Abstract
GWAS in out-crossing perennial crops is typically limited by insufficient marker density to account for population diversity and effects of population structure resulting in high false positive rates. The perennial crop oil palm is the most productive oil crop. We performed GWAS for oil-to-dry-mesocarp content (O/DM) on 2,045 genotyped tenera palms using 200K SNPs that were selected based on the short-range linkage disequilibrium distance, which is inherent with long breeding cycles and heterogeneous breeding populations. Eighty loci were significantly associated with O/DM (p ≤ 10−4) and three key signals were found. We then evaluated the progeny of a Deli x AVROS breeding trial and a 4% higher O/DM was observed amongst those having the beneficial genotypes at two of the three key loci (p < 0.05). We have initiated MAS and large-scale planting of elite dura and pisifera parents to generate the new commercial tenera palms with higher O/DM potential.
Genome-wide association study (GWAS) has emerged as a powerful method and commonly adopted, particularly in human populations to identify a broad range of complex diseases1. The method was then wide implemented in annual plants, including rice2,3,4, foxtail millet5, maize6 and Arabidopsis7 when their reference genomes were successfully sequenced. As for perennial crops, GWAS progress is often hindered due to insufficient marker density and population structure effects8,9. We carried out GWAS on oil palms (Elaeis guineensis Jacq.) as a model to address these limiting factors and to illustrate the potential of marker-assisted selection (MAS) in out-crossing breeding programmes. The oil palm with the highest yield per hectare of all oil crops, it accounts for a quarter of vegetable oil traded worldwide annually, despite occupying only 5% of the global oil planting acreage10. Only five to six generations of selection and breeding have been completed since plantations were established in the 1920s and 1930s11, primarily due to long phenotyping cycles (typically twelve years per cycle).
In recent decades, molecular markers have been employed to identify quantitative trait loci (QTL) for traits of importance in oil palm. By determining the allelic variation present, palms that possess particular combinations of desired QTL can be selected at the nursery stage. Markers could significantly reduce the conventional phenotyping cycles and also enrich the best combinations of alleles in palms planted from each cross. Most marker discovery programmes in oil palm are still mainly based on controlled cross-based linkages using various marker systems with modest density, including restriction fragment length polymorphisms (RFLPs)12,13, amplified fragment length polymorphisms (AFLPs)14 and simple sequence repeats (SSRs)15. To further increase marker density, single nucleotide polymorphisms (SNPs) that distribute abundantly throughout a genome, were developed for oil palm16. Nevertheless, the application of SNPs was still mainly deployed for linkage map construction16 and followed by localisation of fruit form trait with Mendelian inheritance10 and stem height trait17. While the family-based mapping approach and high-density markers are reasonably powerful for detecting the major QTLs, the mapping resolution, particularly for complex traits, like oil yield is always constrained by small population size. Typically, seed numbers from a single breeding cross exceeds 1,000, but currently only 16–96 palms are randomly selected for assessment.
The publication of the oil palm genome consisting of 1.535 gigabase (Gb) of assembled sequence10, the independent assembly of Sime Darby’s oil palm genome and development of high throughput SNP genotyping technologies has enabled us to produce a pipeline for marker-assisted breeding in oil palm, from discovery to the planting of marker-selected breeding materials in the field. We here report the first comprehensive GWAS in oil palm, validation of the results for oil-to-dry mesocarp (O/DM) and the application of the results in a field level selection programme. This is, to our knowledge, the largest SNP array-based GWAS analysis in any tree species and provides a test case for the application of such analysis to address the unique characteristics of perennial food crop species. Such step-wise changes are needed to address the major challenge of food security facing the world.
