Abstract
This study was to determine the association between several single nucleotide polymorphisms (SNPs) in the dedicator of cytokinesis 7 (DOCK7), proprotein convertase subtilisin/kexin type 9 (PCSK9) and polypeptide N-acetylgalactosaminyltransferase 2 (GALNT2) and serum lipid levels. Genotyping of 9 SNPs was performed in 881 Jing subjects and 988 Han participants. Allele and genotype frequencies of the detected SNPs were different between the two populations. Several SNPs were associated with triglyceride (TG, rs10889332, rs615563, rs7552841, rs1997947, rs2760537, rs4846913 and rs11122316), high-density lipoprotein (HDL) cholesterol (rs1997947), low-density lipoprotein (LDL) cholesterol (rs1168013 and rs7552841), apolipoprotein (Apo) A1 (rs1997947), ApoB (rs10889332 and rs7552841), and ApoA1/ApoB ratio (rs7552841) in Jing minority; and with TG (rs10889332, rs615563, rs7552841, rs11206517, rs1997947, rs4846913 and rs11122316), HDL cholesterol (rs11206517 and rs4846913), LDL cholesterol (rs1168013), ApoA1 (rs11206517 and rs4846913), ApoB (rs7552841), and ApoA1/ApoB ratio (rs4846913) in Han nationality. Strong linkage disequilibria were noted among the SNPs. The commonest haplotype was G-C-G-C-T-G-C-C-G (>10%). The frequencies of C-C-G-C-T-G-T-C-G, G-C-A-C-T-G-C-C-G, G-C-G-C-T-A-C-C-A, G-C-G-C-T-G-C-C-A, G-C-G-C-T-G-T-C-A haplotypes were different between the two populations. Haplotypes could explain much more serum lipid variation than any single SNP alone especially for TG. Differences in lipid profiles between the two populations might partially attribute to these SNPs and their haplotypes.
Cardiovascular disease (CVD) remains as the leading cause of morbidity and mortality worldwide, and its prevalence is expected to increase further, which exerts a significant economic burden1,2. Most of the current prevention strategies are focused on identifying and managing the established risk factors including hyperlipidemia3 that can be effectively addressed for individuals and populations suffering from atherosclerosis. It is generally agreed that dyslipidemia is complex and the result of the interactions4,5 of multiple genes6,7,8 and multiple environmental factors9,10. Statins are highly effective for lowering low-density lipoprotein (LDL) cholesterol levels and, consequently, cardiovascular event rates. However, statins do not eliminate cardiovascular risk. High triglyceride (TG) level is a significant risk factor for independent cardiovascular disease and is a marker for atherogenic remnant lipoproteins, such as very low-density lipoprotein (VLDL) cholesterol. Additionally, with elevated TG levels, a combination of LDL cholesterol with VLDL cholesterol in the measure of non-high-density lipoprotein (HDL) cholesterol may be a better predictor of cardiovascular risk than LDL cholesterol alone. Therefore, improved understanding of TG-related loci may optimize patient management strategies, provide potential new targets for future individual therapy, and thereby improve patients’ chances for survival.
Candidate gene and genome-wide association studies (GWASs)11,12,13 have identified a number of sequence variants that explain some of the individual variation in the susceptibility for high TG levels. The dedicator of cytokinesis 7 (DOCK7; Gene ID: 85440; MIM: 615730) formerly known as ZIR2 and EIEE23, is located on chromosome 1p31.3 and the protein encoded by this gene is a guanine nucleotide exchange factor (GEF) that plays a role in axon formation and neuronal polarization. The encoded protein displays GEF activity toward RAC1 and RAC3 Rho small GTPases but not toward CDC42. Several transcript variants encoding different isoforms have been found for this gene. The proprotein convertase subtilisin/kexin type 9 (PCSK9; Gene ID: 255738; MIM: 607786) gene, also known as FH3, PC9, NARC1, LDLCQ1, NARC-1 and HCHOLA3, is located on 1p32.3 and this gene encodes a member of the subtilisin-like proprotein convertase family, which includes proteases that process protein and peptide precursors trafficking through regulated or constitutive branches of the secretory pathway. The encoded protein undergoes an autocatalytic processing event with its prosegment in the ER and is constitutively secreted as an inactive protease into the extracellular matrix and trans-Golgi network. It is expressed in liver, intestine and kidney tissues and escorts specific receptors for lysosomal degradation. It plays a role in cholesterol and fatty acid metabolism. Mutations in this gene have been associated with autosomal dominant familial hypercholesterolemia. Alternative splicing can result in multiple transcript variants. The polypeptide N-acetylgalactosaminyltransferase 2 (GALNT2; Gene ID: 2590; MIM: 602274) gene, formerly known as GalNAc-T2, is located on chromosome 1q41-q42 and encodes a member of the glycosyltransferase 2 protein family. Members of this family initiate mucin-type O-glycoslation of peptides in the Golgi apparatus. The encoded protein may be involved in O-linked glycosylation of the immunoglobulin A1 hinge region. This gene may influence TG levels, and may be involved type 2 diabetes, as well as several types of cancer. Alternative splicing can also result in multiple transcript variants (http://www.ncbi.nlm.nih.gov/gene/).
Human genetic studies of lipid levels can identify targets for new therapies for cholesterol management and prevention of heart disease especially monoclonal anti-PCSK9 antibodies are already on the market to significantly reduced levels of LDL cholesterol when added to statin therapy administered at the maximum tolerated dose14. For comparison with the nonsynonymous single nucleotide polymorphisms (SNPs) in known drug therapies genes, we scored point mutations at synonymous point mutations in housekeeping genes or genes of unknown function on the approximate locations of the chromosome 1. The exact positions of the PCSK9 SNPs were located in the similar position area of the DOCK7 and GLANT2 SNPs (http://hapmap.ncbi.nlm.nih.gov/). Several genetic variants in the DOCK7, PCSK9 and GLANT2 have been associated with serum lipid parameters, especially with TG in Western populations15, e.g. the SNPs of DOCK7 rs1167998, rs1088935316, PCSK9 rs1159114717 and GALNT2 rs484691413 were associated with TG levels in European and PCSK9 rs50515118 and GALNT2 rs2144300 and rs484691419 in the Asian populations. However, the association of the DOCK7 (rs1168013 and rs10889332), PCSK9 (rs615563, rs7552841 and rs1126517) and GALNT2 (rs1997947, rs2760537, rs4846913 and rs11122316) SNPs and serum lipid levels has not been previously reported. Since ancient times China is a multi-ethnic country. Among 56 nationalities in China, the Han nationality is the biggest one. Jing is one of the smallest population of ethnic minorities in southern China with a population of 22,517 (in 2000 the fifth national census statistics of China), China’s only a coastal fishery ethnic minority, and China’s only national ocean at the same time20. Jing populations live in Dongxing city, Guangxi Zhuang Autonomous Region. Diet to rice is given priority to, fresh fish and shrimp more, like to use fish sauce to taste. The history of Jing ethnic minority shows Jing nationality is a relatively conservative and isolated minority, and preserves their custom of intra-ethnic marriage21. Thus, their genetic background may be less heterogeneous within the population. Little is known about the association of SNPs and lipid phenotypes in the Jing population. Therefore, this research was undertaken to detect the association of the DOCK7 rs1168013, DOCK7 rs10889332, PCSK9 rs615563, PCSK9 rs7552841, PCSK9 rs11206517, GALNT2 rs1997947, GALNT2 rs2760537, GALNT2 rs4846913, and GALNT2 rs11122316 SNPs and lipid profiles in the two ethnic groups.
Results
Demographic and clinical characteristics
Table 1 summarized the value of weight, waist circumference, body mass index (BMI), and total cholesterol (TC) and TG levels which were higher and the % of participants who consumed alcohol and the ratio of apolipoprotein (Apo) A1 to ApoB were lower in Jing ethnic minority than in Han nationality (P < 0.01–0.001). However, no such difference in the levels of HDL cholesterol, LDL cholesterol, ApoA1 and ApoB between the two populations (P > 0.05 for all).
Table 1. Lipid profiles and clinical characteristics in the two ethnic groups.
| Characteristics | Jing | Han | test-statistic | P-value |
|---|---|---|---|---|
| Number (n) | 881 | 988 | ||
| Gender (Male/Female) | 456/425 | 536/452 | 1.161 | 0.281 |
| Age (years) | 56.69 ± 13.391 | 56.18 ± 12.85 | −0.726 | 0.468 |
| Height (cm) | 156.51 ± 7.67 | 156.03 ± 7.79 | 1.133 | 0.257 |
| Weight (kg) | 57.62 ± 9.86 | 55.66 ± 9.37 | 3.768 | 0.000 |
| Body mass index (kg/m2) | 23.46 ± 3.25 | 22.82 ± 3.23 | 3.642 | 0.000 |
| Waist circumference (cm) | 79.98 ± 9.04 | 77.49 ± 8.94 | 5.115 | 0.000 |
| SBP (mmHg) | 131.65 ± 22.03 | 132.66 ± 41.95 | −0.550 | 0.582 |
| DBP (mmHg) | 80.37 ± 10.54 | 80.84 ± 10.17 | −0.848 | 0.397 |
| Pulse pressure (mmHg) | 51.28 ± 17.54 | 51.82 ± 10.15 | −0.315 | 0.753 |
| Cigarette smoking [n (%)] | ||||
| Nonsmoker | 775 (87.9) | 846 (85.6) | ||
| ≤20 Cigarettes/day | 27 (3.1) | 36 (3.6) | 1.634 | 0.442 |
| >20 Cigarettes/day | 79 (9.0) | 106 (10.8) | ||
| Alcohol consumption [n (%)] | ||||
| Nondrinker | 788 (89.4) | 834 (84.4) | ||
| ≤25 g/day | 50 (5.7) | 35 (3.5) | 25.016 | 0.000 |
| >25 g/day | 43 (4.9) | 119 (12.1) | ||
| Blood glucose level (mmol/L) | 6.71 ± 1.71 | 6.63 ± 1.08 | 0.983 | 0.326 |
| Total cholesterol (mmol/L) | 5.13 ± 0.93 | 4.89 ± 0.87 | 4.834 | 0.000 |
| Triglyceride (mmol/L) | 1.41 (1.12)2 | 1.32 (1.09) | −2.890 | 0.004 |
| HDL cholesterol (mmol/L) | 1.79 ± 0.52 | 1.80 ± 0.45 | −0.231 | 0.817 |
| LDL cholesterol (mmol/L) | 2.85 ± 0.44 | 2.82 ± 0.44 | 1.314 | 0.189 |
| Apolipoprotein (Apo) A1 (g/L) | 1.30 ± 0.23 | 1.32 ± 0.20 | −1.592 | 0.112 |
| ApoB (g/L) | 1.06 ± 0.25 | 1.03 ± 0.24 | 1.837 | 0.066 |
| ApoA1/ApoB | 1.30 ± 0.38 | 1.35 ± 0.37 | −2.465 | 0.014 |
SBP, systolic blood pressure; DBP, diastolic blood pressure; HDL, high density lipoprotein; LDL, low density lipoprotein;
1Mean ± SD determined by t-test.
2Median (interquartile range) tested by the Wilcoxon-Mann-Whitney test.
Genotyping
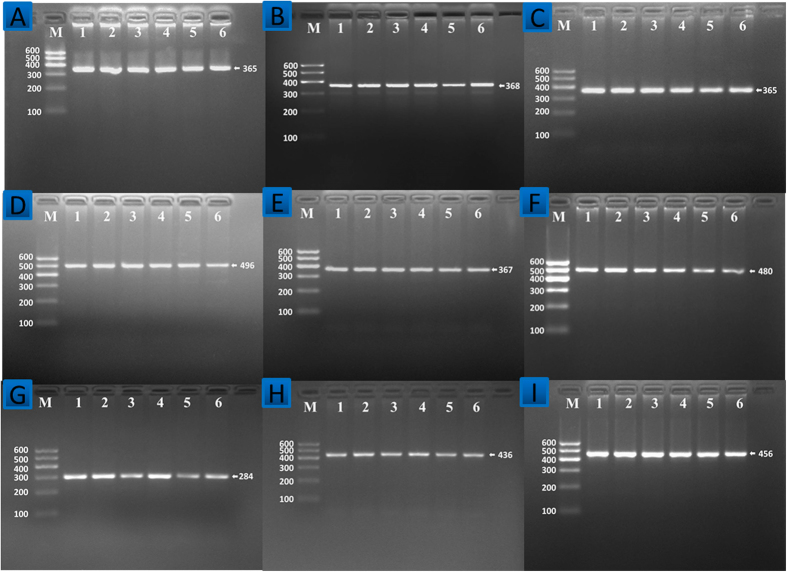
Polymerase chain reaction (PCR) products of DOCK7 rs1168013, DOCK7 rs10889332, PCSK9 rs615563, PCSK9 rs7552841, PCSK9 rs1126517, GALNT2 rs1997947, GALNT2 rs2760537, GALNT2 rs4846913 and GALNT2 rs11122316 SNPs were 365-, 368-, 365-, 496-, 367-, 480-, 284-, 436- and 456-bp nucleotide sequences after electrophoresis; respectively (Fig. 1). After restriction fragment length polymorphism (RFLP) reaction and then imaged by 2% agarose gel electrophoresis, the genotypes of the SNPs identified were labeled according to the presence and absence of the enzyme restriction sites (Fig. 2).
