Abstract
In plasma membranes, flippases translocate aminophospholipids such as phosphatidylserine and phosphatidylethanolamine from the extracellular to the cytoplasmic leaflet. Mammalian ATP11C, a type IV P-type ATPase, acts as a flippase at the plasma membrane. Here, by expressing 12 human type IV P-type ATPases in ATP11C-deficient cells, we determined that ATP8A2 and ATP11A can also act as plasma membrane flippases. As with ATP11C, ATP8A2 and ATP11A localized to the plasma membrane in a CDC50A-dependent manner. ATP11A was cleaved by caspases during apoptosis, and a caspase-resistant ATP11A blocked apoptotic PtdSer exposure. In contrast, ATP8A2 was not cleaved by caspase, and cells expressing ATP8A2 did not expose PtdSer during apoptosis. Similarly, high Ca2+ concentrations inhibited the ATP11A and ATP11C PtdSer flippase activity, but ATP8A2 flippase activity was relatively resistant to Ca2+. ATP11A and ATP11C were ubiquitously expressed in human and mouse adult tissues. In contrast, ATP8A2 was expressed in specific tissues, such as the brain and testis. Thus, ATP8A2 may play a specific role in translocating PtdSer in these tissues.
Keywords: apoptosis, calcium, caspase, phosphatidylserine, plasma membrane, P4-ATPase, flippase
Introduction
The plasma membrane of eukaryotic cells is composed of an inner and an outer leaflet. Phospholipids, a major component of plasma membranes, are asymmetrically distributed between the leaflets (1); aminophospholipids such as phosphatidylserine (PtdSer)2 and phosphatidylethanolamine (PtdEtn) are confined to the inner leaflet, whereas the outer leaflet is rich in phosphatidylcholine (PtdCho) and sphingomyelin (SM) (1, 2). This asymmetrical distribution of phospholipids is disrupted in various biological processes. Cells undergoing apoptosis expose PtdSer on the cell surface as an “eat me” signal that is recognized by phagocytes, which respond by engulfing the apoptotic cell (3, 4). Activated platelets expose PtdSer as a scaffold for blood-clotting factors (5). Three types of membrane proteins regulate the distribution of phospholipids in the plasma membrane: aminophospholipid translocases (flippases) specifically transport PtdSer and PtdEtn from the outer to the inner leaflet in an ATP-dependent manner (1, 2); floppases transport phospholipids (PtdCho in particular) from the inner to the outer leaflet in an ATP-dependent manner, and phospholipid scramblases nonspecifically scramble phospholipids between the inner and outer leaflets in an energy-independent manner (1).
Scramblase systems can be Ca2+-activated or caspase-activated (6, 7). In the transmembrane protein (TMEM)16 family, which has 10 transmembrane regions (8), five of the 10 human family members (TMEM16C, D, F, G, and J) elicit Ca2+-activated phospholipid scrambling (7, 9). Caspase-activated scrambling is executed by members of the Xkr (Xk-related protein) family. The human Xkr family consists of nine membrane proteins having six transmembrane segments; three of these proteins (Xkr4, Xkr8, and Xkr9) scramble phospholipids in apoptotic cells (6, 10). These Xkr members carry a caspase recognition site in the C-terminal tail, and this site must be cleaved by caspase to support phospholipid scrambling (6, 10).
Several Type IV P-type ATPases (P4-ATPases) have been shown to be phospholipid flippases (11–14), but it remains unclear which human P4-ATPases function as flippases at the plasma membrane. Recently, we identified ATP11C, a member of the P4-ATPase family, as a flippase at the plasma membrane (15). Like other P4-ATPases (16–18), ATP11C requires chaperoning by CDC50A to localize to the plasma membrane. CDC50A-deficient cells almost completely lose the ability to translocate PtdSer from the outer to the inner leaflet and constitutively expose PtdSer. Although ATP11C-deficient cells lose most of their flippase activity, they retain sufficient flippase ability to maintain an asymmetrical phospholipid distribution and do not expose PtdSer (15). These results suggested that other P4-ATPases might have compensated for the lack of ATP11C in these cells. In fact, ATP8A2, ATP8B1, and ATP11A were proposed to have PtdSer flippase activity at plasma membranes (11, 19, 20). However, how they are regulated in various biological processes and whether other members of the P4-ATPase family have the plasma membrane flippase activity is not clear.
The human P4-ATPase family has 14 members (16, 17). In this study, we expressed 12 human P4-ATPases in an ATP11C-deficient cell line and found that only ATP8A2, ATP11A, and ATP11C had a strong ability to flip PtdSer at the plasma membrane. These proteins localized to the plasma membrane in the presence of CDC50A. Similar to ATP11C, ATP11A carried two caspase recognition sites in its cytoplasmic region that were cleaved during apoptosis. In contrast, ATP8A2 was not cleaved or inactivated during apoptosis, and cells expressing ATP8A2 did not expose PtdSer during apoptosis. Although high Ca2+ concentrations inhibited ATP11A and ATP11C ATPase activity, ATP8A2 was relatively resistant to Ca2+-mediated inhibition. ATP11A and ATP11C were ubiquitously expressed in various human and mouse tissues, whereas ATP8A2 was specifically expressed in the brain and testis. These results suggest that in most mammalian cells, the major flippases at the plasma membrane are ATP11A and ATP11C. ATP8A2 may have a specific role in flipping phospholipids in the brain and testis.
Experimental Procedures
Mice, Cell Lines, Plasmids, Recombinant Proteins, Antibodies, and Reagents
C57BL/6J mice were purchased from Japan Clea. All mouse studies were approved by the Animal Care and Use Committee of the Research Institute for Microbial Diseases and the Immunology Frontier Research Center of Osaka University. W3 cells were transformed from mouse T-cell lymphoma (WR19L) to express mouse Fas (21). W3-Ildm cells, derived from WR19L cells, expressed Fas and a caspase-resistant form of the inhibitor of caspase-activated DNase (22). W3 and W3-derived cells were cultured in RPMI 1640 containing 10% FCS. ATP11CED22 is an ATP11C-null W3 cell line; CDC50AED29 is a CDC50A-null W3-Ildm cell line (15). HEK293T cells were cultured in DMEM containing 10% FCS. The pMXs-puro retrovirus vector and pGag-pol-IRES-bsr packaging plasmid (23) were provided by Dr. T. Kitamura (Institute of Medical Science, University of Tokyo, Tokyo, Japan). The pCMV-VSV-G plasmid was provided by Dr. H. Miyoshi (Riken Bioresource Center, Riken). We purchased pAdVAntage from Thermo Fisher.
Leucine-zippered human Fas ligand (FasL) was produced in COS7 as previously described (24). Recombinant caspase 3 was produced in Escherichia coli and purified as previously described (25). Rabbit mAb against activated caspase 3 (clone 5A1E), which detects the caspase 3 cleaved at amino acid position Asp175, was purchased from Cell Signaling Technology. Mouse anti-GFP mAb (Clone JL8) was obtained from Takara Bio. HRP-conjugated goat anti-mouse Igs and goat anti-rabbit Igs were purchased from Dako Agilent Technologies. Cy5-labeled annexin V was purchased from Biovision. DRAQ5 (a membrane-permeable DNA-staining dye) and Sytox Blue (a membrane-impermeable DNA-staining dye) were purchased from BioStatus and Thermo Fisher, respectively. We obtained A23187 from Sigma-Aldrich, and LysoTracker from Thermo Fisher. We purchased 1-oleoyl-2-{6-[(7-nitro-2–1,3-benzoxadiazol-4-yl) amino]hexanoyl}-sn-glycero-3-phosphoserine (NBD-PS), 1-oleoyl-2-{6-[(7-nitro-2–1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphoethanolamine (NBD-PE), 1-oleoyl-2-{6-[(7-nitro-2–1,3-benzoxadiazol-4-yl) amino]hexanoyl}-sn-glycero-3-phosphocholine (NBD-PC), N-[6-[(7-nitro-2–1,3-benzoxadiazol-4-yl)amino]hexanoyl]-sphingosine-1-phosphocholine (NBD-SM), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (POPS), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), and N-stearoyl-d-erythro-sphingosylphosphorylcholine (SM) from Avanti Polar Lipids. Polyethylenimine was obtained from Polysciences.
Expression Plasmids
Human cDNA for CDC50A (NCBI: NM_018247.3), ATP8A1 (NM_001105529.1), ATP8B2 (XM_005245356.2), ATP9A (NM_006045.1), and ATP11A (NM_015205.2, carrying SNP rs11616795 and rs368865) was prepared by RT-PCR with RNA from KBM7 cells. Because cDNA carrying the native sequences of ATP8A2 (NM_016529.4), ATP8B1 (NM_005603.4), ATP8B3 (NM_138813.3), ATP8B4 (NM_024837.3), ATP9B (NM_001306085), ATP10A (NM_024490.3), ATP10B (NM_025153.2), ATP10D (NM_020453.3), ATP11B (NM_014616.2), and ATP11C (XM_005262405.1) produced low protein levels in mouse WR19L cells, a sequence to enhance mRNA stability and translational efficiency was custom-designed and synthesized at GENEART.