Result and Discussion
The Deli dura palms are a breeding population of restricted origin (BPRO) derived from the four original palms planted at Bogor Botanical Garden in 184818. Developed in different breeding programmes with different selection objectives, this led to important sub-populations19, such as the Ulu Remis and Johore Labis materials20. The Deli origin exhibits thick mesocarp, high bunch number and high O/DM and is the most important oil palm planting material. Shell thickness is inversely related to mesocarp/fruit (hence oil yield) and is controlled by a single gene with two co-dominant alleles, sh + and sh−21. Planters in Southeast Asia in the 1960s switched from planting Deli dura (sh + sh+) to planting tenera (sh + sh−) derived from Deli x AVROS, realizing a 30% increment in oil yield/hectare22,23. However, this complicated breeding selection by requiring separate development of maternal (dura) and paternal (pisifera) breeding lines, followed by extensive progeny testing. Concerns of a narrow genetic base limiting future breeding progress has also led many oil palm breeders to evaluate other sources of dura germplasm. For this reason, the semi-wild Nigerian dura crossed with AVROS pisifera was also evaluated in Sime Darby.
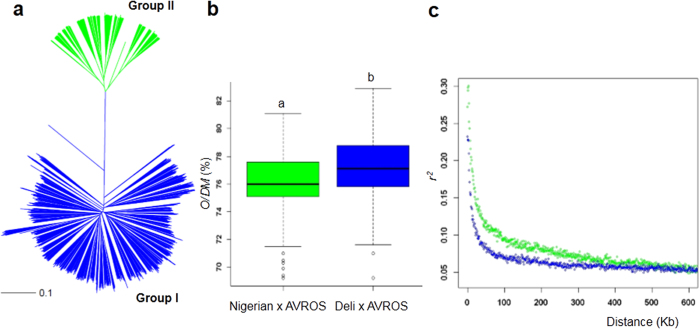
One hundred and thirty-two oil palm representing 59 breeding origins (Supplementary Table 1) were sampled for whole genome re-sequencing, yielding ~900 million raw reads. Approximately 60% of the raw reads were mappable to the published oil palm reference genome10. We identified 7.8 million potential SNPs, of which 200,000 (with minor allele frequency, MAF >0.05) were selected for array design (see Methods). The array was used to genotype 2,045 palms with existing phenotype data (including oil yield collected over 7 years). A clustering analysis based on Hamming distance (Fig. 1a) showed that the palms could be assigned to two divergent groupings i.e. (I) commercially important Deli x AVROS (n = 1,459) palms and (II) semi-wild Nigerian x AVROS (n = 586) palms. On average, group I yielded 1% O/DM more than the semi-wild group II (p < 0.001) (Fig. 1b) (Supplementary Table 2). An increase of 1% in mesocarp oil content (or oil extraction rate, OER) could translate to approximately 500,000 tons of additional oil production annually in Malaysia24, without the need of new plantings.
Figure 1. Genetic stratification of 2,045 oil palm samples representing Deli x AVROS and Nigerian x AVROS.
(a) Neighbor-joining tree (NJ) constructed from Hamming distances for all SNPs. The two divergent groups, Group I and Group II are shown in blue and green, respectively. The scale bar indicates the Hamming distance. (b) The boxplots represent median values, percentile 25–75 and outliers for the oil-to-dry mesocarp (O/DM) of the two divergent groups (color as in a). Statistical significance for each group was determined by a Student-t test at p < 0.001. (c) Genome-wide average LD decay rate estimated from 1,459 Deli x AVROS (Group I) and 586 Nigerian x AVROS (Group II) palms (color as in a).
The linkage disequilibrium (LD) decay rates of Deli x AVROS and Nigerian x AVROS were 25Kb and 20Kb at 0.12 and 0.15 of average pairwise correlation coefficients (r2) (Fig. 1c). The LD in the commercially important population derived from a narrow genetic base of Deli founders only decayed slightly slower than the semi-wild population. The long-range LD in Deli dura might be broken down by the genetic recombination introduced from AVROS pisifera to increase oil yield through heterosis in tenera. The LD of the oil palm populations decays considerably faster than in cultivated rice and foxtail millet. In both cereal species, the LD decay ranged from 100Kb to 200Kb2,3,4,5 accumulated from a long history of reproductive isolation in self-fertilization breeding systems. Overall, the 200K SNP array with an average sampling of one locus for every 11Kb based on the reference genome size10, was selected to provide sufficient genomic resolution for GWAS.
