Abstract
In response to environmental cues, the mitogen-activated protein kinase Sty1-driven signaling cascade activates hundreds of genes to induce a robust anti-stress cellular response in fission yeast. Thus, upon stress imposition Sty1 transiently accumulates in the nucleus where it up-regulates transcription through the Atf1 transcription factor. Several regulators of transcription and translation have been identified as important to mount an integral response to oxidative stress, such as the Spt-Ada-Gcn5-acetyl transferase or Elongator complexes, respectively. With the aim of identifying new regulators of this massive gene expression program, we have used a GFP-based protein reporter and screened a fission yeast deletion collection using flow cytometry. We find that the levels of catalase fused to GFP, both before and after a threat of peroxides, are altered in hundreds of strains lacking components of chromatin modifiers, transcription complexes, and modulators of translation. Thus, the transcription elongation complex Paf1, the histone methylase Set1-COMPASS, and the translation-related Trm112 dimers are all involved in full expression of Ctt1-GFP and in wild-type tolerance to peroxides.
Keywords: catalase, gene expression, Schizosaccharomyces pombe, stress response, transfer RNA (tRNA)
Introduction
Cells adapt to changing environments by activating signaling pathways and modifying the cellular gene expression programs. Often, those signals are stressful and transiently or permanently halt cell growth. In particular, reactive oxygen species such as hydrogen peroxide (H2O2) trigger what is known as oxidative stress. In fission yeast, >550 types of transcripts are up-regulated >2-fold in response to peroxides, and the expression of up to 450 genes is down-regulated also by >2-fold (1). This severe change in the Schizosaccharomyces pombe transcriptome is ruled by the mitogen-activated protein kinase Sty1-driven signaling cascade, which becomes activated upon different stress signals such as H2O2 to allow adaptation and survival.
Upon activation, Sty1 accumulates in the nucleus and orchestrates this massive change in the cell gene expression program. The downstream transcription factor Atf1 is important for this response. Atf1 is phosphorylated by Sty1 in a stress-dependent manner, and transcription of genes occurs. We and others have demonstrated that the SAGA4 complex is recruited to stress genes in a Sty1- and Atf1-dependent manner (2, 3) and participate in chromatin remodeling along stress open reading frames to allow RNA polymerase II (Pol II) initiation and elongation.
For this dramatic change in the gene expression program to occur, not only the Pol II transcriptional machinery but also the translation process of the newly induced stress mRNAs has to be coordinated (4) and also be robust. Thus, we recently described that mutations in Elongator components impair wild-type tolerance to peroxides, probably by decreasing the amount of proteins synthesized from the stress mRNAs (5). Elongator introduces a modification of the anticodon loop of the low copy number tRNAUUULys as well as in other two types of tRNAs with a uracil 34 (U34) in the 5′ position of the anticodon (6, 7). Modification of tRNAUUULys by Elongator (introducing an acetyl group at position 5 of U34 (cm5U34)) and by Ctu1-Ctu2 complex (catalyzing the thiolation at carbon 2 of U34 (s2U34)) is required for the efficient recognition by this tRNA of the codon AAA and, therefore, for optimal performance under situations requiring a robust translation process, such as abrupt changes in the gene expression program during adaptation to stress (5).
Here, we have performed a genome-wide search of S. pombe genes required for wild-type adaptation to stress. We recently demonstrated that overexpression of catalase (Ctt1) is essential for the cellular response to oxidative stress, as ectopic expression of only this protein totally or partially suppresses the absence of components of the Sty1 or Pap1 cascades (8). Using an optimized protocol combining green fluorescent protein (GFP) fluorescence and flow cytometry, we have isolated all genes affecting the levels of expression of Ctt1-GFP expressed from the ctt1 locus. Besides confirming the role of the stress Pap1 and Sty1 cascades as well as of SAGA on Ctt1-GFP expression, we also isolated complexes promoting robust transcription, such as Mediator, Set1-COMPASS, and Paf1 complexes. We centered our attention on the characterization of Trm112, a protein reported in Saccharomyces cerevisiae to heterodimerize and regulate the activity of methylases such as Trm9, Mtq2, Trm11, and Bud23. As will be shown here, the combination of Trm112 with Trm9 and Mtq2 will contribute to fission yeast tolerance to peroxides, probably by promoting efficient translation; the dimer Trm112-Trm9 introduces the last modification at U34 of the tRNAUUULys, a methyl group, to yield mcm5s2U34; Trm112-Mtq2 may methylate a translation release factor, eRF1, to promote efficient translation termination. Our experiments demonstrate that hundreds of genes are required to coordinate the transcription and translation machineries for the acquisition of tolerance to oxidative stress.
Experimental Procedures
Growth Conditions, Plasmids, and Yeast Strains
Cells were grown in rich medium (YES) or synthetic minimal medium as described previously (9). Origins and genotypes of strains used in this study are outlined in Table 1. To tag ctt1 with GFP, we transformed wild-type strain (975, h+) with a linear fragment containing 3′ end of ctt1-GFP::natMX6, obtained by PCR amplification using ctt1 specific primers and the plasmid pFA6a-GFP::natMX6 (10), yielding strain PG102. To generate the strain PG111 used to cross the deletion collection, we crossed the strain PG102 with P392 (11); this strategy (PEM-2, or pombe epistatic mapping) takes advantage of a recessive, cycloheximide-resistance mutation that eases the high throughput double mutant generation between our reporter strain and the Bioneer deletion collection (11). The specific gene deletions of the strains selected in our screen and used thereafter (see below; Paf1, Set1, Trm112 complexes) were confirmed by PCR. To construct S. pombe strains without nutritional requirements and with or without the chimeric ctt1-GFP gene, selected deletion mutants from our genetic screen (see below) were crossed with 972 (wild type) in minimal medium plates without nitrogen followed by germination in YES medium and haploid selection in plates containing G-418 (to select the specific gene deletion) or nourseothricin (NAT) (to select or not for ctt1-GFP) and minimal medium (to select for the absence of requirements). We used integrative plasmids p428 and p428AAG to express HA-Atf1 and HA-Atf1AAG, where all 11 lysine AAA codons or the atf1 open reading frame are replaced by AAG (5). The two plasmids were inserted at the leu1–32 loci of different strain backgrounds, yielding strains JF91, JF94, PG136, PG138, PG139, and PG140. We used the following episomal plasmids containing or not tRNA coding genes: pREP.42x, p465 (expressing tRNAUUULys) and p466 (expressing tRNACUULys) (5).
TABLE 1.
