Abstract
Vibrio cholerae is a human pathogen and the causative agent of cholera. The persistence of this bacterium in aquatic environments is a key epidemiological concern, as cholera is transmitted through contaminated water. Predatory protists, such as amoebae, are major regulators of bacterial populations in such environments. Therefore, we investigated the interaction between V. cholerae and the amoeba Acanthamoeba castellanii at the single-cell level. We observed that V. cholerae can resist intracellular killing. The non-digested bacteria were either released or, alternatively, established a replication niche within the contractile vacuole of A. castellanii. V. cholerae was maintained within this compartment even upon encystment. The pathogen ultimately returned to its aquatic habitat through lysis of A. castellanii, a process that was dependent on the production of extracellular polysaccharide by the pathogen. This study reinforces the concept that V. cholerae is a facultative intracellular bacterium and describes a new host–pathogen interaction.
Introduction
Cholera, a severe diarrheal disease caused by the pathogenic bacterium Vibrio cholerae, remains a serious health threat. Because V. cholerae is an aquatic bacterium and is primarily encountered in coastal waters, estuaries, ponds and rivers, its re-emergence is thought to be climate-related (Colwell, 1996; Morens et al., 2004). In its environmental reservoir, V. cholerae is found not only in a free-living state but also in association with phytoplankton, zooplankton and potentially also with additional environmental hosts.
Until recently, a lot of the research on V. cholerae has focused directly on the clinical aspects. However, the insidious and complex nature of the pathogen still requires more investigations covering the environmental lifestyle of V. cholerae. In an environmental context, predation by protists and particularly amoebae represents a major selective pressure that shapes bacterial populations (Matz and Kjelleberg, 2005; Pernthaler, 2005; Snelling et al., 2006). Moreover, amoebae are widely used as non-mammalian model organisms in research of host–microbe interactions. Previous studies have addressed the killing behavior of few specific V. cholerae isolates towards the social amoeba Dictyostelium discoideum (Pukatzki et al., 2006). However, as D. discoideum is primarily a soil-living amoeba, recurrent encounters with V. cholerae are unlikely. The free-living amoeba Acanthamoeba castellanii, however, is frequently found in aquatic environments (Martinez and Visvesvara, 1997; Khan, 2006, 2015). These amoebae prey on bacteria and have been described as a reservoir of pathogenic bacteria (Fouque et al., 2012). A. castellanii is characterized by a biphasic life cycle; the ‘trophozoite' form is metabolically active and feeding, and the ‘cyst' form is stress-induced, dormant and resistant to various environmental hazards (Khan, 2006). Successive steps of encystment occur in the progression from the trophozoite to the mature cyst, including global cell rounding and increased cell-wall synthesis (Bowers and Korn, 1968; Fouque et al., 2012; Chavez-Munguia et al., 2013). As a result, cysts harbor a thickened cell wall, which enhances their resistance to disinfection treatments.
Though an impact of predatory flagellates on V. cholerae populations has been established (Matz et al., 2005), much less is known about the interaction of V. cholerae with free-living amoebae and the molecular mechanisms underlying such an interaction. Notably, V. cholerae and A. castellanii were detected in the same environmental habitats (Shanan et al., 2011) and a study by Thom et al. (1992) showed that V. cholerae survived better in the presence of the amoebae Acanthamoeba polyphaga and Naegleria gruberi compared with that in their absence. Follow-up work by the Sandström group confirmed the increased survival of V. cholerae in the presence of Acanthamoebae (Abd et al., 2005, 2007; Sandström et al., 2010; Shanan et al., 2011). Notably, these and other studies (Sun et al., 2013) focused primarily on population measurements including the enumeration of colony-forming units, whereas limited information was provided at the single-cell level.
This study aimed at deciphering the interaction of V. cholerae and amoeba using imaging-based approaches. We hypothesized that the pathogen could grow intracellulary within an environmental non-human host. The rationale for these speculations was based on two observations: (i) Nielsen et al. (2010) provided evidence of intracellular growth of V. cholerae within extruded epithelial cells, a phenotype that was observed in a rabbit ileal loop model of cholera; and (ii) the association of the pathogen with so-called ‘ghost cells' was recently reported based on the evaluation of stool samples from cholera patients (Nelson et al., 2007). The authors defined such ghost cells as DAPI (4′,6-diamidino-2-phenylindole) stain-negative cells with a size range of 20–80 μm, which were most likely derived from the mucosal epithelium (Nelson et al., 2007).
Considering the primary niche of this pathogen, it is tempting to speculate that aquatic amoebae could serve as environmental hosts for V. cholerae and might select for pathogenic traits, a phenomenon that has been suggested for other human pathogens such as Legionella pneumophila (Solomon and Isberg, 2000; Swanson and Hammer, 2000; Greub and Raoult, 2004; Hoffmann et al., 2014). In this study, we hypothesized that the putative intracellular occurrence of V. cholerae (in rabbits and humans; Nelson et al., 2007; Nielsen et al., 2010) might represent an adaptive trait of V. cholerae acquired in aquatic reservoirs. We therefore investigated the interaction of V. cholerae and A. castellanii at the single-cell level. We show that V. cholerae bacteria can (i) resist intracellular killing by A. castellanii; (ii) be released from trophozoites by exocytosis; (iii) establish an intracellular proliferation niche; (iv) maintain their niche upon amoebal encystment; and (v) promote the lysis of A. castellanii cysts. Moreover, we demonstrate that a functional quorum sensing (QS) circuit and the Vibrio polysaccharide (VPS) are of importance for this host–pathogen interaction. Our data reinforce the concept that V. cholerae is a facultative intracellular pathogen and strongly suggest that this intracellular lifestyle enhances environmental survival and proliferation.
Materials and methods
Amoebal and bacterial strains and plasmids
The Vibrio cholerae strains used in this study are derived from O1 El Tor strain A1552 (Yildiz and Schoolnik, 1998) and are listed in Table 1 (together with the plasmids). To delete the vpsA (VC0917) or hapR (VC0583) genes from the parental wild-type (WT) strain (A1552), a gene disruption method based on natural transformation and FLP recombination was used (TransFLP method; De Souza Silva and Blokesch, 2010; Marvig and Blokesch, 2010; Blokesch, 2012a; Borgeaud and Blokesch, 2013). The site-directed stable insertion of the mini-Tn7-derived transposons (mTn7-GFP (green fluorescent protein), mTn7-dsRed and mTn7-rrnBP1-gfp(ASV)) into the chromosome of the indicated strains was accomplished through triparental mating (Bao et al., 1991). Escherichia coli strain S17-1λpir/pGP704-mTn7-dsRed served as a non-pathogenic control. Acanthamoeba castellanii Neff strain, genotype T4 (American Type Culture Collection 30010 (ATCC30010)) was used in all amoebal experiments.
Table 1. V. cholerae strains and plasmids used in this study.
| Genotype/descriptiona | Reference | |
|---|---|---|
| Strains | ||
| A1552 (WT) | Wild-type, O1 El Tor Inaba, RifR | Yildiz and Schoolnik (1998) |
| A1552-GFP | A1552 with mTn7-gfp; RifR, GentR | Blokesch (2012b) |
| A1552-dsRed | A1552 with mTn7-dsRed.T3[DNT]; RifR, GentR | Borgeaud et al. (2015) |
| A1552-TnrrnBP1-gfp(ASV) | A1552 with mTn7-rrnBP1-gfp(ASV); RifR, GentR; growth reporter strain | This study |
| ΔhapR | A1552 deleted for hapR (TransFLP); RifR | Borgeaud et al. (2015) |
| ΔhapR-GFP | ΔhapR with mTn7-gfp; RifR, GentR | This study |
| ΔhapR-dsRed | ΔhapR with mTn7-dsRed.T3[DNT]; RifR, GentR | This study |
| ΔhapRΔvpsA | A1552 deleted for hapR and vpsA (TransFLP); RifR | This study |
| ΔhapRΔvpsA-GFP | ΔhapRΔvpsA with mTn7-gfp; RifR, GentR | This study |
| ΔhapRΔvpsA-dsRed | ΔhapRΔvpsA with mTn7-dsRed.T3[DNT]; RifR, GentR | This study |
| ΔvpsA | A1552 deleted for vpsA (TransFLP); RifR | This study |
| ΔvpsA-GFP | ΔvpsA with mTn7-gfp; RifR, GentR | This study |
| Plasmids | ||
| pUX-BF13 | oriR6K, helper plasmid with Tn7 transposition function; AmpR | Bao et al. (1991) |
| pGP704::Tn7-GFP | pGP704 with mini-Tn7 harboring a constitutively expressed gfp cassette; AmpR | Müller et al. (2007) |
| pGP704-mTn7-dsRed | pGP704 with mini-Tn7 carrying dsRed.T3[DNT]; AmpR | Borgeaud et al. (2015) |
| pGP704Sac28-mTn7- rrnBP1-gfp(ASV) | pGP704 with mini-Tn7 carrying growth reporter rrnBP1-gfp(ASV); AmpR | This study |
TransFLP method according to De Souza Silva and Blokesch (2010) and Blokesch (2012a).