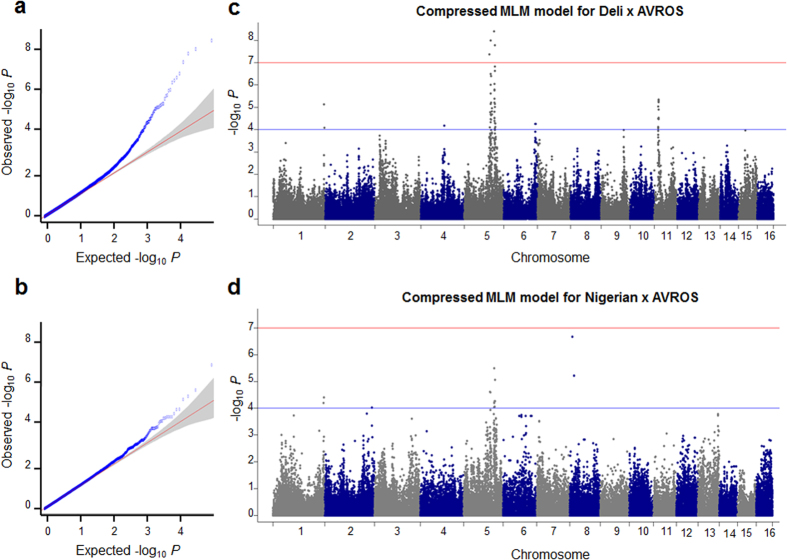
The O/DM of individual palms was measured over 7 years of field planting (summarized in Supplementary Table 2) to perform a phenotype-genotype association analysis using a simple linear model. We discovered inflated false-positive signals (Supplementary Fig. 1) in Deli x AVROS (Genomic inflation factor, GIF = 3.66) and, particularly, in Nigerian x AVROS (GIF = 11.9) (Supplementary Fig. 2). The result suggested that the model failed to account for the recent common ancestry of small groups of individuals, defined as cryptic relatedness25,26. General linear model (GLM)-based methods, including structure association27, genome control26 and family-based28 tests of association are widely used to address population stratification. We adopted a compressed mixed linear model (MLM) with population parameters previously determined (P3D) to address the problem of genomic inflation that was based on a principle component analysis and a group kinship matrix29. This method greatly reduced false positives in Deli x AVROS (GIF = 1.1) and Nigerian x AVROS (GIF = 1.9) as illustrated in Quantile-Quantile plots (Fig. 2a,b).
Figure 2. Genome-wide association studies results for oil-to-dry mesocarp (O/DM).
(a) Quantile-Quantile plot of compressed MLM for Deli x AVROS. (b) Quantile-Quantile plot of compressed MLM for Nigerian x AVROS. (c) Manhattan plots of Compressed MLM model for Deli x AVROS. Negative log10-transformed p values from a genome-wide scan are plotted against position on each of the 16 chromosomes. The blue and red horizontal lines indicates the genome-wide significance cut-off and Bonferroni cut-off. (d) Manhattan plots of Compressed MLM model for Deli x AVROS, as in (c).
