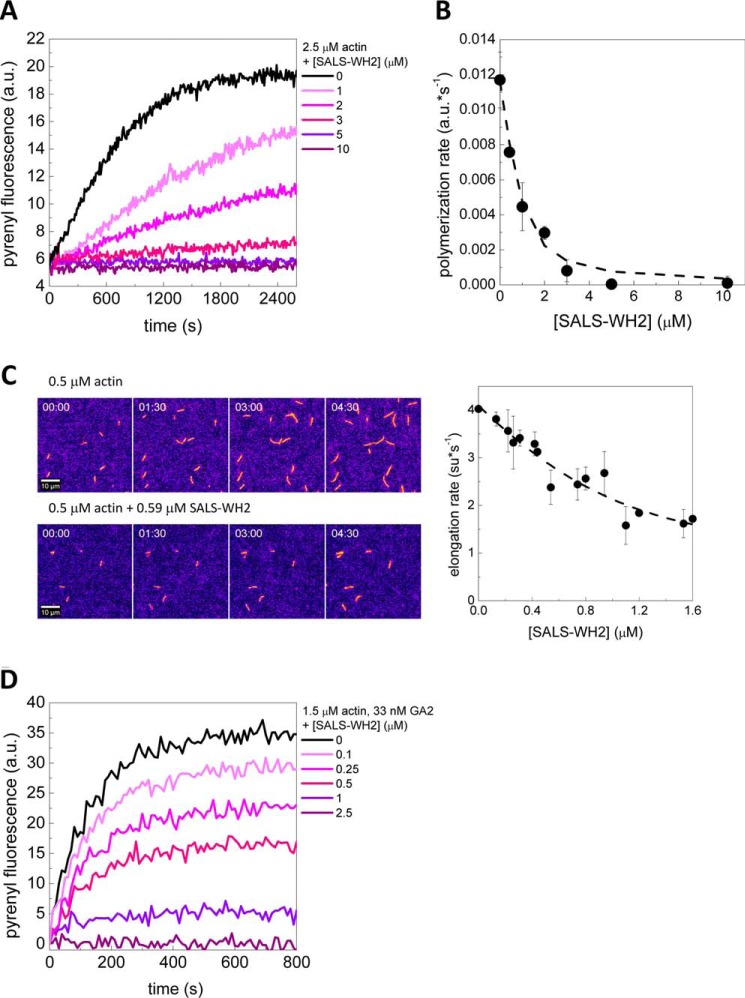
To address whether actin binding by SALS-WH2 can influence actin polymerization, the effects of SALS-WH2 on actin assembly were investigated by using pyrenyl-actin (2.5 μm). We found that the WH2 domains of SALS slowed down actin assembly in a concentration-dependent manner (Fig. 3A). At high SALS-WH2 concentration (>5 μm), the polymerization was completely inhibited. Notably, the extent of actin assembly (i.e. the plateau values of the pyrenyl traces corresponding to the steady-state regime) decreased with increasing concentrations of SALS-WH2 (Fig. 3A). This could not result from the quenching of pyrenyl fluorescence as it is demonstrated by using label-free approaches (e.g. Fig. 4). Analysis of the concentration dependence of the initial polymerization rate (by using Equation 4) revealed that half-inhibition was reached at 0.3 ± 0.13 μm SALS-WH2 (Fig. 3B). This value is close to that derived from steady-state fluorescence measurements (Fig. 2, A and C), suggesting that the polymerization inhibiting activity of SALS-WH2 arises from monomer binding.
FIGURE 3.
SALS-WH2 inhibits actin assembly. A, representative polymerization kinetics of actin (2.5 μm, containing 5% pyrenyl-actin) in the absence and presence of increasing amounts of SALS-WH2, as indicated. a.u., arbitrary units. B, rate of actin assembly determined from the initial slope of the pyrenyl traces as a function of SALS-WH2 concentration. Dashed line corresponds to the fit of the data using Equation 4, and the fit gave half-inhibition at 0.24 ± 0.1 μm SALS-WH2. Error bars, standard deviation. C, left panel, representative montages of TIRFM movies of actin assembly (0.5 μm, containing 10% Alexa488NHS-actin) recorded in the absence and presence of SALS-WH2 (0.59 μm), as indicated. Scale bar, 10 μm, elapsed time = min/s, look-up table: fire. Right panel, SALS-WH2 concentration dependence of the elongation rate of actin filaments, derived from TIRFM experiments. Dashed line corresponds to the fit of the data using Equation 4, and the fit gave half-inhibition at 0.38 ± 0.3 μm SALS-WH2. Error bars, standard deviation. D, representative polymerization kinetics of actin assembly from gelsolin/actin (GA2) seeds (1.5 μm, containing 5% pyrenyl-actin, [GA2] = 33 nm) in the absence and presence of increasing amounts of SALS-WH2, as indicated.
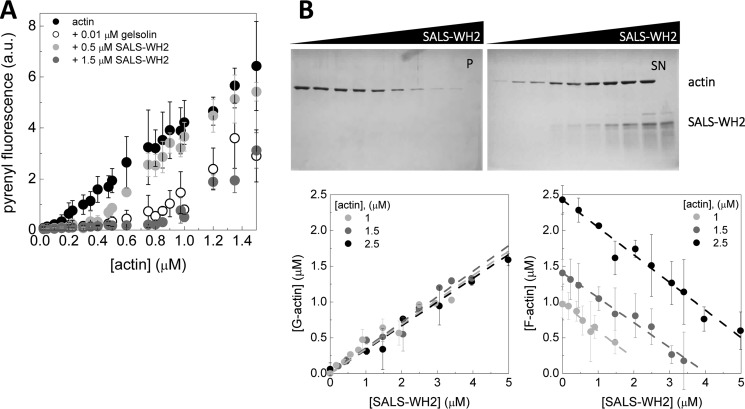
FIGURE 4.