Media and growth conditions
Uninfected amoebae were grown in peptone yeast glucose medium (ATCC medium 712). Bacterial cultures were grown in LB medium at 30 °C under shaking conditions. Defined artificial seawater medium at a 0.5-fold concentration and buffered with 50 mm HEPES (Meibom et al., 2005) served as the infection medium. Antibiotics were supplemented when required at the following concentrations: gentamicin, 50 μg ml−1; ampicillin, 100 μg ml−1; streptomycin, 100 μg ml−1.
Amoebal infections
For the evaluation of bacterial uptake by the amoebae, an infection protocol was implemented for confocal microscopy imaging, in which adherent A. castellanii trophozoites (1 × 105 cells per ml) were infected with the bacteria (multiplicity of infection of 1000) in 35 mm μ-Dish devices (IBIDI, Martinsried, Germany). The incubation occurred statically at 24 °C. Single-cell quantification (contractile vacuole (CV) colonization in trophozoites, CV colonization in cysts, cytosol colonization in cysts and colonized lysed cysts) was carried out at 20 h after post primary contact (p.p.c.). The evaluation was based on at least 1200 counted amoebae per experiment and the data show averages from three independent biological replicates.
For the enumeration of surviving bacteria, the amoebal and bacterial cells were detached from the IBIDI device using a cell scraper. The homogenized amoebae were lysed by three cycles (30 s on, 30 s off) of bead beating (glass beads of 0.75–1 mm diameter; Retsch GmbH, Haan, Germany) using a Precellys 24 homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France) at a speed of 5000 r.p.m. This method allowed the lysis of both trophozoites and cysts of A. castellanii as visually inspected. Recovered bacteria were serial diluted, spotted onto agar plates and enumerated the following day (given as colony-forming units (CFUs) per ml).
Confocal laser scanning microscopy
Confocal laser scanning microscopy images were obtained using an inverted Zeiss LSM 700 confocal microscope (Zeiss, Feldbach, Switzerland) equipped with four laser lines, two confocal channels and a highly sensitive photomultiplier for the detection of transmission signals. For time-lapse imaging, pictures were taken every 30 s in one plane.
To follow the phago-/endosomal pathway of the amoebae, the infection medium was supplemented with Alexa Fluor 647-labeled dextran (size: 10 000 MW; Molecular Probes and purchased via Life Technologies, Zug, Switzerland) at a final concentration of 100 μg ml−1 and fluorescence was monitored in the far-red region (excitation and emission wavelengths are 650 and 668, respectively). One-micrometer Fluoresbrite Yellow Green (YG) microspheres (Polysciences Inc., Eppelheim, Germany) were used at a concentration of ~107 particles per ml.
Fluorescence recovery after photobleaching
Fluorescence recovery after photobleaching was used to test for bacterial protein synthesis and therefore bacterial viability within the CV of infected amoebae. V. cholerae were subjected to 20 iterations of bleaching throughout the full Z-direction of the amoeba using the 488 nm laser line at 100% power. Epifluorescence microscopy images were taken to confirm the complete bleaching of the CV. Fluorescence recovery was monitored after 30, 60 and 90 min using the confocal laser scanning option with the same scanning settings. Quantification of the fluorescent signal was performed using ImageJ (National Institute of Mental Health, Bethesda, MD, USA).
Electron microscopy
Infected amoebae were initially fixed in a solution of 1% acrolein and 1% glutaraldehyde in 0.1m cacodylate buffer using a microwave (Pelco BioWave; Ted Pella, Redding, CA, USA; 30 s at 300, 350 and 400 W) to ensure rapid preservation. Samples were then left for 60 min in the same fixative before being pelleted by gentle centrifugation. The supernatant was replaced by 4% low melting-point agarose (w v−1) in 0.1 m cacodylate and the pellet was resuspended. After cooling on ice, the agarose was cut into small cubes (~0.5 mm each side length) with a razor blade. These cubes were again fixed using the same fixative as above, washed in 0.1 m cacodylate buffer and then stained using 1% osmium (w v−1) in 0.1 m cacodylate buffer followed by 1% uranyl acetate (w v−1) in 0.1 m cacodylate, for 40 min each. The samples were dehydrated in increasing concentrations of ethanol, transferred to propylene oxide and infiltrated for 24 h with epon resin. The samples were embedded between two glass slides and placed in a 65 °C oven for 24 h. The embedded samples were visualized using a dissecting microscope and blocks were prepared from these regions. Fifty nanometer-thick sections were cut using an ultramicrotome (Leica EM UC7; Leica Microsystems AG, Heerbrugg, Switzerland) and collected on formvar-coated slot grids. Finally, the samples were stained with 1% uranyl acetate (w v−1) followed by 3% lead citrate (w v−1). Images were collected in a transmission electron microscope (Tecnai Spirit; FEI Company, Eindhoven, Netherlands) at 80 KV using a digital camera (Eagle; FEI Company).
Statistical analysis
Two-way analysis of variance with Tukey's post-test was performed using GraphPad Prism version 6.00 for Mac (GraphPad Software, San Diego, CA, USA).
Results
Release of viable V. cholerae from the amoeba A. castellanii
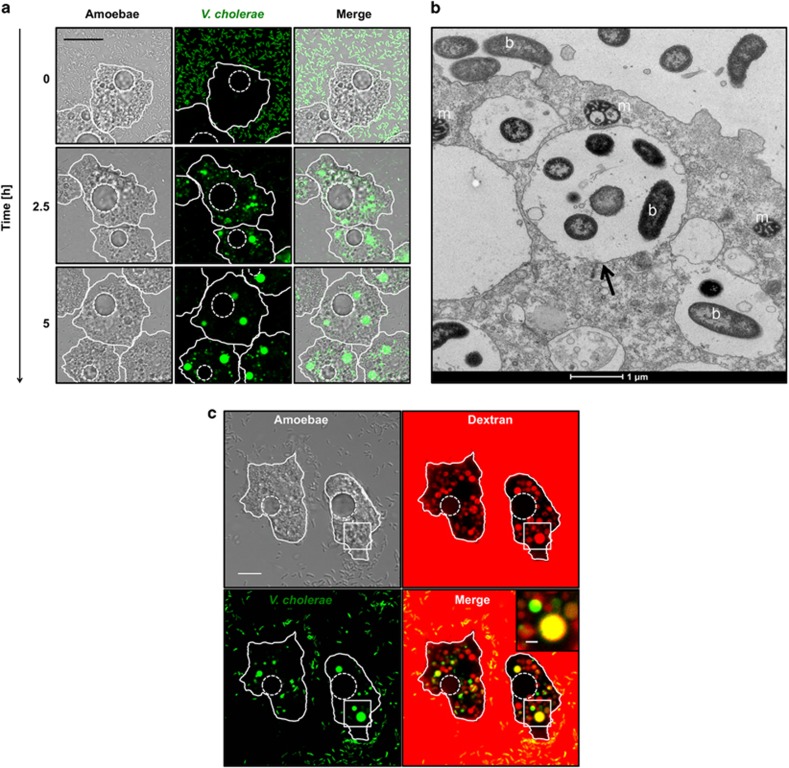
Even though outside of blooms (Gilbert et al., 2012) Vibrios represent on average only a few percent of total bacteria (with 103–104 bacteria per ml), they can contribute significantly to the biomass in aquatic environments (Yooseph et al., 2010). Such balanced biomass is based on rapid growth combined with high predation (Takemura et al., 2014). As predatory free-living amoebae and V. cholerae occur in the same ecological niche (Thom et al., 1992), it is plausible that the two organisms have coevolved based on frequent encounters. We therefore aimed at better understanding the interaction between A. castellanii and V. cholerae at the single-cell level. GFP-producing V. cholerae bacteria were monitored by time-lapse imaging, allowing observation of active internalization of the bacteria by amoebae (Figure 1a and Supplementary Movie S1). Such bacterial uptake resulted in small, bacteria-filled vacuoles, which eventually fused to form larger compartments. Based on the retained morphology observed by electron microscopy (Figure 1b) and the release of non-digested bacteria (see below), we concluded that at least a fraction of the internalized bacteria were not digested within these vacuoles. In contrast to previous studies (Abd et al., 2005, 2007), we did not observe V. cholerae cells within the cytosol of the trophozoites even though mitochondria were readily detectable (see Discussion). These data suggest that undigested V. cholerae are present within intracellular vacuoles of A. castellanii after phagocytosis.