Figure 1. Agarose gel electrophoresis (2%) of PCR products of the DOCK7, PCSK9 and GALNT2 SNPs.
Lane M: DNA ladder 100bp; PCR amplicon of (A) DOCK7 rs1168013, (B) DOCK7 rs10889332, (C) PCSK9 rs615563, (D) PCSK9 rs7552841, (E) PCSK9 rs11206517, (F) GALNT2 rs1997947, (G) GALNT2 rs2760537, (H) GALNT2 rs4846913 and (I) GALNT2 rs11122316 SNPs were 365-, 368-, 365-, 496-, 367-, 480-, 284-, 436- and 456-bp nucleotide sequences; respectively.
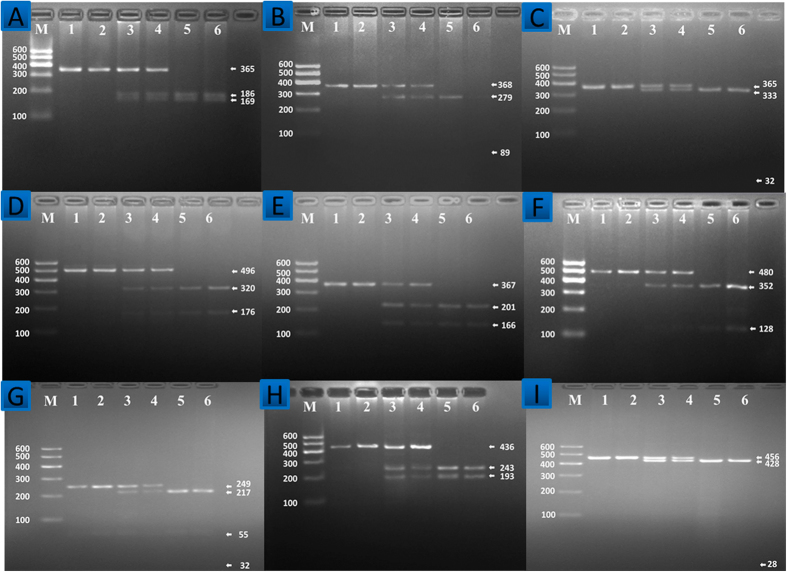
Figure 2. Agarose gel electrophoresis (2%) of genotyping of the DOCK7, PCSK9 and GALNT2 SNPs. Lane M: DNA ladder100bp.
The genotypes of 9 SNPs were as follow: (A) DOCK7 rs1168013: CC (Lanes 1 and 2, 365-bp); CG (lanes 3 and 4, 365-, 186- and 169-bp); and GG genotype (lanes 5 and 6, 186- and 169-bp). (B) DOCK7 rs10889332: TT (lanes 1 and 2, 368-bp); CT (lanes 3 and 4, 368-, 279- and 89-bp); and CC genotype (lanes 5 and 6, 279- and 89-bp). (C) PCSK9 rs615563: AA (lanes 1 and 2, 365-bp); AG (lanes 3 and 4, 365-, 333-, 32-bp); and GG genotype (lanes 5 and 6, 333- and 32-bp). (D) PCSK9 rs7552841: TT (lanes 1 and 2, 496-bp); CT (lanes 3 and 4, 496-, 320- and 176-bp); and CC genotype (lanes 5 and 6, 320- and 176-bp). (E) PCSK9 rs11206517: TT (lanes 1 and 2, 367-bp); GT (lanes 3 and 4, 367-, 201- and 166-bp); and GG (lanes 5 and 6, 201- and 166-bp). (F) GALNT2 rs1997947: AA (lanes 1 and 2, 480-bp); AG (lanes 3 and 4, 480-, 352- and 128-bp); and GG genotype (lanes 5 and 6, 352- and 128-bp). (G) GALNT2 rs2760537: TT (lanes 1 and 2, 249- and 55-bp); CT (lanes 3 and 4, 249-, 217-, 55- and 32-bp); and CC genotype (lanes 5 and 6, 217-, 55- and 32-bp). (H) GALNT2 rs4846913: AA (lanes 1 and 2, 436-bp); AC (lanes 3 and 4, 436-, 243-and 193-bp); and CC genotype (lanes 5 and 6, 243- and 193-bp). (I) GALNT2 rs11122316: AA (lanes 1 and 2, 456-bp); AG (lanes 3 and 4, 456-, 428- and 28-bp); and GG genotype (lanes 5 and 6, 428- and 28-bp). The less than 90-bp fragment was invisible in the gel owing to its fast migration speed.
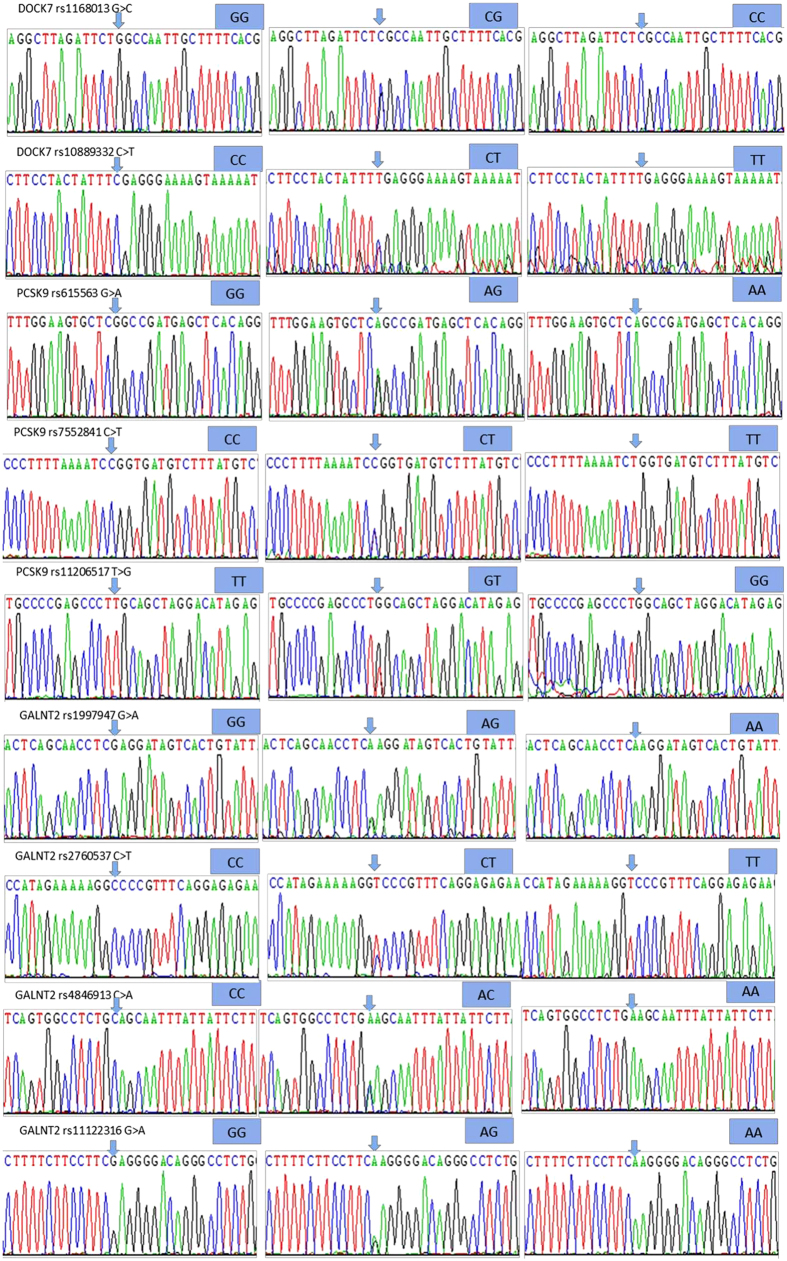
Results of sequencing
The genotypes shown in Fig 2 by PCR-RFLP, the genotypes were also confirmed by the nucleotide direct sequencing (Fig. 3); respectively.
Figure 3. The parts of the nucleotide direct sequencing results of the DOCK7, PCSK9 and GALNT2 SNPs.
DOCK7: dedicator of cytokinesis 7, PCSK9: proprotein convertase subtilisin/kexin type 9 and GALNT2: polypeptide N-acetylgalactosaminyltransferase 2.
Allelic and genotypic frequencies
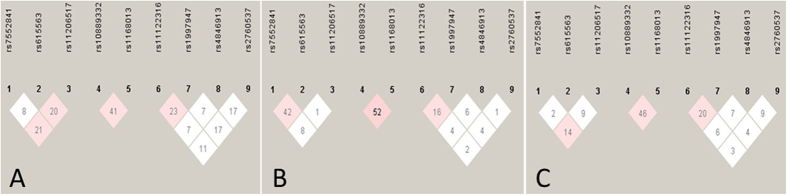
Tables 2 and 3 describe the allelic and genotypic frequencies of the detected SNPs which were different between the two ethnic groups (P < 0.05 for all). All of the detected SNPs were in the Hardy-Weinberg equilibrium (P > 0.05) except DOCK7 rs10889332 (P < 0.05). Linkage disequilibria (LD) were found between PCSK9 rs6165563 and PCSK9 rs11206517, PCSK9 rs7552841 and PCSK9 rs11206517, DOCK7 rs1168013 and DOCK7 rs10889332 and GALNT2 rs11122316 and GALNT2 rs1997947 in Jing, and PCSK9 rs7552841 and PCSK9 rs615563, DOCK7 rs1168013 and DOCK7 rs10889332 and GALNT2 rs11122316 and GALNT2 rs1997947 in Han (P < 0.01 for all; Fig. 4).
Table 2. Prevalence of genotype frequencies in the different populations [n (%)].
| SNP | Genotype | Jing (n = 881) | Han (n = 988) | X2 | P-value |
|---|---|---|---|---|---|
| DOCK7 rs1168013 G > C | GG | 367 (41.65) | 482 (48.75) | 7.457 | 0.024 |
| CG | 409 (46.40) | 413 (41.81) | |||
| CC | 105 (11.95) | 93 (9.44) | |||
| HWE (P) | 0.581 | 0.739 | |||
| DOCK7 rs10889332 C > T | CC | 447 (50.69) | 575 (58.19) | 10.851 | 0.004 |
| CT | 341 (38.74) | 347 (35.14) | |||
| TT | 93 (10.57) | 66 (6.67) | |||
| HWE (P) | 0.023 | 0.168 | |||
| PCSK9 rs615563 G > A | GG | 536 (60.80) | 664 (67.22) | 6.223 | 0.045 |
| AG | 294 (33.38) | 279 (28.20) | |||
| AA | 51 (5.82) | 45 (4.58) | |||
| HWE (P) | 0.208 | 0.271 | |||
| PCSK9 rs7552841 C > T | CC | 571 (64.78) | 689 (69.72) | 6.063 | 0.048 |
| CT | 264 (30.01) | 269 (27.22) | |||
| TT | 46 (5.21) | 30 (3.06) | |||
| HWE (P) | 0.359 | 0.549 | |||
| PCSK9 rs11206517 T > G | TT | 723 (82.08) | 859 (86.95) | 6.357 | 0.042 |
| GT | 146 (16.54) | 121 (12.22) | |||
| GG | 12(1.38) | 8 (0.83) | |||
| HWE (P) | 0.142 | 0.108 | |||
| GALNT2 rs1997947 G > A | GG | 529 (60.03) | 661 (66.94) | 7.586 | 0.023 |
| AG | 297 (33.69) | 283 (28.61) | |||
| AA | 55 (6.28) | 44 (4.45) | |||
| HWE (P) | 0.129 | 0.056 | |||
| GALNT2 rs2760537 C > T | CC | 344 (39.05) | 446 (45.14) | 6.291 | 0.043 |
| CT | 406 (46.10) | 427 (43.19) | |||
| TT | 131 (14.85) | 115 (11.67) | |||
| HWE (P) | 0.531 | 0.408 | |||
| GALNT2 rs4846913 C > A | CC | 554 (62.94) | 685 (69.30) | 6.292 | 0.043 |
| AC | 281 (31.85) | 263 (26.67) | |||
| AA | 46 (5.21) | 40 (4.03) | |||
| HWE (P) | 0.188 | 0.232 | |||
| GALNT2 rs11122316 G > A | GG | 320 (36.29) | 410 (41.53) | 6.541 | 0.038 |
| AG | 429 (48.70) | 468 (47.36) | |||
| AA | 132 (15.01) | 110 (11.11) | |||
| HWE (P) | 0.546 | 0.171 |
SNP: single nucleotide polymorphism; HDL, high density lipoprotein; LDL, low density lipoprotein; HWE, Hardy-Weinberg equilibrium; DOCK7: Dedicator of cytokinesis 7; PCSK9: Proprotein convertase subtilisin/kexin type 9; GALNT2: N-acetylgalactosaminyltransferase 2.