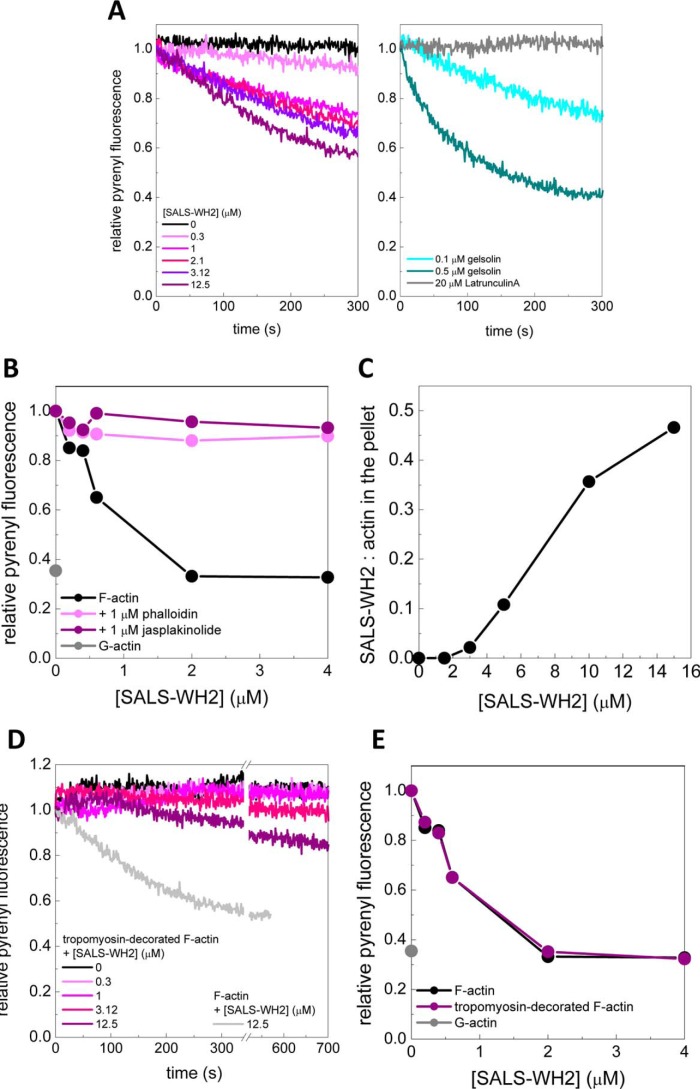
SALS-WH2 sequesters actin monomers. A, critical concentration plots of actin assembly in the absence of any other protein and in the presence of gelsolin (0.01 μm) or SALS-WH2 (0.5 or 1.5 μm), as indicated. Error bars, standard deviation. a.u., arbitrary units. B, upper panel, representative Coomassie-stained gels of pellets (P) and supernatants (SN) from high speed sedimentation experiments. The actin concentration was 2.5 μm from lanes 1 to 9 and 0 μm in lane 10, the SALS-WH2 concentrations were 0, 1, 2, 2.8, 4.2, 5, 6, 9, and 12 μm from lanes 1–9 and 12 μm in lane 10. Lower panel, amount of G-actin and F-actin at steady-state as the function of the SALS-WH2 concentration derived from SDS-PAGE analysis. The actin concentrations were as follows: 1, 1.5, and 2.5 μm. Dashed lines in the corresponding colors show the linear fits to the data yielding average slope of |0.36| ± 0.02. Error bars, standard deviation.
The investigation of the effects of SALS-WH2 on actin polymerization at the level of individual filaments by TIRFM revealed that SALS-WH2 reduces the length of actin filaments (Fig. 3C). Spontaneously growing actin filaments elongated at a rate of 4.1 ± 0.5 subunit·s−1. Considering barbed end critical concentration of cC = 0.15 μm (Fig. 4A) (27, 36–38), this value is in agreement with the well established value of the barbed end association rate constant of Mg2+-ATP-G-actin (k+ ∼11 μm−1·s−1 (Equation 6) (27, 36–38)). We found that SALS-WH2 decreased the elongation rate in a concentration-dependent manner (Fig. 3C, right panel). The analysis of the data gave a dissociation equilibrium constant of ∼0.4 μm (Fig. 3C, right panel, and Equation 4, consistent with the results derived from fluorescence spectroscopy experiments. We also found that the number of actin filaments formed in the presence of SALS-WH2 was reduced significantly, compared with the number of spontaneously formed actin filaments (Fig. 3C, left panel). According to these observations, SALS-WH2 inhibits both the nucleation and the elongation phases of actin assembly.
It was proposed that the full-length protein could facilitate sarcomeric actin filament assembly at the pointed ends (18). To test this, we monitored the effect of SALS-WH2 in a gelsolin·actin (GA2) seeded polymerization assay, which allowed us to measure actin assembly selectively at the pointed ends (27). In contrast to previous reports on the full-length protein (18), we found that SALS-WH2 inhibits actin assembly from GA2 seeds in a similar manner to its effects on spontaneous actin polymerization (Fig. 3D). These results show that the WH2 domains of SALS are not sufficient to promote pointed end assembly.
WH2 Domains of SALS Sequester Actin Monomers
Although the inhibitory effect of SALS-WH2 on actin nucleation and elongation can be due to sequestration that inhibits both filament formation and lengthening at both ends of the filaments, the inhibition of elongation can also arise from barbed end capping. To distinguish these activities, the effects of SALS-WH2 on steady-state actin dynamics were characterized.