Strains used in this study
| Strain | Genotype | Origin |
|---|---|---|
| 972 | h− | (38) |
| 975 | h+ | (38) |
| IV16 | h+ elp3/sin3::natMX6 | (5) |
| JF77 | h− elp3/sin3::kanMX6 leu1-32 ura4D-18 | (5) |
| AV18 | h− sty1::kanMX6 | (39) |
| EP198 | h+ ctt1::natMX6 | (8) |
| PG102 | h+ ctt1-GFP:natMX6 | This work |
| ES10 | h? ctt1-GFP:natMX6 sty1::kanMX6 leu1-32 | This work |
| P392 | h− ade6-M210 leu1-32 ura4-D18 mat1_m-cyhS smt0 rpl42::cyhR (sP56Q) | (11) |
| PG111 | h− ctt1-GFP::natMX6 mat1_m-cyhS smt0 rpl42::cyhR (sP56Q) | This work |
| MS98 | h− atf1::natMX6 | (5) |
| JE21 | h+ prf1/rtf1::kanMX6 | This work |
| JE22 | h+ leo1::kanMX6 | This work |
| JE23 | h− mtq2::kanMX6 | This work |
| JE24 | h+ lys9::kanMX6 | This work |
| JE25 | h− trm9::kanMX6 | This work |
| JE26 | h− paf1::kanMX6 | This work |
| JE27 | h− shg1::kanMX6 | This work |
| JE28 | h− set1::kanMX6 | This work |
| JE29 | h− swd1::kanMX6 | This work |
| JE30 | h− swd3::kanMX6 | This work |
| JE31 | h− spf1::kanMX6 | This work |
| JE32 | h+ ash2::kanMX6 | This work |
| JE33 | h− trm11::kanMX6 | This work |
| JE34 | h+ trm112::kanMX6 | This work |
| PG141 | h− bud23::kanMX6 | This work |
| JE37 | h+ mtq2::kanMX6 ctt1-GFP::natMX6 | This work |
| JE38 | h− lys9::kanMX6 ctt1-GFP::natMX6 | This work |
| JE39 | h− trm9::kanMX6 ctt1-GFP::natMX6 | This work |
| JE41 | h− shg1::kanMX6 ctt1-GFP::natMX6 | This work |
| JE42 | h− set1::kanMX6 ctt1-GFP::natMX6 | This work |
| JE43 | h− swd1::kanMX6 ctt1-GFP::natMX6 | This work |
| JE44 | h− swd3::kanMX6 ctt1-GFP::natMX6 | This work |
| JE45 | h− spf1::kanMX6 ctt1-GFP::natMX6 | This work |
| JE46 | h+ ash2::kanMX6 ctt1-GFP::natMX6 | This work |
| JE47 | h− trm11::kanMX6 ctt1-GFP::natMX6 | This work |
| JE48 | h− trm112::kanMX6 ctt1-GFP::natMX6 | This work |
| PG163 | h− bud23::kanMX6 ctt1-GFP::natMX6 | This work |
| JE49 | h− trm9::kanMX6 ura4-D18 | This work |
| JE52 | h+ trm112::kanMX6 ura4-D18 | This work |
| JE55 | h+ trm9::kanMX6 sin3::natMX6 | This work |
| JE57 | h− trm112::kanMX6 sin3::natMX6 | This work |
| JF91 | h− leu1-32 sty1′-HA-atf1::leu1 | (5) |
| JF94 | h− leu1-32 sty1′-HA-atf1AAG::leu1 | (5) |
| PG139 | h+ trm9::kanMX6 leu1-32 sty1'-HA-atf1::leu1 | This work |
| PG140 | h− trm112::kanMX6 leu1-32 sty1'::HA-atf1::leu1 | This work |
| PG136 | h− trm9::kanMX6 leu1-32 sty1'::HA-atf1AAG::leu1 | This work |
| PG138 | h− trm112::kanMX6 leu1-32 sty1'::HA-atf1AAG::leu1 | This work |
H2O2 Sensitivity Assay
For survival on solid plates, S. pombe strains were grown, diluted, and spotted on YES plates containing or not H2O2 at the indicated concentrations as described previously (3).
ctt1-GFP Deletion Library Construction and Flow Cytometry Analysis
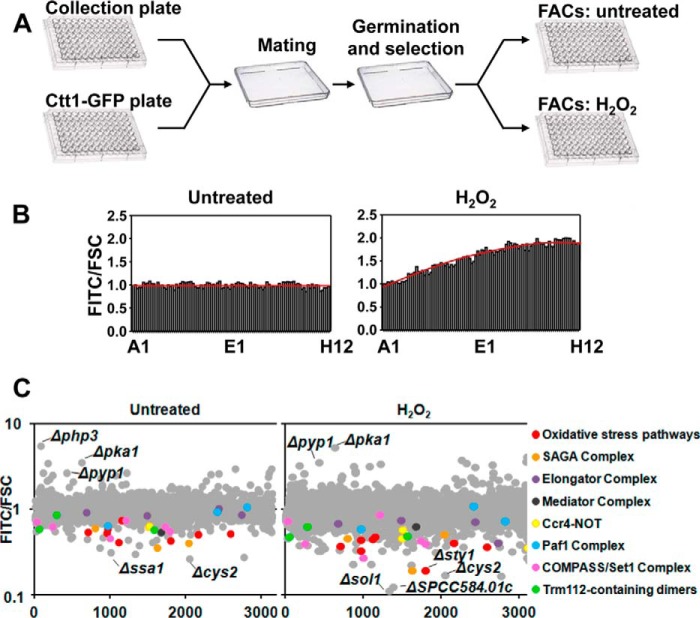
We crossed strain PG111 (ctt1-GFP) with the haploid deletion collection by Bioneer Corp. (version 2), which contains 3004 haploid deletion strains comprising ∼71% of non-essential fission yeast genes, following the protocol proposed by Roguev et al. (11). Briefly, the Bioneer deletion library [h+ leu1–32 ura4-D18 ade6-M210 (or M216) Δgene::kanMX4], which was frozen at −80 °C in 96-well plates, was first transferred to solid YES plates using a 96 pin replicator and incubated at 30 °C for 48 h. The deletion library was then transferred to a minimal medium plate without nitrogen (to promote mating) and mixed with the NAT-marked ctt1-GFP (PG111) and allowed to cross for 4 days at 25 °C followed by germination in YES and haploid selection in plates containing NAT (to select ctt1-GFP), G-418 (to select deletion genes) and cycloheximide (to select haploid cells). The resulting library was transferred using a 96 pin replicator to 96-well microtiter plates containing 200 μl of liquid YES and incubated at 30 °C for 24 h. Then the plates were diluted and incubated overnight to perform flow cytometry analysis during logarithmic growth in untreated condition or after 1 h with 1 mm H2O2 at 30 °C. Flow cytometry analysis was performed using FACScanto equipped with 488- and 633-nm lasers (BD Biosciences), and all flow cytometry data were analyzed with FACSDiva software (BD Biosciences). Ctt1-GFP fluorescence was detected using the FITC green channel, and the final data obtained were expressed as a FITC/FSC (forward scatter) normalized using the mean of each plate. Due to the fact that Ctt1 fluorescence increased after H2O2 treatment during the 96-well plate reading, we normalized the results using the equation y = −0.00009x2 + 0.02x + 0.9137, obtained from flow cytometry analysis of a plate containing the control strain PG111 (ctt1-GFP) in all wells after 60 min of H2O2 treatment (Fig. 2B). The STRING database (12) was used to identify different protein complexes reveled in our genetic screen. STRING is a database that includes known and predicted protein associations from different sources and which assigns normalized confidence scores to many different types of protein associations: some from experiments (physical and genetic protein interactions) or derived from co-expression and others either inferred by literature annotation or transferred from homology.
FIGURE 2.
Flow cytometry-based screening of a fission yeast deletion collection using fluorescence of Ctt1-GFP as a reporter. A, schematic representation of the construction and flow cytometry analysis of the Ctt1-GFP-expressing library of deletion strains. B, rich media cultures of strain PG102 (ctt1-GFP), either untreated or treated with 1 mm H2O2 for 60 min, were distributed in the 96 wells of a flow cytometry plate, and fluorescence along the complete plate was determined. Results are expressed as FITC/FCS ratio, normalized to the values at well number 1, with an assigned value of 1. A trend line is drawn in red. C, graphs show Ctt1-GFP FITC/FCS ratio corresponding to all deletion collection strains in a logarithmic scale, either untreated (left) or treated with 1 mm H2O2 for 60 min (right). Components of some complexes are differentially colored. The names of some deletion strains with the lowest or highest levels of Ctt1-GFP fluorescence are indicated.
RNA Analysis
Total RNA from S. pombe YES cultures was obtained, processed, transferred to membranes, and hybridized with radioactivity-labeled probes as described previously (13). Quantification of the levels of tRNA overexpression in strains carrying episomal tRNA plasmids was performed as described before (5).