In total, 62 and 18 significant association signals for the O/DM phenotype (Supplementary Table 3) were mainly clustered on Chromosomes 5 and Chromosome 11 in Deli x AVROS (Fig. 2c) and Nigerian x AVROS (Fig. 2d) guided by a whole-genome significance cutoff (p) at 10−4 and a Bonferroni cutoff at 10−7. The number of significant signals was smaller in latter group, possibly due to smaller population size at only half of Deli x AVROS group. Higher statistical power will be achievable with larger population size. We identified three significant SNPs i.e. SD_SNP_000010418 (pDeli x AVROS = 2.39 × 10−6; pNigerian x AVROS = 2.45 × 10−5), SD_SNP_000019529 (pDeli x AVROS = 1.51 × 10−7; pNigerian x AVROS = 6.13 × 10−5) and SD_SNP_000002370 (pDeli x AVROS = 9.48 × 10−6; pNigerian x AVROS = 6.39 × 10−5) on Chromosome 5, which were commonly present in both groups and situated near gene models (Supplementary Table 4). Compared to the previous controlled-crossed linkage studies15,30, the significant QTLs for O/DM were located on Chromosome 3 and Chromosome 6 in multi-parent progeny from Deli dura x Yangambi/La Mé pisifera (299 palms) and a progeny from Topi Deli dura x Yangambi pisifera (69 palms). Interestingly, we also observed one of the QTLs located on Chromosome 6 between 41,545,028bp–41,557,789bp (Supplementary Table 3). The major association peak on Chromosome 5 as reported in this study was however not previously observed. We further analyzed the homologies of candidate genes in or close to the three significant SNPs, which might be influencing the phenotype, using known comparative gene and pathway analyses in oil palm and date palm31. For example, SD_SNP_000002370 (physical position: 40,396,733bp; Chromosome 5) located at 2-Kb away from the Pyruvate kinase (PK) gene was found to be associated to O/DM. The expression of PK gene in oil palm was reported to be responsible for an increased flux through to pyruvate for fatty acid biosynthesis31,32 leading to more oil. Nevertheless, we are currently further fine mapping these loci with higher SNP density.
Oil palm breeders deploy a variant of reciprocal recurrent selection (RRS) or family and individual selection (FIS), producing thick-shelled dura maternal and shell-less pisifera paternal pools23. Elite dura and pisifera with good combining ability evaluated via progeny testing are crossed for thin-shelled tenera commercial production. Within the Deli x AVROS group (1,459 palms), the homozygous G/G palms for SD_SNP_000019529 yielded a mean of 77.1% O/DM, which was significantly higher than the 75.8% observed amongst the heterozygous G/A palms (Fig. 3). The same SNP effect was observed in the Nigerian x AVROS group (586 palms) (O/DM: G/G = 76.3%, G/A = 75.1%, A/A = 74.9%) and further, was successfully validated in a small independent Deli x AVROS breeding trial (46 palms) (Fig. 3). The validation of SD_SNP_000002370 was also proven using the same approach (Supplementary Fig. 3). However, SD_SNP_000010418 did not significantly explain the phenotype variation in the breeding trial, possibly due to insufficient statistical power (Supplementary Fig. 4). In addition, we also observed additive effect when combining the three important SNPs in both groups. The O/DM yield significantly increased along with the stacking of positive alleles in individual palms (Supplementary Fig. 5 and Supplementary Fig. 6).
Figure 3. Boxplots of the oil-to-dry-mesocarp (O/DM) trait, grouped according to SNP polymorphism revealed by SD_SNP_000019529.
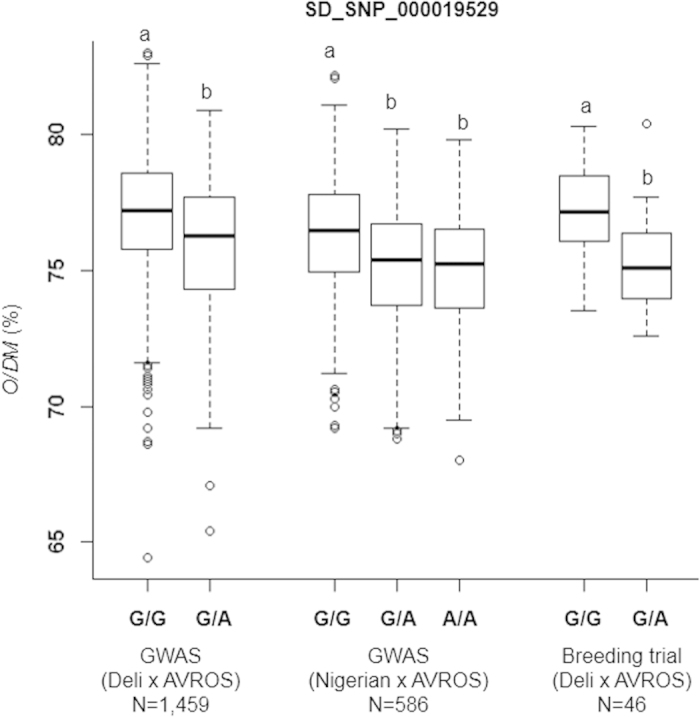
Statistical significance for each genotype in GWAS discovery populations i.e. Deli x AVROS and Nigerian × AVROS was determined by a compressed MLM model at p = 1.51 × 10−7 and p = 6.13 × 10−5, respectively. Statistical significance for differences between genotypes in the Deli x AVROS breeding trial was determined by one-way ANOVA at p < 0.005.