First, to measure whether SALS-WH2 can influence the steady-state amount of unassembled actin, critical concentration plots of actin assembly (J(c)) were constructed (Fig. 4A). In the absence of any other protein, the steady-state amount of unassembled actin is established by the pool of free G-actin, i.e. the critical concentration corresponding to the breaking point of the J(c) plot. It is dominated by barbed end dynamics, and its value is ∼0.15 μm (Fig. 4A, black circles) (36–38). Barbed end capping (by gelsolin, for example) results in a shift in the critical concentration up to ∼0.6 μm that is characteristic to the pointed ends (Fig. 4A, empty circles) (39). The J(c) plots obtained in the presence of SALS-WH2 were parallel to the plot obtained for actin. The breaking point of the plots in the presence of 0.5 and 1.5 μm SALS-WH2 were shifted to higher values of ∼ 0.3 and ∼ 0.8 μm, respectively. According to Equation 5, the measured amount of unassembled actin in the presence of SALS-WH2 is consistent with the sum of the concentration of the SALS-WH2·G-actin complex, as calculated from the dissociation equilibrium constant of the complex (∼ 0.4 μm; Fig. 2), and the barbed end critical concentration. Note that barbed end capping, i.e. pointed end dynamics, would have resulted in breaking points at significantly higher actin concentrations of ∼0.9 and ∼1.5 μm in the presence of 0.5 or 1.5 μm SALS-WH2, respectively. These observations show that SALS-WH2 increases the steady-state pool of unassembled actin in agreement with the decrease in the extent of actin assembly detected in polymerization assays (Fig. 3A).
Based on the above experiments, we reasoned that SALS-WH2 is likely to increase the amount of unpolymerized actin by sequestration. To further test this possibility, increasing amounts of SALS-WH2 were added to pre-formed actin filaments at constant concentrations (either 1, 1.5, or 2.5 μm). The samples were ultracentrifuged, and the protein content of the pellets (P) and supernatants (SN) was analyzed by SDS-PAGE (Fig. 4B). We found that the steady-state amount of monomeric actin increased with increasing the concentration of SALS-WH2 (Fig. 4B, lower left panel), and in parallel, the steady-state amount of filamentous actin decreased (Fig. 4B, lower right panel). These observations again are in agreement with a decrease in the steady-state amount of F-actin, as detected in fluorescence spectroscopy measurements (Fig. 3A). At different actin concentrations, both the increase in the amount of G-actin and the decrease in the amount of F-actin followed a linear tendency and were characterized by parallel straight lines (Fig. 4B, lower panels). The linear fit to the data resulted in the same absolute value of the slopes, 0.36 ± 0.02 (Fig. 4B, lower panels). This value matches well with the dissociation equilibrium constant of the SALS-WH2·G-actin complex and the barbed end critical concentration (Equation 5).
In conclusion, the analyses of the effects of SALS-WH2 on the steady-state distribution of monomeric and filamentous actin suggest that SALS-WH2 does not influence barbed end dynamics. Consequently, its inhibitory effect on actin assembly results solely from sequestration and not from capping of the filament barbed ends.
WH2 Domains of SALS Enhance Actin Filament Disassembly
Sequestration leads to a decrease in the steady-state amount of F-actin (Fig. 4B). However, this activity affects filament disassembly only passively as it is determined by spontaneous actin dynamics. Actin monomers dissociating spontaneously from the filament ends are sequestered, which results in their removal from subsequent actin dynamics. This leads to a stepwise decrease in the concentration of uncomplexed actin and thus the amount of actin filaments.
To address whether the WH2 domains of SALS can actively accelerate actin disassembly, dilution-induced depolymerization assays were performed in the absence and presence of SALS-WH2 (Fig. 5A). The results revealed that SALS-WH2 accelerated the disassembly of actin filaments in a concentration-dependent manner (Fig. 5A, left panel). As controls, we monitored the effects of latrunculin A as a sole sequestrator (33) and gelsolin as a severing protein (39) on actin filament disassembly (Fig. 5A, right panel). We found that latrunculin A did not alter the kinetics of actin disassembly, although gelsolin enhanced disassembly in a concentration-dependent manner. The magnitude of the effect of SALS-WH2 on the disassembly kinetics fell into the range that is characteristic for severing. These results support the theory that the WH2 domains of SALS can actively enhance filament disassembly by severing.
FIGURE 5.
SALS-WH2 binds and enhances the disassembly of actin filaments, which is influenced by actin-binding drugs and tropomyosin. A, left panel, kinetics of actin disassembly (0.02 μm, containing 50% pyrenyl-actin) in the absence and presence of different amount of SALS-WH2, as indicated. Right panel, as controls, actin disassembly kinetics in the presence of latrunculin A (20 μm) or gelsolin (0.1 and 0.5 μm) are shown. B, relative pyrenyl fluorescence of F-actin (1 μm) in the presence of increasing amounts of SALS-WH2, in the absence and presence of phalloidin (1 μm) or jasplakinolide (1 μm), as indicated. The relative pyrenyl fluorescence corresponding to G-actin is shown as a gray dot. C, ratio of SALS-WH2 and actin in the pellet as the function of SALS-WH2 concentration determined from high speed sedimentation experiments using actin filaments (2.5 μm) stabilized by equimolar amount of phalloidin. D, kinetics of actin disassembly (0.02 μm, containing 50% pyrenyl-actin) in the presence of tropomyosin (4.5 μm) and in the absence and presence of different amount of SALS-WH2, as indicated. For easier comparison, gray line shows the pyrenyl trace obtained in the absence of tropomyosin and in the presence of SALS-WH2 (12.5 μm) (same data is shown on A). E, relative pyrenyl fluorescence of F-actin (1 μm) and tropomyosin (4.5 μm) decorated F-actin in the presence of increasing amounts of SALS-WH2, as indicated. The relative pyrenyl fluorescence corresponding to G-actin is shown as a gray dot.