Figure 1.
Uptake of V. cholerae by A. castellanii. (a) Time-lapse confocal microscopy imaging experiment to monitor the interaction of A. castellanii and V. cholerae. The amoebae are visible in the transmitted-light channel (labeled Amoebae) and the GFP-producing bacteria in the green channel (labeled V. cholerae). The merged signals are shown on the right (labeled Merge). The time p.p.c. is indicated on the left (in h). Amoebae are delineated by solid white lines, whereas the CVs are marked by dashed white lines (valid for all figures). Scale bar: 20 μm. See also Supplementary Movie S1. (b) Representative transmission electron micrograph showing undigested bacteria (b) surrounded by a vacuolar membrane (black arrow) compared with cytosolic mitochondria (m) at 3 h p.p.c. (c) Undigested V. cholerae localize to food vacuoles. Colocalization of GFP-producing bacteria (labeled V. cholerae) and Alexa Fluor 647-conjugated dextran (labeled Dextran) within vacuolar compartments of A. castellanii at 3 h p.p.c. (amoebal structures are visible in the transmission-light channel; labeled Amoebae). A merged image of the two fluorescent channels is also depicted (labeled Merge). Scale bar: 10 μm. The inset shows a magnification of the white-boxed area in the merged image (scale bar: 2 μm).
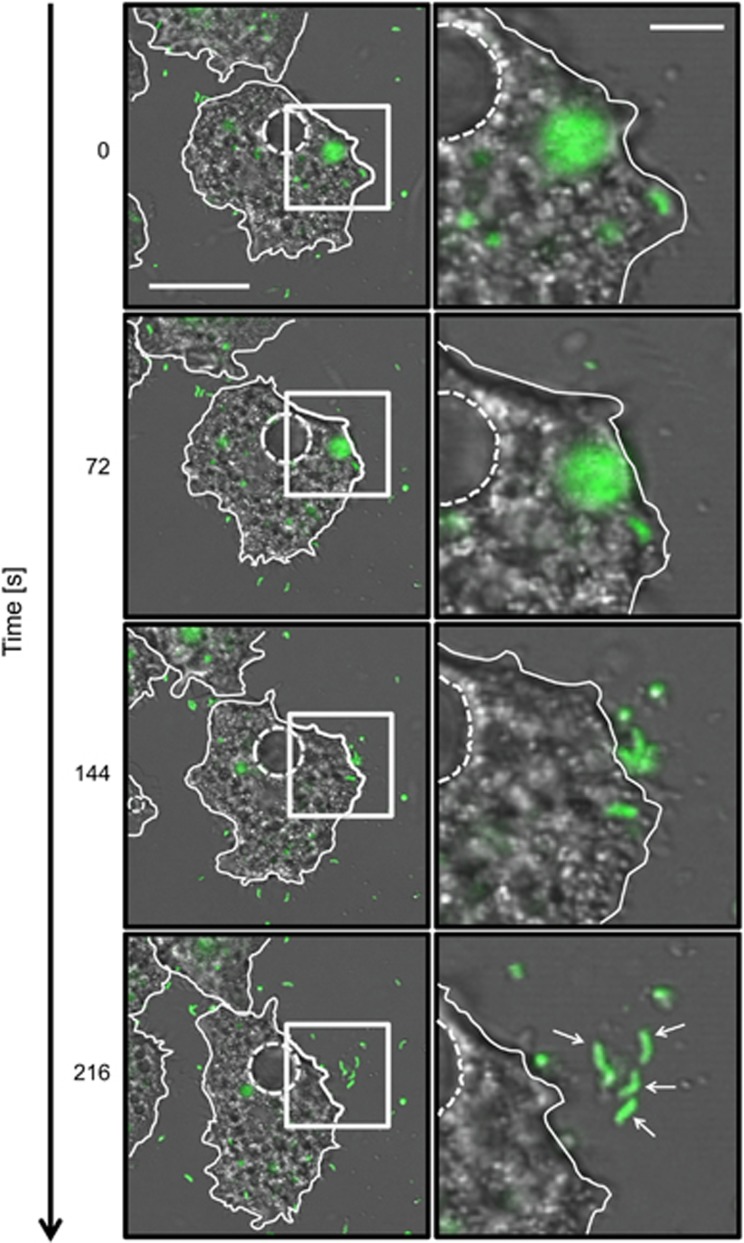
To identify the compartment containing intracellular V. cholerae, we followed the course of the phago-/endosomal pathway by feeding amoebae undigestible fluorescently labeled high-molecular-weight dextran, and tracked its localization (Aubry et al., 1993). Similar to other cellular waste including digested bacteria, the accumulated dextran is ultimately released into the environment by exocytosis. At early time points (0–5 h p.p.c.), phagocytosed bacteria colocalized with the dextran, indicating that intracellular V. cholerae reside within food vacuoles (Figure 1c and quantified in Supplementary Figure S1). In addition, live-cell imaging provided evidence that such pathogen-containing compartments fused to form larger vacuoles, which the amoeba ultimately excreted by exocytosis, thereby releasing live bacteria back into the environment (Figure 2 and Supplementary Movie S2). Such release of undigested bacteria was not observed in the control experiments using non-pathogenic E. coli (Supplementary Figure S2B). We therefore conclude that, upon phagocytosis, a portion of the V. cholerae population resisted intracellular killing and returned to the environment.
Figure 2.
Release of V. cholerae from digestive vacuoles. Time-lapse confocal microscopy image series showing the release of V. cholerae from digestive vacuoles. GFP-producing bacteria were incubated with A. castellanii for 4.5 h and then monitored over a period of 216 s (relative time shown on the left). Images represent merged transmitted-light (amoebae) and GFP channels (V. cholerae). The right row shows magnification of the white-boxed areas highlighted in the left row. The white arrows point to undigested V. cholerae released from the food vacuole. Scale bars: 20 and 5 μm for image in the left and right row, respectively. See also Supplementary Movie S2.