Expression plasmids for ATP11A mutants were prepared by recombinant PCR using ATP11A cDNA as a template with the following primers, which carried mutated nucleotides (the sequence for the restriction enzyme site is underlined): ATP11A, 5′-GGCTTAATTAAGGAGGAGCCATGGACTGCAG-3′ and 5′-GGCGAATTCGAAACTCAGGCTGCTGGAAGTC-3′; Mut1, 5′-CGCTGGGGGACGAGGCAATCATGGCGATTCCTGA C-3′ and 5′-GTCAGGAATCGCCATGATTGCCTCGTCCCCCAGCG-3′; and Mut2, 5′-GATTTCCTGGGGCCGGCTACGCTGGCATCGTCTTTC-3′ and 5′-GAAAGACGATGCCAGCGTAGCCGGCCCCAGGAAATC-3′. The cDNAs were verified by sequencing, Flag- or GFP-tagged at the C terminus, and inserted into the pMXs-puro vector. To produce P4-ATPase, cDNAs of Flag-tagged ATP8A2, 11A, 11C, or 11A mutants and of GFP-tagged CDC50A were introduced into pEF-BOS vector.
Transformation of Mouse Cell Lines
W3 cells and their derivatives were transformed using a retroviral vector carrying VSV-G envelope genes. The cDNAs were inserted into the pMXs-puro vector and introduced into HEK293T cells using FuGENE 6 (Promega) together with pGag-pol-IRES-bsr, pCMV-VSV-G, and pAdVAntage. The resultant retrovirus was concentrated by centrifugation at 6,000 × g for 16 h at 4 °C and used to infect cells; the infected cells were cultured in the presence of 1 μg/ml puromycin. If necessary, GFP-positive cells were sorted using a FACSAria II (BD Biosciences). To examine the subcellular localization of human P4-ATPases, CDC50AED29 cells or CDC50AED29 transformants expressing hCDC50A were infected by a retrovirus carrying GFP-tagged P4-ATPase, and the GFP-positive transformants were sorted using a FACSAria II. The sorted cells (2 × 105) were suspended in phenol red-free DMEM containing 10% FCS and 12.5 μm DRAQ5, plated on a glass-bottomed dish (Iwaki, AGC Techno Glass), incubated for 7 min at 37 °C, and observed by a confocal microscope (FV1000-D; Olympus).
FasL-induced Apoptosis, and Detection of Phosphatidylserine
Cells were treated with 33 units/ml FasL at 37 °C to induce apoptosis. To detect exposed PtdSer, the cells were incubated at 25 °C for 5 min or 4 °C for 15 min with Cy5-annexin V diluted 1:2,000 in 10 mm HEPES-KOH buffer (pH 7.4) containing 140 mm NaCl and 2.5 mm CaCl2. The cells were then incubated with 200 nm Sytox Blue and analyzed using a FACSAria II or FACSCanto II.
Purification of P4-ATPase
To produce recombinant P4-ATPase, pEF-BOS vectors bearing Flag-tagged P4-ATPase (ATP8A2, 11A, 11C, or 11A mutants) and GFP-tagged CDC50A were introduced into HEK293T cells using a previously described polyethylenimine-mediated transfection method (26). After 42 h, cells were scraped, washed with chilled 50 mm Tris-HCl buffer (pH 7.4) containing 150 mm NaCl, frozen in liquid nitrogen, and stored at −80 °C. The cells were thawed on ice and lysed by rotating for 3–4 h at 4 °C in lysis buffer (40 mm MES/Tris buffer, pH 7.0, containing 20 mm magnesium acetate, 150 mm NaCl, 1 mm ATP, 0.5% 2,2-didecylpropane-1,3-bis-β-d-maltopyranoside (LMNG; Anatrace), and a mixture of protease inhibitors (cOmplete, Mini, EDTA-free; Roche Diagnostics)). Insoluble materials were removed by centrifugation at 12,000 × g for 20 min, and the supernatant was mixed with anti-FLAG M2 Magnetic Beads (Sigma-Aldrich) and incubated for 12 h at 4 °C with rotation. The beads were then washed with chilled washing buffer (40 mm MES/Tris buffer, pH 7.0, 5% glycerol, 150 mm NaCl, and 0.05% LMNG), and the protein was eluted with washing buffer containing 160 μg/ml 3× FLAG Peptide (Sigma-Aldrich). The purified proteins were separated by SDS-PAGE on a 7.5% gel (Bio Craft), visualized by staining with Coomassie Brilliant Blue, and analyzed with an ImageQuant LAS4000 Biomolecular Imager (GE Healthcare). The protein concentration was determined with ImageQuant TL software (GE Healthcare) using bovine serum albumin as a standard.
To treat with caspase, the affinity-purified ATP11A or its mutants (100 ng) were incubated at 20 °C for 1 h with 200 ng of caspase 3 in 10 μl of 40 mm MES/Tris buffer (pH 7.0) containing 5% glycerol, 7 mm DTT, 150 mm NaCl, 0.05% LMNG. After caspase 3-treatment, aliquots were used for the ATPase assay or separated with SDS-PAGE on a 7.5% gel, visualized by SYPRO® Ruby protein gel stain (Thermo Fisher) and analyzed with an ImageQuant LAS4000 Biomolecular Imager (GE Healthcare).
ATPase Assay
The ATPase activity of P4-ATPase was determined as described (27–31) with slight modifications. In brief, the purified protein (10 ng) was preincubated for 10 min at room temperature in 40 μl of the reaction mixture (40 mm MES/Tris buffer (pH 7.0), 150 mm NaCl, 5% glycerol, 5 mm MgCl2, 7 mm DTT, 600 μm ATP, and 0.05% LMNG) and incubated for 20 min at 37 °C. After adding 70 μl of stop solution (1.7 n H2SO4 containing 0.5% (w/v) ammonium hepta molybdate tetrahydrate (Fluka)), the mixture was incubated at room temperature for 20 min, 20 μl of 0.035% (w/v) malachite green oxalate (Merck Japan) and 0.35% (w/v) polyvinylalcohol (Sigma-Aldrich) was added, and the mixture was further incubated for 5 min. The malachite green-molybdate phosphate complex was detected at 610 nm using a microplate reader (Infinite M200; TECAN).
Western Blotting
Cells were lysed in radioimmune precipitation assay buffer (50 mm HEPES-NaOH buffer, pH 8.0, 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate, 150 mm NaCl, and a mixture of protease inhibitors). Insoluble materials were removed by centrifugation, and the lysates were mixed 2:1 with 3× SDS sample buffer (200 mm Tris-HCl buffer, pH 6.8, 6% SDS, 25% glycerol, 15% β-mercaptoethanol, and 0.05% bromophenol blue) and incubated at room temperature for 20 min. Proteins were separated by SDS-PAGE on a 7.5% or 10–20% gradient gel (Bio Craft) and transferred to a PVDF membrane (Millipore). The membranes were probed with HRP-conjugated mouse anti-Flag M2 mAb, mouse anti-GFP mAb, or rabbit anti-active caspase 3 mAb and incubated with HRP-conjugated goat anti-mouse Igs or goat anti-rabbit Igs. Peroxidase activity was detected by a Western Lightning-ECL system (PerkinElmer).
NBD Lipid Internalization Assay
NBD lipid internalization was assayed essentially as described previously (15, 32). In brief, 7 × 105 cells were incubated with 1 μm NBD-PS, NBD-PE, NBD-PC, or NBD-SM at 20 °C in 200 μl of Hanks' balanced salt solution (HBSS) containing 1 mm MgCl2 and 2 mm CaCl2. The cells were collected by centrifugation, resuspended in HBSS containing 5 mg/ml fatty acid-free BSA to extract nonincorporated lipids, and analyzed by a FACSAria II or FACSCanto II. In some cases, 2 × 105 cells in 200 μl of HBSS containing 1 mm CaCl2 and1 μm NBD-PS were stimulated at 20 °C for 4 min with A23187 Ca2+ ionophore and analyzed as above. To determine the apoptotic scramblase activity, 5 × 105 cells were treated with FasL, incubated at 25 °C for 3 min with 1 μm NBD-PC, suspended in HBSS containing 5 mg/ml BSA, and analyzed by a FACSCanto II.
Real Time RT-PCR
Total RNA prepared from various mouse tissues using an Isogen RNA isolation kit (Nippon Gene) and the RNeasy mini kit (Qiagen) was reverse-transcribed with a high capacity RNA to cDNA kit (Thermo Fisher). Human cDNA panels for various tissues (human MTC panel I and II) were purchased from Takara Bio and used as templates for real time RT-PCR. The cDNA was amplified using LightCycler 480 SYBR Green I Master (Roche Diagnostics). Primers for real time RT-PCR were as previously described (15).