Severing activity requires direct interactions with the side of actin filaments. Therefore, to further support the conclusion from the dilution-induced depolymerization experiments, we tested the ability of SALS-WH2 to bind directly to filamentous actin. Interestingly, we found that SALS-WH2 failed to disassemble actin filaments stabilized by equimolar amounts of phalloidin or jasplakinolide, as revealed by steady-state pyrenyl fluorescence measurements (Fig. 5B). Using this observation, we assayed the F-actin binding ability of SALS-WH2 in high speed sedimentation assays. Consistent with the steady-state fluorescence spectroscopic experiments (Fig. 5B), the amount of phalloidin-stabilized F-actin did not change with increasing concentrations of SALS-WH2. Additionally, SALS-WH2 appeared in the pellet with actin in a concentration-dependent manner (data not shown). Control experiments showed that this did not result from the self-pelleting of the protein (e.g. Fig. 4B, upper panel, 1th0 lane). Thus, these results indicate that SALS-WH2 can directly bind to phalloidin-bound actin filaments. The amount of SALS-WH2 detected in the pellet was much higher than the one expected due to filament end binding, which suggests that the WH2 domains of SALS interact with the side of actin filaments. The SALS-WH2/F-actin ratio in the pellet derived from these experiments reached saturation at ∼0.5 (Fig. 5C). This stoichiometry indicates that both WH2 domains of SALS bind to actin, consistent with the results obtained in steady-state anisotropy measurements using Alexa488NHS-G-actin (Fig. 2D, right panel). Based on the above results, we propose that, besides sequestration, SALS-WH2 can facilitate actin filament disassembly upon direct binding to F-actin.
Tropomyosin Can Modulate the Depolymerizing Activity of SALS-WH2
Because sarcomeric actin filaments are decorated by tropomyosin, we tested whether muscle tropomyosin influences the interaction of SALS-WH2 with actin filaments. The dilution-induced depolymerization experiments were repeated using tropomyosin-bound actin filaments. The pyrenyl transients reflected significantly slower disassembly kinetics in the presence of both SALS-WH2 and tropomyosin (Fig. 5D) than those observed in the absence of tropomyosin (Fig. 5, A, left panel, and D, gray line). However, at a longer time scale SALS-WH2 completely disassembled tropomyosin-decorated actin filaments in a similar manner to the bare filaments, as shown by steady-state pyrenyl fluorescence measurements (Fig. 5E). These observations suggest that tropomyosin can decrease but cannot inhibit completely the depolymerizing activity of SALS-WH2.
Profilin Does Not Affect the Activities of the WH2 Domains of SALS
Upstream to the WH2 domains, SALS possesses a proline-rich motif (Fig. 1B). In some actin-regulatory proteins, such as formins, the proline-rich regions can interact with profilin, and this interaction can influence their effects on actin dynamics (25, 27, 40, 41). To study this issue, we investigated the biochemical activities of a construct containing both the proline-rich sequence element and the WH2 domains (SALS-Pro-WH2). We found that in the absence of profilin, SALS-Pro-WH2 influenced actin dynamics similar to SALS-WH2. SALS-Pro-WH2 bound to G-actin, inhibited actin assembly, and enhanced actin filament disassembly (Fig. 6, A–C). To address whether the proline-rich region of SALS can bind profilin, the steady-state anisotropy of Alexa568C-profilin (2 μm) was measured in the absence and presence of increasing concentrations of SALS-Pro-WH2. There was no detectable change in the value of steady-state anisotropy of Alexa568C-profilin (0.15 ± 0.01) up to 20 μm SALS-Pro-WH2. We also assayed the effect of profilin on the SALS-Pro-WH2-mediated actin assembly using pyrenyl-actin (Fig. 6D). As expected, profilin slowed down actin assembly due to the inhibition of nucleation and the reduction of the association rate constant of actin monomers to filaments (42, 43). We found that SALS-Pro-WH2 inhibited actin assembly to the same extent in both the absence and presence of profilin (Fig. 6D). These observations show that the proline-rich motif of SALS is unlikely to bind profilin and does not influence the investigated activities of the WH2 domains of SALS.
FIGURE 6.
Proline-rich region does not affect the sequestering and depolymerizing activities of the WH2 domains. A, steady-state anisotropy of Alexa488NHS-Mg2+-ATP-G-actin (1 μm) as a function of SALS-Pro-WH2 concentration. Dashed line shows the fit to the data, according to Equation 3. Dissociation equilibrium constant of 0.4 ± 0.2 μm was obtained. Error bars, standard deviation. B, polymerization kinetics of actin (2.5 μm, containing 5% pyrenyl-actin) in the absence and presence of different amounts of SALS-Pro-WH2, as indicated. C, kinetics of actin disassembly (0.02 μm, containing 50% of pyrenyl-actin) in the absence and presence of SALS-WH2 or SALS-Pro-WH2 (12 μm), as indicated. D, polymerization kinetics of actin (2.5 μm, containing 5% pyrenyl-actin) in the absence and presence of profilin (7.3 μm) and/or SALS-Pro-WH2 (6.1 μm), as indicated. a.u., arbitrary units.
Discussion
In this work, we investigated the biochemical activities of the WH2 domains and the proline-rich motif of D. melanogaster SALS required for sarcomeric actin assembly. Our analysis revealed that the isolated tandem WH2 domains of SALS, as bona fide WH2 domains, are multifunctional regulators of actin dynamics, and their activities are manifested through their interactions with monomeric and filamentous actin.