Table 3. Prevalence of allele frequencies in the different populations [n (%)].
| SNP | Allele | Jing (n = 881) | Han (n = 988) | X2 | P-value |
|---|---|---|---|---|---|
| DOCK7 rs1168013 | G/C | 1143 (64.85)/619 (35.15) | 1376 (69.65)/600 (30.35) | 7.173 | 0.007 |
| DOCK7 rs10889332 | C/T | 1234 (70.06)/528 (29.94) | 1497 (75.76)/479 (24.24) | 11.313 | 0.001 |
| PCSK9 rs615563 | G/A | 1365 (77.49)/397 (22.51) | 1607 (81.32)/369 (18.68) | 6.167 | 0.013 |
| PCSK9 rs7552841 | C/T | 1406 (79.79)/356 (20.21) | 1647 (83.33)/329 (16.67) | 5.752 | 0.016 |
| PCSK9 rs11206517 | T/G | 1592 (90.35)/170 (9.65) | 1839 (93.06)/137 (6.94) | 6.627 | 0.010 |
| GALNT2 rs1997947 | G/A | 1355 (76.88)/407 (23.12) | 1606 (81.25)/370 (18.75) | 7.945 | 0.005 |
| GALNT2 rs2760537 | C/T | 1094 (62.10)/668 (37.90) | 1319 (66.74)/657 (33.26) | 6.437 | 0.011 |
| GALNT2 rs4846913 | C/A | 1390 (78.87)/372 (21.13) | 1633 (82.64)/343 (17.36) | 6.293 | 0.012 |
| GALNT2 rs11122316 | G/A | 1068 (60.64)/694 (39.36) | 1289 (65.21)/687 (34.79) | 6.126 | 0.013 |
SNP: single nucleotide polymorphism; DOCK7: Dedicator of cytokinesis 7; PCSK9: Proprotein convertase subtilisin/kexin type 9; GALNT2: N-acetylgalactosaminyltransferase 2.
Figure 4. The linkage disequilibrium (LD) of the DOCK7, PCSK9 and GALNT2 SNPs.
LD among the (1) PCSK9 rs7552841, (2)PCSK9 rs615563, (3) PCSK9 rs11206517, (4)DOCK7 rs10889332, (5) DOCK7 rs1168013, (6) GALNT2 rs11122316, (7) GALNT2 rs1997947, (8) GALNT2 rs4846913 and (9) GALNT2 rs2760537 SNPs in the Jing (A), Han (B) and combined Jing and Han populations (C). The LD status is expounded by the r2 value.
Haplotype frequencies
The haplotype frequencies are listed in Table 4. The commonest haplotype was G-C-G-C-T-G-C-C-G (in the order of DOCK7 rs1168013, DOCK7 rs10889332, PCSK9 rs615563, PCSK9 rs7552841, PCSK9 rs11206517, GALNT2 rs1997947, GLANT2 rs2760537, GLANT2 rs4846913 and GLANT2 rs11122316; >10% of the samples). The frequencies of the C-C-G-C-T-G-T-C-G, G-C-A-C-T-G-C-C-G, G-C-G-C-T-A-C-C-A, G-C-G-C-T-G-C-C-A, and G-C-G-C-T-G-T-C-A haplotypes were also different between the Jing and Han populations (P < 0.05 − 0.001).
Table 4. Frequencies of haplotypes among 9 SNPs of the DOCK7, PCSK9 and GALNT2 genes in the two ethnic groups [n (%)].
| Haplotype | Jing | Han | X2 | P-value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | ||||
| C | C | G | C | T | G | C | C | G | 61 (3.1) | 47 (2.7) | 0.011 | 0.916 |
| C | C | G | C | T | G | T | C | G | 5 (0.2) | 59 (3.3) | 63.522 | 1.66 × 10−15 |
| G | C | A | C | T | G | C | C | G | 92 (4.7) | 47 (2.7) | 6.786 | 0.009 |
| G | C | G | C | T | A | C | C | A | 4 (0.2) | 55 (3.1) | 60.831 | 6.52 × 10−15 |
| G | C | G | C | T | G | C | C | A | 133 (6.8) | 66 (3.7) | 11.968 | 0.001 |
| G | C | G | C | T | G | C | C | G | 262 (13.3) | 187 (10.6) | 1.908 | 0.167 |
| G | C | G | C | T | G | T | C | A | 80 (4.1) | 42 (2.4) | 5.642 | 0.018 |
| G | C | G | C | T | G | T | C | G | 105 (5.3) | 86 (4.9) | 0.088 | 0.767 |
| Rare Hap (frequency <3%) in both Jing & Han populations has been dropped | ||||||||||||
A, DOCK7 rs1168013; B, DOCK7 rs10889332; C, PCSK9 rs615563; D, PCSK9 rs7552841; E, PCSK9 rs11206517; F, GALNT2 rs1997947; G, GLANT2 rs2760537; H, GLANT2 rs4846913; I, GLANT2 rs11122316; DOCK7, Dedicator of cytokinesis 7; PCSK9, Proprotein convertase subtilisin/kexin type 9; GALNT2, N-acetylgalactosaminyltransferase 2.
Genotypes and lipid parameters
Table 5 shows that the levels of TG (DOCK7 rs10889332, PCSK9 rs615563, PCSK9 rs7552841, GALNT2 rs1997947, GALNT2 rs2760537, GALNT2 rs4846913 and GALNT2 rs11122316), HDL cholesterol (GALNT2 rs1997947), LDL cholesterol (DOCK7 rs1168013 and PCSK9 rs7552841), ApoA1 (GALNT2 rs1997947), ApoB (DOCK7 rs10889332 and PCSK9 rs7552841) and the ratio of ApoA1 to ApoB (PCSK9 rs7552841) in the Jing ethnic minority were different among the three genotypes (P < 0.005–0.001), whereas the levels of TG (DOCK7 rs10889332, PCSK9 rs615563, PCSK9 rs7552841, PCSK9 rs11206517, GALNT2 rs1997947, GALNT2 rs4846913 and GALNT2 rs11122316), HDL cholesterol (PCSK9 rs11206517 and GALNT2 rs4846913), LDL cholesterol (DOCK7 rs1168013), ApoA1 (PCSK9 rs11206517 and GALNT2 rs4846913), ApoB (PCSK9 rs7552841) and the ratio of ApoA1 to ApoB (GALNT2 rs4846913) in the Han nationality were different among the genotypes (P < 0.005–0.001). When both minor homozygous and heterozygous were combined to enhance the power, the levels of TC (DOCK7 rs1168013 and PCSK9 rs7552841), TG (DOCK7 rs10889332, PCSK9 rs7552841, PCSK9 rs11206517, GALNT2 rs1997947, GALNT2 rs2760537, GALNT2 rs4846913 and GALNT2 rs11122316), HDL cholesterol (PCSK9 rs11206517, GALNT2 rs1997947), LDL cholesterol (DOCK7 rs1168013 and PCSK9 rs7552841), ApoA1 (GALNT2 rs1997947), ApoB (DOCK7 rs10889332 and PCSK9 rs7552841) and the ratio of ApoA1 to ApoB (PCSK9 rs7552841) in the Jing ethnic minority were found to be different between the two genotypes (P < 0.005–0.001); whereas the levels of TG (PCSK9 rs615563 and GALNT2 rs4846913), HDL cholesterol (PCSK9 rs11206517, GALNT2 rs1997947 and GALNT2 rs4846913), ApoA1 (PCSK9 rs11206517, GALNT2 rs1997947 and GALNT2 rs4846913) and the ratio of ApoA1 to ApoB (DOCK7 rs10889332, GALNT2 rs1997947 and GALNT2 rs4846913) in the Han nationality were different between the genotypes (P < 0.005–0.001).
Table 5. Lipid profiles according to genotypes for the two ethnic groups.
| Genotype | n | Total cholesterol (mmol/L) | Triglyceride (mmol/L) | HDL cholesterol (mmol/L) | LDL cholesterol (mmol/L) | Apolipoprotein (Apo) A1 (g/L) | Apolipoprotein (Apo) B (g/L) | ApoA1/ ApoB |
|---|---|---|---|---|---|---|---|---|
| DOCK7 rs1168013 G > C | ||||||||
| Jing | ||||||||
| GG | 367 | 4.97 ± 0.92 | 1.40(1.10) | 1.81 ± 0.47 | 2.67 ± 0.55 | 1.31 ± 0.21 | 1.04 ± 0.24 | 1.31 ± 0.37 |
| CG | 409 | 5.13 ± 0.94 | 1.41 (1.14) | 1.80 ± 0.42 | 2.83 ± 0.42 | 1.30 ± 0.25 | 1.06 ± 0.28 | 1.29 ± 0.39 |
| CC | 105 | 5.17 ± 0.91 | 1.51 (1.18) | 1.71 ± 0.45 | 2.84 ± 0.42 | 1.29 ± 0.19 | 1.07 ± 0.25 | 1.29 ± 0.37 |
| F | 4.430 | 3.504 | 1.840 | 6.747 | 0.225 | 3.360 | 1.390 | |
| P | 0.012 | 0.031 | 0.160 | 0.001 | 0.775 | 0.035 | 0.250 | |
| Han | ||||||||
| GG | 482 | 4.80 ± 0.87 | 1.27 (1.04) | 1.86 ± 0.54 | 2.80 ± 0.44 | 1.35 ± 0.21 | 1.00 ± 0.24 | 1.37 ± 0.39 |
| CG | 413 | 4.95 ± 0.87 | 1.31 (1.06) | 1.75 ± 0.51 | 2.87 ± 0.44 | 1.30 ± 0.21 | 1.04 ± 0.25 | 1.36 ± 0.36 |
| CC | 93 | 5.09 ± 0.72 | 1.60 (1.26) | 1.65 ± 0.45 | 2.97 ± 0.41 | 1.30 ± 0.19 | 1.14 ± 0.23 | 1.18 ± 0.29 |
| F | 2.894 | 3.458 | 2.436 | 5.794 | 1.491 | 3.561 | 1.960 | |
| P | 0.056 | 0.032 | 0.088 | 0.003 | 0.226 | 0.029 | 0.142 | |
| DOCK7rs10889332 C > T | ||||||||
| Jing | ||||||||
| CC | 447 | 5.05 ± 0.93 | 1.31 (1.06) | 1.83 ± 0.43 | 2.78 ± 0.43 | 1.34 ± 0.26 | 1.02 ± 0.22 | 1.34 ± 0.39 |
| CT | 341 | 5.15 ± 0.88 | 1.49 (1.20) | 1.78 ± 0.45 | 2.83 ± 0.45 | 1.30 ± 0.21 | 1.06 ± 0.24 | 1.28 ± 0.37 |
| TT | 93 | 5.38 ± 1.02 | 1.91 (1.28) | 1.69 ± 0.54 | 2.94 ± 0.46 | 1.29 ± 0.25 | 1.21 ± 0.33 | 1.17 ± 0.32 |
| F | 2.624 | 6.928 | 1.226 | 3.628 | 1.202 | 10.896 | 3.207 | |
| P | 0.073 | 0.001 | 0.294 | 0.027 | 0.301 | 0.000 | 0.041 | |
| Han | ||||||||
| CC | 575 | 4.85 ± 0.83 | 1.27 (1.07) | 1.81 ± 0.54 | 2.83 ± 0.42 | 1.34 ± 0.20 | 1.02 ± 0.23 | 1.39 ± 0.42 |
| CT | 347 | 4.88 ± 0.91 | 1.33 (1.10) | 1.79 ± 0.50 | 2.84 ± 0.47 | 1.32 ± 0.24 | 1.03 ± 0.26 | 1.34 ± 0.34 |
| TT | 66 | 5.27 ± 0.88 | 1.73 (1.34) | 1.70 ± 0.60 | 3.02 ± 0.38 | 1.31 ± 0.20 | 1.12 ± 0.21 | 1.22 ± 0.34 |
| F | 2.271 | 5.660 | 0.364 | 3.109 | 0.879 | 4.625 | 5.033 | |
| P | 0.104 | 0.004 | 0.695 | 0.045 | 0.416 | 0.010 | 0.007 | |
| PCSK9rs615563 G > A | ||||||||
| Jing | ||||||||
| GG | 536 | 5.07 ± 0.88 | 1.34 (1.10) | 1.87 ± 0.64 | 2.80 ± 0.43 | 1.33 ± 0.24 | 1.03 ± 0.23 | 1.33 ± 0.38 |
| AG | 294 | 5.18 ± 0.94 | 1.51 (1.21) | 1.81 ± 0.42 | 2.83 ± 0.45 | 1.31 ± 0.24 | 1.09 ± 0.26 | 1.24 ± 0.37 |
| AA | 51 | 5.41 ± 1.18 | 1.58 (1.26) | 1.76 ± 0.45 | 2.98 ± 0.46 | 1.29 ± 0.22 | 1.12 ± 0.33 | 1.27 ± 0.38 |
| F | 1.544 | 5.479 | 1.668 | 2.125 | 1.065 | 1.906 | 1.598 | |