Results
Plasma Membrane Flippase Activity of P4-ATPases
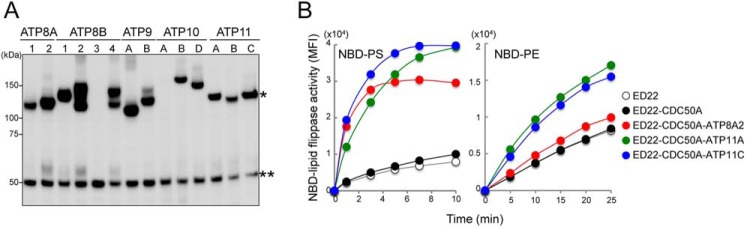
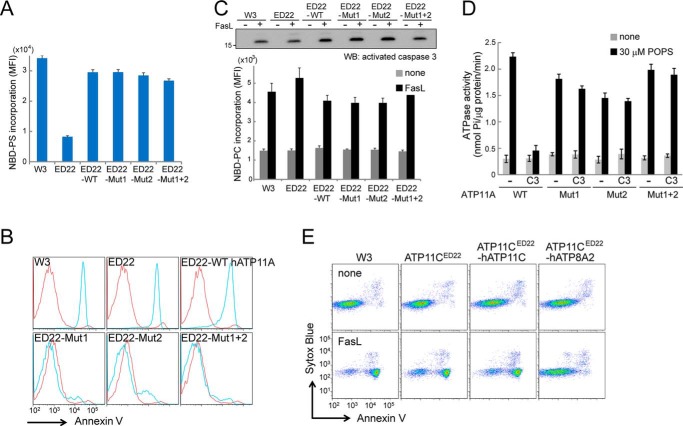
The phospholipid flippase activity is severely reduced in ATP11C-null W3 cells (ATP11CED22) (15). To examine the flippase activity of other human P4-ATPases, each P4-ATPase was Flag-tagged and introduced, along with Flag-tagged human CDC50A, into ATP11CED22 cells using retrovirus-based expression vectors. Stable transformants were analyzed by Western blotting with an anti-Flag mAb. Because the native cDNA sequences of ATP8A2, ATP8B1, ATP8B3, ATP8B4, ATP9B, ATP10A, ATP10B, ATP10D, ATP11B, and ATP11C produced only low levels of these proteins in ATP11CED22 cells, their nucleotide sequences were edited to optimize translational efficiency by considering mRNA stability, codon usage, and secondary structures. As a result, the transformants expressed comparable protein levels for all of the family members except ATP8B3 and ATP10A (Fig. 1A). Because ATP8B3 is exclusively expressed in the testis (33, 34), it might require the testis-specific chaperone CDC50C for stable expression. However, ATP10A is expressed in various mouse tissues, so it is not clear why its expression was low in ATP11CED22 cells.
FIGURE 1.
Phospholipid flippase activity of human P4-ATPases. A, expression of 14 human P4-ATPases. Each was Flag-tagged and introduced, along with Flag-tagged human CDC50A, into mouse ATP11C-deficient ATP11CED22 cells. Stable transformants were analyzed by Western blotting with an anti-Flag mAb. *, bands representing P4-ATPase. **, bands of ∼50 kDa representing CDC50A. B, the flippase activity of human P4-ATPases. ATP11CED22 cells and their transformants expressing the indicated P4-ATPase and/or CDC50A were incubated at 20 °C with 1 μm NBD-PS or NBD-PE for the indicated times and treated with fatty acid-free BSA. The incorporated fluorescence was measured by flow cytometry. MFI, mean fluorescent intensity.
We examined the flippase activity of 12 ATPases (ATP8A1, 8A2, 8B1, 8B2, 8B4, 9A, 9B, 10B, 10D, 11A, 11B, and 11C) by incubating the ATP11CED22 transformants with a fluorescence-conjugated phospholipid (NBD-PS, -PE, -PC, or -SM). As shown in Fig. 1B, the ATP11C-deficient ATP11CED22 cells slowly incorporated NBD-PS; its incorporation was strongly enhanced in cells transformed with ATP8A2, ATP11A, or ATP11C. In these cells, the incorporation reached its maximum within 6 min. In Table 1, the rate of flippase activity (measured by the incorporated NBD-PS at 2 min) was compared among transformants expressing the various ATPases. Considering the protein expression levels (Fig. 1A), ATP8A2, 11A, and 11C appeared to have similar PtdSer flipping activities. ATP8A1 weakly enhanced the NBD-PS incorporation, but little or no enhancement was observed with the other P4-ATPases. ATP11CED22 cells significantly incorporated NBD-PE, and the rate of incorporation was enhanced 2.4–2.7-fold by the cell transformation with ATP11A or ATP11C but not with other P4-ATPases (Fig. 1B and Table 1). On the other hand, none of the P4-ATPases examined, including ATP8A2, ATP11A, and ATP11C, enhanced the incorporation of NBD-PC or NBD-SM (Table 1), confirming that they were aminophospholipid-specific.
TABLE 1.
Flippase activity of human P4-ATPases
ATP11CED22 cells and transformants expressing CDC50A alone or CDC50A with the indicated P4-ATPases were incubated at 20 °C with 1 μm NBD-PS, NBD-PE, NBD-PC, or NBD-SM for 2, 10, 15, and 15 min, respectively. The incorporated NBD-phospholipids were quantified by flow cytometry, and the rate compared with that of the NBD-phospholipid incorporation into ATP11CED22 cells was determined. The values shown are the averages with S.E. (n = 3).
| P4-ATPase | CDC50A | NBD-PS | NBD-PE | NBD-PC | NBD-SM |
|---|---|---|---|---|---|
| − | − | 1.00 ± 0.02 | 1.00 (±0.09) | 1.00 (±0.04) | 1.00 (±0.11) |
| − | + | 1.17 ± 0.03 | 1.02 (±0.08) | 1.26 (±0.07) | 1.02 (±0.10) |
| ATP8A1 | + | 2.53 ± 0.03 | 1.28 (±0.05) | 0.97 (±0.01) | 1.01 (±0.06) |
| ATP8A2 | + | 8.90 ± 0.07 | 1.50 (±0.06) | 1.35 (±0.02) | 0.97 (±0.04) |
| ATP8B1 | + | 1.33 ± 0.03 | 1.13 (±0.07) | 1.05 (±0.05) | 0.82 (±0.02) |
| ATP8B2 | + | 1.23 ± 0.01 | 1.21 (±0.08) | 1.29 (±0.02) | 0.89 (±0.05) |
| ATP8B4 | + | 1.24 ± 0.06 | 1.12 (±0.09) | 1.29 (±0.02) | 0.88 (±0.02) |
| ATP9A | + | 1.25 ± 0.02 | 1.19 (±0.08) | 0.94 (±0.01) | 0.89 (±0.04) |
| ATP9B | + | 1.05 ± 0.01 | 1.28 (±0.07) | 1.09 (±0.01) | 0.99 (±0.05) |
| ATP10B | + | 1.22 ± 0.04 | 1.20 (±0.05) | 0.87 (±0.05) | 0.98 (±0.06) |
| ATP10D | + | 1.06 ± 0.02 | 1.21 (±0.19) | 0.91 (±0.02) | 0.95 (±0.07) |
| ATP11A | + | 4.75 ± 0.09 | 2.67 (±0.02) | 1.50 (±0.02) | 0.87 (±0.07) |
| ATP11B | + | 1.38 ± 0.03 | 1.42 (±0.10) | 1.29 (±0.02) | 0.93 (±0.07) |
| ATP11C | + | 7.68 ± 0.17 | 2.38 (±0.10) | 0.99 (±0.01) | 1.06 (±0.07) |
ATPase Activity of the Three Flippases
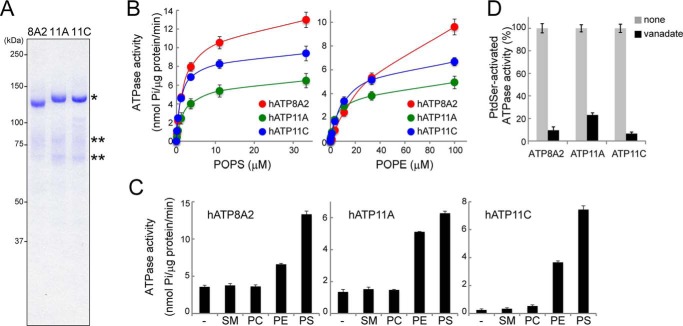
The ATPase activity of P4-ATPases is known to be activated by phospholipids (12, 30). To examine the ATPase activity of human ATP8A2, 11A, and 11C, the proteins were Flag-tagged, produced in 293T cells together with human CDC50A, and affinity-purified. The purified ATPases (ATP8A2, 11A, and 11C) were of the expected size on SDS-PAGE and were associated with proteins of ∼78 and 72 kDa (Fig. 2A), which were probably differently glycosylated CDC50As fused to GFP. The ATPase activity of ATP8A2, 11A, and 11C was strongly activated by POPS (Fig. 2B). A Michael-Menten kinetics analysis indicated that the Km and Vmax values of the three ATPases for POPS: 2.1 μm and 13 nmol Pi/μg protein/min, 1.9 μm and 6.5 nmol Pi/μg protein/min, and 1.3 μm and 8.9 nmol Pi/μg protein/min for ATP8A2, 11A, and 11C, respectively. POPE also activated these ATPases, but the Km for POPE was 3–15 times higher than that for POPS and differed significantly among the three ATPases (29.2 μm (Vmax: 9.6 nmol Pi/μg protein/min), 6.1 μm (Vmax: 4.9 nmol Pi/μg protein/min), and 11.0 μm (Vmax: 6.7 nmol Pi/μg protein/min) for ATP8A2, 11A, and 11C, respectively). POPC and sphingomyelin had little if any effect on the ATPase activity of ATP8A2, 11A, or 11C (Fig. 2C). As reported for the membrane ATPases (35), the POPS-stimulated ATPase activity of ATP8A2, 11A, and 11C was inhibited by orthovanadate (Fig. 2D). The significant POPE-induced ATPase activity of ATP8A2 was inconsistent with its apparently low PtdEtn-flippase activity (Table 1), but the latter finding might have been due to the high background incorporation of NBD-PE into ATP11CED22 cells (Fig. 1).