Growth Curves
Yeast cells were grown in YES from an initial absorbance (A600) of 0.1 with or without the addition of 2 mm H2O2 using an assay based on automatic measurements of optical densities as previously described (14). For each strain, we calculated the “arrest after H2O2” by subtracting the minutes required to reach an A600 of 0.5 between 2 mm H2O2-treated and untreated cultures.
S. pombe Trichloroacetic Acid (TCA) Extracts and Immunoblot Analysis
Modified TCA extracts were prepared as previously described (15). Atf1 and Ctt1-GFP were immunodetected with polyclonal anti-Atf1 (3) and anti-GFP (16) antisera. HA-Atf1 was detected with in-house monoclonal anti-HA antibody. Anti-Sty1 polyclonal antibodies (17) and monoclonal anti-tubulin (Sigma) were used as loading controls. Relative quantification of protein levels in Western blots was performed using the Image StudioTM 4.0 software.
Results
Characterization of a Reporter of the Sty1-Atf1 Cascade
We designed a global approach to isolate all S. pombe genes affecting the change in gene expression program driven to survive against oxidative stress. We chose accumulation of catalase, encoded by the ctt1 gene, as an indicator of both basal and H2O2-dependent levels. Indeed, the levels of ctt1 mRNA are increased by 35-fold 60 min after stress imposition (0.5 mm H2O2 in the growth media) (18). Furthermore, the basal expression of ctt1 in cells lacking the mitogen-activated protein kinase Sty1 or its downstream transcription factor Atf1 are 4–5-fold times lower than in wild-type cells (18). We inserted a GFP-coding sequence downstream of the chromosomal ctt1 locus (Fig. 1A) and determined that the cells expressing the protein chimera did not display any oxidative stress defect (Fig. 1B), suggesting that the protein is fully functional. Accumulation of the mRNA was similar to that of untagged ctt1 (Fig. 1C). The steady-state levels of Ctt1-GFP protein increased with time after stress imposition as determined in extracts by Western blot (Fig. 1D). Fluorescence also increased in live cells after H2O2 stress in a wild-type background as determined by flow cytometry, whereas basal and H2O2-induced fluorescence was clearly diminished in cells lacking Sty1 (Fig. 1E).
FIGURE 1.
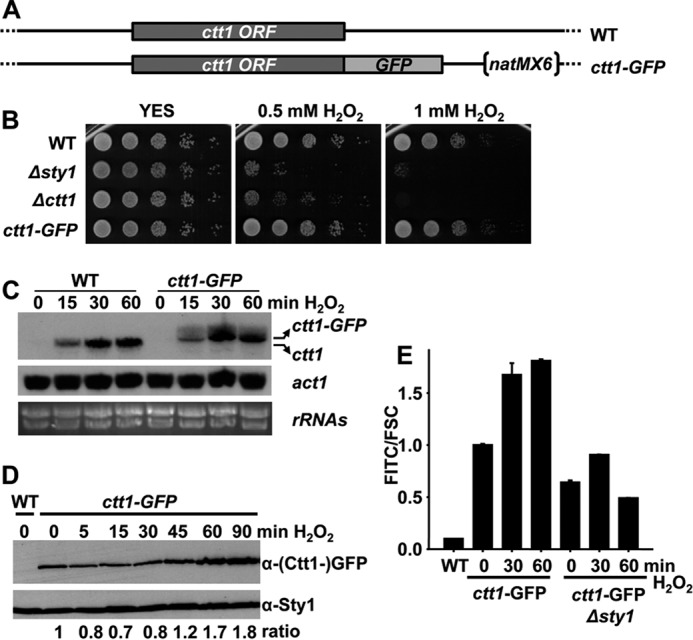
Ctt1-GFP is a good reporter of the oxidative stress response. A, schematic representation of chromosomal ctt1 tagged with GFP. B, ctt1-GFP strain is not sensitive to oxidative stress. Serial dilutions from cultures of strains 972 (WT), AV18 (Δsty1), EP198 (Δctt1), and PG102 (ctt1-GFP) were spotted onto rich plates without (YES) or with H2O2. C, relative levels of ctt1 mRNA by Northern blot. Total RNA from strains 972 (WT) and EP102 (ctt1-GFP), either untreated (0) or treated with 1 mm H2O2 for the indicated times, was analyzed by Northern blot with probes for ctt1. rRNA and act1 are shown as loading controls. D, rich media cultures of strains 972 (WT) and PG102 (ctt1-GFP), either untreated (0) or treated with 1 mm H2O2 for the indicated times, were analyzed to determine Ctt1-GFP protein levels by Western blot using polyclonal antibody against GFP. Sty1 is shown as a loading control. E, rich media cultures of strains 972 (WT), PG102 (ctt1-GFP), and ES10 (ctt1-GFP Δsty1), either untreated (0) or treated with 1 mm H2O2 for the indicated times, were analyzed to determine Ctt1-GFP fluorescence levels by flow cytometry. Results are expressed as FITC/FCS ratio, normalized to the levels of untreated wild-type ctt1-GFP cells, with an assigned value of 1. S.D. were calculated from biological duplicates.
Flow Cytometry-based Screening of a Fission Yeast Deletion Collection Using Ctt1-GFP as a Reporter
To isolate all fission yeast genes that affect Ctt1-GFP levels, we constructed a deletion library crossing the Bioneer deletion collection with PG111, an S. pombe strain carrying the ctt1-GFP allele as a reporter in a P392 background, which take advantage of a recessive cycloheximide-resistance mutation to facilitate selection of haploid cells (11). Briefly, the PG111 strain was crossed with the deletion library in minimal medium plates without nitrogen followed by germination and haploid selection (Fig. 2A). We failed to obtain the targeted segregants for 165 deletion mutants. Once the ctt1-GFP deletion library was constructed, each plate was analyzed by flow cytometry to determine the fluorescence level of Ctt1-GFP treated or not with H2O2. We used a FITC filter to measure GFP fluorescence, and the results are expressed as FITC/FCS ratio, FCS being proportional to cell surface area or size. We observed that the Ctt1 protein level increases with time after stress imposition at least until 90 min of treatment (Fig. 1D). Accordingly, Ctt1 fluorescence increased after H2O2 treatment during the analysis by flow cytometry of the 96-well plates, which took ∼40 min. To solve this problem we analyzed the fluorescence by flow cytometry of a plate containing the control strain PG111 (ctt1-GFP) in all 96 wells in untreated and stress conditions (Fig. 2B). We normalized the data, giving the value 1 to the first position of the plate, and obtained a trend line (Fig. 2B, red line) whose equation for the H2O2 plate was used to normalize the results to abolish reading differences due to the position occupied by each strain in the plate. Our flow cytometry-based screen revealed that >100 mutants are Ctt1-GFP up-regulated and 88 are down-regulated >1.5-fold under untreated conditions. After stress, numbers increased to reach >160 up-regulated and >280 down-regulated also by >1.5-fold. Top 150 genes with the lowest values for Ctt1-GFP fluorescence were selected from the screen and loaded in STRING database to search for protein complexes (12). Gene deletions coding for most known subunits of the oxidative stress pathway or of the SAGA complex were identified as well as different components of Ccr4-NOT, Mediator, Set1-COMPASS, and Paf1 complexes as well as of the Trm112-containing dimers (Fig. 2C).
The SAGA, Paf1, and Set1 Complexes Are Revealed in the Genetic Screen and Regulate Transcription of Stress Genes
As described above, most components of the Sty1 and Pap1 cascades were highlighted by our flow cytometry-based screen. Similarly, deletion of genes coding for members of the SAGA complex such as Gcn5, Ada2, or Ngg1 also displayed lower levels of Ctt1-GFP fluorescence both under treated and untreated conditions (Table 2). Among others, deletion of genes coding for components of the Paf1 complex also had lower levels of fluorescence as shown in Table 2.
