Abstract
Autism Spectrum Disorders (ASDs) are a group of neurodevelopmental disorders characterized by ritualistic-repetitive behaviors and impaired verbal and non-verbal communication. Objectives were to determine the contribution of genetic variation to ASDs in the Lebanese. Affymetrix Cytogenetics Whole-Genome 2.7 M and CytoScan™ HD Arrays were used to detect CNVs in 41 Lebanese autistic children and 35 non-autistic, developmentally delayed and intellectually disabled patients. 33 normal participants were used as controls. 16 de novo CNVs and 57 inherited CNVs, including recognized pathogenic 16p11.2 duplications and 2p16.3 deletions were identified. A duplication at 1q43 classified as likely pathogenic encompasses RYR2 as a potential ASD candidate gene. This previously identified CNV has been classified as both pathogenic, and, of uncertain significance. A duplication of unknown significance at 10q11.22, proposed as a modulator for phenotypic disease expression in Rett syndrome, was also identified. The novel potential autism susceptibility genes PTDSS1 and AREG were uncovered and warrant further genetic and functional analyses. Previously described and novel genetic targets in ASD were identified in Lebanese families with autism. These findings may lead to improved diagnosis of ASDs and informed genetic counseling, and may also lead to untapped therapeutic targets applicable to Lebanese and non-Lebanese patients.
Autism Spectrum Disorders (ASD) are a group of non-progressive neurodevelopmental disorders characterized by repetitive/restrictive behaviors and impaired language/social behavior that set in between 16 to 36 months of age. Deficits are noted within the areas of communicative, interactive and behavioral core domains. These may be coupled with mental retardation (70%)1 and seizures (20–25%)2,3. Prevalence estimates of autism in the general population have gradually increased over the last 20 years. Recent figures from the Center for Disease Control and Prevention state that 1 of 68 children were diagnosed with autism in the USA in 20104. In the Eastern Mediterranean region, small scale studies such as the one in Saudi Arabia states that 12.5% of patients ≤to 18 years who attended child psychiatric clinics have ASD5. In Oman, the prevalence is lower at 1.4 per 10,000 in children less than 14 years of age6. In Lebanon, formal estimates for prevalence of ASD in the greater Beirut and Mount Lebanon areas are now published7, and are 1/66 children.
ASD is a complex disorder with clinical variability, genetic heterogeneity and high de novo mutation rates that can also be impacted by epigenetic factors. The concordance rate for autism has been estimated to be 88.1% in monozygotic and 30.5% in dizygotic twin pairs8. Yet, several factors hinder identification of culprit gene(s)9,10,11,12. The fact that affected individuals seldom bear children, makes it difficult to perform linkage studies searching for ASD loci. A few recurrent aberrations have been reported and include maternally derived duplication of chromosome band 15q11.2-13. This was identified in 0.5–3% of ASD cases13. In addition, autistic features often occur in single gene disorders such as tuberous sclerosis, Fragile X syndrome, and Rett syndrome, but these disorders only explain around 2–5% of patients presenting with autism symptoms14,15,16. These disorders are no longer classified as being on the autism spectrum. Both copy number variations (CNVs) and mutations in more than 100 genes including SHANK3, NLGN3, NLGN4, and NRXN1, have been reported in rare cases of ASD17,18,19.
Recently, next generation sequencing in large ASD cohorts has added multiple risk-conferring genes. One of the most successful findings is the discovery of the CDH8 gene. Multiple independent studies have reported the contribution of de novo deleterious mutations in chromo-domain helicase DNA-binding protein 8 (CDH8) in ASD20,21,22,23. Subjects with CDH8 mutations share common features including macrocephaly, distinct facial features and gastrointestinal difficulties, which define a distinct ASD subtype24. Later, CDH8 was found to be involved in a highly interconnected gene network with numerous previously identified ASD candidate genes25.
The unique genetic makeup of the Lebanese makes them an ideal population to study and identify novel autism-related genes. The Lebanese population is genetically distinguished by significant shared ancestry26,27, a distinctive blend of DNA recombination events occurring over millennia and high rates of consanguinity28,29,30. The specific genetic fingerprint of the Lebanese population is ideally suited for Comparative Genomic Hybridization or CGH microarray studies, homozygosity mapping and copy number variation (CNV) analysis for discovery of novel as well as previously described ASD susceptibility genes. In fact, in the Lebanese population, male to female ASD cases are 1:1.05, unlike the reported 1:4 ratio reported elsewhere, underscoring the important contribution of unique genetic characteristics to the incidence of ASD in Lebanon7.
Array chromosomal genomic hybridization has increased identification of putative ASD genes, and raised up to 25% the percentage of children for whom an ASD-related genetic change can be identified31. CGH microarray analysis allows identification of normal benign CNVs, pathogenic CNVs associated with disease, and novel CNVs classified as unknown, but that may be associated with autism.
In this work, the objective was to determine the potential contribution of genomic variation in ASD in the Lebanese population. Affymetrix Cytogenetics Whole-Genome 2.7 M and CytoScan™ HD Arrays were used to identify CNVs across families of ASD children. We identified loci and genes that have been previously detected in the West. Additionally, we discovered new potential candidate ASD susceptibility loci and several candidate genes that code for proteins that function within the synapse. A systems biology approach was utilized to study the implications and contributions of these genes to the pathology of autism.
Results
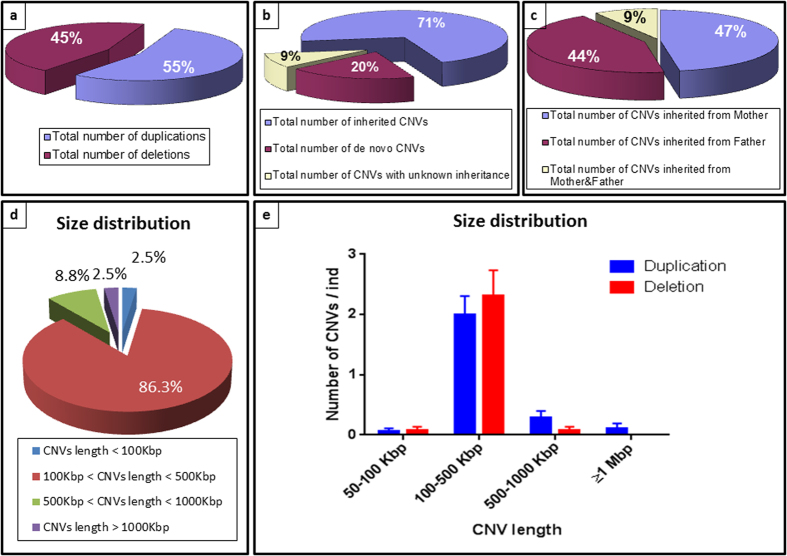
The chromosomal microarray analysis of 41 children with autism revealed 80 CNVs (44 gains, 36 losses) (Fig. 1a). Of these, 16 CNVs are de novo (20%) and 57 CNVs are inherited (71%) (Fig. 1b).