V. cholerae escapes the endosomal pathway through the water discharge system of A. castellanii
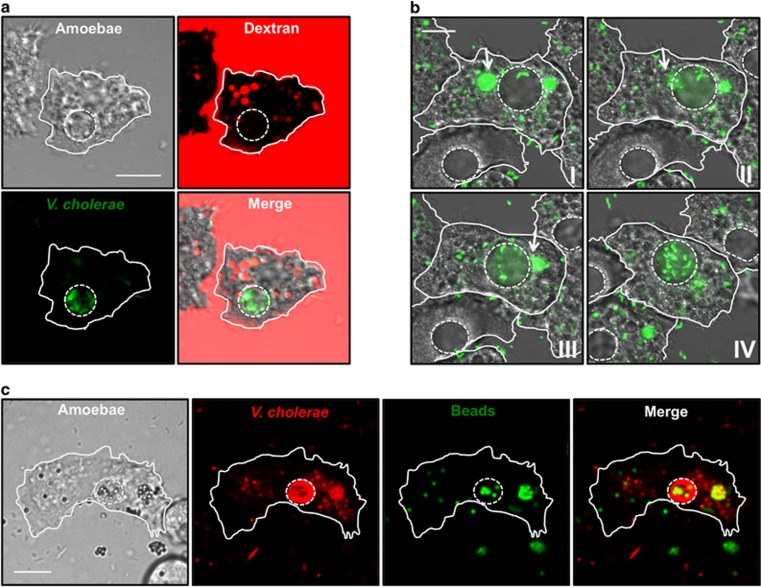
After we had established the intracellular survival phenotype, we were curious to determine whether all ingested V. cholerae follow the same fate. Interestingly, quantification of the images showed that even though the vast majority of A. castellanii contained internalized V. cholerae within the phagosomal pathway, a small but consistently observed fraction of the amoebal population contained fluorescently labeled bacteria that did not colocalize with the fluorescent dextran (Figure 3a and quantified in Supplementary Figure S1). Instead, we witnessed the presence of V. cholerae accumulated inside the CV of A. castellanii (Figure 3a). The CV is a highly dynamic organelle involved in osmoregulation (Doberstein et al., 1993), which is readily visible in transmission-light images. It regulates osmotic pressure by collecting cytosolic water and actively discharging excess water by means of contraction (Allen 2000; Khan 2015). Upon closer inspection, we observed that colonization of the CV was nearly coincident with fusion of small food vacuoles (e.g., ~4 h p.p.c.). In accordance with this observation, we were able to image the entry of V. cholerae into the CV upon vacuolar fusion (Figure 3b and Supplementary Movie S3). Notably, we never observed colonization of the CV by E. coli (quantified in Supplementary Figure S1), suggesting that V. cholerae promotes these fusion events in an unknown (direct or indirect) manner. To confirm this, we first fed A. castellanii indigestible fluorescent microspheres. Upon phagocytosis, such beads were excluded from the CV and instead colocalized with the dextran, indicating passage through the regular phago-/endosomal pathway (Supplementary Figure S1B and quantified in Supplementary Table S1). However, upon coinoculation with V. cholerae, such beads became readily detectable within the CV (Figure 3c and quantified in Supplementary Table S1). Again, the same phenomenon was never observed when E. coli was used in place of V. cholerae (Supplementary Table S1). These data are indicative of a V. cholerae-promoted CV colonization process.
Figure 3.
V. cholerae colonizes the CV of A. castellanii. (a) The amoebae were coincubated with gfp-expressing bacteria before confocal laser scanning microscopy (CLSM) imaging. The image composition is as described for Figure 1c, except that the merged image also contains the transmitted-light channel. (b) Vacuolar fusion leads to CV colonization. GFP-producing V. cholerae (green) were incubated with A. castellanii and followed over 6 min starting at 5h07 p.p.c. The relative time is I, 0 s; II, 30 s; III, 60 s; and IV, 360 s. The time series shows the fusion between V. cholerae-containing food vacuoles (depicted by the white arrows) and the CV leading to the release of bacteria into the CV. The image composition is as described for Figure 2. See also Supplementary Movie S3. (c) Colocalization between dsRed-expressing V. cholerae (red) and fluorescent beads (green) within the CV. Scale bar for all panels: 10 μm.
CV-infected amoebae maintain the pathogen upon encystment followed by CV destruction and cyst lysis
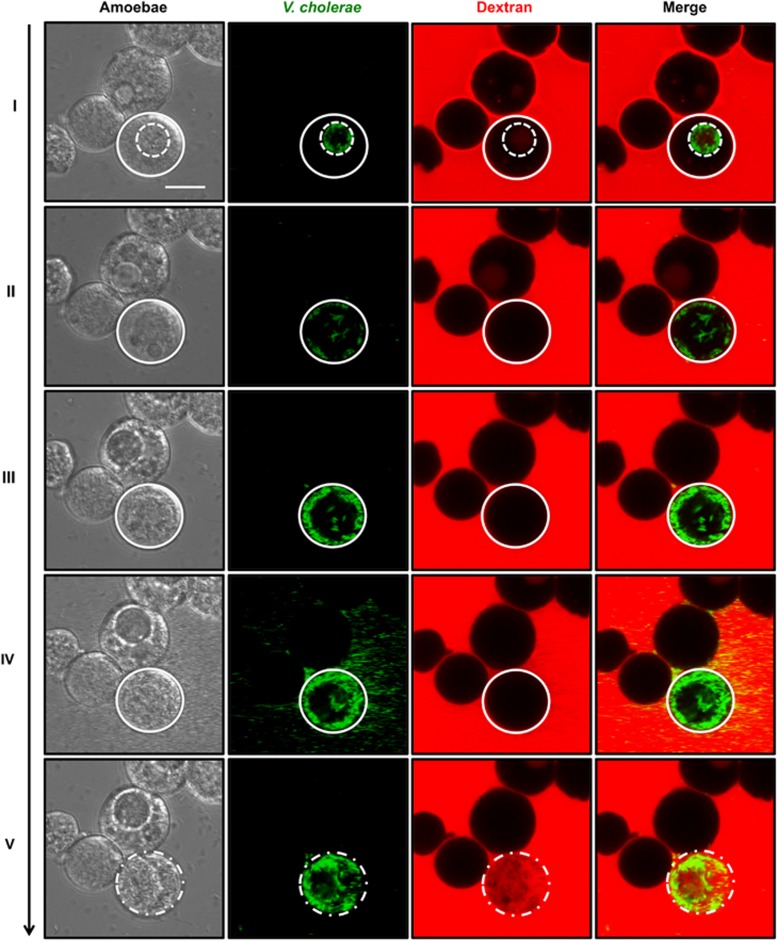
We followed the fate of the CV-infected A. castellanii over time (Supplementary Movie S4–S6). Under unfavorable conditions, the metabolically active amoebal trophozoites undergo a differentiation leading to the generation of cysts (Bowers and Korn, 1968; Bowers and Korn, 1969; Fouque et al., 2012; Siddiqui and Khan, 2012). The major structural changes accompanying encysting trophozoites are (i) the retraction of the pseudopodia; (ii) cell rounding; and (iii) the exocytosis of food vacuole content (Bowers and Korn, 1968; Stewart and Weisman, 1972; Chavez-Munguia et al., 2013). During encystment of infected amoebae, however, V. cholerae remained within the CV in contrast to the content of the food vacuoles (Figure 4). Moreover, despite encystation, V. cholerae grew intracellularly (see below) and lysed the CV, thereby ensuring its release into the cyst's cytosol (Figure 4 and Supplementary Movie S4 and S5). Ultimately, the cysts themselves were lysed, as visualized by the release of fluorescently labeled motile bacteria and influx of dextran through the compromised plasma membrane (Figure 4 and Supplementary Movie S6). Based on these observations, we conclude that lysis of the CV, followed by the lysis of the whole cyst, forms a sequential two-step process allowing V. cholerae to return to the environment. Moreover, this behavior defines V. cholerae as a pathogen for A. castellanii as CV-infected amoebae consistently lysed upon encystment.
Figure 4.
Cyst lysis occurs after disruption of the V. cholerae-colonized CV. Pathogen-containing CVs (I; relative t=0 h, corresponding to 16 h p.p.c.) are lysed within the cyst, which allows the spread of gfp-expressing V. cholerae into the cyst's cytosol (II; t=3.22 h; Supplementary Movie S4) followed by bacterial proliferation (III; t=5.33 h; Supplementary Movie S5). This leads to the disintegration of the amoebal cell as noted by the escape of V. cholerae from the cyst (IV; t=5.93 h; Supplementary Movie S6) and the influx of dextran into the lysed cyst (V; t=6.07 h; Supplementary Movie S6). Scale bar: 10 μm.
Intracellular proliferation of V. cholerae within the CV
To test the viability of the V. cholerae bacteria within the CV, we used fluorescence recovery after photobleaching. We observed that GFP was resynthesized by the intracellular bacteria, demonstrating their ability for protein synthesis, an indicator of their sustained viability (Supplementary Figure S3). Notably, the recovery kinetics indicated in Supplementary Figure S3 might be an underestimation, as partial killing of bacteria through the iterations of bleaching cannot be excluded.