TABLE 2.
Complexes with low levels of Ctt1-GFP under basal and H2O2-treated conditions
| Name of the complex or pathway | Deletion mutants revealed in the screen | Levels of Ctt1-GFP (unt./H2O2)a |
|---|---|---|
| Oxidative stress pathways | Δsty1 | 0.44/0.19 |
| Δpap1 | 0.75/0.48 | |
| Δcsx1 | 0.41/0.45 | |
| Δatf1 | 0.53/0.37 | |
| Δwin1 | 0.60/0.45 | |
| Δwis4 | 0.54/0.33 | |
| Δpcr1 | 0.51/0.41 | |
| Δmcs4 | 0.56/0.38 | |
| SAGA complex | Δgcn5 | 0.41/0.51 |
| Δada2 | 0.60/0.47 | |
| Δngg1 | 0.36/0.19 | |
| Elongator complex | Δelp3 | 0.85/0.75 |
| Δelp6 | 0.87/0.40 | |
| Δelp4 | 1.02/0.73 | |
| Δiki3 | 0.93/0.69 | |
| Mediator complex | Δsep11/pmc6 | 0.54/0.63 |
| Paf1 complex | Δpaf1 | 1.07/0.74 |
| Δrtf1/prf1 | 0.64/0.59 | |
| Δleo1 | 0.94/1.09 | |
| COMPASS/Set1 complex | Δset1 | 0.47/0.28 |
| Δspf1 | 0.56/0.40 | |
| Δswd1 | 0.64/0.44 | |
| Δswd3 | 0.64/0.40 | |
| Δshg1 | 0.73/0.74 | |
| Δash2 | 0.75/0.88 | |
| Trm112-containing dimers | Δtrm112 | 0.60/0.59 |
| Δmtq2 | 0.58/0.50 | |
| Δtrm9 | 0.87/0.64 |
a Levels of Ctt1-GFP fluorescence in Δgene cells under untreated/H2O2-treated conditions normalized to wild-type cells, with an assigned value at both untreated and treated conditions of 1.
The Pol II-associated Paf1 complex (polymerase associated factors), linked to active transcription, serves as a platform to coordinate the association between the transcriptional machinery with transcription factors, histone modifiers, elongating complexes, and termination/cleavage/3′ processing (for review, see Ref. 19). The Paf1 complex associates with phosphorylated and unphosphorylated Pol II, and it is found near promoters and along the ORFs of genes and detaches near the poly(A) site. It has been suggested that the Paf1 complex couples histone modifications to efficient transcription elongation. In particular, during transcription elongation Paf1 directly or indirectly facilitates recruitment of enzymes that mediate Pol II phosphorylation at Ser2 and Set2-dependent methylation of H3K36 (20), ubiquitination of H2B, and Set1-dependent methylation of H3 at lysine 4 (21–24).
In fission yeast, deletion of either prf1/rtf1, paf1, or leo1 genes decreases tolerance to oxidative stress on solid plates (Fig. 3A). We also determined the growth curves of wild-type and mutant cells in liquid cultures in the presence or absence of peroxides and determined for each strain the time of arrest after H2O2 as described under “Experimental Procedures.” We used cells lacking the transcription factor Atf1 as a control of cells with a pronounced arrest under H2O2 conditions. As shown in Fig. 3B, cells lacking Prf1, Paf1, or Leo1 display elongated time arrest after H2O2 stress compared with wild-type cells. This seems to indicate that the Paf1 complex exerts a positive role on transcription. Of the three deletion strains tested (lacking Prf1/Rtf1, Paf1, or Leo1-coding genes), the loss of Prf1/Rtf1 affects tolerance to H2O2 to a lower extent than a lack of the other two components, consistent with a recent report indicating that this protein may not belong to the core of the Paf1 complex in S. pombe (25).
FIGURE 3.
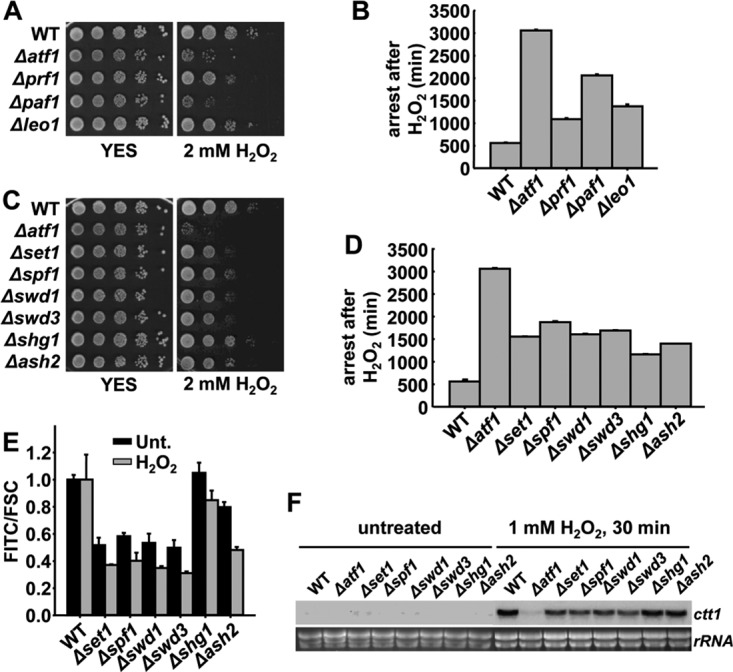
Paf1 and Set1 complexes regulate the levels of ctt1-GFP mRNA. A, deletion strains of Paf1 complex subunits are sensitive to oxidative stress. Serial dilutions from cultures of strains 972 (WT), MS98 (Δatf1), JE21 (Δprf1), JE23 (Δpaf1), and JE22 (Δleo1) were spotted onto rich plates without (YES) or with 2 mm of H2O2. B, arrest in the growth of the same strains as in A was calculated by subtracting the time required to reach an A600 of 0.5 in the presence or absence of 2 mm of H2O2. C, deletion strains of Set1 complex subunits are sensitive to oxidative stress. Serial dilutions from cultures of strains 972 (WT), MS98 (Δatf1), JE28 (Δset1), JE31 (Δspf1), JE29 (Δswd1), JE30 (Δswd3), JE27 (Δshg1), and JE32 (Δash2) were spotted onto rich plates without (YES) or with 2 mm of H2O2. D, arrest in the growth of the same strains as in C was calculated as in B. E, rich media cultures of strains PG111 (WT ctt1-GFP), JE42 (Δset1 ctt1-GFP), JE45 (Δspf1 ctt1-GFP), JE43 (Δswd1 ctt1-GFP), JE44 (Δswd3 ctt1-GFP), JE41 (Δshg1 ctt1-GFP), and JE46 (Δash2 ctt1-GFP) either untreated (Unt.) or treated with 1 mm H2O2 for 60 min were analyzed to determine Ctt1-GFP fluorescence levels by flow cytometry. Results are expressed as FITC/FCS ratio, normalized to the levels of untreated or treated wild-type cells, with an assigned value of 1. F, relative levels of ctt1 mRNA by Northern blot. Total RNA from strains as in C was analyzed by Northern blot with probes for ctt1. rRNA is shown as loading control. B, D, and E, S.D. were calculated from biological duplicates.