Figure 1. Comparative CNV landscape identified in Lebanese autism patients.
Charts summarize (a) Number of duplications versus deletions; (b) Mode of inheritance of CNVs; (c) Number of CNVs inherited from father versus mother; (d) Size distribution of total CNVs; (e) Size distribution of deletions versus duplications.
2 CNVs (2.5%) were less than 100 Kbp, 69 CNVs (86.3%) ranged between 100 Kbp and 500 Kbp, 7 CNVs (8.8%) ranged between 500 Kbp and 1000 Kbp and 2 CNVs (2.5%) were above 1000 Kbp (Fig. 1d). Duplications and deletions were not distributed equally, and, deletions tended to be smaller than duplications (Fig. 1e).
A CNV (duplication at 1q43) was validated using quantitative PCR (qPCR) to ensure reliability of the CNV detection method employed (see Supplementary figure 2).
De novo CNVs
Sixteen de novo CNVs (11 gains and 5 losses) were detected in 10 children with autism from 9/35 (25%) of families fully analyzed. The frequency of de novo CNVs in ASD patients was 27% (10 patients out of 37 analyzed cases showed at least one de novo CNV). The rate of de novo CNVs per child was 1.6 (16 de novo CNVs were observed in 10 patients). Table 1 and Supplementary table 1 list the 16 de novo CNVs and their gene content.
Table 1. De novo CNVs identified in patients with ASD classified as pathogenic, likely pathogenic or of unknown significance.
| ID | Cytoband | Type | Location | Size (kb) | Genes | Classification |
|---|---|---|---|---|---|---|
| LAS17 | 16p11.2 | Gain | 29396001-30190676 | 795 | YPEL3, PRRT2, SULT1A4, GDPD3, C16orf53, LOC440356, BOLA2, PPP4C, ZG16, CDIPT, HIRIP3, LOC388242, TAOK2, ASPHD1, KCTD13, INO80E, SPN, QPRT, LOC606724, SEZ6L2, MAZ, TMEM219, LOC440354, ALDOA, C16orf54, KIF22, C16orf92, SLC7A5P1, TBX6, MAPK3, MVP | Pathogenic |
| CLIN19 | 1q42.3 | Gain | 236270756-236557162 | 286 | ERO1LB, GPR137B | Unknown significance |
| 3q24 | Gain | 147147614-147082426 | 67 | ZIC1, ZIC4 | Unknown significance | |
| CLIN27 | 10q26.13 | Gain | 124808072-124919262 | 111 | HMX3, HMX2, BUB3 | Unknown significance |
| CLIN29 | 4q13.3 | Loss | 75405778-75521036 | 115 | AREG | Unknown significance |
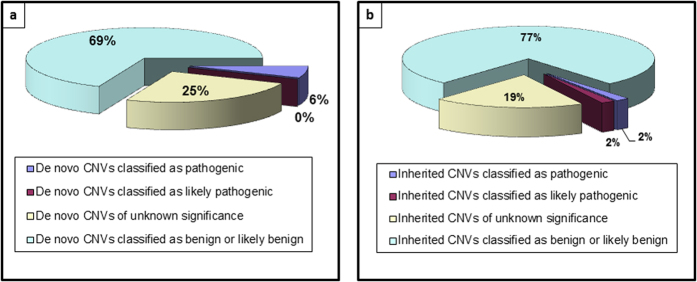
We identified one duplication located on chromosome 16 (16p11.2), which is classified as pathogenic, and 4 CNVs of unclear clinical significance located on chromosomes 1, 3, 4 and 10, respectively (1q42.3, 3q24, 4q13.3, 10q26.13) (Fig. 2a).
Figure 2. Pie charts summarizing the classification of (a) de novo and (b) inherited CNVs in autism patients.
Inherited CNVs
57 CNVs (26 gains and 31 losses) inherited from at least one of the parents were detected in 26 of 35 families fully analyzed (two families had 2 affected children each making the total number of cases analyzed 37 out of 41). Table 2 and Supplementary table 2 list the 57 inherited CNVs and their gene content. Twenty seven out of 57 CNVs are inherited from the mother (47%) and 25 from the father (44%). Five CNVs were inherited from both the mother and father (Fig. 1c).
Table 2. Inherited CNVs identified in patients with ASD classified as pathogenic, likely pathogenic or of unknown significance.
| ID | Cytoband | Type | Location | Size (kb) | Genes | Classification | Inheritance |
|---|---|---|---|---|---|---|---|
| SES3 | 10q11.22 | Gain | 46776777-47939116 | 1162 | GPRIN2, FAM35B2, ANTXRL, FAM35B, SYT15, AGAP9, ANXA8L2, PPYR1, FAM21B, FAM25C, LOC642826, ANXA8, LOC643650 | Unknown significance | Mother |
| LAS9 | 10q11.22 | Gain | 46950168-47939116 | 989 | FAM21B/25B/35B2, SYT15, ANXA8L2, ANTXRL, PPYR1, AGAP9, LOC643650/642826, GPRIN2, FAM25G | Unknown significance | Father |
| 7q22.3 | Gain | 105890353-106064794 | 174 | NAMPT | Unknown significance | Father & Mother | |
| LAS17 | 2p16.3 | Loss | 51166006-51302668 | 137 | NRXN1 | Pathogenic | Father |
| CLIN22 | 14q12 | Gain | 31642542-31778371 | 136 | HECTD1, HEATR5A | Unknown significance | Mother |
| CLIN24 | 20p11.23 | Loss | 20441027-20573017 | 132 | RALGAPA2 | Unknown significance | Mother |
| BAK41 | Yp11.31 | Gain | 2867178-3222952 | 356 | LINC00278 | Unknown significance | Father |
| Yp11.2 | Gain | 3973124-4271373 | 298 | No genes | Unknown significance | Father | |
| BAK42 | 4q28.3 | Loss | 138092729-138196101 | 103 | No genes | Unknown significance | Mother |
| 5q35.2 | Loss | 175438045-175638617 | 201 | FAM153B, LOC100507387, LOC643201 | Unknown significance | Father | |
| 8q22.1 | Gain | 97151500-97303937 | 152 | GDF6, MTERFD1, PTDSS1, UQCRB | Unknown significance | Mother | |
| BAK45 | 1q43 | Gain | 237517711-238090569 | 573 | RYR2, LOC100130331, ZP4 | Likely pathogenic | Mother |
| Xp22.33 | Gain | 1841391-2303322 | 462 | DHRSX | Unknown significance | Father |
A deletion of 137 Kbps inherited from the father was identified. It is classified as pathogenic. It is located on 2p16.3 and encompasses the gene NRXN1 (deletion from exon1 to exon 6). Another duplication affecting 1q43 and inherited from the mother was classified as likely pathogenic. Eleven CNVs of unclear clinical significance were also identified at 4q28.3, 5q35.2, 7q22.3, 8q21.1, 10q11.22, 14q12, 20p11.23, Xp22.33, Yp11.31 and Yp11.2 (Fig. 2b).