We found that SALS-WH2 binds actin monomers with sub-micromolar affinity (KD ∼0.4 μm) and sequesters them into non-polymerizable complexes (Figs. 2–4). The binding was accompanied by marked changes in the fluorescence emission of IAEDANS attached to Cys374 in subdomain 1 at the barbed face of G-actin (44), as reported previously to other WH2/Thymosinβ4 proteins(Fig. 2A) (12, 45). This implies that the binding of the WH2 domains of SALS affects the local milieu of the fluorophore, resulting in an altered potentially more solvent-exposed environment in the complex. This observation supports that SALS-WH2 adapts to the characteristic G-actin-binding mode of WH2 domains by inserting its N-terminal amphipathic α-helix into the hydrophobic cleft between subdomains 1 and 3 of G-actin (8). In contrast to IAEDANS, no changes were observed in the fluorescence emission of Alexa488NHS coupled to Lys328 in subdomain 3 of G-actin (Fig. 2B). This is again in agreement with the known structural features of the WH2·actin complexes, according to which this region is not included in the binding site. Importantly, we found that the SALS-WH2 domain/G-actin ratio in the complex depended on the fluorophore (Fig. 2D). Cys374 is located at a site that is involved in the WH2 domain/G-actin interaction, as well as in longitudinal intersubunit contacts between actin protomers in oligomers, whereas Lys328 is not. Considering this, IAEDANS can interfere with the tandem G-actin binding by WH2 domains resulting in lower stoichiometry than is characteristic to a label-free system. We propose that Alexa488NHS is a more accurate probe to derive the WH2 domain·G-actin stoichiometry in the complexes. The fact that the stoichiometry of SALS-WH2·G-actin depends on the location of the fluorophore, highlights the importance of the careful use of fluorescent probes in deriving binding ratios of proteins in their complexes with actin.
Comparative sequence analysis of WH2 domains from different proteins supports the conclusion that both WH2 domains of SALS bind to G-actin and predict a sequestering mechanism (Fig. 1A) (7). The N-terminal part of both WH2 domains of SALS possesses a conserved hydrophobic amino acid doublet/triplet composed of Met, Leu, Val, or Ile. These amino acids with hydrophobic side chains are important in stabilizing the interaction between the N terminus of the WH2 domain and the hydrophobic cleft of G-actin. In addition, the WH2 domains of SALS contain a lysine/isoleucine as the central amino acid of the FXXXK linker preceding the LKK(T/V) motif, as well as an arginine as the 5th residue after the consensus LKK(T/V) motif. In the WH2 domains of WIP (Wiskott-Aldrich syndrome protein-interacting protein) and Ciboulot, these amino acids were shown to form salt bridges with actin residues Glu-334 and Glu-93, respectively (7). These interactions can restrict the dynamics of the C-terminal half of the WH2 domain, which was proposed to endow the domain with sequestration activity. Despite possessing two WH2 domains, SALS-WH2 can only sequester actin monomers but cannot promote nucleation. However, the WH2 domains of SALS are similar to those found in Spire, Cordon Bleu, N-WASP or in VopL/F proteins, which sequester actin monomers, and their efficient nucleation promoting activity requires other modules surrounding the WH2 motifs (10, 12–15).
In the initial study on SALS, it was found that the full-length protein slows down the assembly of gelsolin-capped actin filaments, which was interpreted as the association of SALS to filament pointed ends (18). According to our results, SALS-WH2 inhibits spontaneous actin assembly, as well as pointed end growth from gelsolin actin seeds to the same extent (Fig. 3); therefore, it is more likely that the inhibitory activity of SALS-WH2 in both cases results from sequestration and not from pointed end binding.
Besides sequestration, another aspect of the multifunctional nature of SALS-WH2 is its depolymerizing activity. Our results from dilution-induced depolymerization experiments clearly show that SALS-WH2 can actively enhance filament depolymerization at a rate characteristic to severing activity (Fig. 5A). Notably, we found that SALS-WH2 binds to phalloidin- or jasplakinolide-stabilized actin filaments but fails to disassemble them (Fig. 5B). From this observation, we were able to directly observe filament binding by the WH2 domains (Fig. 5C). These drugs bind to F-actin in the inner groove of the filament and stabilize their structure (46, 47). The drug-binding sites do not overlap with the binding interface of the WH2 domains, which is located on the outer surface of F-actin (48–51). This would provide an explanation as to why SALS-WH2 can still bind to phalloidin-stabilized actin but cannot disassemble them (Fig. 5, B and C). Besides these structural features, the filament stabilization by the drugs also results in the arrest of filament end dynamics. This effect is expected to inhibit the sequestration activity of SALS-WH2. Considering the dual effect of the drugs, SALS-WH2 should be unable to induce the accumulation of unassembled actin at steady state, which is entirely consistent with our observations (Fig. 5B). However, our finding that the WH2 domains can still associate with actin filaments in the presence of phalloidin may be a helpful approach to study the structural features of the WH2 domain/F-actin interaction. In agreement with the previous characterization of WH2 domains from other proteins such as Cordon Bleu and Spire (10, 12), our results indicate that the binding and effective destabilization of F-actin requires at least two actin-binding motifs (Fig. 5C).
We found that tropomyosin, a canonical F-actin-binding element in sarcomeres, severely reduces the ability of SALS-WH2 to enhance the disassembly of actin filaments (Fig. 5D). However, at equilibrium the amount of unassembled actin was the same both in the presence and in absence of tropomyosin (Fig. 5E). Tropomyosin maintains filament end dynamics (52, 53), and hence it does not influence the accumulation of unassembled actin due to sequestration. Considering this, we propose that tropomyosin can inhibit only one of the two main effects of SALS-WH2 on actin dynamics. The side binding of tropomyosin interferes with the severing activity of SALS-WH2. However, actin monomers spontaneously dissociated from tropomyosin-bound filaments can still be captured into SALS-WH2·G-actin non-polymerizable complexes, which leads to the accumulation of unassembled actin.