FIGURE 2.
ATPase activity of human ATP8A2, ATP11A, and ATP11C. A, purified human P4-ATPases (400 ng) were separated by SDS-PAGE and stained with Coomassie Brilliant Blue. *, Flag-tagged P4-ATPase. **, GFP-tagged CDC50A. B, ATPase activity of human ATP8A2, ATP11A, and ATP11C, stimulated by PtdSer and PtdEtn. Each purified P4-ATPase (10 ng) was incubated for 20 min at 37 °C with 600 μm ATP in the presence of the indicated concentrations of POPS or POPE, and the released phosphate was measured. To determine the phospholipid-stimulated ATPase activity (nmol released phosphate/μg protein/min), the ATPase activity without POPS or POPE was subtracted, and the result was plotted with S.E. (n = 3). C, effect of various phospholipids on the ATPase activity. Purified ATP8A2, ATP11A, or ATP11C (10 ng) was incubated for 20 min at 37 °C with 600 μm ATP in the presence or absence of 15 μm SM, POPC (PC), POPE (PE), or POPS (PS). The released phosphate was measured, and the ATPase activity (nmol released phosphate/μg protein/min) was plotted with S.E. (n = 3). D, the indicated purified ATPases (10 ng) were incubated for 20 min at 37 °C with 600 μm ATP and 15 μm POPS in the presence or absence (none) of 500 μm orthovanadate. The PtdSer-activated ATPase activity was obtained by subtracting the ATPase activity obtained without POPS and is expressed as the percentage of the activity observed in the absence of orthovanadate with S.E. (n = 3).
Subcellular Localization of P4-ATPases
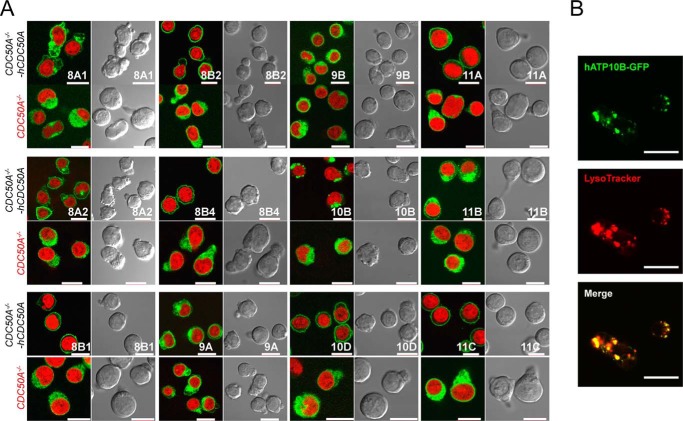
Little or no evidence of flippase activity was found in the remaining eight human P4-ATPases (ATP8B1, 8B2, 8B4, 9A, 9B, 10B, 10D, and 11B). It is possible that these ATPases had low flippase activity because they did not localize to the plasma membrane. To examine this possibility, we tagged the 12 human P4-ATPases with GFP and introduced them into CDC50A-null CDC50AED29 cells or CDC50A-transformed CDC50AED29 cells to establish stable transformants expressing each ATPase. As expected from their flippase activity, ATP8A2, 11A, and 11C localized to the plasma membrane in the presence of CDC50A (Fig. 3). In the absence of CDC50A, these proteins were found intracellularly, indicating that CDC50A was required for their localization at the plasma membrane. ATP8B1, 8B2, 8B4, and 10D also localized to the plasma membrane in a CDC50A-dependent manner. ATP8A1 and 11B were found mainly in intracellular vesicles and partly at the plasma membrane in the presence of CDC50A. As previously reported (36), ATP9A and ATP9B localized predominantly to intracellular vesicles, and the presence of CDC50A did not change their localization. On the other hand, the localization of ATP10B was dramatically changed by the presence of CDC50A; ATP10B-GFP was diffusely present in intracellular vesicles in the absence of CDC50A but appeared as a large patch in the presence of CDC50A (Fig. 3A). The patch-like ATP10B-GFP structure co-localized with LysoTracker (Fig. 3B), indicating that ATP10B localized to the late endosome or lysosome, as previously reported (37).
FIGURE 3.
Cellular P4-ATPase localization. A, cellular localization of 12 P4-ATPases. CDC50A-null (CDC50AED29) or CDC50A-expressing cells (CDC50AED29-CDC50A) were stably transformed with the indicated GFP-tagged human P4-ATPases (ATP8B3 and ATP10A were not used) and observed by confocal microscopy. Nuclei were stained with 12.5 μm DRAQ5 (red). Scale bar, 10 μm. ATP8A2, 8B1, 8B2, 8B4, 10D, 11A, and 11C localized to the plasma membranes when co-expressed with CDC50A. B, ATP10B localized to lysosomes. CDC50AED29 cells were stably transformed with human CDC50A and GFP-tagged human ATP10B. The transformants were stained with 100 μm LysoTracker for 30 min at 37 °C and observed by confocal microscopy. Images show GFP (green), LysoTracker (red), and merged. Scale bar, 10 μm.
Caspase Cleavage of ATP11A
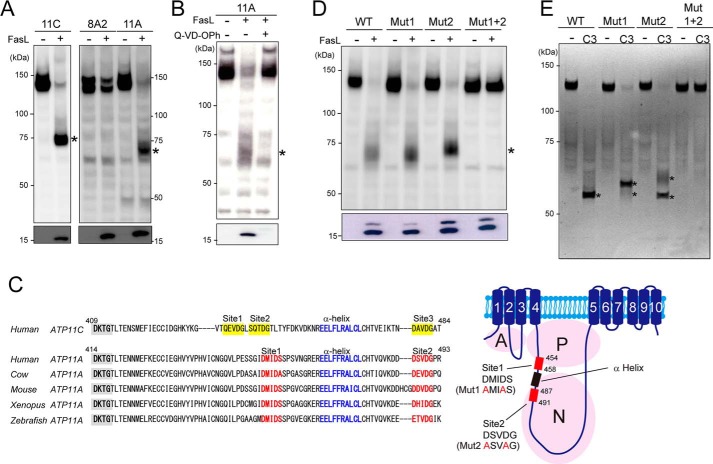
Human ATP11C carries three caspase recognition sites in its nucleotide-binding domain and is cleaved by caspases for apoptotic PtdSer exposure (15). To examine whether other PtdSer-flippases are similarly cleaved during apoptosis, ATP11CED22 transformants expressing GFP-tagged ATP8A2, 11A, or 11C were treated with FasL for 1 h and analyzed by Western blotting with an anti-GFP mAb. In FasL-treated cells, ATP11A was cleaved from 140 to 75 kDa, but ATP8A2 remained intact (Fig. 4A). A caspase inhibitor (Q-VD-OPh) blocked the cleavage of ATP11A in the FasL-treated cells, indicating that the cleavage was caspase-dependent (Fig. 4B).
FIGURE 4.