Comparison of autism patients with controls, and non-autistic developmentally delayed/intellectually disabled (DD/ID) patients
Some autistic patients initially diagnosed as just mentally retarded miss out on early intensive behavioral intervention, which alleviates symptoms and promotes improvement. To assess the difference at the genetic level between these two groups, we compared CNVs found in autism patients with those in the DD/ID group and in healthy controls (Fig. 3).
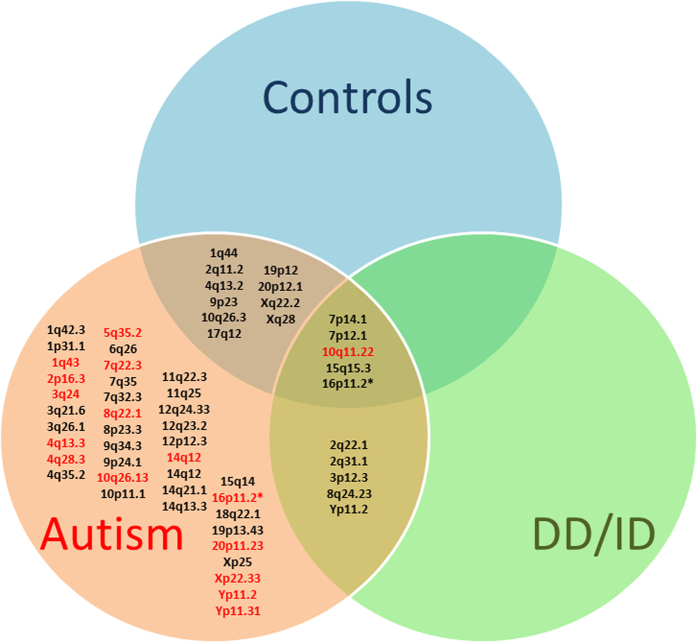
Figure 3. Venn diagram depicting the comparison of CNVs found in autism patients (red circle), healthy controls (blue circle), and those with developmental delay/intellectual disabilities, DD/ID (green circle).
Only CNVs identified in autism patients are listed in the figure. Benign CNVs are indicated in black. Pathogenic CNVs, likely pathogenic CNVs and CNVs of unknown significance are indicated in red. *Two different CNVs are identified at 16p11.2 (pathogenic is in red and benign in black).
All the CNVs classified as pathogenic, likely pathogenic and of unknown significance were restricted to the autistic group, except for one CNV, a duplication of unknown significance at 10q11.22 which was common to all three groups (autistics, DD/ID and normal controls). This duplication of ~1000 Kbp was inherited from one of the parents in 2 different autistic families. In one of these families, we observed this duplication in the autistic boy but not in his autistic sister with mental retardation. It was also found in 3/35 patients of the DD/ID group and in 5/37 healthy controls.
The 20 CNVs that were common to the autistic group and either normal controls and/or the DD/ID groups were all classified as benign except for a duplication of unknown significance at 10q11.22 (Fig. 3). Five CNVs common to the 3 groups were located at 7p14.1, 7p12.1, 10q11.22, 15q15.3 and 16p11.2 (Table 3). Five CNVs were common to the autism group and DD/ID group only and these were located at 2q22.1, 2q31.1, 3p12.3, 8q24.23 and Yp11.2 (Table 4).
Table 3. Common CNVs identified in patients with ASD, developmental and/or intellectual delays, and in healthy controls.
| Cytoband | Type | Location | Size (kb) | Genes | Classification |
|---|---|---|---|---|---|
| 7p14.1 | Loss | 38290298-38402607 | 112 | TARP | Benign |
| 7p12.1 | Loss | 53459876-53591589 | 132 | No genes | Benign |
| 10q11.22 | Gain | 46776777-47939116 | 1162 | GPRIN2, FAM35B2, ANTXRL, FAM35B, SYT15, AGAP9, ANXA8L2, PPYR1, FAM21B, FAM25C, LOC642826, ANXA8, LOC643650 | Unknown significance |
| 15q15.3 | Gain | 43859057-44041632 | 183 | CKMT1B, CATSPER2, CKMT1A, PPIP5K1, PDIA3, STRC | Benign |
| 16p11.2 | Gain | 34449594-34755816 | 306 | LOC283914, LOC146481, LOC100130700 | Benign |
Table 4. Common CNVs identified in patients with ASD and in patients with developmental and/or intellectual delays.
| Cytoband | Type | Location | Size (kb) | Genes | Classification |
|---|---|---|---|---|---|
| 2q22.1 | Loss | 142050109-142166828 | 117 | LRP1B | Benign |
| 2q31.1 | Gain | 176919432-177066433 | 147 | HOXD3/4/8/9/10/11/12/13, EVX2 | Benign |
| 3p12.3 | Gain | 75093330-76311299 | 1218 | ZNF717, FAM86D, FRG2C | Likely benign |
| 8q24.23 | Loss | 137687247-137863344 | 176 | no genes | Benign |
| Yp11.2 | Gain | 9198721-9319924 | 121 | FAM197Y2P, TSPY1, TSPY3, TSPY4 | Benign |
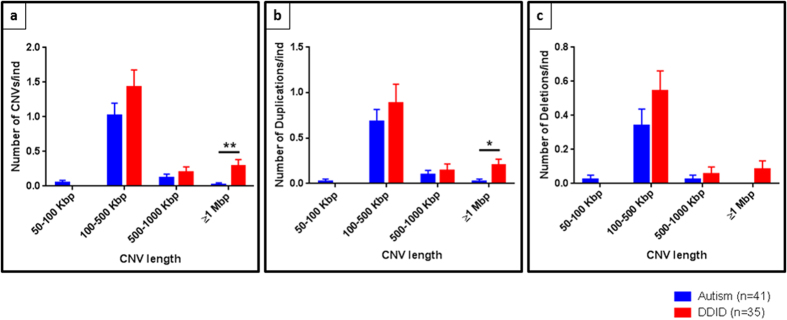
After excluding all CNVs common to healthy controls and autism or DD/ID patients, we compared the length of the remaining CNVs in the autistic group versus the DD/ID group (Fig. 4). The DD/ID group has a greater number of CNVs per individual than autism patients, except for CNVs smaller than 100 Kbp which were slightly increased in ASD (0.05 in autism vs 0 in DD/ID) (Fig. 4a). This increase in CNVs in DD/ID controls is significantly more pronounced for CNVs greater than 1 Mbp (0.02 in autism vs 0.29 in DD/ID, p = 0.0082). The same trend between ASD and DD/ID was observed when duplications (Fig. 4b) or deletions (Fig. 4c) were analyzed separately. Thus, DD/ID individuals have an enrichment in total number of CNVs, especially larger CNVs (>1Mbp), whereas autism patients show an enrichment in the number of smaller deletions (<100 Kb).