Moreover, upon observing the accumulation of V. cholerae within the A. castellanii CV, we wished to determine whether this phenomenon resulted from an accumulation of phagocytosed bacteria (e.g., resulting from a series of vacuolar fusions) or from intracellular growth and proliferation of the pathogen. We therefore infected amoebae with a coculture of gfp- or dsRed-expressing WT V. cholerae strains (at a ratio 1:1; Supplementary Figure S4A). Interestingly, even though dual-colored infected CVs were detectable, single-colored CVs were also observed, indicating that the bacterial population growth resulted from proliferation of one or very few single bacteria (quantified in Table 2). This phenomenon was observed in both the trophozoite and cyst stages (Supplementary Figure S4B).
Table 2. Mixed color CV colonization assay.
| Inoculum GFP:dsRed=1:1 |
Colonized CV |
|||
|---|---|---|---|---|
| Mixed colors | Mixed colors | GFP only | dsRed only | |
| Expected (%)a | 100% | 100% | 0% | 0% |
| Observed (%)b | 100% | 79.7% (±5.5) | 11.7% (±4.1) | 8.6% (±1.6) |
| Supplementary Figure S4A | Supplementary Figure S4B | |||
Expected percentage values are based on the assumption of repeated vacuolar fusions delivering V. cholerae to the CV or single fusion events between one food vacuole containing many bacteria with the CV (e.g., no growth within the CV).
Averages are derived from three independent biological experiments (±s.d.).
To further confirm the intracellular growth phenotype, we genetically engineered a growth reporter strain of V. cholerae in accordance with previous reports (Bartlett and Gourse, 1994; Sternberg et al., 1999; Nielsen et al., 2010). Such growth reporters are widely used and based on a growth-rate-dependent P1 promoter of ribosomal rrnB fused to a reporter gene such as gfp(ASV), which encodes a destabilized version of GFP, which is quickly degraded and suited to study transient gene expression in bacteria (Andersen et al., 1998). It therefore only reports on growing bacteria. This reporter strain allowed us to monitor the growth of cells by observing the accumulation of the fluorescent signal. Indeed, we confirmed the intracellular growth of V. cholerae within the CV of both trophozoites and cysts and within the cytosol of cysts (Figure 5). No such fluorescent signal was observable for extracellular bacteria (except for V. cholerae cells that recently escaped from burst cysts (Supplementary Figure S4C) owing to the lack of a carbon source in the infection medium, which rendered V. cholerae unable to grow extracellularly. These data indicate that V. cholerae proliferates within the CV of infected amoebae and within the cytosol of cysts after CV lysis, effectively transforming these compartments into an intracellular replication niche for the pathogen.
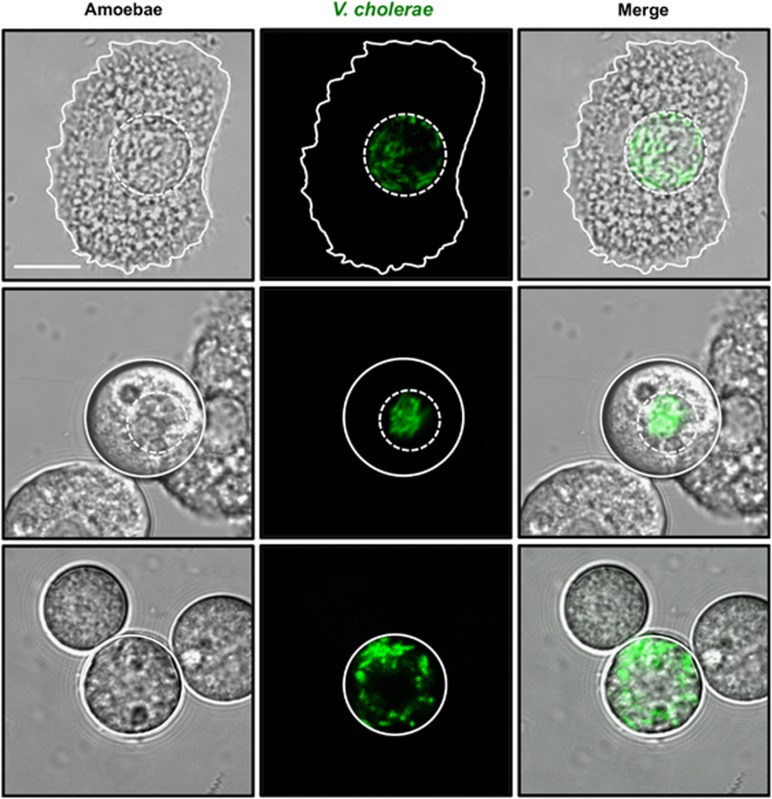
Figure 5.
Evaluation of intracellular growth based on a growth reporter strain. A V. cholerae WT strain harboring a chromosomally encoded growth reporter construct was cocultured with A. castellanii. Growth of bacteria expressing destabilized GFP was detected in the CVs of trophozoites (upper row), the CVs of cysts (middle row) and the cytosol of cysts (lower row). Scale bar valid for all panels: 10 μm.
VPS is required for lysis of the CV
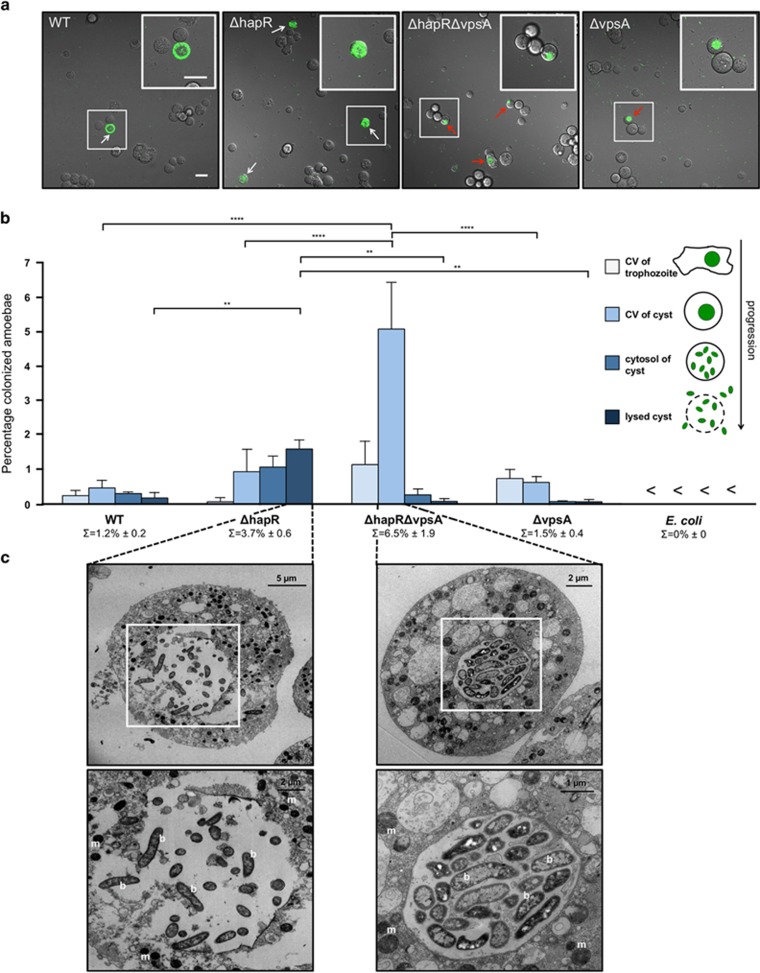
After we had established this intracellular replication niche for V. cholerae, we wondered which bacterial factors and/or signal- transduction cascades might be important to establish this host–pathogen interaction. We first focused on QS, which often has a role in virulence regulation in pathogenic bacteria (Rutherford and Bassler, 2012). QS is a cell-to-cell communication system that involves the production and sensing of small molecules, or ‘autoinducers' within bacterial populations. At high population densities, the concentration of autoinducers is sufficiently elevated to initiate a signaling cascade and to ultimately influence gene expression and bacterial behavior (Platt and Fuqua, 2010). The QS circuit of V. cholerae has been extensively studied by Bassler and co-workers. In V. cholerae, QS involves the production of the major QS regulator HapR at high cell densities (Ng and Bassler 2009). Previous studies had shown that HapR represses the virulence cascade in V. cholerae and decreases the production of extracellular VPS, thereby leading to reduced biofilm formation (Zhu et al., 2002; Zhu and Mekalanos, 2003; Hammer and Bassler, 2003; Teschler et al., 2015). VPS is primarily composed of glucose and galactose combined with 2-acetamido-2-deoxy-α-l-gulopyranosyluronic acid, glycine and N-acetylglucosamine (Yildiz et al., 2014; Teschler et al., 2015). HapR also acts as a positive regulator of natural competence for transformation and of the bacterium's type VI secretion system (T6SS) (Meibom et al., 2005; Zheng et al., 2010; Seitz and Blokesch, 2013; Borgeaud et al., 2015). We therefore wondered whether V. cholerae's QS ability also affected the phenotypes observed in this study. Comparing the dynamics of the intracellular localization of the WT and QS-defective V. cholerae strains revealed several altered behaviors such as (i) an overall increase in CV colonization; (ii) a superior CV destruction dynamics; and (iii) enhanced lysis of the amoebal cyst (Figure 6). Indeed, upon careful quantification of these phenotypes, we observed that the HapR-deficient strain showed a statistically significant enhanced CV-related amoebal colonization compared with the WT (3.7%±0.6 (s.d.) versus 1.2%±0.2 (s.d.); values are averages of three independent experiments and a total of 4200 counted amoebae per strain; **P=0.0061), although CV-colonized trophozoites were rarely detectable (Figure 6b).