| P | 0.214 | 0.004 | 0.190 | 0.120 | 0.345 | 0.150 | 0.203 | |
| Han | ||||||||
| GG | 664 | 4.84 ± 0.85 | 1.26 (1.04) | 1.90 ± 0.40 | 2.82 ± 0.43 | 1.37 ± 0.19 | 1.02 ± 0.24 | 1.37 ± 0.38 |
| AG | 279 | 4.98 ± 0.90 | 1.27 (1.11) | 1.78 ± 0.53 | 2.89 ± 0.45 | 1.32 ± 0.20 | 1.06 ± 0.26 | 1.32 ± 0.27 |
| AA | 45 | 5.05 ± 0.77 | 1.47 (1.15) | 1.78 ± 0.52 | 2.95 ± 0.41 | 1.31 ± 0.20 | 1.07 ± 0.23 | 1.30 ± 0.37 |
| F | 0.825 | 6.574 | 0.533 | 1.105 | 1.101 | 2.041 | 1.674 | |
| P | 0.439 | 0.001 | 0.587 | 0.332 | 0.333 | 0.131 | 0.188 | |
| PCSK9 rs7552841 C > T | ||||||||
| Jing | ||||||||
| CC | 571 | 5.02 ± 0.90 | 1.33 (1.06) | 1.89 ± 0.52 | 2.76 ± 0.43 | 1.31 ± 0.25 | 1.01 ± 0.22 | 1.36 ± 0.39 |
| CT | 264 | 5.30 ± 0.93 | 1.56 (1.26) | 1.81 ± 0.45 | 2.91 ± 0.44 | 1.30 ± 0.21 | 1.14 ± 0.26 | 1.19 ± 0.34 |
| TT | 46 | 5.44 ± 1.04 | 1.58 (1.22) | 1.75 ± 0.44 | 2.96 ± 0.45 | 1.29 ± 0.27 | 1.18 ± 0.30 | 1.16 ± 0.30 |
| F | 3.683 | 8.648 | 1.429 | 6.058 | 0.073 | 17.987 | 12.017 | |
| P | 0.026 | 0.000 | 0.240 | 0.002 | 0.930 | 0.000 | 0.000 | |
| Han | ||||||||
| CC | 689 | 4.86 ± 0.86 | 1.31 (1.07) | 1.89 ± 0.62 | 2.83 ± 0.44 | 1.37 ± 0.19 | 1.02 ± 0.24 | 1.36 ± 0.38 |
| CT | 269 | 4.90 ± 0.84 | 1.32 (1.10) | 1.81 ± 0.59 | 2.85 ± 0.41 | 1.33 ± 0.20 | 1.04 ± 0.24 | 1.34 ± 0.36 |
| TT | 30 | 5.42 ± 1.12 | 1.78 (1.51) | 1.78 ± 0.49 | 3.11 ± 0.59 | 1.31 ± 0.20 | 1.21 ± 0.33 | 1.20 ± 0.33 |
| F | 3.672 | 8.841 | 0.666 | 3.611 | 1.756 | 5.595 | 3.144 | |
| P | 0.026 | 0.000 | 0.514 | 0.028 | 0.174 | 0.004 | 0.044 | |
| PCSK9 rs11206517 T > G | ||||||||
| Jing | ||||||||
| TT | 723 | 5.07 ± 0.90 | 1.40 (1.12) | 1.83 ± 0.45 | 2.80 ± 0.44 | 1.35 ± 0.25 | 1.04 ± 0.23 | 1.31 ± 0.37 |
| GT | 146 | 5.36 ± 0.98 | 1.47 (1.10) | 1.79 ± 0.44 | 2.89 ± 0.43 | 1.29 ± 0.23 | 1.14 ± 0.30 | 1.27 ± 0.41 |
| GG | 12 | 5.61 ± 1.16 | 1.75 (1.35) | 1.41 ± 0.57 | 3.03 ± 0.57 | 1.27 ± 0.26 | 1.30 ± 0.44 | 1.02 ± 0.20 |
| F | 0.356 | 4.366 | 4.202 | 0.164 | 3.059 | 2.062 | 1.692 | |
| P | 0.694 | 0.013 | 0.015 | 0.849 | 0.048 | 0.128 | 0.185 | |
| Han | ||||||||
| TT | 859 | 4.85 ± 0.77 | 1.28 (1.06) | 1.82 ± 0.53 | 2.84 ± 0.38 | 1.34 ± 0.19 | 1.03 ± 0.25 | 1.36 ± 0.37 |
| GT | 121 | 4.89 ± 0.88 | 1.58 (1.24) | 1.59 ± 0.41 | 2.85 ± 0.44 | 1.33 ± 0.21 | 1.05 ± 0.23 | 1.34 ± 0.50 |
| GG | 8 | 4.95 ± 0.38 | 1.95 (1.26) | 1.52 ± 0.42 | 2.91 ± 0.34 | 1.24 ± 0.15 | 1.07 ± 0.24 | 1.24 ± 0.34 |
| F | 0.254 | 6.780 | 5.430 | 0.229 | 7.519 | 0.172 | 1.116 | |
| P | 0.775 | 0.001 | 0.005 | 0.795 | 0.001 | 0.842 | 0.328 | |
| GALNT2 rs1997947 G > A | ||||||||
| Jing | ||||||||
| GG | 529 | 5.07 ± 0.94 | 1.29 (1.06) | 1.84 ± 0.44 | 2.71 ± 0.53 | 1.33 ± 0.24 | 1.04 ± 0.24 | 1.34 ± 0.37 |
| AG | 297 | 5.17 ± 0.84 | 1.55 (1.23) | 1.77 ± 0.46 | 2.80 ± 0.44 | 1.29 ± 0.22 | 1.08 ± 0.27 | 1.26 ± 0.39 |
| AA | 55 | 5.22 ± 0.92 | 1.67 (1.50) | 1.52 ± 0.30 | 2.87 ± 0.42 | 1.14 ± 0.13 | 1.09 ± 0.21 | 1.10 ± 0.31 |
| F | 0.687 | 5.251 | 5.226 | 1.549 | 9.989 | 0.010 | 3.799 | |
| P | 0.503 | 0.005 | 0.005 | 0.213 | 0.000 | 0.990 | 0.023 | |
| Han | ||||||||
| GG | 661 | 4.88 ± 0.87 | 1.27 (1.04) | 1.81 ± 0.45 | 2.84 ± 0.44 | 1.33 ± 0.20 | 1.02 ± 0.25 | 1.37 ± 0.37 |
| AG | 283 | 4.90 ± 0.88 | 1.38 (1.16) | 1.80 ± 0.55 | 2.86 ± 0.45 | 1.31 ± 0.20 | 1.05 ± 0.24 | 1.33 ± 0.39 |
| AA | 44 | 4.99 ± 0.69 | 2.16 (1.71) | 1.42 ± 0.36 | 2.93 ± 0.29 | 1.20 ± 0.15 | 1.12 ± 0.16 | 1.09 ± 0.20 |
| F | 0.369 | 6.376 | 4.391 | 0.501 | 4.573 | 0.873 | 3.144 | |
| P | 0.692 | 0.002 | 0.013 | 0.606 | 0.011 | 0.418 | 0.044 | |
| GALNT2 rs2760537 C > T | ||||||||
| Jing | ||||||||
| CC | 344 | 5.05 ± 0.95 | 1.36 (1.15) | 1.82 ± 0.46 | 2.80 ± 0.42 | 1.31 ± 0.25 | 1.04 ± 0.23 | 1.30 ± 0.35 |
| CT | 406 | 5.15 ± 0.90 | 1.41 (1.08) | 1.78 ± 0.41 | 2.81 ± 0.54 | 1.30 ± 0.22 | 1.06 ± 0.25 | 1.30 ± 0.40 |
| TT | 131 | 5.23 ± 0.96 | 1.54 (1.24) | 1.76 ± 0.52 | 2.83 ± 0.34 | 1.29 ± 0.21 | 1.07 ± 0.28 | 1.29 ± 0.39 |
| F | 2.062 | 5.588 | 0.038 | 0.232 | 0.234 | 0.525 | 0.163 | |
| P | 0.128 | 0.004 | 0.963 | 0.793 | 0.792 | 0.592 | 0.850 | |
| Han | ||||||||
| CC | 446 | 4.86 ± 0.87 | 1.30 (1.03) | 1.82 ± 0.55 | 2.83 ± 0.45 | 1.32 ± 0.20 | 1.02 ± 0.23 | 1.37 ± 0.38 |
| CT | 427 | 4.87 ± 0.84 | 1.31 (1.09) | 1.79 ± 0.54 | 2.85 ± 0.41 | 1.32 ± 0.20 | 1.03 ± 0.24 | 1.34 ± 0.35 |
| TT | 115 | 5.06 ± 0.91 | 1.46 (1.21) | 1.76 ± 0.49 | 2.93 ± 0.47 | 1.31 ± 0.21 | 1.07 ± 0.28 | 1.30 ± 0.42 |
| F | 0.835 | 4.692 | 1.401 | 0.879 | 0.008 | 0.833 | 0.290 | |
| P | 0.434 | 0.009 | 0.247 | 0.416 | 0.992 | 0.435 | 0.749 | |
| GALNT2 rs4846913 C > A | ||||||||
| Jing | ||||||||
| CC | 554 | 5.14 ± 0.94 | 1.37 (1.10) | 1.83 ± 0.45 | 2.81 ± 0.44 | 1.32 ± 0.24 | 1.05 ± 0.26 | 1.32 ± 0.38 |
| AC | 281 | 5.07 ± 0.88 | 1.54 (1.11) | 1.82 ± 0.50 | 2.82 ± 0.41 | 1.27 ± 0.20 | 1.06 ± 0.23 | 1.27 ± 0.37 |
| AA | 46 | 5.30 ± 1.02 | 1.69 (1.49) | 1.73 ± 0.43 | 2.86 ± 0.62 | 1.26 ± 0.24 | 1.09 ± 0.21 | 1.21 ± 0.34 |
| F | 0.378 | 5.388 | 4.764 | 0.406 | 2.828 | 0.207 | 1.254 | |
| P | 0.685 | 0.005 | 0.009 | 0.667 | 0.060 | 0.813 | 0.286 | |
| Han | ||||||||
| CC | 685 | 4.86 ± 0.88 | 1.25 (1.03) | 1.84 ± 0.53 | 2.84 ± 0.44 | 1.34 ± 0.21 | 1.02 ± 0.24 | 1.39 ± 0.37 |
| AC | 263 | 4.95 ± 0.82 | 1.50 (1.16) | 1.77 ± 0.47 | 2.87 ± 0.43 | 1.27 ± 0.17 | 1.05 ± 0.25 | 1.27 ± 0.35 |
| AA | 40 | 5.03 ± 0.88 | 1.54 (1.24) | 1.65 ± 0.48 | 2.93 ± 0.47 | 1.24 ± 0.20 | 1.06 ± 0.24 | 1.24 ± 0.43 |
| F | 0.527 | 5.320 | 8.741 | 0.410 | 9.946 | 0.945 | 5.623 | |
| P | 0.591 | 0.005 | 0.000 | 0.664 | 0.000 | 0.389 | 0.004 | |
| GALNT2 rs11122316 G > A | ||||||||
| Jing | ||||||||
| GG | 320 | 5.06 ± 0.92 | 1.30 (1.03) | 1.87 ± 0.40 | 2.79 ± 0.46 | 1.32 ± 0.21 | 1.06 ± 0.24 | 1.32 ± 0.37 |
| AG | 429 | 5.17 ± 0.95 | 1.49 (1.16) | 1.81 ± 0.52 | 2.84 ± 0.42 | 1.31 ± 0.24 | 1.06 ± 0.25 | 1.31 ± 0.37 |
| AA | 132 | 5.19 ± 0.95 | 1.54 (1.24) | 1.73 ± 0.45 | 2.87 ± 0.43 | 1.28 ± 0.24 | 1.06 ± 0.26 | 1.28 ± 0.39 |
| F | 2.561 | 6.549 | 3.245 | 1.726 | 1.231 | 0.414 | 0.237 | |
| P | 0.078 | 0.002 | 0.040 | 0.179 | 0.293 | 0.661 | 0.789 | |
| Han | ||||||||
| GG | 410 | 4.85 ± 0.84 | 1.25 (1.03) | 1.81 ± 0.55 | 2.83 ± 0.42 | 1.34 ± 0.21 | 1.02 ± 0.23 | 1.36 ± 0.38 |
| AG | 468 | 4.92 ± 0.91 | 1.31 (1.07) | 1.78 ± 0.51 | 2.84 ± 0.46 | 1.32 ± 0.21 | 1.02 ± 0.26 | 1.35 ± 0.37 |
| AA | 110 | 4.97 ± 0.82 | 1.49 (1.32) | 1.73 ± 0.48 | 2.93 ± 0.42 | 1.32 ± 0.20 | 1.09 ± 0.23 | 1.29 ± 0.35 |
| F | 1.172 | 6.503 | 0.902 | 1.414 | 1.041 | 2.670 | 0.920 | |
| P | 0.310 | 0.002 | 0.406 | 0.244 | 0.354 | 0.070 | 0.399 | |
HDL, high density lipoprotein; LDL, low density lipoprotein; The P-value calculated by ANCOVA, using general linear models, and adjusted for age, sex, BMI, smoking status, alcohol use, glucose and hypertension, and less than 0.005 was considered statistically significant after adjusting by Bonferroni correction.
n = sample size.
Haplotypes and lipid profiles
The correlation of the haplotypes and lipid profiles is shown in Table 6. Rare Hap (frequency < 3%) in both Jing and Han populations has been dropped. The carriers of C-C-G-C-T-G-T-C-G haplotype had lower TG and higher HDL cholesterol levels in Jing plus Han populations and lower TG levels in Jing population than the non-carriers of C-C-G-C-T-G-T-C-G haplotype (P < 0.05). There were no differences in lipid parameters between the carriers and non-carriers of C-C-G-C-T-G-T-C-G haplotype in the Han population. Haplotype G-C-A-C-T-G-C-C-G carriers had lower serum TG in the Han populations than the haplotype non-carriers (P < 0.05). Haplotype G-C-G-C-T-A-C-C-A carriers had higher serum TG and lower ApoA1 levels in Jing plus Han population, and higher serum TG and lower HDL cholesterol and ApoA1 in Jing population than the haplotype G-C-G-C-T-A-C-C-A non-carriers (P < 0.05 for each). Haplotype G-C-G-C-T-G-C-C-A carriers had lower TC, TG, LDL cholesterol and ApoB levels in Jing plus Han population, lower TC, TG and ApoB in Jing ethnic minority and lower TG levels than the haplotype G-C-G-C-T-A-C-C-A non-carriers (P < 0.05 for all).