ATP11A but not ATP8A2 was cleaved during apoptosis. A and B, FasL induced the cleavage of ATP11A during apoptosis. ATP11CED22 transformants expressing GFP-tagged ATP8A2, ATP11A, or ATP11C were left untreated or treated with 33 units/ml FasL for 1 h at 37 °C with or without 20 μm Q-VD-OPh. Cell lysates were analyzed by Western blotting with an anti-GFP (upper panels) or anti-active caspase 3 mAb (lower panels). *, cleaved P4-ATPase. C, left panel, amino acid sequence alignment around caspase recognition sites in ATP11A orthologs: human (UniProt; P98196), bovine (UniProt; F1N0J3), mouse (UniProt; P98197), Xenopus (UniProt; F6XT62), and zebrafish (UniProt; F8W2B0). Caspase recognition sequences and the flanking α-helix in ATP11A are shown in red and blue, respectively. The DKTG motif, the conserved phosphorylation site in P-type ATPases, is shaded in gray. The amino acid sequence of the corresponding region of human ATP11C is shown at the top, with caspase recognition sites highlighted in yellow. Right panel, schematic presentation of human ATP11A. Two caspase recognition sites (red boxes) and α-helix (black box) flanked by the caspase recognition sites in the large cytoplasmic loop are indicated. Three cytoplasmic domains (A, actuator; P, phosphorylation; N, nucleotide binding) are shown in pale purple circles, and transmembrane segments are numbered. D, ATP11A with mutations at putative caspase recognition sites was not cleaved during apoptosis. ATP11CED22 transformants expressing WT, single-mutant (Mut1 or Mut2), or double-mutant (Mut1 + 2) ATP11A-GFP were left untreated (−) or treated with FasL (+) at 37 °C for 1 h, and the cell lysates were analyzed by Western blotting with an anti-GFP (upper panel) or anti-active caspase 3 mAb (lower panel). *, cleaved ATP11A. E, ATP11A was cleaved by caspase 3. The purified ATP11A and indicated mutants were incubated with or without caspase 3 at 20 °C for 1 h, separated by SDS-PAGE, and stained with SYPRO® Ruby protein gel stain. C3, recombinant caspase 3. *, cleaved ATP11A.
We searched for caspase recognition sites using Cascleave (59) and identified two putative phylogenetically well conserved caspase 3 recognition sites in human ATP11A at positions similar to those in human ATP11C (Fig. 4C). To determine whether ATP11A is cleaved at these sites during apoptosis, the two caspase recognition sites (site 1, DMIDS; site 2, DSVDG) were mutated singly to AMIAS (Mut1) and ASVAG (Mut2), fused to GFP, and expressed in ATP11CED22 cells. ATP11A with a mutation at only one caspase recognition site (Mut1 or Mut2) could be cleaved during FasL-induced apoptosis, whereas ATP11A with mutations in both caspase recognition sites (Mut1 + 2) resisted cleavage (Fig. 4D). This was confirmed with the purified proteins. That is, ATP11A and its caspase-resistant mutants were Flag-tagged, expressed in 293T cells together with GFP-tagged CDC50A, and purified with anti-Flag antibody. As shown in Fig. 4E, incubating the purified proteins with caspase 3 confirmed that caspase 3 could directly cleave ATP11A at these two recognition sites.
Caspase Regulation of Flippase
We previously found that cells expressing a caspase-resistant ATP11C do not expose PtdSer during apoptosis (15). To examine whether this is also true of ATP11A, ATP11CED22 cells were transformed with wild-type (WT) or mutant (Mut1, Mut2, or Mut1 + 2) ATP11A. All of the transformants expressing a single- or double-mutant ATP11A had flippase activity comparable with that of WT ATP11A (Fig. 5A); thus, the mutations did not affect flippase activity. When these transformants were treated with FasL, cells expressing WT ATP11A exposed PtdSer as efficiently as cells lacking ATP11C. In contrast, FasL treatment could not induce PtdSer exposure in cells expressing Mut1, Mut2, or Mut1 + 2 ATP11A (Fig. 5B). The expression of ATP11A mutants did not affect the FasL-induced caspase 3 activation or phospholipid scrambling, as determined by NBD-PC incorporation assays (Fig. 5C). These results confirmed that ATP11A, like ATP11C, must be inactivated by caspase for efficient PtdSer exposure. The ability of Mut1 and Mut2 to inhibit the PtdSer-exposure suggested that cleavage at one caspase recognition site did not abolish the flippase activity of ATP11A. To verify this, the ATPase activity of caspase 3-treated recombinant WT, Mut 1, Mut 2, and Mut1 + 2 ATP11A (Fig. 4E) was measured. As shown in Fig. 5D, caspase 3 treatment abolished the PtdSer-dependent ATPase activity of WT AT11A, but it had little effect on the ATPase activity of Mut1, Mut 2, and Mut 1 + 2 ATP11A, supporting that cleavage at one position of ATP11A is not sufficient to inactivate the flippase. Because ATP8A2 had no caspase recognition site and was not cleaved during apoptosis (Fig. 4A), it is possible that WT ATP8A2 would inhibit apoptotic PtdSer exposure. In fact, ATP11CED22 transformants expressing ATP11C efficiently exposed PtdSer when treated with FasL, but those expressing ATP8A2 did not expose PtdSer (Fig. 5E). These results confirmed that ATP8A2 flips PtdSer, but unlike ATP11A and 11C, ATP8A2 is not inactivated during apoptosis.
FIGURE 5.
Caspase-mediated ATP11A cleavage elicits apoptotic PtdSer exposure. A, PtdSer-flippase activity of human ATP11A with caspase recognition site mutations. ATP11CED22 transformants expressing WT or mutant (Mut1, Mut2, or Mut1 + 2) ATP11A-GFP were incubated at 25 °C for 3 min with 1 μm NBD-PS, treated with fatty-acid-free BSA, and analyzed by flow cytometry. The mean fluorescent intensity was plotted with S.E. (n = 3). B, very little PtdSer exposure was detected in cells expressing a caspase-resistant form of ATP11A. W3 cells, ATP11CED22 cells, or ATP11CED22 transformants expressing WT or mutant (Mut1, Mut2, or Mut1 + 2) ATP11A-GFP were left untreated (red) or treated (blue) with 33 units/ml FasL for 2 h at 37 °C, stained with Cy5-Annexin V and Sytox blue, and analyzed by flow cytometry. Annexin V-staining profiles in the Sytox Blue-negative population are shown. C, flippase mutations had no effect on apoptotic caspase activation and phospholipid scrambling. W3 cells, ATP11CED22 cells, or ATP11CED22 transformants expressing WT ATP11A-GFP, Mut1-GFP, Mut2-GFP, or Mut1 + 2-GFP were left untreated or treated with 33 units/ml FasL at 37 °C for 1 h (upper panel) or 2 h (lower panel). The cell lysates were analyzed by Western blotting with an anti-active caspase 3 mAb (upper panel). −, untreated; +, treated with FasL. In the lower panel, untreated (gray bars) or FasL-treated (black bars) cells were incubated at 25 °C for 3 min with 1 μm NBD-PC and analyzed by flow cytometry. Phospholipid scrambling activity was expressed as the incorporation of NBD-PC. Experiments were conducted in triplicate, and the average values were plotted with S.E. D, ATPase activity of caspase 3-treated ATP11A and its mutants. Purified ATP11A and indicated mutants were left untreated or treated with caspase 3 at 20 °C for 1 h. Using aliquots of the protein (10 ng), their ATPase activity (nmol released phosphate/μg protein/min) was measured and is plotted with S.E. (n = 3). C3, recombinant caspase 3. E, apoptotic PtdSer exposure was inhibited in ATP8A2-expressing cells. W3 cells, ATP11CED22 cells, or ATP11CED22 transformants expressing Flag-tagged CDC50A with ATP11C or ATP8A2 were left untreated of treated with 33 units/ml of FasL for 2 h at 37 °C, stained with Cy5-annexin V and Sytox blue, and analyzed by flow cytometry. MFI, mean fluorescence intensity.
Calcium Regulates PtdSer Flippase
Increasing intracellular Ca2+ levels in human erythrocytes inhibits their ability to incorporate spin-labeled aminophospholipids (38), suggesting that Ca2+ can regulate flippase activity. To confirm this finding at a molecular level, ATP11CED22 transformants expressing ATP8A2, 11A, or 11C were incubated for 4 min with NBD-PS in the presence of various concentrations of A23187, a calcium ionophore that dose-dependently increases intracellular Ca2+ (39). As shown in Fig. 6A, A23187 dose-dependently inhibited the ATP8A2, ATP11A, and ATP11C PtdSer-flippase activity, with a 3-fold difference in the sensitivity of the three flippases: the A23187 IC50 was 1.54, 0.48, and 0.56 μm for ATP8A2, 11A, and 11C, respectively. To confirm the differing sensitivities of ATP8A2, 11A, and 11C to Ca2+, their PtdSer-activated ATPase activity was measured in the presence of various concentrations of CaCl2. As shown in Fig. 6B, CaCl2 inhibited the PtdSer-stimulated ATPase activity of ATP8A2, 11A, and 11C. Again, sensitivity to Ca2+ differed between the three flippases: the IC50 was 581, 206, and 130 μm for ATP8A2, 11A, and 11C, respectively.
FIGURE 6.