A classical mark of active chromatin, methylation of histone H3 at lysine 4, is introduced by the Set1 complex, which has also been highlighted in our genetic screening (Table 2). As shown above for deletion strains of Paf1 components, cells lacking Set1 or other components of the complex also displayed impaired tolerance to peroxides, both on solid plates (Fig. 3C) and in liquid media (Fig. 3D). At least in S. cerevisiae, recruitment of the Set1-COMPASS complex in the transition from initiation to elongation of Pol II genes is mediated by Paf1 complex and promotes the H3-lysine 4-methylation mark close to the promoters of actively transcribed genes (22, 23, 26). To confirm the results from the genome-wide screening, we measured with regular flow cytometry the ratio of fluorescence to cell size of the individual mutants after eliminating all auxotrophic markers. As shown in Fig. 3E, both untreated and H2O2-treated levels of Ctt1-GFP fluorescence were reduced in cells lacking Set1, Spf1, Swd1, and Swd3, with moderate reductions for strains lacking Shg1 or Ash2. Concomitantly, H2O2-induced levels of ctt1 mRNA in cells lacking Shg1 or Ash2 were only moderately reduced compared with wild-type cells (Fig. 3F).
The Elongator and Trm112 Complexes Are Revealed in the Genetic Screen
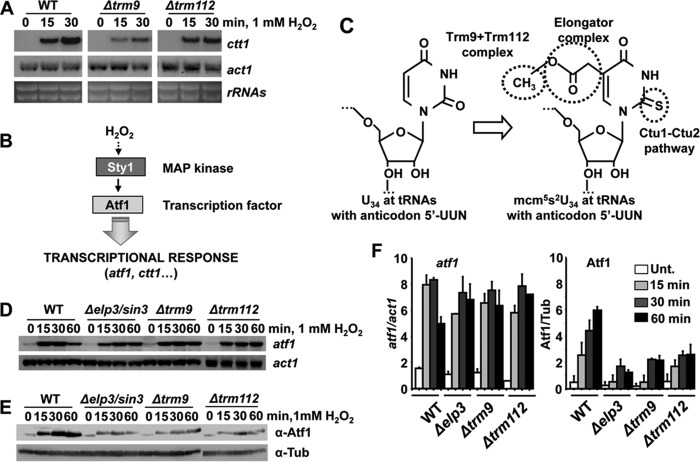
We recently reported that Elongator, a tRNA-modifying complex promoting efficient translation, was required for wild-type tolerance to peroxides (5). Indeed, our genetic screen also indicates that deletion of genes coding for different Elongator components express lower levels of basal or induced Ctt1-GFP (Table 2).
Interestingly, our screening also highlighted that Trm112 and some of its partners (Mtq2 and Trm9, specifically) are also required for wild-type expression of Ctt1-GFP, as shown in Fig. 4A for strains without auxotrophies carrying the indicated gene deletions and the ctt1-GFP chimera. In S. cerevisiae, Trm112 is an auxiliary protein that dimerizes with another subunit to form an active methylase. These two-subunit enzymes modify nucleotides in RNA chains of specific tRNAs (in the case of Trm112-Trm9 or Trm112-Trm11; Fig. 4B), rRNAs (as in the case of Trm112 + Bud23), or in the translation release factor eRF1 (Trm112 + Mtq2; Fig. 4B). As shown in Fig. 4CD, S. pombe strains lacking Trm112 or two of its partners, Mtq2 or Trm9, displayed sensitivity to H2O2 both on solid plates and in liquid cultures (Fig. 4, C and D).
FIGURE 4.
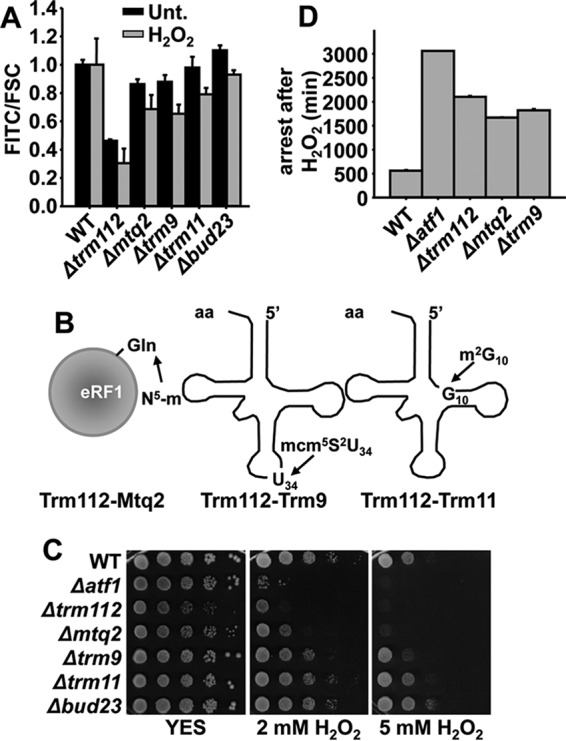
Trm112 and some of its partners display defects in oxidative stress tolerance. A, rich media cultures of strains PG111 (WT ctt1-GFP), JE48 (Δtrm112 ctt1-GFP), JE37 (Δmtq2 ctt1-GFP), JE39 (Δtrm9 ctt1-GFP), JE47 (Δtrm11 ctt1-GFP), and PG163 (Δbud23 ctt1-GFP) either untreated (Unt.) or treated with 1 mm H2O2 for 60 min were analyzed to determine Ctt1-GFP fluorescence level by flow cytometry. Results are expressed as a FITC/FCS ratio, normalized to the levels of untreated or treated wild-type cells, with an assigned value of 1. B, scheme illustrating tRNA modifications that require Trm112-methylase dimers. C, serial dilutions from cultures of strains 972 (WT), MS98 (Δatf1), JE34 (Δtrm112), JE23 (Δmtq2), JE25 (Δtrm9), JE33 (Δtrm11), and PG141 (Δbud23) were spotted onto rich plates without (YES) or with 2 mm and 5 mm of H2O2. D, arrest in the growth of strains 972 (WT), MS98 (Δatf1), JE34 (Δtrm112), JE23 (Δmtq2), and JE25 (Δtrm9) was calculated as described in Fig. 3B. A and D, S.D. were calculated from biological duplicates.
Cells Lacking Trm112, Mtq2, or Trm9 Display Defects in Oxidative Stress Tolerance
Cells lacking Trm112 displayed severe growth defects, as exemplified by spotting these cells on rich media plates (Fig. 4C). Probably, this “hub” protein participates in several cellular processes, many of them linked to translation (27), and the phenotype of Δtrm112 cells is the sum of the lack of different enzymatic activities. Only two of its putative dimers, Mtq2 and Trm9 (not previously characterized in fission yeast) display decreased tolerance to H2O2. The S. cerevisiae Trm112-Mtq2 dimer has been described to introduce an N5 methylation at a glutamine residue of eukaryotic release factor 1 (eRF1) (28). To confirm the possible participation of S. pombe Mtq2 in eRF1/Sup45/SPBC18B5.06 modification, we tried to complement the sensitivity of Δmtq2 cells to grow in the presence of peroxides with a plasmid allowing overexpression of fission yeast eRF1, without success.
We focused our attention of the role of the Trm112-Trm9 dimer in H2O2 tolerance. We first determined that not only the H2O2-dependent expression of the protein Ctt1-GFP but also of the ctt1 mRNA was slightly impaired in cells lacking either Trm9 or Trm112 (Fig. 5A). As described in the Introduction, transcription of the ctt1 gene depends on the mitogen-activated protein kinase Sty1 and its downstream transcription factor Atf1, and the gene coding for this transcription factor is also under the control of the Sty1-driven regulatory cascade (Fig. 5B). At least in S. cerevisiae, the methyl group introduced by Trm112-Trm9 is the third step (after 5-acetylation and 2-thiolation) in the generation of the mcm5S2U34 modification at the tRNAs with an anticodon 5′-UUN, decoding lysine, glutamine, and glutamic acid (Fig. 5C). As shown before for cells lacking the Elongator component Elp3 (5), although the H2O2-dependent mRNA levels of atf1 are not significantly altered in strains Δtmr9 or Δtmr112 (Fig. 5, D and F), protein levels are severely affected (Fig. 5, E and F).