Similarly to many other WH2 domain proteins (such as the activators of the Arp2/3 machinery, VASP, MIM, WIP, Cordon Bleu, and leiomodin) (2), SALS also possesses a region rich in proline upstream to the WH2 domains. In the case of certain Arp2/3 complex activators and VASP, this region collaborates with the WH2 domains by binding profilin to direct profilin·actin to filament ends (6). However, in most of the cases the functional significance of this motif was not addressed experimentally. Our results indicate that this region of SALS does not bind profilin and does not affect the activities of the WH2 domains of SALS either in the absence or presence of profilin (Fig. 6). This suggests that the proline-rich region may function as a flexible structural element in the protein, instead of targeting profilin·actin to the WH2 domains.
Previously, it was found that the purified full-length SALS bound both G- and F-actin but did not affect spontaneous actin assembly kinetics and did not alter the steady-state amount of filamentous actin. In addition, SALS was found to be important for the establishment of proper sarcomeric actin filament lengthening (18). This implies that the full-length SALS protein does not behave as an actin monomer sequestrator or filament depolymerizing factor. In contrast, we found that the combination of multiple activities of isolated WH2 domains of SALS results in the accumulation of monomeric actin. Thus, the activities of the WH2 domains do not reconstitute the effect of the full-length SALS protein on thin filament dynamics. Collectively, these observations imply that other regions in the protein or other sarcomeric elements must be important to modulate the biochemical activities and localization of the WH2 domains. These regions can be other actin-binding motifs like in the case of Lmod (20). Regarding this issue, former pulldown experiments with truncated SALS proteins suggested that the WH2 domains of SALS are sufficient for G-actin binding (18). However, the full-length protein was not mapped for additional G- or F-actin-binding sites. Furthermore, we cannot exclude that other sarcomeric components can also contribute to the biological activity/localization of the full-length protein.
In conclusion, this study established that the WH2 domains of SALS are multifunctional regulators of actin dynamics, possessing two main biochemical activities previously reported for other members of the WH2 domain protein family. The data highlight the importance of the detailed analysis of the activities of other regions of SALS and underline the necessity of the characterization of its interactions with different sarcomeric elements in future investigations. Further studies will help elucidate the molecular mechanisms underlying the function of SALS. It will contribute to a better understanding of the tailoring of the biochemical activities in WH2 domains by other cellular components in the biological context.
Author Contributions
M. A. T., A. T. V., A. K. M., E. M., and B. B. performed the experiments and analyzed the data; J. M. and B. B. designed the study, analyzed the data, interpreted the results, and wrote the paper; M. N. revised the paper critically for important intellectual content. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Tamás Huber for the purification of gelsolin. We thank Éva Hunyadi-Gulyás for the mass spectrometry measurements, which were done in the Proteomics Research Group of the Biological Research Centre (Hungarian Academy of Sciences, Szeged, Hungary).
This work was supported by Hungarian Science Foundation (OTKA) Grants K109689 (to B. B.), K109330 (to J. M.), and NN 107776 (to M. N.), National Innovation Office “Baross Gábor” Program Grant REG-DD-09-1-2009-0009 Tirfm 09 (to B. B.), and by the European Union and the State of Hungary and co-financed by the European Social Fund in the framework of TÁMOP 4.2.4.A/2-11-1-2012-0001 “National Excellence Program.” The authors declare that they have no conflicts of interest with the contents of this article.
- Tmod
- tropomodulin
- WH2
- Wiskott-Aldrich syndrome protein homology 2
- Lmod
- leiomodin
- SALS
- sarcomere length short
- IAEDANS
- 1,5-IAEDANS, 5-((((2-iodoacetyl)amino)ethyl)amino)naphthalene-1-sulfonic acid
- Alexa488NHS
- Alexa Fluor® 488 carboxylic acid succinimidyl ester
- TIRFM
- total internal reflection fluorescence microscopy
- WIP
- Wiskott-Aldrich syndrome protein-interacting protein.