Table 6. Lipid profiles according to haplotypes for the two ethnic groups.
| Haplotype | Group | n | Total cholesterol (mmol/L) | Triglyceride (mmol/L) | HDL cholesterol (mmol/L) | LDL cholesterol (mmol/L) | Apolipoprotein (Apo) A1 (g/L) | Apolipoprotein (Apo) B (g/L) | ApoA1/ ApoB |
|---|---|---|---|---|---|---|---|---|---|
| C-C-G-C-T-G-T-C-G | Jing plus Han | 1869 | |||||||
| Carrier | 64 | 4.98 ± 0.86 | 1.30(1.08) | 1.84 ± 0.50 | 2.82 ± 0.43 | 1.31 ± 0.23 | 1.03 ± 0.24 | 1.34 ± 0.38 | |
| Non-carrier | 1805 | 4.99 ± 0.90 | 1.39 (1.10) | 1.75 ± 0.49 | 2.84 ± 0.43 | 1.31 ± 0.21 | 1.05 ± 0.25 | 1.31 ± 0.38 | |
| F | 0.035 | −2.397 | 5.661 | 0.269 | 0.011 | 0.736 | 0.355 | ||
| P | 0.852 | 0.017 | 0.017 | 0.604 | 0.916 | 0.391 | 0.552 | ||
| Jing | 881 | ||||||||
| Carrier | 5 | 5.11 ± 0.92 | 1.30 (1.06) | 1.83 ± 0.44 | 2.81 ± 0.43 | 1.29 ± 0.26 | 1.04 ± 0.23 | 1.30 ± 0.39 | |
| Non-carrier | 876 | 5.13 ± 0.85 | 1.44 (1.14) | 1.77 ± 0.45 | 2.82 ± 0.42 | 1.30 ± 0.22 | 1.06 ± 0.25 | 1.29 ± 0.38 | |
| F | 0.062 | −2.414 | 1.672 | 0.077 | 0.232 | 0.665 | 0.008 | ||
| P | 0.803 | 0.016 | 0.196 | 0.782 | 0.630 | 0.415 | 0.927 | ||
| Han | 988 | ||||||||
| Carrier | 59 | 4.85 ± 0.85 | 1.29 (1.09) | 1.86 ± 0.55 | 2.83 ± 0.44 | 1.33 ± 0.21 | 1.02 ± 0.24 | 1.37 ± 0.39 | |
| Non-carrier | 929 | 4.88 ± 0.87 | 1.33 (1.07) | 1.74 ± 0.52 | 2.85 ± 0.44 | 1.32 ± 0.20 | 1.04 ± 0.24 | 1.34 ± 0.38 | |
| F | 0.112 | −0.916 | 3.735 | 0.069 | 0.550 | 0.001 | 0.097 | ||
| P | 0.738 | 0.360 | 0.054 | 0.793 | 0.458 | 0.973 | 0.755 | ||
| G-C-A-C-T-G-C-C-G | Jing plus Han | 1869 | |||||||
| Carrier | 139 | 4.98 ± 0.88 | 1.36 (1.06) | 1.79 ± 0.46 | 2.83 ± 0.43 | 1.32 ± 0.21 | 1.04 ± 0.24 | 1.33 ± 0.38 | |
| Non-carrier | 1730 | 5.01 ± 0.94 | 1.37 (1.10) | 1.77 ± 0.50 | 2.84 ± 0.44 | 1.30 ± 0.22 | 1.05 ± 0.24 | 1.31 ± 0.38 | |
| F | 0.015 | −0.926 | 0.112 | 0.009 | 0.925 | 0.011 | 0.387 | ||
| P | 0.901 | 0.354 | 0.738 | 0.924 | 0.336 | 0.915 | 0.534 | ||
| Jing | 881 | ||||||||
| Carrier | 92 | 5.10 ± 0.89 | 1.41 (1.12) | 1.79 ± 0.47 | 2.81 ± 0.43 | 1.30 ± 0.23 | 1.06 ± 0.24 | 1.29 ± 0.39 | |
| Non-carrier | 789 | 5.17 ± 0.96 | 1.51 (1.16) | 1.78 ± 0.45 | 2.82 ± 0.42 | 1.29 ± 0.21 | 1.06 ± 0.25 | 1.28 ± 0.38 | |
| F | 1.009 | −0.756 | 0.183 | 0.105 | 0.157 | 0.152 | 0.309 | ||
| P | 0.315 | 0.450 | 0.669 | 0.746 | 0.692 | 0.697 | 0.578 | ||
| Han | 988 | ||||||||
| Carrier | 47 | 4.87 ± 0.90 | 1.25 (0.98) | 1.79 ± 0.46 | 2.85 ± 0.45 | 1.35 ± 0.20 | 1.03 ± 0.24 | 1.39 ± 0.37 | |
| Non-carrier | 941 | 4.88 ± 0.86 | 1.34 (1.09) | 1.76 ± 0.54 | 2.85 ± 0.43 | 1.31 ± 0.20 | 1.04 ± 0.24 | 1.33 ± 0.38 | |
| F | 0.052 | −2.134 | 0.086 | 0.026 | 3.701 | 0.093 | 1.562 | ||
| P | 0.820 | 0.033 | 0.770 | 0.872 | 0.055 | 0.760 | 0.212 | ||
| G-C-G-C-T-A-C-C-A | Jing plus Han | 1869 | |||||||
| Carrier | 59 | 4.99 ± 0.91 | 1.48 (1.19) | 1.77 ± 0.40 | 2.83 ± 0.44 | 1.28 ± 0.20 | 1.05 ± 0.25 | 1.29 ± 0.38 | |
| Non-carrier | 1810 | 4.97 ± 0.77 | 1.35 (1.07) | 1.77 ± 0.51 | 2.83 ± 0.38 | 1.31 ± 0.22 | 1.04 ± 0.22 | 1.32 ± 0.38 | |
| F | 0.775 | −3.088 | 0.090 | 0.005 | 5.947 | 0.296 | 1.381 | ||
| P | 0.379 | 0.002 | 0.765 | 0.944 | 0.015 | 0.587 | 0.240 | ||
| Jing | 881 | ||||||||
| Carrier | 4 | 5.14 ± 0.93 | 1.52 (1.21) | 1.71 ± 0.38 | 2.82 ± 0.44 | 1.26 ± 0.21 | 1.06 ± 0.25 | 1.27 ± 0.38 | |
| Non-carrier | 877 | 5.03 ± 0.76 | 1.38 (1.09) | 1.80 ± 0.46 | 2.79 ± 0.35 | 1.31 ± 0.23 | 1.04 ± 0.22 | 1.30 ± 0.39 | |
| F | 1.423 | −2.312 | 4.245 | 0.648 | 7.008 | 1.558 | 0.566 | ||
| P | 0.233 | 0.021 | 0.040 | 0.421 | 0.008 | 0.212 | 0.452 | ||
| Han | 988 | ||||||||
| Carrier | 55 | 4.89 ± 0.79 | 1.34 (1.16) | 1.75 ± 0.54 | 2.89 ± 0.41 | 1.32 ± 0.19 | 1.04 ± 0.23 | 1.33 ± 0.38 | |
| Non-carrier | 933 | 4.87 ± 0.88 | 1.31 (1.06) | 1.86 ± 0.41 | 2.84 ± 0.44 | 1.32 ± 0.20 | 1.04 ± 0.25 | 1.34 ± 0.38 | |
| F | 0.021 | −1.490 | 1.910 | 0.876 | 0.231 | 0.198 | 0.616 | ||
| P | 0.885 | 0.136 | 0.167 | 0.350 | 0.631 | 0.657 | 0.433 | ||
| G-C-G-C-T-G-C-C-A | Jing plus Han | 1869 | |||||||
| Carrier | 199 | 4.90 ± 0.86 | 1.30 (1.08) | 1.79 ± 0.49 | 2.80 ± 0.40 | 1.31 ± 0.23 | 1.02 ± 0.22 | 1.34 ± 0.37 | |
| Non-carrier | 1670 | 5.03 ± 0.91 | 1.42 (1.10) | 1.77 ± 0.50 | 2.85 ± 0.45 | 1.31 ± 0.21 | 1.06 ± 0.25 | 1.31 ± 0.39 | |
| F | 8.329 | −4.113 | 0.530 | 4.982 | 0.028 | 9.864 | 2.914 | ||
| P | 0.004 | 0.000 | 0.467 | 0.026 | 0.867 | 0.002 | 0.088 | ||
| Jing | 881 | ||||||||
| Carrier | 133 | 5.00 ± 0.88 | 1.35 (1.12) | 1.79 ± 0.47 | 2.78 ± 0.40 | 1.30 ± 0.22 | 1.03 ± 0.23 | 1.31 ± 0.39 | |
| Non-carrier | 748 | 5.18 ± 0.91 | 1.45 (1.13) | 1.77 ± 0.41 | 2.84 ± 0.43 | 1.29 ± 0.25 | 1.08 ± 0.25 | 1.28 ± 0.38 | |
| F | 7.709 | −2.520 | 0.013 | 3.046 | 0.128 | 6.670 | 2.105 | ||
| P | 0.006 | 0.012 | 0.909 | 0.081 | 0.720 | 0.010 | 0.147 | ||
| Han | 988 | ||||||||
| Carrier | 66 | 4.82 ± 0.83 | 1.26 (1.05) | 1.80 ± 0.55 | 2.82 ± 0.40 | 1.32 ± 0.21 | 1.01 ± 0.22 | 1.36 ± 0.36 | |
| Non-carrier | 922 | 4.90 ± 0.89 | 1.37 (1.09) | 1.75 ± 0.52 | 2.87 ± 0.46 | 1.32 ± 0.20 | 1.05 ± 0.25 | 1.33 ± 0.39 | |
| F | 2.273 | −3.327 | 0.717 | 2.079 | 0.015 | 3.558 | 0.842 | ||
| P | 0.132 | 0.001 | 0.397 | 0.150 | 0.904 | 0.060 | 0.359 | ||
| G-C-G-C-T-G-T-C-A | Jing plus Han | 1869 | |||||||
| Carrier | 122 | 4.94 ± 0.86 | 1.36 (1.16) | 1.81 ± 0.53 | 2.81 ± 0.40 | 1.31 ± 0.23 | 1.03 ± 0.23 | 1.35 ± 0.42 | |
| Non-carrier | 1747 | 5.00 ± 0.90 | 1.37 (1.07) | 1.76 ± 0.48 | 2.84 ± 0.44 | 1.31 ± 0.21 | 1.05 ± 0.25 | 1.31 ± 0.37 | |
| F | 1.882 | −0.431 | 1.209 | 1.204 | 0.015 | 3.363 | 3.350 | ||
| P | 0.170 | 0.666 | 0.272 | 0.273 | 0.903 | 0.067 | 0.067 | ||
| Jing | 881 | ||||||||
| Carrier | 80 | 5.06 ± 0.86 | 1.42 (1.11) | 1.78 ± 0.46 | 2.79 ± 0.41 | 1.30 ± 0.26 | 1.04 ± 0.24 | 1.32 ± 0.44 | |
| Non-carrier | 801 | 5.13 ± 0.91 | 1.44 (1.16) | 1.78 ± 0.43 | 2.82 ± 0.43 | 1.30 ± 0.22 | 1.06 ± 0.25 | 1.28 ± 0.37 | |
| F | 0.922 | −0.637 | 0.179 | 0.859 | 0.144 | 0.798 | 0.554 | ||
| P | 0.337 | 0.524 | 0.673 | 0.354 | 0.704 | 0.372 | 0.457 | ||
| Han | 988 | ||||||||
| Carrier | 42 | 4.83 ± 0.84 | 1.31 (1.04) | 1.83 ± 0.60 | 2.83 ± 0.40 | 1.33 ± 0.20 | 1.01 ± 0.23 | 1.38 ± 0.41 | |
| Non-carrier | 946 | 4.89 ± 0.88 | 1.33 (1.17) | 1.75 ± 0.51 | 2.85 ± 0.45 | 1.32 ± 0.20 | 1.04 ± 0.25 | 1.33 ± 0.37 | |
| F | 1.143 | −1.183 | 2.233 | 0.372 | 0.496 | 2.237 | 2.717 | ||
| P | 0.285 | 0.237 | 0.135 | 0.542 | 0.482 | 0.135 | 0.100 | ||
HDL, high density lipoprotein; LDL, low density lipoprotein.
Correlation between lipid parameters and alleles or genotypes
Table 7 depicts the direction and magnitude of associations between lipid parameters and alleles or genotypes of the 9 SNPs in the Jing and Han populations. Adjusting for age, sex, BMI, smoking status, alcohol use, and exercise, logistic regression analysis showed that several the examined SNPs were significant correlated with lipid parameters.