FIGURE 5.
Trm112-Trm9 is necessary for efficient Atf1 translation upon oxidative stress. A, deletion of trm9 or trm112 affects transcription of ctt1. Cultures of strains 972 (WT), JE25 (Δtrm9), and JE34 (Δtrm112) were treated with 1 mm H2O2 for the indicated times. Total RNA was analyzed by Northern blot with probes for ctt1 and act1. rRNA and act1 were used as loading controls. B, scheme illustrating the activation of the stress gene expression program by Sty1 and Atf1. MAP, mitogen-activated protein. C, Trm9 and Trm112 methyltransferases are responsible for formation of the terminal methyl group of mcm5U34 (5-methoxycarbonylmethyluridine). D, absence of Sin3/Elp3, Trm9, or Trm112 barely affects transcription of the atf1 gene. Cultures of strains 972 (WT), IV16 (Δelp3/sin3), JE25 (Δtrm9), and JE34 (Δtrm112) were treated with 1 mm H2O2 for the indicated times. Total RNA was analyzed by Northern blot with probes for atf1 and act1. E, absence of Sin3/Elp3, Trm9, or Trm112 affects Atf1 protein levels. Strains as in D were analyzed by Western blot using polyclonal anti-Atf1 antibodies. Anti-tubulin was used as the loading control. F, quantification of the relative mRNA (left) and protein levels (right) of the blots in D and E, respectively. Unt., untreated. S.D. were calculated from biological duplicates.
The Stress Phenotypes of Cells Lacking Trm9 Are Suppressed by Overexpression of tRNAUUULys
The mcm5S2U34 modification at the tRNAs with an anticodon 5′-UUN has been proposed to be required for translation efficiency and/or fidelity of mRNAs containing 5′-NAA codons (29, 30). We demonstrated before that the H2O2 sensitivity of cells lacking the Elongator component Elp3 can be alleviated by overexpressing only one of its target tRNAs, specifically tRNAUUULys but not by the control tRNACUULys, which recognizes the other codon for lysine (AAG) and is not modified by Elongator (5) (Fig. 6, A and D). To confirm the role of S. pombe Trm9 in methylation of the same target tRNAs as Elongator, we transformed strains Δtrm9 and Δtrm112 with the same two tRNAs and confirmed that only tRNAUUULys was sufficient to suppress the stress defects of cells lacking Trm9 (Fig. 6, B and E). As expected, the pleiotropic defects of cells lacking the hub protein Trm112, caused by deficiencies of up to five different enzymatic activities, could not be suppressed by overexpressing this tRNA (Fig. 6, C and F).
FIGURE 6.
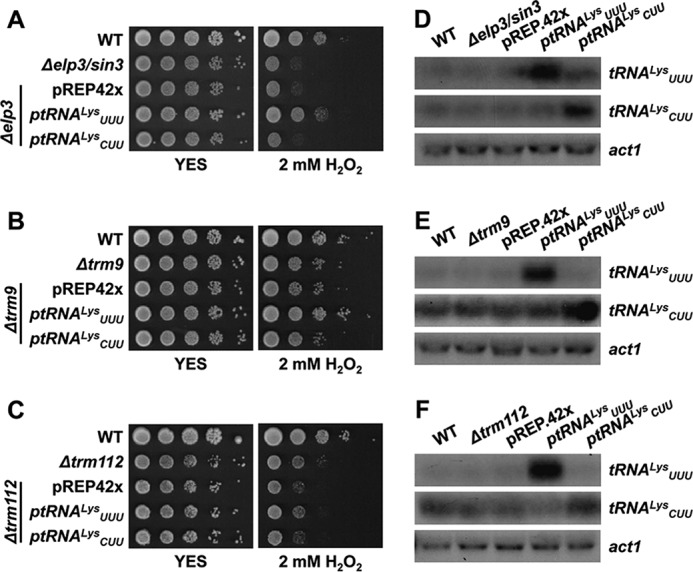
Overexpression of tRNAUUU suppresses the growth defects of Δtrm9 but not of Δtrm112 upon oxidative stress. A, B, and C, serial dilutions from cultures of strains 972 (WT), JF77 (Δelp3/sin3), JE49 (Δtrm9), and JE52 (Δtrm112) transformed with episomal plasmids p465 (ptRNAUUULys), p466 (ptRNACUULys), or the empty vector pREP.42x, were spotted onto rich plates without (YES) or with 2 mm H2O2. D, E, and F, relative levels of tRNA overexpression by Northern blot. Total RNA from the same strains as in A, B, and C was analyzed by Northern blot with probes of dsDNA of the indicated tRNAs labeled at their antisense strand. act1 is shown as a loading control.
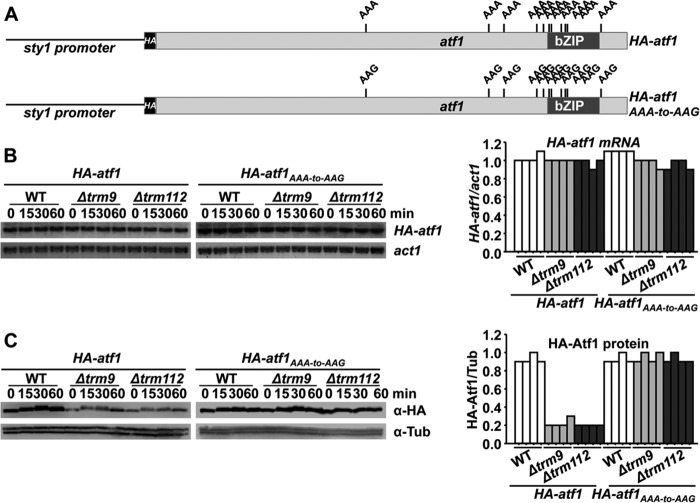
In cells lacking Elongator, to demonstrate that low levels of a stress protein, Atf1, were caused by defective translation of the atf1 mRNA, we used a chimeric, constitutively expressed HA-atf1 gene carrying 11 AAA-to-AAG substitutions so that all lysine codons in the atf1 transcript would not depend on the tRNA modified by Elongator (5) (Fig. 7A). In cells lacking Trm9 or Trm112, expression of both HA-tagged mRNAs was constitutive and identical in both endogenous and mutated atf1 (Fig. 7B). As shown before for Elongator, the defects in Atf1 protein expression displayed by strains Δtrm9 or Δtrm112 were suppressed in cells carrying the HA-atf1AAA-to-AAG chimeric gene (Fig. 7C). We conclude that the Trm9-Trm112 introduces the terminal methyl group of mcm5S2U34 at the anticodon of specific tRNAs, and we propose that the sensitivity to H2O2 of cells lacking Trm9 is caused by defective translation of AAA codons by the tRNAUUULys.
FIGURE 7.