References
- 1.Renault L., Deville C., and van Heijenoort C. (2013) Structural features and interfacial properties of WH2, β-thymosin domains, and other intrinsically disordered domains in the regulation of actin cytoskeleton dynamics. Cytoskeleton 70, 686–705 [DOI] [PubMed] [Google Scholar]
- 2.Carlier M. F., Husson C., Renault L., and Didry D. (2011) Control of actin assembly by the WH2 domains and their multifunctional tandem repeats in Spire and Cordon-Bleu. Int. Rev. Cell Mol. Biol. 290, 55–85 [DOI] [PubMed] [Google Scholar]
- 3.Paunola E., Mattila P. K., and Lappalainen P. (2002) WH2 domain: a small, versatile adapter for actin monomers. FEBS Lett. 513, 92–97 [DOI] [PubMed] [Google Scholar]
- 4.Dominguez R. (2007) The β-thymosin/WH2 fold: multifunctionality and structure. Ann. N.Y. Acad. Sci. 1112, 86–94 [DOI] [PubMed] [Google Scholar]
- 5.Husson C., Cantrelle F. X., Roblin P., Didry D., Le K. H., Perez J., Guittet E., Van Heijenoort C., Renault L., and Carlier M. F. (2010) Multifunctionality of the β-thymosin/WH2 module: G-actin sequestration, actin filament growth, nucleation, and severing. Ann. N.Y. Acad. Sci. 1194, 44–52 [DOI] [PubMed] [Google Scholar]
- 6.Chereau D., Kerff F., Graceffa P., Grabarek Z., Langsetmo K., and Dominguez R. (2005) Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc. Natl. Acad. Sci. U.S.A. 102, 16644–16649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Didry D., Cantrelle F. X., Husson C., Roblin P., Moorthy A. M., Perez J., Le Clainche C., Hertzog M., Guittet E., Carlier M. F., van Heijenoort C., and Renault L. (2012) How a single residue in individual β-thymosin/WH2 domains controls their functions in actin assembly. EMBO J. 31, 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hertzog M., van Heijenoort C., Didry D., Gaudier M., Coutant J., Gigant B., Didelot G., Préat T., Knossow M., Guittet E., and Carlier M. F. (2004) The β-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell 117, 611–623 [DOI] [PubMed] [Google Scholar]
- 9.Boquet I., Boujemaa R., Carlier M. F., and Préat T. (2000) Ciboulot regulates actin assembly during Drosophila brain metamorphosis. Cell 102, 797–808 [DOI] [PubMed] [Google Scholar]
- 10.Husson C., Renault L., Didry D., Pantaloni D., and Carlier M. F. (2011) Cordon-Bleu uses WH2 domains as multifunctional dynamizers of actin filament assembly. Mol. Cell 43, 464–477 [DOI] [PubMed] [Google Scholar]
- 11.Hertzog M., Yarmola E. G., Didry D., Bubb M. R., and Carlier M. F. (2002) Control of actin dynamics by proteins made of β-thymosin repeats: the actobindin family. J. Biol. Chem. 277, 14786–14792 [DOI] [PubMed] [Google Scholar]
- 12.Bosch M., Le K. H., Bugyi B., Correia J. J., Renault L., and Carlier M. F. (2007) Analysis of the function of Spire in actin assembly and its synergy with formin and profilin. Mol. Cell 28, 555–568 [DOI] [PubMed] [Google Scholar]
- 13.Namgoong S., Boczkowska M., Glista M. J., Winkelman J. D., Rebowski G., Kovar D. R., and Dominguez R. (2011) Mechanism of actin filament nucleation by Vibrio VopL and implications for tandem W domain nucleation. Nat. Struct. Mol. Biol. 18, 1060–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pernier J., Orban J., Avvaru B. S., Jégou A., Romet-Lemonne G., Guichard B., and Carlier M. F. (2013) Dimeric WH2 domains in Vibrio VopF promote actin filament barbed-end uncapping and assisted elongation. Nat. Struct. Mol. Biol. 20, 1069–1076 [DOI] [PubMed] [Google Scholar]
- 15.Yu B., Cheng H. C., Brautigam C. A., Tomchick D. R., and Rosen M. K. (2011) Mechanism of actin filament nucleation by the bacterial effector VopL. Nat. Struct. Mol. Biol. 18, 1068–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuchero J. B., Coutts A. S., Quinlan M. E., Thangue N. B., and Mullins R. D. (2009) p53-cofactor JMY is a multifunctional actin nucleation factor. Nat. Cell Biol. 11, 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Renault L., Bugyi B., and Carlier M. F. (2008) Spire and Cordon-bleu: multifunctional regulators of actin dynamics. Trends Cell Biol. 18, 494–504 [DOI] [PubMed] [Google Scholar]
- 18.Bai J., Hartwig J. H., and Perrimon N. (2007) SALS, a WH2-domain-containing protein, promotes sarcomeric actin filament elongation from pointed ends during Drosophila muscle growth. Dev. Cell 13, 828–842 [DOI] [PubMed] [Google Scholar]
- 19.Yuen M., Sandaradura S. A., Dowling J. J., Kostyukova A. S., Moroz N., Quinlan K. G., Lehtokari V. L., Ravenscroft G., Todd E. J., Ceyhan-Birsoy O., Gokhin D. S., Maluenda J., Lek M., Nolent F., Pappas C. T., et al. (2014) Leiomodin-3 dysfunction results in thin filament disorganization and nemaline myopathy. J. Clin. Invest. 124, 4693–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chereau D., Boczkowska M., Skwarek-Maruszewska A., Fujiwara I., Hayes D. B., Rebowski G., Lappalainen P., Pollard T. D., and Dominguez R. (2008) Leiomodin is an actin filament nucleator in muscle cells. Science 320, 239–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nworu C. U., Kraft R., Schnurr D. C., Gregorio C. C., and Krieg P. A. (2015) Leiomodin 3 and tropomodulin 4 have overlapping functions during skeletal myofibrillogenesis. J. Cell Sci. 128, 239–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olson B. J., and Markwell J. (2001) Assays for determination of protein concentration. Curr. Protoc. Protein Sci. 2007, Chapter 3, Unit 3.4 [DOI] [PubMed] [Google Scholar]
- 23.Spudich J. A., and Watt S. (1971) The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246, 4866–4871 [PubMed] [Google Scholar]
- 24.Nag S., Ma Q., Wang H., Chumnarnsilpa S., Lee W. L., Larsson M., Kannan B., Hernandez-Valladares M., Burtnick L. D., and Robinson R. C. (2009) Ca2+ binding by domain 2 plays a critical role in the activation and stabilization of gelsolin. Proc. Natl. Acad. Sci. U.S.A. 106, 13713–13718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perelroizen I., Marchand J. B., Blanchoin L., Didry D., and Carlier M. F. (1994) Interaction of profilin with G-actin and poly(l-proline). Biochemistry 33, 8472–8478 [DOI] [PubMed] [Google Scholar]