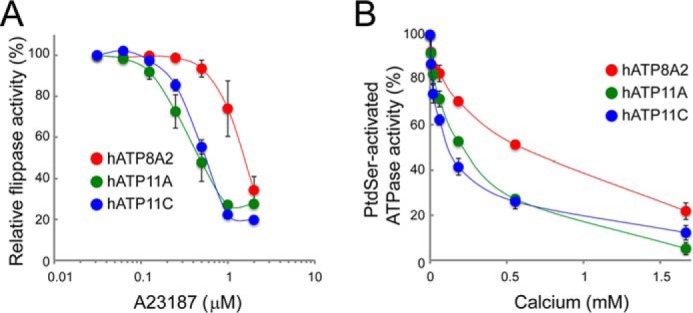
Calcium inhibits the ATP11A and ATP11C flippase activity. A, ATP11CED22 transformants expressing Flag-tagged CDC50A with ATP8A2, ATP11A, or ATP11C were incubated with 1 μm NBD-PS for 4 min at 20 °C in the presence of 1 mm Ca2+ and the indicated concentrations of A23187, then treated with fatty-acid-free BSA, and analyzed by flow cytometry. NBD-PS incorporation is expressed as a percentage of the maximal flippase activity observed without A23187. Experiments were conducted three times, and the average values were plotted with S.E. B, calcium inhibition of ATPase activity. Purified ATP8A2, ATP11A, or ATP11C (10 ng each) was incubated in triplicate for 20 min at 37 °C with 600 μm ATP and 15 μm POPS in the presence of calcium at the indicated concentrations. PtdSer-activated ATPase activity is expressed as the percentage of the activity observed in the absence of calcium with S.E. (n = 3).
Tissue Distribution of P4-ATPases with PtdSer-Flippase Activity
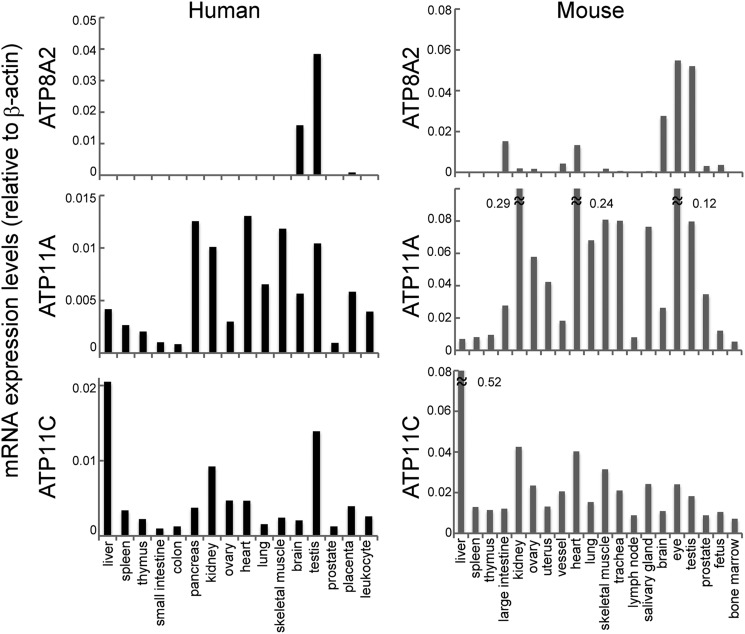
Because our results indicated that ATP8A2, 11A, and 11C act as flippases at the plasma membrane and that ATP8A2 sensitivity to caspase and Ca2+ differed greatly from that of ATP11A and 11C, we next used real time RT-PCR to analyze the distribution of these flippases in human and mouse tissues. As shown in Fig. 7, ATP11A and 11C were ubiquitously expressed in various tissues but with varying preferences. ATP11C was highly expressed in the liver, whereas ATP11A expression was relatively low; this difference was even more pronounced in the mouse. The levels of ATP11A mRNA in the mouse kidney and heart were 6–7 times higher than those of ATP11C mRNA. In contrast, the ATP8A2 expression was rather tissue-specific; ATP8A2 mRNA was abundantly expressed in the human and mouse brain and testis, less abundantly in the mouse large intestine and heart, and hardly at all in other adult human and mouse tissues.
FIGURE 7.
Tissue distribution of ATP8A2, ATP11A, and ATP11C. Using human tissue cDNA panels (human MTC panels I and II) and mouse cDNA prepared from the indicated tissues of C57BL/6 mice aged 10–12 weeks, the mRNA levels for ATP8A2, 11A, and 11C were quantified by real time RT-PCR and expressed relative to β-actin mRNA.
Discussion
Early biochemical studies of membranes from human red blood cells (40) and bovine chromaffin granules (41) showed a specific ATP-dependent inward incorporation of aminophospholipids. An ATPase purified from human erythrocyte membranes was shown to flip aminophospholipids in an ATP-dependent manner in a reconstituted liposome system (42). Cloning of the ATPase cDNA revealed that this ATPase was a member of the large P4-ATPase family (43). Although many groups have since studied the biochemical characteristics of each member of the P4-ATPase family, there is confusion in the literature as to which flippases act at the plasma membrane. We previously showed that most of the PtdSer flippase activity is lost in ATP11C-null human and mouse leukemia cells. In the present study, we expressed 12 human P4-ATPases in ATP11C-null cells and found that ATP11C is not the only P4-ATPase that functions as a plasma membrane flippase; ATP11A and ATP8A2 also localize to the plasma membrane and have PtdSer-flippase activity. Accordingly, the ATPase activity of these flippases was strongly stimulated by PtdSer. The ability of ATP8A2 to flip PtdEtn was low compared with its ability to flip PtdSer. However, the ATPase activity of ATP8A2 was enhanced by high PtdEtn concentrations, suggesting that ATP8A2 may also flip PtdEtn. ATP11A, ATP11C, and ATP8A2 did not flip PtdCho or SM, and their ATPase activity was not stimulated by PtdCho or SM, confirming that they are aminophospholipid translocases.
The flippase activity of ATP11A at plasma membrane agrees with a previous report by Takatsu et al. (19). However, it is controversial where ATP8A2 is localized. One report indicates that ATP8A2 works as a flippase at plasma membranes (20), whereas Lee et al. (13) showed that ATP8A2 localizes to recycling endosomes and works as a flippase for membrane fission. Although we cannot rule out the possibility that ATP8A2 localization depends on cell type, ATP8A2 inhibition of apoptotic PtdSer exposure supports its role as a flippase at the plasma membrane. ATP8B1 is reported to localize to the plasma membrane and to flip PtdSer (11), cardiolipin (44), or PtdCho (19). Although we confirmed ATP8B1 plasma membrane localization, we did not detect any translocase activity for fluorescent-labeled PtdSer or PtdCho. The reason for this difference in findings is not clear. ATP8B1 is specifically expressed in liver, intestinal, and pancreatic cells (45), and it may be necessary to examine its function in these cells. We found three P4-ATPases (ATP8B2, ATP8B4, and ATP10D) that localized to the plasma membrane but did not support the flipping of PtdSer, PtdEtn, PtdCho, or SM. Whether these ATPases are involved in flipping other phospholipids or require different substrates, such as oligosaccharide diphosphate dolichols (46), remains to be studied.
We previously showed that ATP11C is cleaved by caspases and inactivated during apoptosis to expose PtdSer to the cell surface (15); this process is essential for the dead cell to be engulfed. Because ATP11A carries two caspase recognition sites at similar positions to those in ATP11C, we examined whether ATP11A might behave similarly. Apoptosis is a universal process in mammals and occurs in most cells (47). Because the delayed clearance of apoptotic cells is detrimental for animals (4), inactivation of the ubiquitously expressed ATP11A and ATP11C during apoptosis would be reasonable for the prompt engulfment of dead cells. Thus, it was a surprise that ATP8A2 was not cleaved during apoptosis.
ATP8A2 is expressed in photoreceptor cells in the eyes (12, 20). Every day, distal outer segments of these photoreceptor cells are shed, expose PtdSer, and are phagocytosed by adjacent retinal pigment epithelial cells that express PtdSer-recognizing proteins (48, 49). How PtdSer is exposed on the shed outer segments is not known. Within the outer segments, ATP8A2 is present in the disk membrane (12), which is derived from the plasma membrane, but later is disconnected from it (50). It is possible that the role of ATP8A2 in maintaining asymmetrical phospholipid distribution is lost in the outer segments, at least in rod photoreceptor cells, to facilitate PtdSer exposure. ATP8A2 is also widely expressed by neurons in the central nervous system and by spermatids in the testis (51). PtdSer is exposed when neurons undergo apoptosis (52) and probably when axons are pruned (53). PtdSer is also exposed when sperm undergoes capacitation (54). It will be interesting to examine how ATP8A2 is regulated to enable PtdSer exposure in these processes.
PtdSer is exposed in various activated cells, including platelets (5) and lymphocytes (55, 56). When cells are activated, the free Ca2+ concentration is known to locally and transiently increase near or beneath the plasma membrane, reaching to more than 100 μm (57, 58). The Ca2+ in these activated cells would activate Ca2+-dependent phospholipid scramblase and inactivate flippase. This inhibition of flippase by Ca2+ should not be complete, otherwise the activated cells would be engulfed by macrophages. Once the Ca2+ concentrations return to normal levels, scramblases would stop working, and flippase would restart to translocate PtdSer and PtdEtn from outer to inner leaflet of the plasma membrane. Whether Ca2+ directly binds to the flippase or not remains to be studied.