Table 7. Association of the alleles and genotypes of the DOCK7, PCSK9 and GAALNT2 SNPs and serum lipid traits in the Jing and Han populations.
| Lipid | SNP | Affected allele/ Other allele | Affected genotype/ Other genotype | Beta | Std.error | t | P-value |
|---|---|---|---|---|---|---|---|
| Jing plus Han | |||||||
| TC | DOCK7rs10889332 | CC,CT/TT | 0.081 | 0.036 | 3.161 | 0.002 | |
| DOCK7rs10889332 | C/T | 0.057 | 0.047 | 2.231 | 0.026 | ||
| PCSK9rs615563 | GG,AG/AA | 0.068 | 0.039 | 2.634 | 0.009 | ||
| PCSK9rs615563 | G/A | 0.059 | 0.048 | 2.292 | 0.022 | ||
| PCSK9 rs7552841 | CC,CT/TT | 0.108 | 0.041 | 4.234 | 0.000 | ||
| PCSK9 rs7552841 | C/T | 0.101 | 0.049 | 3.941 | 0.000 | ||
| GALNT2rs2760537 | CC,CT/TT | 0.055 | 0.034 | 2.154 | 0.031 | ||
| GALNT2rs11122316 | −0.055 | 0.047 | −2.145 | 0.032 | |||
| TG | DOCK7 rs1168013 | GG,CG/CC | 0.087 | 0.033 | 3.426 | 0.001 | |
| DOCK7rs10889332 | CC,CT/TT | 0.099 | 0.034 | 3.903 | 0.000 | ||
| PCSK9rs615563 | GG,AG/AA | 0.094 | 0.035 | 3.995 | 0.000 | ||
| PCSK9rs7552841 | CC,CT/TT | 0.126 | 0.037 | 5.325 | 0.000 | ||
| PCSK9rs11206517 | T/G | −0.056 | 0.035 | −2.155 | 0.031 | ||
| GALNT2rs1997947 | GG,AG/AA | 0.083 | 0.035 | 3.440 | 0.001 | ||
| GALNT2rs2760537 | CC,CT/TT | 0.114 | 0.030 | 4.811 | 0.000 | ||
| GALNT2rs4846913 | CC,AC/AA | 0.104 | 0.036 | 4.384 | 0.000 | ||
| GALNT2rs4846913 | C/A | −0.109 | 0.027 | −4.235 | 0.000 | ||
| GALNT2rs11122316 | GG,AG/AA | 0.119 | 0.031 | 4.978 | 0.000 | ||
| HDL cholesterol | PCSK9rs615563 | G/A | 0.052 | 0.024 | 1.988 | 0.047 | |
| PCSK9 rs7552841 | C/T | 0.100 | 0.025 | 3.816 | 0.000 | ||
| PCSK9rs11206517 | TT,GT/GG | −0.067 | 0.032 | −2.573 | 0.010 | ||
| GALNT2rs1997947 | GG,AG/AA | −0.081 | 0.021 | −3.111 | 0.002 | ||
| GALNT2rs4846913 | GG,AG/AA | −0.078 | 0.022 | −3.014 | 0.003 | ||
| LDL cholesterol | DOCK7rs10889332 | CC,CT/TT | 0.069 | 0.018 | 2.617 | 0.009 | |
| PCSK9rs615563 | GG,AG/AA | 0.068 | 0.020 | 2.595 | 0.010 | ||
| PCSK9rs615563 | G/A | 0.052 | 0.024 | 1.988 | 0.047 | ||
| PCSK9 rs7552841 | CC,CT/TT | 0.114 | 0.021 | 4.337 | 0.000 | ||
| PCSK9rs7552841 | C/T | 0.100 | 0.025 | 3.816 | 0.000 | ||
| ApoA1 | DOCK7 rs1168013 | G/C | 0.053 | 0.011 | 2.050 | 0.041 | |
| DOCK7rs10889332 | CC,CT/TT | 0.065 | 0.009 | 2.485 | 0.013 | ||
| GALNT2rs1997947 | GG,AG/AA | −0.128 | 0.010 | −4.905 | 0.000 | ||
| GALNT2rs1997947 | G/A | −0.089 | 0.012 | −3.434 | 0.001 | ||
| GALNT2rs4846913 | CC,AC/AA | −0.117 | 0.010 | −4.488 | 0.000 | ||
| GALNT2rs4846913 | G/A | −0.128 | 0.012 | −4.901 | 0.000 | ||
| ApoB | DOCK7rs10889332 | CC,CT/TT | 0.103 | 0.010 | 4.023 | 0.000 | |
| DOCK7rs10889332 | C/T | 0.060 | 0.013 | 2.341 | 0.019 | ||
| PCSK9rs615563 | GG,AG/AA | 0.091 | 0.011 | 3.572 | 0.000 | ||
| PCSK9rs615563 | G/A | 0.093 | 0.013 | 3.598 | 0.000 | ||
| PCSK9rs7552841 | CC,CT/TT | 0.175 | 0.011 | 6.843 | 0.000 | ||
| PCSK9rs7552841 | C/T | 0.164 | 0.014 | 6.386 | 0.000 | ||
| ApoA1/ApoB | PCSK9rs615563 | GG,AG/AA | −0.073 | 0.016 | −2.860 | 0.004 | |
| PCSK9rs615563 | G/A | −0.087 | 0.020 | −3.420 | 0.001 | ||
| PCSK9rs7552841 | CC,CT/TT | −0.115 | 0.017 | −4.543 | 0.000 | ||
| PCSK9rs7552841 | C/T | −0.112 | 0.020 | −4.390 | 0.000 | ||
| GALNT2rs1997947 | GG,AG/AA | −0.089 | 0.016 | −3.443 | 0.001 | ||
| GALNT2rs1997947 | G/A | −0.060 | 0.020 | −2.321 | 0.020 | ||
| GALNT2rs4846913 | CC,AC/AA | −0.088 | 0.017 | −3.483 | 0.001 | ||
| GALNT2rs4846913 | C/A | −0.095 | 0.020 | −3.747 | 0.000 | ||
| Jing | |||||||
| TC | DOCK7 rs1168013 | GG,CG/CC | −0.109 | 0.054 | −2.819 | 0.005 | |
| DOCK7rs10889332 | CC,CT/TT | 0.111 | 0.054 | 2.823 | 0.005 | ||
| PCSK9rs7552841 | CC,CT/TT | 0.116 | 0.058 | 3.159 | 0.002 | ||
| PCSK9rs7552841 | C/T | 0.132 | 0.072 | 3.561 | 0.000 | ||
| GALNT2rs2760537 | CC,CT/TT | 0.079 | 0.049 | 2.138 | 0.033 | ||
| GALNT2rs11122316 | G/A | −0.076 | 0.071 | −2.048 | 0.041 | ||
| TG | DOCK7 rs1168013 | GG,CG/CC | 0.089 | 0.045 | 2.512 | 0.012 | |
| DOCK7 rs1168013 | G/C | 0.115 | 0.060 | 3.335 | 0.001 | ||
| DOCK7rs10889332 | CC,CT/TT | 0.089 | 0.046 | 2.460 | 0.014 | ||
| PCSK9rs615563 | GG,AG/AA | 0.072 | 0.048 | 2.131 | 0.034 | ||
| PCSK9rs7552841 | CC,CT/TT | 0.141 | 0.049 | 4.169 | 0.000 | ||
| PCSK9rs7552841 | C/T | 0.139 | 0.062 | 4.022 | 0.000 | ||
| GALNT2rs1997947 | GG,AG/AA | 0.104 | 0.048 | 3.015 | 0.003 | ||
| GALNT2rs1997947 | G/A | 0.107 | 0.061 | 3.048 | 0.002 | ||
| GALNT2rs2760537 | CC,CT/TT | 0.140 | 0.042 | 4.111 | 0.000 | ||
| GALNT2rs2760537 | C/T | 0.116 | 0.061 | 3.337 | 0.001 | ||
| GALNT2rs4846913 | CC,AC/AA | 0.095 | 0.049 | 2.806 | 0.005 | ||
| GALNT2rs4846913 | C/A | 0.103 | 0.061 | 3.004 | 0.003 | ||
| GALNT2rs11122316 | GG,AG/AA | 0.124 | 0.043 | 3.605 | 0.000 | ||
| GALNT2rs11122316 | G/A | 0.117 | 0.062 | 3.342 | 0.001 | ||
| HDL cholesterol | GALNT2rs1997947 | GG,AG/AA | −0.102 | 0.027 | −2.766 | 0.006 | |
| GALNT2rs11122316 | G/A | −0.106 | 0.034 | −2.916 | 0.004 | ||
| LDL cholesterol | DOCK7 rs1168013 | GG,CG/CC | −0.139 | 0.026 | −3.487 | 0.001 | |
| DOCK7rs10889332 | CC,CT/TT | 0.129 | 0.026 | 3.217 | 0.001 | ||
| PCSK9rs7552841 | CC,CT/TT | 0.145 | 0.028 | 3.836 | 0.000 | ||
| PCSK9rs7552841 | C/T | 0.155 | 0.035 | 4.052 | 0.000 | ||
| ApoA1 | PCSK9rs11206517 | T/G | 0.076 | 0.023 | 1.987 | 0.047 | |
| GALNT2rs1997947 | GG,AG/AA | −0.157 | 0.014 | −4.121 | 0.000 | ||
| GALNT2rs1997947 | G/A | −0.129 | 0.018 | −3.390 | 0.001 | ||
| GALNT2rs4846913 | CC,AC/AA | −0.076 | 0.015 | −2.010 | 0.045 | ||
| GALNT2rs4846913 | C/A | −0.090 | 0.018 | −2.369 | 0.018 | ||
| ApoB | DOCK7 rs1168013 | GG,CG/CC | −0.112 | 0.014 | −2.923 | 0.004 | |
| DOCK7 rs1168013 | G/C | −0.110 | 0.019 | −2.857 | 0.004 | ||
| DOCK7rs10889332 | CC,CT/TT | 0.175 | 0.014 | 4.502 | 0.000 | ||
| DOCK7rs10889332 | C/T | 0.125 | 0.019 | 3.200 | 0.001 | ||
| PCSK9rs7552841 | CC,CT/TT | 0.221 | 0.015 | 6.079 | 0.000 | ||
| PCSK9rs7552841 | C/T | 0.236 | 0.019 | 6.435 | 0.000 | ||
| PCSK9rs11206517 | TT,GT/GG | 0.093 | 0.022 | 2.444 | 0.015 | ||
| PCSK9rs11206517 | T/G | 0.085 | 0.025 | 2.222 | 0.027 | ||
| ApoA1/ApoB | PCSK9rs615563 | G/A | −0.075 | 0.029 | −2.017 | 0.044 | |
| DOCK7rs10889332 | CC,CT/TT | −0.080 | 0.021 | −2.154 | 0.032 | ||
| PCSK9rs7552841 | CC,CT/TT | −0.177 | 0.024 | −4.770 | 0.000 | ||
| PCSK9rs7552841 | C/T | −0.183 | 0.030 | −4.905 | 0.000 | ||
| GALNT2rs1997947 | GG,AG/AA | −0.098 | 0.023 | −2.597 | 0.000 | ||
| GALNT2rs1997947 | G/A | −0.074 | 0.029 | −1.968 | 0.050 | ||
| Han | |||||||
| TC | DOCK7 rs1168013 | GG,CG/CC | 0.101 | 0.047 | 2.831 | 0.005 | |
| DOCK7 rs1168013 | G/C | 0.103 | 0.061 | 2.902 | 0.004 | ||
| PCSK9rs7552841 | CC,CT/TT | 0.076 | 0.058 | 2.143 | 0.032 | ||
| TG | DOCK7 rs1168013 | GG,CG/CC | 0.084 | 0.049 | 2.316 | 0.021 | |
| DOCK7rs10889332 | CC,CT/TT | 0.111 | 0.052 | 3.083 | 0.002 | ||
| DOCK7rs10889332 | C/T | 0.116 | 0.060 | 3.435 | 0.001 | ||
| PCSK9rs615563 | GG,AG/AA | 0.123 | 0.052 | 3.661 | 0.000 | ||
| PCSK9rs615563 | G/A | 0.153 | 0.064 | 4.476 | 0.000 | ||
| PCSK9rs7552841 | CC,CT/TT | 0.119 | 0.056 | 3.509 | 0.000 | ||
| PCSK9rs7552841 | C/T | 0.100 | 0.065 | 2.933 | 0.003 | ||
| PCSK9rs11206517 | TT,GT/GG | 0.105 | 0.079 | 3.121 | 0.002 | ||
| PCSK9rs11206517 | T/G | 0.076 | 0.088 | 2.248 | 0.025 | ||
| GALNT2rs2760537 | CC,CT/TT | 0.090 | 0.043 | 2.717 | 0.007 | ||
| GALNT2rs2760537 | C/T | 0.078 | 0.059 | 2.312 | 0.021 | ||
| GALNT2rs4846913 | CC,AC/AA | 0.120 | 0.052 | 3.640 | 0.000 | ||
| GALNT2rs4846913 | C/A | 0.122 | 0.064 | 3.618 | 0.000 | ||
| GALNT2rs11122316 | GG,AG/AA | 0.121 | 0.044 | 3.666 | 0.000 | ||
| GALNT2rs11122316 | G/A | 0.090 | 0.060 | 2.690 | 0.007 | ||
| HDL cholesterol | DOCK7 rs1168013 | G/C | 0.084 | 0.037 | 2.351 | 0.019 | |
| PCSK9rs11206517 | TT,GT/GG | −0.112 | 0.052 | −3.044 | 0.002 | ||
| PCSK9rs11206517 | T/G | −0.132 | 0.056 | −3.673 | 0.000 | ||
| GALNT2rs4846913 | CC,AC/AA | −0.116 | 0.033 | −3.262 | 0.001 | ||
| GALNT2rs4846913 | C/A | −0.146 | 0.040 | −4.111 | 0.000 | ||
| LDL cholesterol | DOCK7 rs1168013 | GG,CG/CC | 0.107 | 0.024 | 2.965 | 0.003 | |
| DOCK7 rs1168013 | G/C | 0.093 | 0.032 | 2.567 | 0.010 | ||
| ApoA1 | DOCK7 rs1168013 | G/C | 0.109 | 0.014 | 3.094 | 0.002 | |
| DOCK7rs10889332 | CC,CT/TT | 0.082 | 0.012 | 2.315 | 0.021 | ||
| PCSK9rs11206517 | TT,GT/GG | −0.114 | 0.020 | −3.138 | 0.002 | ||
| PCSK9rs11206517 | T/G | −0.125 | 0.022 | −3.448 | 0.001 | ||
| GALNT2rs1997947 | GG,AG/AA | −0.079 | 0.013 | −2.231 | 0.026 | ||
| GALNT2rs4846913 | CC,AC/AA | −0.164 | 0.013 | −4.627 | 0.000 | ||
| GALNT2rs4846913 | C/A | −0.176 | 0.016 | −5.000 | 0.000 | ||
| ApoB | DOCK7 rs1168013 | GG,CG/CC | 0.119 | 0.013 | 3.314 | 0.001 | |
| DOCK7 rs1168013 | G/C | 0.101 | 0.018 | 2.797 | 0.005 | ||
| PCSK9rs615563 | GG,AG/AA | 0.087 | 0.015 | 2.406 | 0.016 | ||
| PCSK9rs615563 | G/A | 0.080 | 0.019 | 2.200 | 0.028 | ||
| PCSK9rs7552841 | CC,CT/TT | 0.104 | 0.017 | 2.868 | 0.004 | ||
| ApoA1/ApoB | DOCK7 rs1168013 | GG,CG/CC | −0.073 | 0.020 | −2.074 | 0.038 | |
| PCSK9rs615563 | G/A | −0.084 | 0.028 | −2.394 | 0.017 | ||
| PCSK9rs11206517 | TT,GT/GG | −0.072 | 0.036 | −2.027 | 0.043 | ||
| GALNT2rs1997947 | GG,AG/AA | −0.110 | 0.023 | −3.139 | 0.002 | ||
| GALNT2rs4846913 | CC,AC/AA | −0.118 | 0.023 | −3.363 | 0.001 | ||
| GALNT2rs4846913 | C/A | −0.136 | 0.028 | −3.908 | 0.000 | ||
HDL, high density lipoprotein; LDL, low density lipoprotein; Association of serum lipid traits and allele and genotypes in Jing, Han and combined the Jing and Han populations were assessed by multivariable linear regression analyses with stepwise modeling.