Figure 4. Size distribution of CNVs in autism patients (in blue) versus the Developmentally Delayed/intellectually disabled (DD/ID) group (in red).
Data are means ± SD. (a) The DD/ID group have more CNVs per individual than autism patients except for CNVs that are smaller than 100Kbp. The trend is similar when we look at (b) duplications and (c) deletions (**p value = 0.0082 ; *p value = 0.00209; Mann-Whitney test).
Gene content analysis
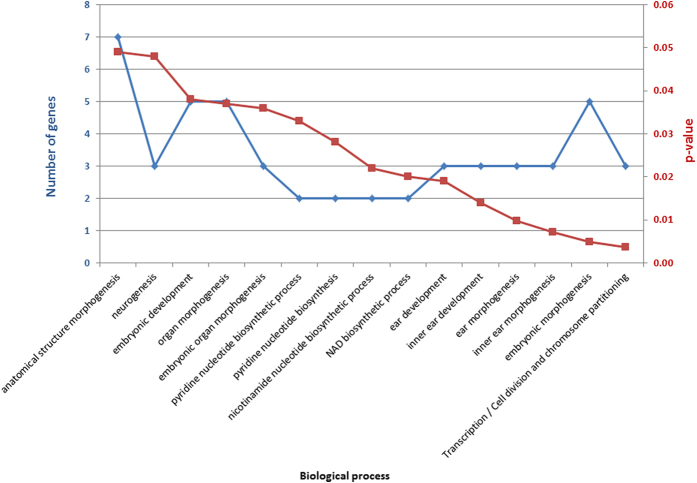
Subsequent analysis focused on the 19 CNVs that were classified as pathogenic, likely pathogenic and of unknown significance. These CNVs were mapped across 13 chromosomes (Supplementary figure 1). Gene content was first analyzed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) to assess statistical enrichment of Gene Ontology (GO) (Supplementary table 3). In order to increase significance, we selected all p-value less than 0.05. Among GO biological processes, the most significant enrichment was in transcription, cell division and chromosome partitioning (3 genes, p = 0.004) (Fig. 5).
Figure 5. Functional categories of CNV-related genes by gene ontology analysis ranked by p-value (Corrected P values < 0.05).
The second bar plot represents gene counts within each GO category.
Genes involved in cell and organ development (including embryonic development, organ morphogenesis, and neurogenesis) were also identified.
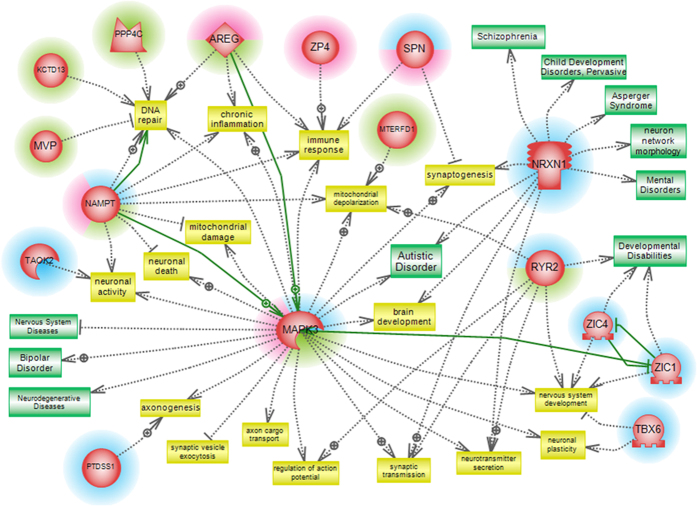
Using Pathway Studio software, a network was generated linking genes with selected functional biological processes and neurological diseases. The network was also used to investigate gene-gene interactions possibly existing among genes (Fig. 6). The functional processes in this network, such as neurogenesis, axonogenesis, DNA repair, mitochondrial damage and immune response where manually chosen for their relevance to identified deficits in ASDs. The network generated included 16 genes and 3 different functional sub-networks. The number of studies and citations used to draw each interaction has been included in Supplementary table 4. The first sub-network includes SPN, NRXN1, RYR2, ZIC4, TBX6, MAPK3, ZIC1, PTDSS1, TAOK2 related to nervous system functions. The second sub-network includes SPN, AREG, NAMPT, ZP4 and MAPK3 related to the immune response. The third sub-network includes NAMPT, PPP4C, MVP, KCTD13, MTERFD1, AREG and MAPK3 related to mitochondrial dysfunction and DNA repair. Of the 16 genes, MAPK3 shows the highest “connectivity” with other components within the 3 sub-networks. MAPK3 is related to autism, neurodegenerative diseases and bipolar disorder, and can exacerbate nervous system diseases. NRXN1 is related to autism, Asperger, schizophrenia, and pervasive developmental disorder.
Figure 6. Pathway Studio analysis of 16 potential candidate genes.
A biological network was created using the Pathway Studio 9.0 program to identify biological functions and disorders associated with potential candidate genes selected from pathogenic and likely pathogenic CNVs, and from CNVs of unknown significance. Out of 88 genes, 16 genes were found to be involved in direct interactions. 3 different functional sub-networks were detected in this pathway analysis: components of the first sub-network related to nervous system functions are highlighted with a blue halo; Components of the second sub-network related to immune response are highlighted with a red halo; Components of the third sub-network related to mitochondrial dysfunction and DNA repair are highlighted with a green halo. Green arrows show possible direct interactions between genes.
Discussion
This is the first genome-wide CNV association study in 41 autism patients in the Lebanese screening the genome for microdeletions and microduplications.