PtdSer is exposed on the cell surface or vesicles not only in the engulfment of dead cells and in blood clotting, but also in many other biological processes. Two ubiquitously expressed scramblases, TMEM16F and Xkr8, are activated by Ca2+ and caspase, respectively (6, 7). We found that two similar, ubiquitously expressed P4-ATPases (ATP11A and ATP11C) are down-regulated by Ca2+ and caspases (15). In addition, we found a caspase- and Ca2+-resistant flippase that is tissue-specifically expressed. There is a tissue-specific Ca2+-dependent scramblase (9) and a caspase-dependent scramblase (10). It will be interesting to investigate how these scramblases and flippases interact to expose PtdSer in various biological processes.
Author Contributions
K. S. and S. N. conceived the study. K. S. and S. K. performed the experiments. K. S. and S. N. wrote the manuscript.
Acknowledgments
We thank Dr. Kazuhiro Abe (Nagoya University) for advice about the ATPase assay and M. Fujii for secretarial assistance.
This work was supported in part by Grants-in-Aid from the Ministry of Education, Science, Sports, and Culture in Japan, and CREST, Japan Science Technology Corporation. The authors declare that they have no conflicts of interest with the contents of this article.
- PtdSer
- phosphatidylserine
- PtdEtn
- phosphatidylethanolamine
- PtdCho
- phosphatidylcholine
- FasL
- Fas ligand
- HBSS
- Hanks' balanced salt solution
- P4-ATPase
- type IV P-type ATPase
- NBD-PS
- 1-oleoyl-2-{6-[(7-nitro-2–1,3-benzoxadiazol-4-yl) amino]hexanoyl}-sn-glycero-3-phosphoserine
- NBD-PE
- 1-oleoyl-2-{6-[(7-nitro-2–1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphoethanolamine
- NBD-PC
- 1-oleoyl-2-{6-[(7-nitro-2–1,3-benzoxadiazol-4-yl) amino]hexanoyl}-sn-glycero-3-phosphocholine
- NBD-SM
- N-[6-[(7-nitro-2–1,3-benzoxadiazol-4-yl)amino]hexanoyl]-sphingosine-1-phosphocholine
- POPS
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine
- POPE
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- SM
- N-stearoyl-d-erythro-sphingosylphosphorylcholine
- LMNG
- 2,2-didecylpropane-1,3-bis-β-d-maltopyranoside
- WT
- wild-type.
References
- 1.Leventis P. A., and Grinstein S. (2010) The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 39, 407–427 [DOI] [PubMed] [Google Scholar]
- 2.van Meer G., Voelker D. R., and Feigenson G. W. (2008) Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fadok V. A., de Cathelineau A., Daleke D. L., Henson P. M., and Bratton D. L. (2001) Loss of phospholipid asymmetry and surface exposure of phosphatidylserine is required for phagocytosis of apoptotic cells by macrophages and fibroblasts. J. Biol. Chem. 276, 1071–1077 [DOI] [PubMed] [Google Scholar]
- 4.Nagata S., Hanayama R., and Kawane K. (2010) Autoimmunity and the clearance of dead cells. Cell 140, 619–630 [DOI] [PubMed] [Google Scholar]
- 5.Lhermusier T., Chap H., and Payrastre B. (2011) Platelet membrane phospholipid asymmetry: from the characterization of a scramblase activity to the identification of an essential protein mutated in Scott syndrome. J. Thromb. Haemost. 9, 1883–1891 [DOI] [PubMed] [Google Scholar]
- 6.Suzuki J., Denning D. P., Imanishi E., Horvitz H. R., and Nagata S. (2013) Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science 341, 403–406 [DOI] [PubMed] [Google Scholar]
- 7.Suzuki J., Umeda M., Sims P. J., and Nagata S. (2010) Calcium-dependent phospholipid scrambling by TMEM16F. Nature 468, 834–838 [DOI] [PubMed] [Google Scholar]
- 8.Brunner J. D., Lim N. K., Schenck S., Duerst A., and Dutzler R. (2014) X-ray structure of a calcium-activated TMEM16 lipid scramblase. Nature 516, 207–212 [DOI] [PubMed] [Google Scholar]
- 9.Suzuki J., Fujii T., Imao T., Ishihara K., Kuba H., and Nagata S. (2013) Calcium-dependent phospholipid scramblase activity of TMEM16 protein family members. J. Biol. Chem. 288, 13305–13316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki J., Imanishi E., and Nagata S. (2014) Exposure of phosphatidylserine by Xk-related protein family members during apoptosis. J. Biol. Chem. 289, 30257–30267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paulusma C. C., Folmer D. E., Ho-Mok K. S., de Waart D. R., Hilarius P. M., Verhoeven A. J., and Oude Elferink R. P. (2008) ATP8B1 requires an accessory protein for endoplasmic reticulum exit and plasma membrane lipid flippase activity. Hepatology 47, 268–278 [DOI] [PubMed] [Google Scholar]
- 12.Coleman J. A., Kwok M. C., and Molday R. S. (2009) Localization, purification, and functional reconstitution of the P4-ATPase Atp8a2, a phosphatidylserine flippase in photoreceptor disc membranes. J. Biol. Chem. 284, 32670–32679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S., Uchida Y., Wang J., Matsudaira T., Nakagawa T., Kishimoto T., Mukai K., Inaba T., Kobayashi T., Molday R. S., Taguchi T., and Arai H. (2015) Transport through recycling endosomes requires EHD1 recruitment by a phosphatidylserine translocase. EMBO J. 34, 669–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yabas M., Teh C. E., Frankenreiter S., Lal D., Roots C. M., Whittle B., Andrews D. T., Zhang Y., Teoh N. C., Sprent J., Tze L. E., Kucharska E. M., Kofler J., Farell G. C., Bröer S., Goodnow C. C., and Enders A. (2011) ATP11C is critical for the internalization phosphatidylserine and differentiation of B lymphocytes. Nat. Immunol. 12, 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segawa K., Kurata S., Yanagihashi Y., Brummelkamp T. R., Matsuda F., and Nagata S. (2014) Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science 344, 1164–1168 [DOI] [PubMed] [Google Scholar]
- 16.Coleman J. A., Quazi F., and Molday R. S. (2013) Mammalian P4-ATPases and ABC transporters and their role in phospholipid transport. Biochim. Biophys. Acta 1831, 555–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanaka K., Fujimura-Kamada K., and Yamamoto T. (2011) Functions of phospholipid flippases. J. Biochem. 149, 131–143 [DOI] [PubMed] [Google Scholar]
- 18.Lopez-Marques R. L., Theorin L., Palmgren M. G., and Pomorski T. G. (2014) P4-ATPases: lipid flippases in cell membranes. Pflügers Arch. 466, 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takatsu H., Tanaka G., Segawa K., Suzuki J., Nagata S., Nakayama K., and Shin H. W. (2014) Phospholipid flippase activities and substrate specificities of human type IV P-type ATPases localized to the plasma membrane. J. Biol. Chem. 289, 33543–33556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu X., Libby R. T., de Vries W. N., Smith R. S., Wright D. L., Bronson R. T., Seburn K. L., and John S. W. (2012) Mutations in a P-type ATPase gene cause axonal degeneration. PLoS Genet. 8, e1002853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogasawara J., Watanabe-Fukunaga R., Adachi M., Matsuzawa A., Kasugai T., Kitamura Y., Itoh N., Suda T., and Nagata S. (1993) Lethal effect of the anti-Fas antibody in mice. Nature 364, 806–809 [DOI] [PubMed] [Google Scholar]
- 22.Sakahira H., Enari M., and Nagata S. (1998) Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature 391, 96–99 [DOI] [PubMed] [Google Scholar]
- 23.Kitamura T., Koshino Y., Shibata F., Oki T., Nakajima H., Nosaka T., and Kumagai H. (2003) Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 31, 1007–1014 [PubMed] [Google Scholar]
- 24.Shiraishi T., Suzuyama K., Okamoto H., Mineta T., Tabuchi K., Nakayama K., Shimizu Y., Tohma J., Ogihara T., Naba H., Mochizuki H., and Nagata S. (2004) Increased cytotoxicity of soluble Fas ligand by fusing isoleucine zipper motif. Biochem. Biophys. Res. Commun. 322, 197–202 [DOI] [PubMed] [Google Scholar]
- 25.Kamens J., Paskind M., Hugunin M., Talanian R. V., Allen H., Banach D., Bump N., Hackett M., Johnston C. G., and Li P. (1995) Identification and characterization of ICH-2, a novel member of the interleukin-1 beta-converting enzyme family of cysteine proteases. J. Biol. Chem. 270, 15250–15256 [DOI] [PubMed] [Google Scholar]