Discussion
In the present study, we showed for the first time the association of the DOCK7 (rs1168013 and rs10889332), PCSK9 (rs615563, rs7552841 and rs1126517) and GALNT2 (rs1997947, rs2760537, rs4846913 and rs11122316) SNPs and some serum lipid parameters; the LD status and the haplotype frequencies of the detected SNPs. In addition, we also successfully replicated the association of DOCK7 rs10889332, PCSK9 rs615563, PCSK9 rs7552841, GALNT2 rs1997947, GALNT2 rs2760537, GALNT2 rs4846913 and GALNT2 rs11122316 SNPs with the levels of serum TG in the Jing ethnic minority; and DOCK7 rs10889332, PCSK9 rs615563, PCSK9 rs7552841, PCSK9 rs11206517, GALNT2 rs1997947, GALNT2 rs4846913 and GALNT2 rs11122316 with serum TG levels in the Han nationality.
The SNPs of rs636523 and rs1213033322 near the DOCK7/ ANGPTL3, PCSK9 rs50515123 and GALNT2 rs484691424,25 have been associated with TG in some previous studies, but the association of the 9 SNPs and other serum lipid parameters has not been reported previously. The genotype and allele frequencies of several SNPs in this study were also not reported previously in different racial/ethnic groups. In the present study, we revealed that the genotypic and allelic frequencies of the DOCK7 rs1168013, DOCK7 rs10889332, PCSK9 rs615563, PCSK9 rs7552841, PCSK9 rs1126517, GALNT2 rs1997947, GALNT2 rs2760537, GALNT2 rs4846913 and GALNT2 rs11122316 SNPs were different between the two ethnic groups. All of the detected SNPs were in the Hardy-Weinberg equilibrium except DOCK7 rs10889332. The minor allele or rare homozygote genotype frequencies of the 9 SNPs in the Han nationality were in close proximity to those of CHB from the international haplotype map (HapMap; http://hapmap.ncbi.nlm.nih.gov/cgi-perl/gbrowse/hapmap24_B36/) data. The minor allele or rare homozygote genotype frequencies of the 9 detected SNPs were also lower in European ancestries than in Asian nationalities from the data. These results suggest that the prevalence of the minor allele or rare homozygote genotype frequencies of the 9 SNPs may have a racial/ethnic-specificity.
In the present reasearch, our findings also showed that there may be a racial/ethnic specific association of the 9 SNPs and lipid parameters. The association of other SNPs near DOCK7, PCSK9 and GALNT2 and lipid profiles has been reported previously. Through fine-mapping, previous study discovered the SNP with significant associations, with consistent effect on TG levels across ancestral groups: rs636523 near DOCK7/ANGPTL3. African LD patterns did not assist in narrowing association signals22. PCSK9 (TG, HDL cholesterol, ApoB and ApoA1/ApoB) was shown interactions with overweight/obesity to influence serum lipid levels23. Our team reported that the correlations of both GALNT2 rs2144300 and GALNT2 rs4846914 SNPs and lipid parameters were different between the two ethnic groups19. However, Several GWASs and candidate gene researches failed to find the association between the GALNT2 polymorphisms and lipid parameters26,27,28. There was no any effect of the GALNT2 rs4846914 on the levels of serum TC or TG reported by Polgár et al. previously26. In Whitehall II, there was a significant correlation of the GALNT2 variants and serum lipoprotein (a) levels. Whereas any of these findings did not confirmed in the previously meta-analysis of six studies28. It could be due to the effects of these SNPs were modest on serum lipid concentrations and/or lower statistical power to determine the correlation was present26,29. In addition, gene-environmental and environmental- environmental factors on lipid parameters remain to be interpreted.
Many GWASs have reported that the association of other variants near DOCK7, PCSK9 and GALNT2 and serum lipid levels is still controversial. Pleiotropic effects on the lipid profile, the potential correspondence was detected for ANGPTL330 and DOCK731 being highly associated with cholesterol and LDL cholesterol levels. Loss-of-function mutations in the ANGPTL3 were associated with decreased levels of LDL cholesterol, HDL cholesterol and TG32. The associations observed for the DOCK7 locus, which is involved in neurogenesis, myelination and axon formation33 but not in lipid metabolism probably reflect the co-localization of this gene with ANGPTL3. As expected, rare variants that contribute to population differences tend to be population specific, exemplified by multiple African-specific variants in PCSK9 associated with LDL cholesterol34. The SNPs in intron 1 of a GalNac transferase (GALNT2) were identified as a novel lipid-associated region from GWAS and subsequent knock-down and overexpression of this gene in mouse liver clearly demonstrated that GALNT2 can influence HDL cholesterol levels35.
The cause of the contradictions in correlation of the detected SNPs with lipid parameters among the different population is not completely understood. This could be because of the differences in genetic background in some degree. Compared to the Han nationality, the Jing ethnic minority had higher the value of weight, BMI, waist circumference, the serum TC and TG levels and the lower percentage of participants who consumed alcohol and the ratio of ApoA1 to ApoB. Among 56 nationalities in China, Han nationality is the largest one. Jing ethnic minority was less population nationality with the population of 22517 according to the China’s fifth national census in 2000. Approximately 90% of the Jing people live in the three islands of Wanwei, Wutou and Shanxin in the Dongxing city, Guangxi, China. About 1511, their ancestors emigrated from Vietnam to China and first settled on the aforementioned three islands. Therefore, some hereditary background and alleles/genotypes of lipid metabolism-related genes in Jing ethnic minority might be somewhat different from those in Han nationality.
Another reason could be because of the ethnic difference in their LD pattern. In this research, we detected that the frequencies of the C-C-G-C-T-G-T-C-G, G-C-A-C-T-G-C-C-G, G-C-G-C-T-A-C-C-A, G-C-G-C-T-G-C-C-A, G-C-G-C-T-G-T-C-A haplotypes were significantly different between the Jing and Han populations. The haplotypes with nine SNPs could explain much more serum lipid variation than any single SNP alone, especially for TG. Therefore, ethnic differences in the LD pattern could partially explain the discrepancy in the correlation of the detected SNPs with lipid parameters among diverse nationalities.
Several environmental factors independently such as hypertension, obesity, physical activity, dietary patterns and lifestyle are related with lipid parameters strongly36,37,38,39,40,41,42. There was association of gender, age, BMI, cigarette smoking, alcohol consumption, blood pressure and lipid levels in both Jing and Han populations. These detected data determined some environmental factors play an important role in determining lipid parameters. For approximately half a century it has been acknowledged that diets of high-fat particularly contain the large quantities of saturated fatty acids raise predispose individuals to hyperlipidemia and CVD43. In the current study, we found that the % of participants who consumed alcohol were lower in Jing than in Han nationality. Although effects of the alcohol consumption on lipid parameters appear to vary by types of specific patient or the alcohol intake patterns, and perhaps by sex and population, the subject research has been the focus of so much current studies44,45,46.
GWASs have identified many loci that will harbor genes relevant to the biology of lipid levels. The results show that significant associations can be identified by studying a relatively small number of subjects with extreme values of a quantitative lipoprotein trait. Re-sequencing genes at GWAS loci may reveal new rare loss-of-function mutations, creating potential new therapeutic targets for decreasing the prevalence of heart disease. Until recently, most genome-wide efforts have used genotyping arrays and imputation to assay most of the common variation across the genome. Recent technological advances have enabled whole genome sequencing approaches, which hold the promise of discovery of novel rare variants with large effects on lipid levels and heart disease risk.
There are several potential limitations in the present study. Firstly, the sample size is a bit small as compared with many previous GWASs. Hence, further studies with larger sample sizes are needed to confirm our results. Secondly, interactions of gene-environmental or environmental-environmental factors on serum lipid traits remain to be detected. Thirdly, there are no independent haplotypes of each gene considered instead. Phasing of the 9 SNPs is so far away on chromosome 1 (~40MB). There will be many recombination events that will be missed due to the lack of information across the region. Finally, although we have detected the effects of 9 SNPs in the PCSK9, DOCK7 and GALNT2 on serum lipid levels in this study, there are still many lipid-related SNPs and the interactions of SNP-SNP and/or SNP-environmental factors. What’s more, the relevance of this finding has to be defined in further high caliber of studies including incorporating the genetic information of the DOCK7, PCSK9 and GALNT2 SNPs and their haplotypes and in vitro functional studies to confirm the impact of a variant on a molecular level.
In summary, several SNPs in the present study were associated with TG (rs10889332, rs615563, rs7552841, rs1997947, rs2760537, rs4846913 and rs11122316), HDL cholesterol (rs1997947), LDL cholesterol (rs1168013 and rs7552841), ApoA1 (rs1997947), ApoB (rs10889332 and rs7552841), and the ApoA1/ApoB ratio (rs7552841) in the Jing population, whereas they were associated with TG (rs10889332, rs615563, rs7552841, rs11206517, rs1997947, rs4846913 and rs11122316), HDL cholesterol (rs11206517 and rs4846913), LDL cholesterol (rs1168013), ApoA1 (rs11206517 and rs4846913), ApoB (rs7552841), and the ApoA1/ApoB ratio (rs4846913) in the Han participants. The frequencies of several haplotypes among the 9 SNPs were also different between the Jing and Han populations. The differences in lipid parameters between the Jing and Han populations might result from different SNPs and their haplotypes partially.
Materials and methods
Subjects and research design
For the current study, 881 unrelated subjects (456 males, 51.76% and 425 females, 48.24%) of Jing and 988 (536 men, 54.25% and 452 women, 45.75%) unrelated individuals of Han from Dongxing city, Guangxi Zhuang Autonomous Region, China were selected randomly from our randomized, stratified samples. The subjects’ age ranged from 15 to 80 years and with the average age of 56.69 ± 13.39 years in Jing and 56.18 ± 12.85 years in Han. All participants were healthy with no disease history of atherosclerosis, CVD, diabetes, thyroid and/or kidney. When blood samples were taken, none of them used lipid-modulating therapy such as fibrates or statins. The study design was approved by the Ethics Committee of the First Affiliated Hospital, Guangxi Medical University. Written informed consent was obtained from all participants. All experiments were performed in accordance with relevant guidelines and regulations47,48,49,50.