Compared to healthy controls, patients show a decreased number of CNVs (1.9 CNV per individual in autistics versus 3.5 CNV per individual in normal controls; p-value = 0.0003). This finding contradicts previous studies claiming larger number of rare de novo CNVs32,33 and common genetic variants34 in autism. Here, the frequency of de novo CNVs in ASD patients was 27% and the rate of de novo CNVs per child was 1.6. This is much higher than what has been previously described (frequency of 5.5 to 10%32,33,35,36,37, and a rate of 1.135,36). These discrepancies may represent specific characteristics of the Lebanese population, but may also be due to the small number of cases analyzed.
Eighty six percent (86.3%) of CNVs were 100–500 kbp in size. When comparing CNV length, duplications were enriched for the larger size (>500 kbp), while deletions were enriched for the smaller 50–500 kbp size. Our findings reproduce results of large-scale genome-wide surveys on ASD individuals of European ancestry32,38. Large CNVs lead to significant effects as they perturb dosage of many genes. Multiple small events may hit critical genes simultaneously and have an overall significant effect.
Two pathogenic CNVs (duplication 16p11.2 and deletion 2p16.3) and a likely pathogenic duplication (1q43) were identified in 2 (4.8%) of the ASD patients (Tables 1 and 2).
Two pathogenic CNVs identified in the same patient (Family LAS17) were located at previously reported ASD loci, including a de novo duplication at 16p11.2, and a deletion inherited from the father at 2p16.3. The region encompassing 16p11.2 (MIM 614671) is a genomic and variably expressed hotspot that is associated with neurological diseases39,40. CNVs at 16p11.2 are genetically linked to 1% of autism-related disorders39. This interval described by us and in the literature, contains several genes suspected to have a role in autism, especially the MAPK3 gene, which expresses the MAP kinase protein (ERK1). The 2p16.3 deletion described by us and inherited from the father implicates exons 1 to 6 of the neurexin gene (NRXN1). This deletion is associated with neuropsychiatric and neurodevelopmental disorders, including ASD36,41,42,43,44, schizophrenia45,46, and dysmorphic features. The NRXN1 gene is a presynaptic neuronal protein and is required for synaptic formation and efficient neurotransmission47.
We classified a maternally inherited duplication at 1q43 as likely pathogenic. It encompasses the genes RYR2, LOC100130331 and ZP4. It overlaps CNVs found in 3 cases from the International Standards for Cytogenomic Arrays (ISCA) consortium with developmental delay and/or other significant developmental or morphological phenotypes48. One case from ISCA has a duplication (nssv578551 at 1:237418348-239615755) and is classified as pathogenic without validation. This duplication encompasses the genes RYR2, LOC100130331, ZP4 and CHRM3. Two other ISCA cases bear a duplication of uncertain significance (nssv583285 at 1:237606586-23794780 and nssv1415541 at 1:237910201-237994811) that involves only the RYR2 gene. The duplication described at 1q43 overlaps a duplication (1:237694745-237824100) within the RYR2 gene found in a patient (DECIPHER ID. 248533) with autism, intellectual disability and an unspecified morphological phenotype. RYR2 protein is a member of the family of intracellular ryanodine receptor Ca2 +-release channels implicated in several disorders49,50. RYR2 is expressed in the brain and contributes to regulation of cytosolic calcium dynamics. A recent study reveals a SNP association highlighting RYR2 as a potential ASD risk gene51. Evidence supports a role for RYR2-mediated control of calcium homeostasis in stress-induced defects in cognitive function and postsynaptic plasticity in hippocampal neurons52.
Also, 12/41 of patients (29%) had aberrations of unknown significance with no reports in databases or the literature. Among those CNVs a duplication was found at 10q11.22 in 2/41 in autism patients from two families (SES3 and LAS9). This duplication overlaps benign CNVs in the DGV database. In DECIPHER, this duplication overlaps CNVs found in patients with autism and/or intellectual disabilities. A similar duplication at 10q11.22 in two patients with Rett syndrome that includes two candidate modifier genes (GPRIN2 and PPYR1) is reported53. GPRIN2 is highly expressed in the cerebellum and regulates neurite outgrowth. PPYR1, known as neuropeptide Y receptor or pancreatic polypeptide 1, is a key regulator of energy homeostasis, and is directly involved in the regulation of food intake. We found this same 10q11.22 duplication in 3/35 patients from the DD/ID group with heterogeneous phenotypes. One patient had mental retardation, the second had moderate mental retardation, language delay, stereotypies and features of autism, and the third patient had a deletion at 22q13.31q13.33 causing Phelan-McDermid syndrome (referred for mild developmental delay, dysmorphism and speech delay). This duplication was identified in 5/37 of controls, indicating this variant could be a common CNV or a pathogenic CNV with incomplete penetrance. Taken together, these facts imply that this CNV, which is common to different neurodevelopmental disorders, may modulate the phenotypic expression of disease, instead of being a single causative factor.
We performed gene ontology (GO) analysis by querying the gene content of all CNVs classified as pathogenic, likely pathogenic and of unknown significance into the records of the GO database (Fig. 5 and Supplementary table 3). Functional processes related to DNA repair, organ development, and neurogenesis, were found. Using Pathway Studio 9, functional processes impacting deficits in ASD were chosen and used to build a literature-mined network based on CNV gene content. The network generated included 16 genes of interest (Fig. 6).
Among these genes, the phosphatidylserine synthase 1 gene (PTDSS1) provides for a plausible ASD candidate gene and was identified in family BAK42 within a maternally inherited 152 Kbp duplication classified as ‘of unknown significance’. PTDSS1 encodes for the phosphatidylserine synthase 1 protein that synthesizes phosphatidylserine. Phosphatidylserine is present in all membrane phospholipids in mammalian tissues and cells54,55. In addition to its roles in apoptosis, internalization of viruses and coagulation, phosphatidylserine participates in intracellular processes by interacting with key signaling proteins, including the Ras and Rho family of GTPases and protein kinase C56. Heterozygous missense gain-of-function mutations in PTDSS1 were found in patients with Lenz-Majewski hyperostotic dwarfism57,58. This syndrome manifests sclerosing bone dysplasia, distinct craniofacial, dental, cutaneous, distal-limb anomalies, and intellectual deficits. Other membrane phospholipid abnormalities have been reported in many psychiatric and behavioral disorders including schizophrenia, dyslexia, and dyspraxia59,60,61. Membrane phospholipids, a prime target for reactive oxygen species (ROS) damage are altered in autism62,63. Phosphatidylserine levels were increased in erythrocyte membranes from children with autism compared to unaffected siblings64. Phosphatidylserine metabolism in the nervous system of individuals with autism may alter axonogenesis, neuronal activity, and brain development.