- 26.Smillie L. B. (1982) Preparation and identification of α- and β-tropomyosins. Methods Enzymol. 85, 234–241 [DOI] [PubMed] [Google Scholar]
- 27.Barkó S., Bugyi B., Carlier M. F., Gombos R., Matusek T., Mihály J., and Nyitrai M. (2010) Characterization of the biochemical properties and biological function of the formin homology domains of Drosophila DAAM. J. Biol. Chem. 285, 13154–13169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bugyi B., Papp G., Hild G., Lõrinczy D., Nevalainen E. M., Lappalainen P., Somogyi B., and Nyitrai M. (2006) Formins regulate actin filament flexibility through long range allosteric interactions. J. Biol. Chem. 281, 10727–10736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bugyi B., Didry D., and Carlier M. F. (2010) How tropomyosin regulates lamellipodial actin-based motility: a combined biochemical and reconstituted motility approach. EMBO J. 29, 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hertzog M., and Carlier M. F. (2005) Functional characterization of proteins regulating actin assembly. Curr. Protoc. Cell Biol. Chapter 13, Unit 13.16 [DOI] [PubMed] [Google Scholar]
- 31.Pollard T. D. (2010) A guide to simple and informative binding assays. Mol. Biol. Cell 21, 4061–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanson J., and Lowy J. (1963) The structure of F-actin and of actin filaments isolated from muscle. J. Mol. Biol. 6, 46–60 [Google Scholar]
- 33.Coué M., Brenner S. L., Spector I., and Korn E. D. (1987) Inhibition of actin polymerization by latrunculin A. FEBS Lett. 213, 316–318 [DOI] [PubMed] [Google Scholar]
- 34.Nyitrai M., Szent-Györgyi A. G., and Geeves M. A. (2003) Interactions of the two heads of scallop (Argopecten irradians) heavy meromyosin with actin: influence of calcium and nucleotides. Biochem. J. 370, 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pollard T. D., and De La Cruz E. M. (2013) Take advantage of time in your experiments: a guide to simple, informative kinetics assays. Mol. Biol. Cell 24, 1103–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pollard T. D. (1986) Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J. Cell Biol. 103, 2747–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pollard T. D. (2007) Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 36, 451–477 [DOI] [PubMed] [Google Scholar]
- 38.Bugyi B., and Carlier M. F. (2010) Control of actin filament treadmilling in cell motility. Annu. Rev. Biophys. 39, 449–470 [DOI] [PubMed] [Google Scholar]
- 39.Ghoshdastider U., Popp D., Burtnick L. D., and Robinson R. C. (2013) The expanding superfamily of gelsolin homology domain proteins. Cytoskeleton 70, 775–795 [DOI] [PubMed] [Google Scholar]
- 40.Romero S., Le Clainche C., Didry D., Egile C., Pantaloni D., and Carlier M. F. (2004) Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell 119, 419–429 [DOI] [PubMed] [Google Scholar]
- 41.Dominguez R. (2009) Actin filament nucleation and elongation factors–structure-function relationships. Crit. Rev. Biochem. Mol. Biol. 44, 351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Didry D., Carlier M. F., and Pantaloni D. (1998) Synergy between actin depolymerizing factor/cofilin and profilin in increasing actin filament turnover. J. Biol. Chem. 273, 25602–25611 [DOI] [PubMed] [Google Scholar]
- 43.Gutsche-Perelroizen I., Lepault J., Ott A., and Carlier M. F. (1999) Filament assembly from profilin-actin. J. Biol. Chem. 274, 6234–6243 [DOI] [PubMed] [Google Scholar]
- 44.Miki M., dos Remedios C. G., and Barden J. A. (1987) Spatial relationship between the nucleotide-binding site, Lys-61 and Cys-374 in actin and a conformational change induced by myosin subfragment-1 binding. Eur. J. Biochem. 168, 339–345 [DOI] [PubMed] [Google Scholar]
- 45.De La Cruz E. M., Ostap E. M., Brundage R. A., Reddy K. S., Sweeney H. L., and Safer D. (2000) Thymosin-β(4) changes the conformation and dynamics of actin monomers. Biophys. J. 78, 2516–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kudryashov D. S., Phillips M., and Reisler E. (2004) Formation and destabilization of actin filaments with tetramethylrhodamine-modified actin. Biophys. J. 87, 1136–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oda T., Namba K., and Maéda Y. (2005) Position and orientation of phalloidin in F-actin determined by x-ray fiber diffraction analysis. Biophys. J. 88, 2727–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ducka A. M., Joel P., Popowicz G. M., Trybus K. M., Schleicher M., Noegel A. A., Huber R., Holak T. A., and Sitar T. (2010) Structures of actin-bound Wiskott-Aldrich syndrome protein homology 2 (WH2) domains of Spire and the implication for filament nucleation. Proc. Natl. Acad. Sci. U.S.A. 107, 11757–11762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zahm J. A., Padrick S. B., Chen Z., Pak C. W., Yunus A. A., Henry L., Tomchick D. R., Chen Z., and Rosen M. K. (2013) The bacterial effector VopL organizes actin into filament-like structures. Cell 155, 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rebowski G., Boczkowska M., Hayes D. B., Guo L., Irving T. C., and Dominguez R. (2008) X-ray scattering study of actin polymerization nuclei assembled by tandem W domains. Proc. Natl. Acad. Sci. U.S.A. 105, 10785–10790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Avvaru B. S., Pernier J., and Carlier M. F. (2015) Dimeric WH2 repeats of VopF sequester actin monomers into non-nucleating linear string conformations: an x-ray scattering study. J. Struct. Biol. 190, 192–199 [DOI] [PubMed] [Google Scholar]
- 52.Broschat K. O. (1990) Tropomyosin prevents depolymerization of actin filaments from the pointed end. J. Biol. Chem. 265, 21323–21329 [PubMed] [Google Scholar]
- 53.Kis-Bicskei N., Vig A., Nyitrai M., Bugyi B., and Talián G. C. (2013) Purification of tropomyosin Br-3 and 5NM1 and characterization of their interactions with actin. Cytoskeleton 70, 755–765 [DOI] [PubMed] [Google Scholar]