- 26.Boussif O., Lezoualc'h F., Zanta M. A., Mergny M. D., Scherman D., Demeneix B., and Behr J. P. (1995) A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. U.S.A. 92, 7297–7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abe K., Tani K., Friedrich T., and Fujiyoshi Y. (2012) Cryo-EM structure of gastric H+,K+-ATPase with a single occupied cation-binding site. Proc. Natl. Acad. Sci. U.S.A. 109, 18401–18406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Veldhoven P. P., and Mannaerts G. P. (1987) Inorganic and organic phosphate measurements in the nanomolar range. Anal. Biochem. 161, 45–48 [DOI] [PubMed] [Google Scholar]
- 29.Zhou X., Sebastian T. T., and Graham T. R. (2013) Auto-inhibition of Drs2p, a yeast phospholipid flippase, by its carboxyl-terminal tail. J. Biol. Chem. 288, 31807–31815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paterson J. K., Renkema K., Burden L., Halleck M. S., Schlegel R. A., Williamson P., and Daleke D. L. (2006) Lipid specific activation of the murine P4-ATPase Atp8a1 (ATPase II). Biochemistry 45, 5367–5376 [DOI] [PubMed] [Google Scholar]
- 31.Coleman J. A., and Molday R. S. (2011) Critical role of the beta-subunit CDC50A in the stable expression, assembly, subcellular localization, and lipid transport activity of the P4-ATPase ATP8A2. J. Biol. Chem. 286, 17205–17216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki J., and Nagata S. (2014) Phospholipid scrambling on the plasma membrane. Methods Enzymol. 544, 381–393 [DOI] [PubMed] [Google Scholar]
- 33.Gong E. Y., Park E., Lee H. J., and Lee K. (2009) Expression of Atp8b3 in murine testis and its characterization as a testis specific P-type ATPase. Reproduction 137, 345–351 [DOI] [PubMed] [Google Scholar]
- 34.Wang L., Beserra C., and Garbers D. L. (2004) A novel aminophospholipid transporter exclusively expressed in spermatozoa is required for membrane lipid asymmetry and normal fertilization. Develop. Biol. 267, 203–215 [DOI] [PubMed] [Google Scholar]
- 35.Bowman B. J., and Slayman C. W. (1979) The effects of vanadate on the plasma membrane ATPase of Neurospora crassa. J. Biol. Chem. 254, 2928–2934 [PubMed] [Google Scholar]
- 36.Takatsu H., Baba K., Shima T., Umino H., Kato U., Umeda M., Nakayama K., and Shin H. W. (2011) ATP9B, a P4-ATPase (a putative aminophospholipid translocase), localizes to the trans-Golgi network in a CDC50 protein-independent manner. J. Biol. Chem. 286, 38159–38167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naito T., Takatsu H., Miyano R., Takada N., Nakayama K., and Shin H. W. (2015) Phospholipid flippase ATP10A translocates phosphatidylcholine and is involved in plasma membrane dynamics. J. Biol. Chem. 290, 15004–15017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bitbol M., Fellmann P., Zachowski A., and Devaux P. F. (1987) Ion regulation of phosphatidylserine and phosphatidylethanolamine outside-inside translocation in human erythrocytes. Biochim. Biophys. Acta 904, 268–282 [DOI] [PubMed] [Google Scholar]
- 39.Korenaga R., Ando J., Ohtsuka A., Sakuma I., Yang W., Toyo-oka T., and Kamiya A. (1993) Close correlation between cytoplasmic Ca++ levels and release of an endothelium-derived relaxing factor from cultured endothelial cells. Cell Struct. Funct. 18, 95–104 [DOI] [PubMed] [Google Scholar]
- 40.Seigneuret M., and Devaux P. (1984) ATP-dependent asymmetric distribution of spin-labeled phospholipids in the erythrocyte membrane: relation to shape changes. Proc. Natl. Acad. Sci. U.S.A. 81, 3751–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zachowski A., Henry J. P., and Devaux P. F. (1989) Control of transmembrane lipid asymmetry in chromaffin granules by an ATP-dependent protein. Nature 340, 75–76 [DOI] [PubMed] [Google Scholar]
- 42.Auland M. E., Roufogalis B. D., Devaux P. F., and Zachowski A. (1994) Reconstitution of ATP-dependent aminophospholipid translocation in proteoliposomes. Proc. Natl. Acad. Sci. U.S.A. 91, 10938–10942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang X., Halleck M. S., Schlegel R. A., and Williamson P. (1996) A subfamily of P-type ATPases with aminophospholipid transporting activity. Science 272, 1495–1497 [DOI] [PubMed] [Google Scholar]
- 44.Ray N. B., Durairaj L., Chen B. B., McVerry B. J., Ryan A. J., Donahoe M., Waltenbaugh A. K., O'Donnell C. P., Henderson F. C., Etscheidt C. A., McCoy D. M., Agassandian M., Hayes-Rowan E. C., Coon T. A., Butler P. L., Gakhar L., Mathur S. N., Sieren J. C., Tyurina Y. Y., Kagan V. E., McLennan G., and Mallampalli R. K. (2010) Dynamic regulation of cardiolipin by the lipid pump Atp8b1 determines the severity of lung injury in experimental pneumonia. Nat. Med. 16, 1120–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bull L. N., van Eijk M. J., Pawlikowska L., DeYoung J. A., Juijn J. A., Liao M., Klomp L. W., Lomri N., Berger R., Scharschmidt B. F., Knisely A. S., Houwen R. H., and Freimer N. B. (1998) A gene encoding a P-type ATPase mutated in two forms of hereditary cholestasis. Nat. Genet. 18, 219–224 [DOI] [PubMed] [Google Scholar]
- 46.Sanyal S., Frank C. G., and Menon A. K. (2008) Distinct flippases translocate glycerophospholipids and oligosaccharide diphosphate dolichols across the endoplasmic reticulum. Biochemistry 47, 7937–7946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuchs Y., and Steller H. (2011) Programmed cell death in animal development and disease. Cell 147, 742–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruggiero L., Connor M. P., Chen J., Langen R., and Finnemann S. C. (2012) Diurnal, localized exposure of phosphatidylserine by rod outer segment tips in wild-type but not Itgb5−/− or Mfge8−/− mouse retina. Proc. Natl. Acad. Sci. U.S.A. 109, 8145–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kevany B. M., and Palczewski K. (2010) Phagocytosis of retinal rod and cone photoreceptors. Physiology 25, 8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young R. W. (1967) The renewal of photoreceptor cell outer segments. J. Cell Biol. 33, 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Halleck M. S., Lawler JF, J. R., Blackshaw S., Gao L., Nagarajan P., Hacker C., Pyle S., Newman J. T., Nakanishi Y., Ando H., Weinstock D., Williamson P., and Schlegel R. A. (1999) Differential expression of putative transbilayer amphipath transporters. Physiol. Genomics 1, 139–150 [DOI] [PubMed] [Google Scholar]
- 52.Kim Y. E., Chen J., Chan J. R., and Langen R. (2010) Engineering a polarity-sensitive biosensor for time-lapse imaging of apoptotic processes and degeneration. Nat. Methods 7, 67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schuldiner O., and Yaron A. (2015) Mechanisms of developmental neurite pruning. Cell. Mol. Life Sci. 72, 101–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flesch F. M., and Gadella B. M. (2000) Dynamics of the mammalian sperm plasma membrane in the process of fertilization. Biochim. Biophys. Acta 1469, 197–235 [DOI] [PubMed] [Google Scholar]
- 55.Dillon S. R., Mancini M., Rosen A., and Schlissel M. S. (2000) Annexin V binds to viable B cells and colocalizes with a marker of lipid rafts upon B cell receptor activation. J. Immunol. 164, 1322–1332 [DOI] [PubMed] [Google Scholar]
- 56.Fischer K., Voelkl S., Berger J., Andreesen R., Pomorski T., and Mackensen A. (2006) Antigen recognition induces phosphatidylserine exposure on the cell surface of human CD8+ T cells. Blood 108, 4094–4101 [DOI] [PubMed] [Google Scholar]
- 57.Marsault R., Murgia M., Pozzan T., and Rizzuto R. (1997) Domains of high Ca2+ beneath the plasma membrane of living A7r5 cells. EMBO J. 16, 1575–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rizzuto R., Brini M., Murgia M., and Pozzan T. (1993) Microdomains with high Ca2+ close to IP3-sensitive channels that are sensed by neighboring mitochondria. Science 262, 744–747 [DOI] [PubMed] [Google Scholar]
- 59.Song J., Tan H., Shen H. B., Mahmood K., Boyd S. E., Webb G. I., Akutsu T., and Whisstock J. C. (2010) Cascleave: towards more accurate prediction of caspase substrate cleavage sites. Bioinformatics 26, 752–760 [DOI] [PubMed] [Google Scholar]