A second ASD candidate gene of interest generated by Pathway Studio 9 analysis is the amphiregulin gene (AREG), and was identified in family CLIN29 within a de novo 115 Kbp deletion classified as of unknown significance. Amphiregulin is a member of the epidermal growth factor family (EGF) that includes EGF, transforming growth factor-alpha (TGF-alpha), heparin-binding EGF, betacellulin and epiregulin. Amphiregulin binds to ErbB1 receptor or epidermal growth factor receptor (EGFR)65. Plasma EGFR levels are significantly higher in autistic children compared to neurotypic controls66. Amphiregulin can act as an autocrine mitogen on Schwann cells in the sciatic nerve67, and a survival factor for sensory neurons that stimulate axonal outgrowth via the EGF receptor68. Amphiregulin, similarly to FGF-2 or EGF69, is a mitogen for neural stem cells in culture, and promotes survival and differentiation of neuronal precursor PC12 cells70,71. Genetic studies in autism have identified mutations and copy number variations in many genes involved in the MAPK/ERK signaling pathway. A gain of function mutation in the CACNA1C gene, deletions and disruption of the SYNGAP1 gene, the latter a CNV encompassing the MAPK3 gene, all indicate that in some autism patients the ERK cascade is inappropriately activated72. Recent analyses clearly link the MAPK pathway to functional gene networks affected in ASDs73. It is proposed that AREG is a novel autism candidate gene, supporting the theory that alteration of MAPK/ERK signaling may be common to many autistic patients74.
Conclusion
The study of Lebanese autism patients adds additional support for the contribution of known CNVs to ASD. Also, shared ancestry and high rates of consanguinity in the Lebanese led to identification of new variants and previously unrecognized susceptibility candidate genes for ASD in a relatively small number of Lebanese ASD children.
Methods
Subject material
We studied three different groups. Group 1 includes a total of 41 autistic children (38 boys and 3 girls). 37/41were from 35 fully analyzed families (33 with one affected and 2 with 2 affected each). The DNA from parents of 4 autistic children was not available for analysis (Supplementary table 5). Autistic children were 3–18 years of age at the time of recruitment, and fulfilled the Diagnostic and Statistical Manual of Mental Disorders V criteria for autism; group 2 includes a control cohort of 37 normal participants (19 males and 18 females); group 3 includes 35 individuals referred to our clinical neurogenetics service with non-syndromic developmental delay and intellectual disabilities (DD/ID), or rare neurological syndromes. All patients selected had a normal karyotype and were negative for fragile-X syndrome. Informed written consent was obtained in accordance with an AUBMC IRB approved protocol (ID: Bioch.RB.06), and all methods were conducted in accordance with approved guidelines and regulations.
DNA samples from all participants were extracted from peripheral blood using QIAamp® blood midi kit (Qiagen, Inc., Valencia, CA). Affymetrix Cytogenetics 2.7M arrays and CytoScan™ HD Arrays were used for CNV screening.
Chromosomal Microarray Analysis
Chromosomal microarray analysis was performed using the Affymetrix Cytogenetics 2.7M and CytoScan™ HD arrays, according to the manufacturer’s instruction (Affymetrix Inc., Santa Clara, CA). These are chips with more than 2.6 million probes capable of detecting known and novel chromosome aberrations across the entire human genome consisting of 25 kb or larger copy number variations (CNVs). Briefly, 100–250 ng of genomic DNA is amplified, fragmented, labeled then hybridized to the arrays for 16–18 h. Arrays were then washed and stained on the GeneChip® Fluidics Station 450 and DAT images acquired using the GeneChip® Scanner 3000 7G (Affymetrix Inc.). Data analysis was performed using the Affymetrix Chromosome Analysis Suite (CHAS). Families were analyzed in trios to determine the de novo or inherited type of each CNV detected in the autistic patients.
Functional classification of loci and genes
The genomic variants were evaluated using DECIPHER (http://decipher.sanger.ac.uk/), Database Genomic Variants (http://dgv.tcag.ca/dgv/app/home), and the International Collaboration for Clinical Genomics-International Standards Cytogenomic Arrays Database Search (https://www.iscaconsortium.org/) and classified into five groups: pathogenic, likely pathogenic, unknown significance, likely benign and benign. CNVs that were clearly found related to Autism in databases were classified as pathogenic. CNVs reported in several autism case studies without direct evidence of pathogenicity were classified as likely pathogenic. CNVs with no reported information, or incomplete and/or contradictory information were classified as of unknown significance. CNVs reported in databases at high frequency in control data were classified as benign or likely benign according to the degree of uncertainty.
The genes in CNV regions were classified according to biological processes using the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 b (http://david.abcc.ncifcrf.gov/)75,76. Gene ontology options (GOTERM_BP_ALL, GOTERM_CC_ALL, GOTERM_MF_ALL), and functional categories (COG_ONTOLOGY, and SP_PIR_KEYWORDS) were selected and a functional annotation chart generated. A maximum p-value of 0.05 was chosen to select only significant categories. For gene-enrichment analysis DAVID uses the “Ease score statistics”, an alternative name of Fisher Exact statistics, referring to the one-tail Fisher Exact test. P-values were corrected by the Benjamini correction which controls the False Discovery Rate (FDR). Network analysis was performed by Pathway Studio software version 9.0 (Ariadne Genomics, Rockville, Md., USA).
Quantitative real-time PCR validation
Quantitative real-time PCR reactions were performed in triplicate in 96-well plates using specific primers (TIB MOLBIOL) and the iQTM SYBR® Green Supermix (BioRad) as a fluorescent detection dye, in CFX96TM Real-Time PCR (BioRad), in a final volume of 12.5 μl. The input amount of genomic DNA used in each well was 35 ng. To characterize generated amplicons and to control contamination by unspecific by-products, melt curve analysis is applied. All results were normalized to GAPDH level and calculated using the ΔΔCT method. Primer sequences are GAPDH Fwd: CGAGATCCCTCCAAAATCAA; GAPDH Rev: AGGCATTGCTGCAAAGAAAG; RYR2 Fwd: TGCGTGCTGGCTACTATGAC; RYR2 Rev: TGCTTCAAGTCCTCGTTGTG.
Additional Information
How to cite this article: Soueid, J. et al. RYR2, PTDSS1 and AREG genes are implicated in a Lebanese population-based study of copy number variation in autism. Sci. Rep. 6, 19088; doi: 10.1038/srep19088 (2016).