Expression of a synthetic AAA-to-AAG atf1 gene renders wild-type Atf1 protein levels in cell lacking Trm9 or Trm112. A, schematic representation of HA-Atf1 and HA-Atf1AAA-to-AAG proteins, expressed from the constitutive sty1 promoter. The relative positions of the 11 AAA and AAG lysine codons are indicated. B and C, vectors carrying the wild-type (HA-atf1) or mutated atf1 genes (HA-atf1AAA-to-AAG) under the control of a constitutive promoter were integrated in the chromosomes of wild type and Δtrm9 and Δtrm112 strains. Rich media cultures of strains JF91 (sty1′::HA-atf1), JF94 (sty1′::HA-atf1AAA-to-AAG), PG136 (Δtrm9 sty1′::HA-atf1AAA-to-AAG), PG138 (Δtrm112 sty1′::HA-atf1AAA-to-AAG), PG139 (Δtrm9 sty1′::HA-atf1), and PG140 (Δtrm112 sty1′::HA-atf1) either untreated (0) or treated with 1 mm H2O2 for the indicated times were analyzed to determine HA-atf1 mRNA levels by Northern blot using an anti-HA probe (B) or HA-Atf1 protein levels by Western blot using monoclonal antibody against HA (C). The graphs on the right panels indicate the relative levels of HA-atf1/act1 mRNAs (B) or HA-Atf1/tubulin protein levels (C).
Discussion
Upon oxidative stress, the Sty1-Atf1 pathway triggers a major change in the gene expression program, as mitotic cell growth and general metabolism is halted and >500 genes are up-regulated >2-fold to promote survival (18). We have performed here a global screening of genes required to allow such a strong shift in the gene expression program using expression of the antioxidant protein Ctt1-GFP as a reporter of efficient adaptation. We have isolated hundreds of genes required to promote this shift, demonstrating that for optimal performance not only complex chromatin modifications, but also efficient transcription and translation are required to mount a survival response. Future work will be required to determine the contribution of many of those protein functions in the development of the stress program.
So far we have shown that Paf1 and Set1-COMPASS components contribute to Ctt1-GFP expression and to wild-type tolerance to peroxides. The role of the Paf1 complex on efficient Pol-II-dependent transcription elongation and histone modification by Set1 complex has been widely characterized in other eukaryotes (22, 23). However, two recent reports have established connections between the Paf1 complex and the nucleation and spreading of heterochromatin in S. pombe. In the first, Bühler and co-workers (31) demonstrate that fission yeast can trigger heterochromatin formation at a specific locus by expressing a synthetic RNA hairpin homologous to that region but only if the Paf1 complex is disrupted, which suggests that Paf1 represses small RNA-mediated gene repression, probably by speeding up transcription elongation. Second, the group of Bayne has shown that the Paf1 complex in fission yeast has a role in maintaining the boundaries, limiting the spreading of heterochromatin out of the telomeres, centromeres, and mating loci. In particular, cells lacking the Paf1 complex component Leo1 were isolated in a genetic screen meant to search for mutants displaying repression of euchromatic genes placed just flanking the boundary from centromeric heterochromatin (32). Because many stress genes are subtelomeric, repression of the ctt1-GFP chimera in Paf1 mutants could be due to heterochromatin spreading from the telomeres. In fact, we have constructed another stress gene chimera, hsp9-GFP, that also depends on Sty1-Atf1 for expression. Because the hsp9 gene is located >250 kb away from one end of chromosome I and ctt1 is only 55 kb away from one end of chromosome III, loss of strict heterochromatin boundaries by a lack of Paf1 complex components would affect expression of ctt1 more than hsp9. We have determined that although the basal levels of expression of the Hsp9-GFP protein are reduced in cells lacking Set1 or Trm9, in the absence of Paf1 the expression levels are similar to wild-type cells.
In this work we have also identified the hub protein Trm112 and some of its catalytic partners as important for wild-type tolerance to peroxides. Trm112 activates not only the tRNA methyltransferase Trm9, but also the methylase catalytic subunit Mtq2 (in eukaryotes, the Mtq2-Trm112 holoenzyme methylates the glutamine of an essential GGQ motif at class I translation termination factors) and Trm11 (the heterodimer introduces in this case the m2G10 modification at some tRNAs) (33, 34). Furthermore, Trm112 can also form a complex with the methylase Bud23, which modifies the 18S rRNA subunit, but only with stabilization purposes (34). According to our genetic data, only Trm9 and Mtq2, but not Bud23 or Trm11, contribute to wild-type tolerance to oxidative stress. Apparently, the stress phenotypes of cells lacking Mtq2 are not suppressed by overexpression of eRF1. Future work will be required to decipher the role of Mtq2 in adaptation to stress.
Regarding Trm9, we have confirmed here that it contributes to S. pombe stress tolerance by similar means as Elongator: modification of the U34 of the anticodon loop of the tRNAUUULys to generate the mcm5S2U34 modification is important for stress survival. Similar to other modifications, it is not clear whether transcription efficiency or fidelity or both are affected when the codon AAA has to be decoded by unmodified tRNAUUULys; although Begley et al. (35) described that S. cerevisiae Trm9-mediated tRNA modifications participate in the DNA damage response by enhancing translation elongation of general and specific stress proteins, they also reported later that those tRNA modifications lead to translation infidelity of specific transcripts, misfolding of the resulting proteins, and activation of the unfolded protein response pathway (36). Similarly, it has recently been suggested that Elongator mutants in budding yeast display basal proteostasis defects due to inefficient or incorrect translation, and this basal phenotype may be exacerbated in the presence of stress conditions (37). Our study demonstrates that cells lacking Trm9 express low levels of Atf1 but not of Atf1AAA-to-AAG, so inefficient translation of stress proteins seems clear. Whether basal or stress-induced proteostasis defects are also present in S. pombe Δtrm9 or Δelp3 strains and whether those defects contribute to the stress phenotype of these mutants will have to be elucidated.
Author Contributions
P. G. and J. E. D. conducted most of the experiments. J. A. designed the flow cytometry-based screening, P. G., J. E. D., J. A., and E. H. designed the experiments and analyzed the results. E. H. wrote most of the paper.
Acknowledgments
We are very thankful to Nevan Krogan for providing strain P392 and to Pilar Pérez and Laura Gaspa for advice in the use of the deletion collection. We thank Rafael Carazo-Salas for advice in the generation of the ctt1-GFP reporter strain.
This work was supported by the Spanish Ministry of Science and Innovation (BFU2012-32045 (to E. H.) and BFU2012-31939 (to J. A.), PLAN E and FEDER, and by 2014-SGR-154 from Generalitat de Catalunya (Spain) (to E. H. and J. A.). The authors declare that they have no conflicts of interest with the contents of this article.
- SAGA
- Spt-Ada-Gcn5-acetyl transferase
- Pol II
- RNA polymerase I
- YES
- rich medium
- NAT
- nourseothricin
- eRF1
- eukaryotic release factor 1.