Data collection
Epidemiological investigation and measurements of biochemical markers
Participants participated in baseline examination conducted in the study center by trained staff following standardized protocols, which included anthropometric, blood pressure measurements, height, weight (without shoes) and waist circumference parameters (in cm was measured at the midpoint between the lower ribs and the iliac crest), a blood sample collection as well as a personal interview on medical history, a sociodemographic, socioeconomic status and lifestyle questionnaire and a self-administered food frequency questionnaire; BMI was calculated as the ratio of weight (kg) to squared height (m2). During the blood sample collection, 5 ml of venous blood were drawn, rapidly processed and serum lipids, lipoproteins, and apolipoproteins were measured by enzymatic methods with commercially available kits: RANDOX Laboratories Ltd., Ardmore, Diamond Road, Crumlin Co. Antrim, United Kingdom, BT29 4QY and Daiichi Pure Chemicals Co., Ltd., Tokyo, Japan, and the immunoturbidimetric assay using a commercial kit: RANDOX Laboratories Ltd. in the Clinical Science Experiment Center of the First Affiliated Hospital, Guangxi Medical University with an autoanalyzer: Type 7170A; Hitachi Ltd., Tokyo, Japan. The normal values of serum lipid phenotypes in our Clinical Science Experiment Center were as follows: TC, 3.10–5.17 mmol/L; TG, 0.56–1.70 mmol/L; HDL cholesterol, 1.16–1.42 mmol/L; LDL cholesterol, 2.70–3.10 mmol/L; ApoA1, 1.20–1.60 g/L; ApoB, 0.80–1.05 g/L; and the ratio of ApoA1 to ApoB, 1.00–2.50; respectively50.
SNPs selection
Nine TG-related loci in the DOCK7, PCSK9 and GALNT2 were selected by three criteria encompass (i) Tag SNPs, which were established by Haploview (Broad Institute of MIT and Harvard, USA, version 4.2) and functional/missense SNPs in functional area of the gene fragments (http://www.ncbi.nlm.nih.gov/SNP/snp); (ii) a known minor allele frequency higher than 1% in European ancestry from the Human Genome Project Database; and (iii) the target SNP region should be adequately replicated by PCR, and the polymorphic site should have a commercially available restriction endonuclease enzyme cleavage site to be genotyped with RFLP.
Genotyping
DNA was isolated from blood samples using DNA Blood Midi kits (Qiagen, Hilden, Germany) following the protocol recommended by the vendor. We identified 9 SNPs genotyping by PCR-RFLP. The characteristics of each SNP and the details of each primer pair, annealing temperature, length of the PCR products and corresponding restriction enzyme used for genotyping are summarized in supplemental Tables 1 and 2. The PCR products of the samples (two samples of each genotype) were sequenced with an ABI Prism 3100 (Applied Biosystems, international Equipment Trading Ltd., Vernon Hill, IL, USA) in Shanghai Sangon Biological Engineering Technology & Services Co., Ltd., China.
Statistical methods
The statistical analyses were done with the statistical software package SPSS 17.0 (SPSS Inc., Chicago, Illinois). Data were presented as the mean ± SD for those, that are normally distributed, and the medians and interquartile ranges for TG, which is not normally distributed. Clinical characteristics between the Jing and Han populations were compared by Student’s unpaired t-test. The allele, genotype and haplotype distribution between the Jing and Han populations were analyzed by the chi-squared test; and the standard goodness-of-fit verified the test Hardy-Weinberg equilibrium. Haploview (Broad Institute of MIT and Harvard, USA, version 4.2) analyzed the haplotype frequencies and pair-wise LD among the detected SNPs. The correlation of genotypes and lipid profiles was calculated by ANCOVA. Any SNPs associated with the lipid profiles at the value of P < 0.005 (corresponding to P < 0.05 after adjusting for 9 independent tests by the Bonferroni correction) were considered statistically significant. Unconditional logistic regression was used to assess the assocation between lipid parameters and genotypes (common homozygote genotype = 1, heterozygote genotype = 2, rare homozygote genotype = 3) or alleles (the minor allele non-carrier = 1, the minor allele carrier = 2). Gender, age, BMI, alcohol consumption, cigarette smoking and hypertension were adjusted for statistical analysis. Two-sided P value of less than 0.05 was considered statistically significant.
Additional Information
How to cite this article: Guo, T. et al. Association between the DOCK7, PCSK9 and GALNT2 Gene Polymorphisms and Serum Lipid levels. Sci. Rep. 6, 19079; doi: 10.1038/srep19079 (2016).
Supplementary Material
Acknowledgments
The current study was supported by the Innovation Project of Guangxi Graduate Education and the National Natural Science Foundation of China (No: 81160111).
Footnotes
Author Contributions T.G. participated in the research design, carried out the epidemiological investigation, collected the samples, undertook genotyping, performed statistical analyses, and drafted the manuscript. R.-X.Y. conceived the study, participated in the research design, carried out the epidemiological investigation, collected samples, helped to draft the manuscript and edited the final manuscript. F.H., L.-M.Y., W.-X.L. and S.-L.P. carried out the epidemiological investigation, and collected samples. All authors had read and approved the final manuscript.
References
- Murray C. J. L. & Lopez A. D. Alternative projections of mortality and disability by cause 1990–2020: Global Burden of Disease Study. Lancet. 349, 98–1504 (1997). [DOI] [PubMed] [Google Scholar]
- Mathers C. D. & Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 3, e442 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson R., Frohlich J., Fodor G. & Genest J. & Canadian Cardiovascular Society.Canadian Cardiovascular Society position statement–recommendations for the diagnosis and treatment of dyslipidemia and prevention of cardiovascular disease. Can J Cardiol. 22, 913–27 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitziel N. O. et al. Exome sequencing in suspected monogenic dyslipidemias. Circ Cardiovasc Genet. 8, 343–50 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musunuru K. Atherogenic dyslipidemia: cardiovascular risk and dietary intervention. Lipids. 45, 907–914 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen C. T. et al. LipidSeq: a next-generation clinical resequencing panel for monogenic dyslipidemias. J Lipid Res. 55, 765–772 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay G. J. et al. Association Analysis of Dyslipidemia-Related Genes in Diabetic Nephropathy. PLoS One. 8, e58472 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S. et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 41, 56–65 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel C. J., Cullen M. R., Ioannidis J. P. & Butte A. J. Systematic evaluation of environmental factors: persistent pollutants and nutrients correlated with serum lipid levels. Int J Epidemiol. 41, 828–43 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir J. M. et al. Plasma lipid profiling in a large population-based cohort. J Lipid Res. 54, 2898–2908 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer C. J. et al. Discovery and Refinement of Loci Associated with Lipid Levels11. Nat Genet. 45, 2797 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folkert W. et al. Large-Scale Gene-Centric Meta-analysis across 32 Studies Identifies Multiple Lipid Loci. Am J Hum Genet. 91, 823–838 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanya M. et al. Biological, Clinical, and Population Relevance of 95 Loci for Blood Lipids. Nature. 466, 707–713 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. G. et al. Efficacy and safety of alirocumab in reducing lipids and cardiovascular events. N Engl J Med. 372, 1489–1499 (2015). [DOI] [PubMed] [Google Scholar]
- Jeemon P., Pettigrew K., Sainsbury C., Prabhakaran D. & Padmanabhan S. Implications of discoveries from genome-wide association studies in current cardiovascular practice. World J Cardiol. 3, 230–247 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko Y. S. et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 41, 47–55 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasman D. I. et al. Forty-three loci associated with plasma lipoprotein size, con-centration, and cholesterol content in genome-wide analysis. PLoS Genet. 5, e1000730 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung L. H. et al. The proprotein convertase subtilisin/kexin type 9 gene E670G polymorphism and serum lipid levels in the Guangxi Bai Ku Yao and Han populations. Lipids Health Dis. 10, 5 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. et al. Association of the GALNT2 gene polymorphisms and several environmental factors with serum lipid levels in the Mulao and Han populations. Lipids Health Dis. 10, 160 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C. et al. Prevalence of chronic kidney disease in Jing adults in China: a village-based study. Clin Nephrol. 79, 50–56 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang J. P. et al. Mutant thiopurine S-methyltransferase alleles among Jing Chinese in Guangxi province. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 20, 303–306 (2003). [PubMed] [Google Scholar]
- Keebler M. E. et al. Fine-mapping in African Americans of 8 recently discovered genetic loci for plasma lipids: the Jackson Heart Study. Circ Cardiovasc Genet. 3, 358–64 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R. X. et al. Several genetic polymorphisms interact with overweight/obesity to influence serum lipid levels. Cardiovasc Diabetol. 11, 123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C. B. et al. Adiposity significantly modifies genetic risk for dyslipidemia. J Lipid Res. 55, 2416–22 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegele R. A. et al. A polygenic basis for four classical Fredrickson hyperlipoproteinemia phenotypes that are characterized by hypertriglyceridemia. Hum Mol Genet. 18, 4189–4194 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polgár N. et al. Triglyceride level modifying functional variants of GALTN2 and MLXIPL in patients with ischaemic stroke. Eur J Neurol. 17, 1033–1039 (2010). [DOI] [PubMed] [Google Scholar]
- Reynolds C. A. et al. Analysis of lipid pathway genes indicates association of sequence variation near SREBF1/TOM1L2/ATPAF2 with dementia risk. Hum Mol Genet. 19, 2068–2078 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabaneh D. et al. Meta-analysis of candidate gene variants outside the LPA locus with Lp(a) plasma levels in 14, 500 participants of six White European cohorts. Atherosclerosis. 217, 447–451 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasman D., Pare G. & Ridker P. Population-based genomewide genetic analysis of common clinical chemistry analytes. Clin Chem. 55, 39–51 (2009). [DOI] [PubMed] [Google Scholar]
- Arianna M. et al. A genome-wide association analysis for porcine serum lipid traits reveals the existence of age-specific genetic determinants. BMC Genomics. 15, 758 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth D. M. et al. Genetic variants influencing circulating lipid levels and risk of coronary artery disease. Arterioscler Thromb Vasc Biol. 30, 2264–2276 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musunuru K. et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med. 363, 2220–2227 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. T., Wang C. L. & Van Aelst L. DOCK7 interacts with TACC3 to regulate interkinetic nuclear migration and cortical neurogenesis. Nat Neurosci. 15, 1201–1210 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coram M. A. et al. Genome-wide characterization of shared and distinct genetic components that influence blood lipid levels in ethnically diverse human populations. Am J Hum Genet. 92, 904–916 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer C. J. & Mohlke K. L. F. inding genes and variants for lipid levels after genome-wide association analysis. Curr Opin Lipidol. 23, 98–103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R. X. et al. Effects of demographic, dietary and other lifestyle factors on the prevalence of hyperlipidemia in Guangxi Hei Yi Zhuang and Han populations. Eur J Cardiovasc Prev Rehabil. 13, 977–84 (2006). [DOI] [PubMed] [Google Scholar]
- Yin R. X. et al. Comparison of demography, diet, lifestyle, and serum lipid levels between the Guangxi Bai Ku Yao and Han populations. J Lipid Res. 48, 2673–81 (2007). [DOI] [PubMed] [Google Scholar]
- Garcia-Palmieri M. R. et al. Nutrient intake and serum lipids in urban and ruralPuerto Rican men. Am J Clin Nutr. 30, 2092–2100 (1977). [DOI] [PubMed] [Google Scholar]
- Sola R. et al. Effect of a traditional Mediterranean diet on apolipoproteins B, A-I,and their ratio: a randomized, controlled trial. Atherosclerosis. 218, 174–180 (2011). [DOI] [PubMed] [Google Scholar]
- Valente E. A. et al. The effect of the addition of resistance training to a dietary education intervention on apolipoproteins and diet quality in overweight andobese older adults. Clin Interv Aging. 6, 235–241 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield E., McPherson R. & Koski K. G. Diet and waist-to-hip ratio: importantpredictors of lipoprotein levels in sedentary and active young men with no evidence of cardiovascular disease. J Am Diet Assoc. 99, 1373–1379 (1999). [DOI] [PubMed] [Google Scholar]
- Joffe Y. T., Collins M. & Goedecke J. H. The relationship between dietary fatty acids and inflammatory genes on the obese phenotype and serum lipids. Nutrients. 5, 1672–1705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu-Poth S. et al. Effects of National Cholesterol Education Program’s Step I and Step II dietary intervention programs on cardiovascular disease risk factors: a meta-analysis. Am J Clin Nutr. 69, 632–46 (1999). [DOI] [PubMed] [Google Scholar]
- Brinton E. A. Effects of ethanol intake on lipoproteins and atherosclerosis. Curr Opin Lipidol. 21, 346–51 (2010). [DOI] [PubMed] [Google Scholar]
- Perissinotto E. et al. ILSA Working Group. Alcohol consumption and cardiovascular risk factors in older lifelong wine drinkers: the Italian Longitudinal Study on Aging. Nutr Metab Cardiovasc Dis. 20, 647–55 (2010). [DOI] [PubMed] [Google Scholar]
- Onat A. et al. Associations of alcohol consumption with blood pressure, lipoproteins, and subclinical inflammation among Turks. Alcohol. 42, 593–601 (2008). [DOI] [PubMed] [Google Scholar]
- Lin Q. Z. et al. Sex-specific association of the peptidase D gene rs731839 polymorphism and serum lipid levels in the Mulao and Han populations. Int J Clin Exp Pathol. 7, 4156–4172 (2014). [PMC free article] [PubMed] [Google Scholar]
- Guo T. et al. Polymorphism of rs873308 near the transmembrane protein 57 gene is associated with serum lipid levels. Biosci Rep. 34, e00095 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T. et al. Sex-specific association of the SPTY2D1 rs7934205 polymorphism and serum lipid levels. Int J Clin Exp Pathol. 8, 665–681 (2015). [PMC free article] [PubMed] [Google Scholar]
- Aung L. H. et al. Association between the MLX interacting protein-like, BUD13 homolog and zinc finger protein 259 gene polymorphisms and serum lipid levels. Sci Rep. 4, 5565 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.