Supplementary Material
Acknowledgments
We thank all the families and staff from the Lebanese Autism Society (LAS), SESOBEL, and the AUBMC Special Kids Clinic, who participated in this study. The work is supported by a generous grant to RMB from OpenMinds and from AUBMC (Program Projects in Biomedical Research).
Footnotes
Author Contributions J.S. performed experiments, analyzed data, interpreted the results, and wrote the manuscript. S.K. performed experiments, analyzed data and contributed to writing of the manuscript. N.M. generated and helped analyze the array-based data. J.M. acquired, participated in processing patient samples and assisted in the statistical analysis of the data. S.G. aided in the experimental setup, and generated preliminary array-based data. S.H. assisted in the study design, and analyzed preliminary array-based data. F.K. assisted in the analysis of the data, and reviewed the manuscript. R.M.B. conceived the study, performed clinical diagnoses, obtained funding for the study, designed experiments and reviewed data and analyses, revised and edited the manuscript. All authors read and approved the final manuscript.
References
- Kanner L. Autistic disturbances of affective contact Nerv. Child 2, 217–250 (1943). [PubMed] [Google Scholar]
- Steer C. D., Golding J. & Bolton P. F. Traits contributing to the autistic spectrum. PLoS One 5, e12633 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canitano R. Epilepsy in autism spectrum disorders. Eur Child Adolesc Psychiatry 16, 61–6 (2007). [DOI] [PubMed] [Google Scholar]
- Developmental Disabilities Monitoring Network Surveillance Year Principal, I., Centers for Disease, C. & Prevention. Prevalence of autism spectrum disorder among children aged 8 years-autism and developmental disabilities monitoring network, 11 sites, United States, 2010. MMWR Surveill Summ 63, 1–21 (2014). [PubMed] [Google Scholar]
- Al-Habeeb A. A., Qureshi N. A. & Al-Maliki T. A. Pattern of child and adolescent psychiatric disorders among patients consulting publicly-funded child psychiatric clinics in Saudi Arabia. East Mediterr Health J 18, 112–9 (2012). [DOI] [PubMed] [Google Scholar]
- Al-Farsi Y. M. et al. Brief report: Prevalence of autistic spectrum disorders in the Sultanate of Oman. J Autism Dev Disord 41, 821–5 (2011). [DOI] [PubMed] [Google Scholar]
- Chaaya M., Saab D., Maalouf F. T. & Boustany R. M. Prevalence of Autism Spectrum Disorder in Nurseries in Lebanon: A Cross Sectional Study. J Autism Dev Disord (2015). 10.1007/s10803-015-2590-7 [DOI] [PubMed] [Google Scholar]
- Rosenberg R. E. et al. Characteristics and concordance of autism spectrum disorders among 277 twin pairs. Arch Pediatr Adolesc Med 163, 907–14 (2009). [DOI] [PubMed] [Google Scholar]
- Abrahams B. S. & Geschwind D. H. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet 9, 341–55 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A. et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med 25, 63–77 (1995). [DOI] [PubMed] [Google Scholar]
- Steffenburg S. et al. A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J Child Psychol Psychiatry 30, 405–16 (1989). [DOI] [PubMed] [Google Scholar]
- Jorde L. B. et al. Complex segregation analysis of autism. Am J Hum Genet 49, 932–8 (1991). [PMC free article] [PubMed] [Google Scholar]
- Hogart A., Wu D., LaSalle J. M. & Schanen N. C. The comorbidity of autism with the genomic disorders of chromosome 15q11.2-q13. Neurobiol Dis 38, 181–91 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker P., Piven J. & Sato Y. Autism and tuberous sclerosis complex: prevalence and clinical features. J Autism Dev Disord 28, 279–85 (1998). [DOI] [PubMed] [Google Scholar]
- Kielinen M., Rantala H., Timonen E., Linna S. L. & Moilanen I. Associated medical disorders and disabilities in children with autistic disorder: a population-based study. Autism 8, 49–60 (2004). [DOI] [PubMed] [Google Scholar]
- Hatton D. D. et al. Autistic behavior in children with fragile X syndrome: prevalence, stability, and the impact of FMRP. Am J Med Genet A 140A, 1804–13 (2006). [DOI] [PubMed] [Google Scholar]
- Jamain S. et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet 34, 27–9 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumonnier F. et al. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet 74, 552–7 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching M. S. et al. Deletions of NRXN1 (neurexin-1) predispose to a wide spectrum of developmental disorders. Am J Med Genet B Neuropsychiatr Genet 153B, 937–47 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale B. M. et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485, 242–5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak B. J. et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338, 1619–22 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak B. J. et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–50 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders S. J. et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–41 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier R. et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell 158, 263–76 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson B. et al. The autism-associated gene chromodomain helicase DNA-binding protein 8 (CHD8) regulates noncoding RNAs and autism-related genes. Transl Psychiatry 5, e568 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sibai M. et al. Geographical structure of the Y-chromosomal genetic landscape of the Levant: a coastal-inland contrast. Ann Hum Genet 73, 568–81 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber M. et al. Genome-wide diversity in the levant reveals recent structuring by culture. PLoS Genet 9, e1003316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour B. & Salameh P. Consanguinity in Lebanon: prevalence, distribution and determinants. J Biosoc Sci 41, 505–17 (2009). [DOI] [PubMed] [Google Scholar]
- El-Kheshen G. & Saadat M. Prevalence of consanguineous marriages among shi’a populations of Lebanon. J Biosoc Sci 45, 675–82 (2013). [DOI] [PubMed] [Google Scholar]
- Hamamy H. et al. Consanguineous marriages, pearls and perils: Geneva International Consanguinity Workshop Report. Genet Med 13, 841–7 (2011). [DOI] [PubMed] [Google Scholar]
- Miles J. H. Autism spectrum disorders-a genetics review. Genet Med 13, 278–94 (2011). [DOI] [PubMed] [Google Scholar]
- Pinto D. et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466, 368–72 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J. et al. Strong association of de novo copy number mutations with autism. Science 316, 445–9 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S. et al. Global increases in both common and rare copy number load associated with autism. Hum Mol Genet 22, 2870–80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D. et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron 70, 886–97 (2011). [DOI] [PubMed] [Google Scholar]