Figure 6.
The VPS contributes to CV lysis. (a) V. cholerae strains lacking VPS are severely impaired in amoebal CV lysis. Confocal imaging of amoebal cysts formed in the presence of V. cholerae WT strain (WT) or mutant V. cholerae strains lacking hapR (ΔhapR), vpsA (ΔvpsA) or hapR and vpsA (ΔhapRΔvpsA). White arrows depict V. cholerae within the cytosol of the cysts after CV lysis and red arrows indicate colonized but intact CVs. Shown are merged images of the transmitted light and the GFP channels taken at 20 h p.p.c. The insets show magnifications of the boxed regions. Scale bars: 20 μm. (b) Quantification of specific events during amoebal colonization. A. castellanii cells harboring V. cholerae inside the CV of the trophozoite, the CV of the cyst and within the cytosol of the cyst before and after cyst lysis were counted. Values represent averages from three independent experiments (4200 total amoebae counted for each strain). <, below the detection limit of 0.02%. Statistically significant differences are indicated (****P<0.0001; **P<0.01). ∑ indicates the total percentage of colonized CVs per V. cholerae strain (±s.d.). (c) Representative transmission electron micrographs showing colonized compartments of A. castellanii. V. cholerae strains ΔhapR and ΔhapRΔvpsA were cocultured with A. castellanii for 20 h before infected cysts were visualized by electron microscopy. The lower images are magnifications of the boxed regions in the upper images. b, bacteria; m, mitochondria. V. cholerae strains used for all panels were producing fluorescent proteins.
Because HapR-deficient strains are known to develop rugose colony morphology and to form enhanced biofilms upon overproduction of VPS (Yildiz and Schoolnik, 1999; Zhu and Mekalanos, 2003), we wondered whether the loss of HapR per se or the overproduction of VPS contributed to amoebal colonization and CV/cyst lysis. We therefore tested V. cholerae strains that lacked the vpsA gene, which encodes UDP-N-acetylglucosamine 2-epimerase (Fong et al., 2010). This enzyme is required for the production of VPS sugar precursors; thus, vpsA-negative (ΔvpsA) strains exhibit reduced VPS production and biofilm formation (Fong et al., 2010). In our amoebal infection model, we noticed that the ΔhapRΔvpsA strain still exhibited a hyper CV colonization phenotype (6.5%±1.9 (s.d.) versus 1.2%±0.2 (s.d.) for the WT; averages of three independent experiments and a total of 4200 counted amoebae per strain; *P=0.03) and that ΔvpsA strains were severely impaired in CV lysis, regardless of the presence or absence of hapR (Figure 6). We therefore conclude that VPS has a critical role in CV lysis. Although the ΔhapR strain often localized to the cytosol of intact or recently lysed cysts, the ΔhapRΔvpsA strain was severely impaired in CV lysis, which was also visible upon inspection of EM images (Figure 6c).
Taken together, these data indicate that QS influences the dynamics of this host–pathogen interaction, that the absence of HapR leads to an increased ability of V. cholerae to colonize the CV and that VPS is involved in the CV lysis process.
Discussion
Cholera is a water-borne disease caused by V. cholerae. Understanding the environmental lifestyle of this facultative pathogen is therefore of prime importance as such studies might elucidate the evolutionary development of virulence factors and the mechanisms through which aquatic bacteria transform into human pathogens.
In this study, we examined the interaction of V. cholerae with the amoeba A. castellanii at the single-cell level. We report a previously unrecognized interaction mode and therefore propose a novel model of V. cholerae's life cycle in cohabitation with free-living amoebae (Figure 7). The pathogen enters the trophozoite's phago-/endosomal pathway via phagocytosis. However, in contrast to other bacteria (e.g., E. coli), a fraction of the V. cholerae population resists amoebal digestion. Instead, the bacteria are either exocytosed alive (Figure 2) or enter the CV (Figure 3b). Within this organelle, V. cholerae establishes a replication niche, which the pathogen maintains throughout the amoebal encystment process. Bacterial proliferation occurs first within the CV; then, V. cholerae escapes into the cytosol of the cyst via CV lysis (Figure 4 and Supplementary Movie S4). The CV and the cyst cytosol offer a protective environment for the pathogen's growth (Figure 4 and Supplementary Movie S5) until a cyst-lysis event releases the bacteria back into the environment (Figure 4 and Supplementary Movie S6).
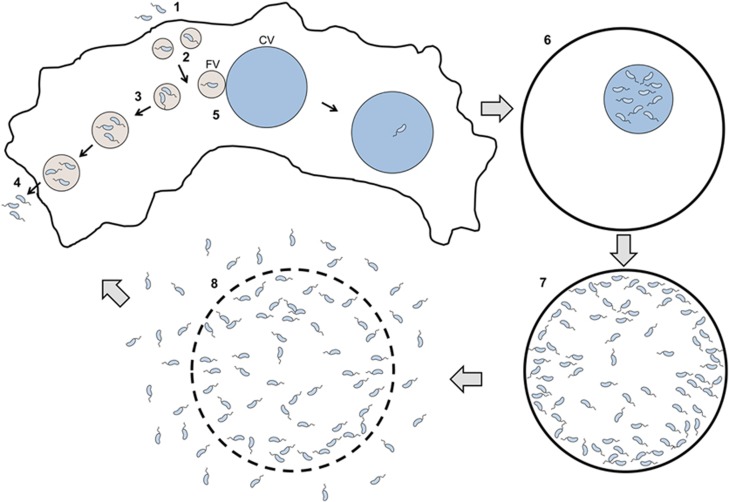
Figure 7.
Scheme summarizing the interaction between V. cholerae and A. castellanii. Upon phagocytosis of free V. cholerae cells (1) by amoebal trophozoites, the bacteria are contained in endosomal vacuoles (2). Fusion of endosomal vacuoles leads to the generation of large vacuoles (3). The content of small and large vacuoles is frequently emptied by A. castellanii (4), thereby releasing live bacteria back into the environment. At times endosomal vacuoles can also shuffle V. cholerae cells into the CV (5), which allows the bacteria to establish a replication niche. When amoebae containing such colonized CVs undergo encystment, the digestive vacuoles are emptied but V. cholerae remains within the cyst's CV (6). V. cholerae proliferates in this protective environment, ultimately leading to CV lysis (7). This process is dependent on VPS. Finally, weakening of the cyst cell wall occurs (8), leading to the release of the intracellular proliferating V. cholerae cells into the extracellular environment.