References
- 1.Chen D., Toone W. M., Mata J., Lyne R., Burns G., Kivinen K., Brazma A., Jones N., and Bähler J. (2003) Global transcriptional responses of fission yeast to environmental stress. Mol. Biol. Cell 14, 214–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnsson A., Xue-Franzén Y., Lundin M., and Wright A. P. (2006) Stress-specific role of fission yeast Gcn5 histone acetyltransferase in programming a subset of stress response genes. Eukaryot. Cell 5, 1337–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sansó M., Gogol M., Ayté J., Seidel C., and Hidalgo E. (2008) Transcription factors Pcr1 and Atf1 have distinct roles in stress- and Sty1-dependent gene regulation. Eukaryot. Cell 7, 826–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lackner D. H., Schmidt M. W., Wu S., Wolf D. A., and Bähler J. (2012) Regulation of transcriptome, translation, and proteome in response to environmental stress in fission yeast. Genome Biol. 13, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernández-Vázquez J., Vargas-Pérez I., Sansó M., Buhne K., Carmona M., Paulo E., Hermand D., Rodríguez-Gabriel M., Ayté J., Leidel S., and Hidalgo E. (2013) Modification of tRNA(Lys) UUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet. 9, e1003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang B., Johansson M. J., and Byström A. S. (2005) An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 11, 424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esberg A., Huang B., Johansson M. J., and Byström A. S. (2006) Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell 24, 139–148 [DOI] [PubMed] [Google Scholar]
- 8.Paulo E., García-Santamarina S., Calvo I. A., Carmona M., Boronat S., Domènech A., Ayté J., and Hidalgo E. (2014) A genetic approach to study H2O2 scavenging in fission yeast: distinct roles of peroxiredoxin and catalase. Mol. Microbiol. 92, 246–257 [DOI] [PubMed] [Google Scholar]
- 9.Alfa C., Fantes P., Hyams J., McLeod M., and Warbrick E. (1993) Experiments with Fission Yeast: A Laboratory Course Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 10.Bähler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A. 3rd, Steever A. B., Wach A., Philippsen P., and Pringle J. R. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 [DOI] [PubMed] [Google Scholar]
- 11.Roguev A., Wiren M., Weissman J. S., and Krogan N. J. (2007) High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nat. Methods 4, 861–866 [DOI] [PubMed] [Google Scholar]
- 12.Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., von Mering C., and Jensen L. J. (2013) STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vivancos A. P., Castillo E. A., Jones N., Ayté J., and Hidalgo E. (2004) Activation of the redox sensor Pap1 by hydrogen peroxide requires modulation of the intracellular oxidant concentration. Mol. Microbiol. 52, 1427–1435 [DOI] [PubMed] [Google Scholar]
- 14.Calvo I. A., Gabrielli N., Iglesias-Baena I., García-Santamarina S., Hoe K. L., Kim D. U., Sansó M., Zuin A., Pérez P., Ayté J., and Hidalgo E. (2009) Genome-wide screen of genes required for caffeine tolerance in fission yeast. PLoS ONE 4, e6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vivancos A. P., Castillo E. A., Biteau B., Nicot C., Ayté J., Toledano M. B., and Hidalgo E. (2005) A cysteine-sulfinic acid in peroxiredoxin regulates H2O2-sensing by the antioxidant Pap1 pathway. Proc. Natl. Acad. Sci. U.S.A. 102, 8875–8880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calvo I. A., García P., Ayté J., and Hidalgo E. (2012) The transcription factors Pap1 and Prr1 collaborate to activate antioxidant, but not drug tolerance, genes in response to H2O2. Nucleic Acids Res. 40, 4816–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jara M., Vivancos A. P., Calvo I. A., Moldón A., Sansó M., and Hidalgo E. (2007) The peroxiredoxin Tpx1 is essential as a H2O2 scavenger during aerobic growth in fission yeast. Mol. Biol. Cell 18, 2288–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen D., Wilkinson C. R., Watt S., Penkett C. J., Toone W. M., Jones N., and Bähler J. (2008) Multiple pathways differentially regulate global oxidative stress responses in fission yeast. Mol. Biol. Cell 19, 308–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomson B. N., and Arndt K. M. (2013) The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states. Biochim. Biophys. Acta 1829, 116–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krogan N. J., Kim M., Tong A., Golshani A., Cagney G., Canadien V., Richards D. P., Beattie B. K., Emili A., Boone C., Shilatifard A., Buratowski S., and Greenblatt J. (2003) Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23, 4207–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng H. H., Dole S., and Struhl K. (2003) The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278, 33625–33628 [DOI] [PubMed] [Google Scholar]
- 22.Ng H. H., Robert F., Young R. A., and Struhl K. (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11, 709–719 [DOI] [PubMed] [Google Scholar]
- 23.Krogan N. J., Dover J., Wood A., Schneider J., Heidt J., Boateng M. A., Dean K., Ryan O. W., Golshani A., Johnston M., Greenblatt J. F., and Shilatifard A. (2003) The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11, 721–729 [DOI] [PubMed] [Google Scholar]
- 24.Wood A., Schneider J., Dover J., Johnston M., and Shilatifard A. (2003) The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278, 34739–34742 [DOI] [PubMed] [Google Scholar]
- 25.Mbogning J., Nagy S., Pagé V., Schwer B., Shuman S., Fisher R. P., and Tanny J. C. (2013) The PAF complex and Prf1/Rtf1 delineate distinct Cdk9-dependent pathways regulating transcription elongation in fission yeast. PLoS Genet. 9, e1004029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pokholok D. K., Harbison C. T., Levine S., Cole M., Hannett N. M., Lee T. I., Bell G. W., Walker K., Rolfe P. A., Herbolsheimer E., Zeitlinger J., Lewitter F., Gifford D. K., and Young R. A. (2005) Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122, 517–527 [DOI] [PubMed] [Google Scholar]
- 27.Liger D., Mora L., Lazar N., Figaro S., Henri J., Scrima N., Buckingham R. H., van Tilbeurgh H., Heurgué-Hamard V., and Graille M. (2011) Mechanism of activation of methyltransferases involved in translation by the Trm112 “hub” protein. Nucleic Acids Res. 39, 6249–6259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heurgué-Hamard V., Champ S., Mora L., Merkulova-Rainon T., Merkoulova-Rainon T., Kisselev L. L., and Buckingham R. H. (2005) The glutamine residue of the conserved GGQ motif in Saccharomyces cerevisiae release factor eRF1 is methylated by the product of the YDR140w gene. J. Biol. Chem. 280, 2439–2445 [DOI] [PubMed] [Google Scholar]
- 29.Ashraf S. S., Sochacka E., Cain R., Guenther R., Malkiewicz A., and Agris P. F. (1999) Single atom modification (O → S) of tRNA confers ribosome binding. RNA 5, 188–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krüger M. K., Pedersen S., Hagervall T. G., and Sørensen M. A. (1998) The modification of the wobble base of tRNAGlu modulates the translation rate of glutamic acid codons in vivo. J. Mol. Biol. 284, 621–631 [DOI] [PubMed] [Google Scholar]
- 31.Kowalik K. M., Shimada Y., Flury V., Stadler M. B., Batki J., and Bühler M. (2015) The Paf1 complex represses small-RNA-mediated epigenetic gene silencing. Nature 520, 248–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verrier L., Taglini F., Barrales R. R., Webb S., Urano T., Braun S., and Bayne E. H. (2015) Global regulation of heterochromatin spreading by Leo1. Open Biol. 5, 150045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Towns W. L., and Begley T. J. (2012) Transfer RNA methytransferases and their corresponding modifications in budding yeast and humans: activities, predications, and potential roles in human health. DNA Cell Biol. 31, 434–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guy M. P., and Phizicky E. M. (2014) Two-subunit enzymes involved in eukaryotic post-transcriptional tRNA modification. RNA Biol. 11, 1608–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Begley U., Dyavaiah M., Patil A., Rooney J. P., DiRenzo D., Young C. M., Conklin D. S., Zitomer R. S., and Begley T. J. (2007) Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell 28, 860–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patil A., Chan C. T., Dyavaiah M., Rooney J. P., Dedon P. C., and Begley T. J. (2012) Translational infidelity-induced protein stress results from a deficiency in Trm9-catalyzed tRNA modifications. RNA Biol. 9, 990–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nedialkova D. D., and Leidel S. A. (2015) Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell 161, 1606–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leupold U. (1970) Genetical methods for Schizosaccharomyces pombe. in Methods in Cell Physiology (Prescott D. M.), pp. 169–177, Vol. 4, Academic Press, New York and London [Google Scholar]
- 39.Zuin A., Vivancos A. P., Sansó M., Takatsume Y., Ayté J., Inoue Y., and Hidalgo E. (2005) The glycolytic metabolite methylglyoxal activates Pap1 and Sty1 stress responses in Schizosaccharomyces pombe. J. Biol. Chem. 280, 36708–36713 [DOI] [PubMed] [Google Scholar]