- Marshall C. R. et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet 82, 477–88 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders S. J. et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron 70, 863–85 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia, C. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 455, 237–41 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss L. A. et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med 358, 667–75 (2008). [DOI] [PubMed] [Google Scholar]
- Shinawi M. et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet 47, 332–41 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniowiecka-Kowalnik B. et al. Intragenic rearrangements in NRXN1 in three families with autism spectrum disorder, developmental delay, and speech delay. Am J Med Genet B Neuropsychiatr Genet 153B, 983–93 (2010). [DOI] [PubMed] [Google Scholar]
- Kim H. G. et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet 82, 199–207 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner J. T. et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 459, 569–73 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucan M. et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet 5, e1000536 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T. et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science 320, 539–43 (2008). [DOI] [PubMed] [Google Scholar]
- Kirov G. et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet 17, 458–65 (2008). [DOI] [PubMed] [Google Scholar]
- Dabell M. P. et al. Investigation of NRXN1 deletions: clinical and molecular characterization. Am J Med Genet A 161A, 717–31 (2013). [DOI] [PubMed] [Google Scholar]
- Miller D. T. et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 86, 749–64 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priori S. G. et al. Clinical and molecular characterization of patients with catecholaminergic polymorphic ventricular tachycardia. Circulation 106, 69–74 (2002). [DOI] [PubMed] [Google Scholar]
- Medeiros-Domingo A. et al. The RYR2-encoded ryanodine receptor/calcium release channel in patients diagnosed previously with either catecholaminergic polymorphic ventricular tachycardia or genotype negative, exercise-induced long QT syndrome: a comprehensive open reading frame mutational analysis. J Am Coll Cardiol 54, 2065–74 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. T. & Cantor R. M. Allowing for sex differences increases power in a GWAS of multiplex Autism families. Mol Psychiatry 17, 215–22 (2012). [DOI] [PubMed] [Google Scholar]
- Liu X. et al. Role of leaky neuronal ryanodine receptors in stress-induced cognitive dysfunction. Cell 150, 1055–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artuso R. et al. Investigation of modifier genes within copy number variations in Rett syndrome. J Hum Genet 56, 508–15 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventis P. A. & Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys 39, 407–27 (2010). [DOI] [PubMed] [Google Scholar]
- Tomohiro S., Kawaguti A., Kawabe Y., Kitada S. & Kuge O. Purification and characterization of human phosphatidylserine synthases 1 and 2. Biochem J 418, 421–9 (2009). [DOI] [PubMed] [Google Scholar]
- Vance J. E. & Tasseva G. Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim Biophys Acta 1831, 543–54 (2013). [DOI] [PubMed] [Google Scholar]
- Sousa S. B. et al. Gain-of-function mutations in the phosphatidylserine synthase 1 (PTDSS1) gene cause Lenz-Majewski syndrome. Nat Genet 46, 70–6 (2014). [DOI] [PubMed] [Google Scholar]
- Whyte M. P. et al. Lenz-Majewski hyperostotic dwarfism with hyperphosphoserinuria from a novel mutation in PTDSS1 encoding phosphatidylserine synthase 1. J Bone Miner Res 30, 606–14 (2015). [DOI] [PubMed] [Google Scholar]
- Horrobin D. F. The membrane phospholipid hypothesis as a biochemical basis for the neurodevelopmental concept of schizophrenia. Schizophr Res 30, 193–208 (1998). [DOI] [PubMed] [Google Scholar]
- Richardson A. J., Cox I. J., Sargentoni J. & Puri B. K. Abnormal cerebral phospholipid metabolism in dyslexia indicated by phosphorus-31 magnetic resonance spectroscopy. NMR Biomed 10, 309–14 (1997). [DOI] [PubMed] [Google Scholar]
- Richardson A. J. & Ross M. A. Fatty acid metabolism in neurodevelopmental disorder: a new perspective on associations between attention-deficit/hyperactivity disorder, dyslexia, dyspraxia and the autistic spectrum. Prostaglandins Leukot Essent Fatty Acids 63, 1–9 (2000). [DOI] [PubMed] [Google Scholar]
- Chauhan A. & Chauhan V. Oxidative stress in autism. Pathophysiology 13, 171–81 (2006). [DOI] [PubMed] [Google Scholar]
- Chauhan A., Chauhan V., Brown W. T. & Cohen I. Oxidative stress in autism: increased lipid peroxidation and reduced serum levels of ceruloplasmin and transferrin-the antioxidant proteins. Life Sci 75, 2539–49 (2004). [DOI] [PubMed] [Google Scholar]
- Chauhan V., Chauhan A., Cohen I. L., Brown W. T. & Sheikh A. Alteration in amino-glycerophospholipids levels in the plasma of children with autism: a potential biochemical diagnostic marker. Life Sci 74, 1635–43 (2004). [DOI] [PubMed] [Google Scholar]
- Maher P. A. & Schubert D. Schwannoma-derived growth factor interacts with the epidermal growth factor receptor. J Neurochem 65, 1895–8 (1995). [DOI] [PubMed] [Google Scholar]
- Russo A. J. Increased Epidermal Growth Factor Receptor (EGFR) Associated with Hepatocyte Growth Factor (HGF) and Symptom Severity in Children with Autism Spectrum Disorders (ASDs). J Cent Nerv Syst Dis 6, 79–83 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Fischer W. H. & Schubert D. Structure, expression and function of a schwannoma-derived growth factor. Nature 348, 257–60 (1990). [DOI] [PubMed] [Google Scholar]
- Nilsson A. & Kanje M. Amphiregulin acts as an autocrine survival factor for adult sensory neurons. Neuroreport 16, 213–8 (2005). [DOI] [PubMed] [Google Scholar]
- Falk A. & Frisen J. Amphiregulin is a mitogen for adult neural stem cells. J Neurosci Res 69, 757–62 (2002). [DOI] [PubMed] [Google Scholar]
- Kimura H. & Schubert D. Schwannoma-derived growth factor promotes the neuronal differentiation and survival of PC12 cells. J Cell Biol 116, 777–83 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenouchi T., Kadosaka M., Shin S. Y. & Munekata E. Biological actions of the epidermal growth factors-like domain peptides of mouse schwannoma-derived growth factor and human amphiregulin. J Pept Res 53, 120–5 (1999). [DOI] [PubMed] [Google Scholar]
- Kalkman H. O. Potential opposite roles of the extracellular signal-regulated kinase (ERK) pathway in autism spectrum and bipolar disorders. Neurosci Biobehav Rev 36, 2206–13 (2012). [DOI] [PubMed] [Google Scholar]
- Pinto D. et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet 94, 677–94 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels I. S., Saitta S. C. & Landreth G. E. MAP’ing CNS development and cognition: an ERKsome process. Neuron 61, 160–7 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T. & Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- Dennis G. Jr. et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4, P3 (2003). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.