At this stage, it is unclear whether the encystment, CV destruction and/or amoebal lysis are actively induced by the pathogen or whether colonization of the CV hinders the normal osmoregulatory function of the organelle, which ultimately results in passive CV and cyst lysis. However, we provide evidence that the bacterial VPS is of prime importance for the lysis of the CV (Figure 6). We therefore hypothesize that the extracellular polysaccharide might hamper CV contraction-dependent osmoregulation of A. castellanii, potentially through an agglutination effect, which leads to lysis of the organelle and ultimately the whole amoeba. Such VPS-mediated agglutination is consistent with an impaired intracellular motility that was observed for the VPS-overproducing HapR-deficient strain (compare the even distribution of ΔhapR bacteria to the motility-driven circular localization of WT in the cytosol of the cysts; Figure 6a and Supplementary Movie S5 and S6). This study therefore suggests that selective pressure exerted on bacterial pathogens in their natural environments can lead to unique pathogenic traits such as the production of polysaccharide. Indeed, the majority of V. cholerae cells that lacked VPS were unable to exit their amoebal host (Figure 6), and this characteristic would therefore be counter-selected in nature. Intriguingly, the production of VPS is also important for intestinal colonization, as VPS-mutant strains of V. cholerae show a colonization defect in the infant mouse model compared with their WT parental strain (Fong et al., 2010).
Previous studies have suggested that survival of V. cholerae O1 strains (classical and El Tor) and A. castellanii is enhanced in cocultivation experiments performed in rich peptone yeast glucose medium (ATCC medium 712) (Abd et al., 2007). Moreover, the authors reported intracellular V. cholerae growth for up to 14 days (Abd et al., 2007), which is not consistent with the forced release of the bacteria that we observed in this study after intracellular proliferation (Figure 4 and Supplementary Movie S4–S6). Notably, when we repeated the experiments described by Abd et al., 2007 we observed massive extracellular proliferation of V. cholerae within the rich medium, which fully sustained bacterial growth. Thus, in agreement with the high numbers of extracellular V. cholerae detected by Abd et al. (2007, 2011) in the coculture experiments (allegedly up to 2 × 1011 CFU ml-1 as stated in Abd et al. (2007) and Abd et al. (2011)), it is possible that the authors scored recent phagocytosis events as ‘intracellularly growing bacteria'. Indeed, extracellular bacteria were not killed throughout the experiment but solely at the time of sampling (Abd et al., 2007). To avoid such extracellular proliferation of the pathogen in the current study, we used an infection medium that does not sustain bacterial growth. Moreover, in addition to scoring CFUs, we directly imaged the intracellular behavior and growth of V. cholerae. Using these methods, combined with electron microscopy imaging, we provide evidence for the absence of free V. cholerae within the cytosol of trophozoites, contrary to previous claims (Abd et al., 2005, 2007, 2011), even though mitochondria were easily detected (Figure 1b) (which can be misled for cross-sectioned bacteria if the mitochondrial cristae structures are not well preserved or overstained). Instead, we primarily uncovered V. cholerae in small and large food vacuoles as part of the phago-/endosomal pathway and frequently observed exocytosis of undigested bacteria (Figure 2). Most importantly, we consistently detected a small percentage of the amoebal population with proliferating V. cholerae cells contained within their CV (Figures 3, 4, 5). To our knowledge, this is the first report showing active colonization of the CV by a pathogen through vacuolar fusion, followed by proliferation, encystment of the host and CV lysis. Although, a previous study showed that rare CV colonization by Salmonella occurred when A. polyphaga encountered a high (packing) density of surrounding bacteria (specifically when amoebae were spotted onto bacterial lawns formed on agar plates and observed after 2–4 days p.p.c.) (Gaze et al., 2003). Gaze et al. (2003) hypothesized that the Salmonella entered the CV via reflux through the pore during contraction-dependent fluid discharge and concluded that ‘the fate of the amoebae and bacteria following this interaction is unknown'. Based on the data presented for V. cholerae in this study, we conclude that the CV represents a new replication niche in which the bacterium is protected against external stresses (e.g., phages, competing bacteria) and intracellular stresses (e.g., vacuole acidification, anti-microbial compounds). The proposed bacterial strategy avoids destruction of the entire predator population, which can be favorable for environmental purposes and would result in a long-term equilibrium between amoebae and V. cholerae. A similar scenario occurs for cholera as a human disease, in which the majority of infected persons do not show symptoms (WHO, 2011). Instead, only a small fraction of infected people suffer from severe diarrhea, which is accompanied by massive bacterial proliferation and the release of the pathogen back into the environment.
Acknowledgments
We thank Sandrine Borgeaud for technical assistance and Gilbert Greub and Sebastien Aeby for providing us with the Acanthamoeba castellanii strain and for valuable methodological advice. We are indebted to Graham Knott from the Bioelectron Microscopy Core Facility for discussions on the project. We also acknowledge Thierry Soldati for advice and helpful comments on the manuscript and John McKinney, Pierre Cosson, Matthieu Delincé, and Ana T. López-Jiménez for fruitful discussions. This work was supported by grants from the European Research Council (309064-VIR4ENV) and the Swiss National Science Foundation (31003A_143356) to MB.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abd H, Saeed A, Weintraub A, Nair GB, Sandström G. (2007). Vibrio cholerae O1 strains are facultative intracellular bacteria, able to survive and multiply symbiotically inside the aquatic free-living amoeba Acanthamoeba castellanii. FEMS Microbiol Ecol 60: 33–39. [DOI] [PubMed] [Google Scholar]
- Abd H, Shanan S, Saeed A, Sandström G. (2011). Survival of Vibrio cholerae inside Acanthamoeba and detection of both microorganisms from natural water samples may point out the amoeba as a protozoal host for V. cholerae. Bacteriol ParasitolS1-003; doi:10.4172/2155-9597.S1-003.
- Abd H, Weintraub A, Sandström G. (2005). Intracellular survival and replication of Vibrio cholerae O139 in aquatic free-living amoebae. Environ Microbiol 7: 1003–1008. [DOI] [PubMed] [Google Scholar]
- Allen RD. (2000). The contractile vacuole and its membrane dynamics. BioEssays 22: 1035–1042. [DOI] [PubMed] [Google Scholar]
- Andersen JB, Sternberg C, Poulsen LK, Bjorn SP, Givskov M, Molin S. (1998). New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol 64: 2240–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry L, Klein G, Martiel JL, Satre M. (1993). Kinetics of endosomal pH evolution in Dictyostelium discoideum amoebae. Study by fluorescence spectroscopy. J Cell Sci 105: 861–866. [DOI] [PubMed] [Google Scholar]
- Bao Y, Lies DP, Fu H, Roberts GP. (1991). An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of Gram-negative bacteria. Gene 109: 167–168. [DOI] [PubMed] [Google Scholar]
- Bartlett MS, Gourse RL. (1994). Growth rate-dependent control of the rrnB P1 core promoter in Escherichia coli. J Bacteriol 176: 5560–5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokesch M. (2012. a). TransFLP—a method to genetically modify V. cholerae based on natural transformation and FLP-recombination. J Vis Exp 68: e3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokesch M. (2012. b). Chitin colonization, chitin degradation and chitin-induced natural competence of Vibrio cholerae are subject to catabolite repression. Environ Microbiol 14: 1898–1912. [DOI] [PubMed] [Google Scholar]
- Borgeaud S, Blokesch M. (2013). Overexpression of the tcp gene cluster using the T7 RNA polymerase/promoter system and natural transformation-mediated genetic engineering of Vibrio cholerae. PLoS One 8: e53952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgeaud S, Metzger LC, Scrignari T, Blokesch M. (2015). The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347: 63–67. [DOI] [PubMed] [Google Scholar]
- Bowers B, Korn ED. (1968). The fine structure of Acanthamoeba castellanii. I. The trophozoite. J Cell Biol 39: 95–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers B, Korn ED. (1969). The fine structure of Acanthamoeba castellanii (Neff strain). II. Encystment. J Cell Biol 41: 786–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez-Munguia B, Salazar-Villatoro L, Lagunes-Guillen A, Omana-Molina M, Espinosa-Cantellano M, Martinez-Palomo A. (2013). Acanthamoeba castellanii cysts: new ultrastructural findings. Parasitol Res 112: 1125–1130. [DOI] [PubMed] [Google Scholar]
- Colwell RR. (1996). Global climate and infectious disease: the cholera paradigm. Science 274: 2025–2031. [DOI] [PubMed] [Google Scholar]
- De Souza Silva O, Blokesch M. (2010). Genetic manipulation of Vibrio cholerae by combining natural transformation with FLP recombination. Plasmid 64: 186–195. [DOI] [PubMed] [Google Scholar]
- Doberstein SK, Baines IC, Wiegand G, Korn ED, Pollard TD. (1993). Inhibition of contractile vacuole function in vivo by antibodies against myosin-I. Nature 365: 841–843. [DOI] [PubMed] [Google Scholar]
- Fong JC, Syed KA, Klose KE, Yildiz FH. (2010). Role of Vibrio polysaccharide (vps genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiology 156: 2757–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouque E, Trouilhe MC, Thomas V, Hartemann P, Rodier MH, Hechard Y. (2012). Cellular, biochemical, and molecular changes during encystment of free-living amoebae. Eukaryot Cell 11: 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaze WH, Burroughs N, Gallagher MP, Wellington EM. (2003). Interactions between Salmonella typhimurium and Acanthamoeba polyphaga, and observation of a new mode of intracellular growth within contractile vacuoles. Microb Ecol 46: 358–369. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Steele JA, Caporaso JG, Steinbruck L, Reeder J, Temperton B et al. (2012). Defining seasonal marine microbial community dynamics. ISME J 6: 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greub G, Raoult D. (2004). Microorganisms resistant to free-living amoebae. Clin Microbiol Rev 17: 413–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer BK, Bassler BL. (2003). Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol 50: 101–104. [DOI] [PubMed] [Google Scholar]
- Hoffmann C, Harrison CF, Hilbi H. (2014). The natural alternative: protozoa as cellular models for Legionella infection. Cell Microbiol 16: 15–26. [DOI] [PubMed] [Google Scholar]
- Khan NA. (2006). Acanthamoeba: biology and increasing importance in human health. FEMS Microbiol Rev 30: 564–595. [DOI] [PubMed] [Google Scholar]
- Khan NA. (2015) Acanthamoeba—Biology and Pathogenesis, 2nd edn. Caister Academic Press: Norfolk, UK. [Google Scholar]
- Martinez AJ, Visvesvara GS. (1997). Free-living, amphizoic and opportunistic amebas. Brain Pathol 7: 583–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvig RL, Blokesch M. (2010). Natural transformation of Vibrio cholerae as a tool-optimizing the procedure. BMC Microbiol 10: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz C, Kjelleberg S. (2005). Off the hook—how bacteria survive protozoan grazing. Trends Microbiol 13: 302–307. [DOI] [PubMed] [Google Scholar]
- Matz C, McDougald D, Moreno AM, Yung PY, Yildiz FH, Kjelleberg S. (2005). Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc Natl Acad Sci USA 102: 16819–16824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meibom KL, Blokesch M, Dolganov NA, Wu C-Y, Schoolnik GK. (2005). Chitin induces natural competence in Vibrio cholerae. Science 310: 1824–1827. [DOI] [PubMed] [Google Scholar]
- Morens DM, Folkers GK, Fauci AS. (2004). The challenge of emerging and re-emerging infectious diseases. Nature 430: 242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Miller MC, Nielsen AT, Schoolnik GK, Spormann AM. (2007). vpsA- and luxO-independent biofilms of Vibrio cholerae. FEMS Microbiol Lett 275: 199–206. [DOI] [PubMed] [Google Scholar]
- Nelson EJ, Chowdhury A, Harris JB, Begum YA, Chowdhury F, Khan AI et al. (2007). Complexity of rice-water stool from patients with Vibrio cholerae plays a role in the transmission of infectious diarrhea. Proc Natl Acad Sci USA 104: 19091–19096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Bassler BL. (2009). Bacterial quorum-sensing network architectures. Annu Rev Genet 43: 197–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AT, Dolganov NA, Rasmussen T, Otto G, Miller MC, Felt SA et al. (2010). A bistable switch and anatomical site control Vibrio cholerae virulence gene expression in the intestine. PLoS Pathogen 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernthaler J. (2005). Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol 3: 537–546. [DOI] [PubMed] [Google Scholar]
- Platt TG, Fuqua C. (2010). What's in a name? The semantics of quorum sensing. Trends Microbiol 18: 383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA 103: 1528–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, Bassler BL. (2012). Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harbor Perspect Med 2: a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandström G, Saeed A, Abd H. (2010). Acanthamoeba polyphaga is a possible host for Vibrio cholerae in aquatic environments. Exp Parasitol 126: 65–68. [DOI] [PubMed] [Google Scholar]
- Seitz P, Blokesch M. (2013). Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol Rev 37: 336–363. [DOI] [PubMed] [Google Scholar]
- Shanan S, Abd H, Hedenström I, Saeed A, Sandström G. (2011). Detection of Vibrio cholerae and Acanthamoeba species from same natural water samples collected from different cholera endemic areas in Sudan. BMC Res Notes 4: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui R, Khan NA. (2012). Biology and pathogenesis of Acanthamoeba. Parasites Vectors 5: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelling WJ, Moore JE, McKenna JP, Lecky DM, Dooley JS. (2006). Bacterial-protozoa interactions; an update on the role these phenomena play towards human illness. Microbes Infect 8: 578–587. [DOI] [PubMed] [Google Scholar]
- Solomon JM, Isberg RR. (2000). Growth of Legionella pneumophila in Dictyostelium discoideum: a novel system for genetic analysis of host-pathogen interactions. Trends Microbiol 8: 478–480. [DOI] [PubMed] [Google Scholar]
- Sternberg C, Christensen BB, Johansen T, Toftgaard Nielsen A, Andersen JB, Givskov M et al. (1999). Distribution of bacterial growth activity in flow-chamber biofilms. Appl Environ Microbiol 65: 4108–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JR, Weisman RA. (1972). Exocytosis of latex beads during the encystment of Acanthamoeba. J Cell Biol 52: 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Kjelleberg S, McDougald D. (2013). Relative contributions of Vibrio polysaccharide and quorum sensing to the resistance of Vibrio cholerae to predation by heterotrophic protists. PLoS One 8: e56338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MS, Hammer BK. (2000). Legionella pneumophila pathogesesis: a fateful journey from amoebae to macrophages. Annu Rev Microbiol 54: 567–613. [DOI] [PubMed] [Google Scholar]
- Takemura AF, Chien DM, Polz MF. (2014). Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front Microbiol 5: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschler JK, Zamorano-Sanchez D, Utada AS, Warner CJ, Wong GC, Linington RG et al. (2015). Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat Rev Microbiol 13: 255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thom S, Warhurst D, Drasar BS. (1992). Association of Vibrio cholerae with fresh water amoebae. J Med Microbiol 36: 303–306. [DOI] [PubMed] [Google Scholar]
- WHO. (2011) Fact sheet No.107. WHO: Geneva, Switzerland.
- Yildiz F, Fong J, Sadovskaya I, Grard T, Vinogradov E. (2014). Structural characterization of the extracellular polysaccharide from Vibrio cholerae O1 El-Tor. PLoS One 9: e86751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Schoolnik GK. (1998). Role of rpoS in stress survival and virulence of Vibrio cholerae. J Bacteriol 180: 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Schoolnik GK. (1999). Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci USA 96: 4028–4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yooseph S, Nealson KH, Rusch DB, McCrow JP, Dupont CL, Kim M et al. (2010). Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature 468: 60–66. [DOI] [PubMed] [Google Scholar]
- Zheng J, Shin OS, Cameron DE, Mekalanos JJ. (2010). Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc Natl Acad Sci USA 107: 21128–21133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Mekalanos JJ. (2003). Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev Cell 5: 647–656. [DOI] [PubMed] [Google Scholar]
- Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ. (2002). Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA 99: 3129–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.