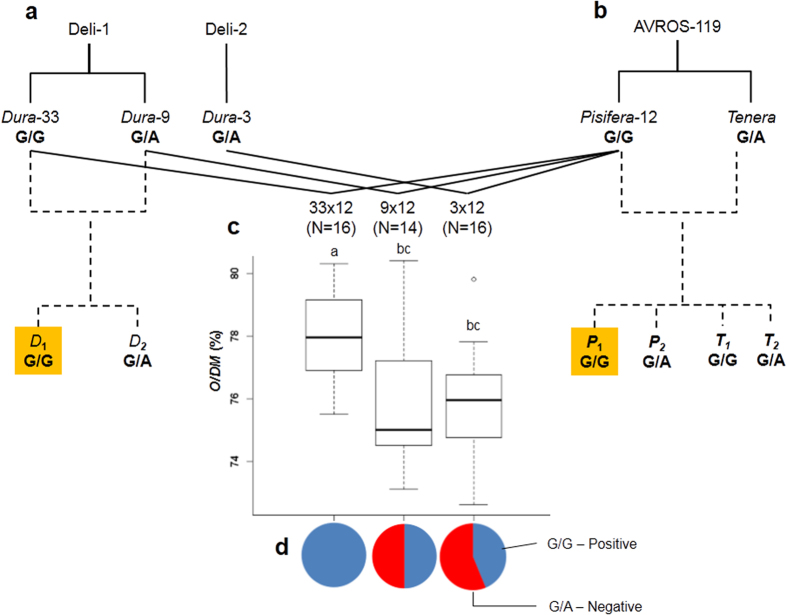
The most significant locus, SD_SNP_000019529 was selected to illustrate the marker application in an oil palm breeding programme. In the Deli x AVROS breeding trial (Fig. 4), the best combining ability of the Dura-33 and Pisifera-12 cross (Fig. 4a,b) was associated with the parent carrying the homozygous G/G SD_SNP_000019529; thus, each parent contributed the positive G allele to form a 100% homozygous G/G tenera progeny (Dura -33 x Pisifera-12; n = 16). The O/DM mean for this progeny was 4% higher than the other two progeny (p < 0.005; one-way Analysis of Variance) (Fig. 4c), while the poorer combining ability of the remaining dura parents was due to transmission of the A alleles (Fig. 4d). Based on these results, we implemented a MAS programme through screening of large number of seedlings for Deli dura and AVROS pisifera parents derived from Dura-33 x Dura-9 and Pisifera-12 x Tenera, respectively, using SD_SNP_000019529. Only the homozygous G/G dura and pisifera seedlings were planted.
Figure 4. Marker-assisted selection (MAS) of Deli dura palms using SD_SNP_000019529.
(a) The pedigree of Deli dura with known genotypes. A new Deli population which consists of G/G (D1) and G/A (D2) genotypes was generated from Dura-33 x Dura-9. Only D1palms (highlighted in orange color) were field planted. D–dura palm. (b) The pedigree of AVROS-199 with known genotypes. A new AVROS population which consists of G/G (P1 and T1) and G/A (P2 and T2). Only P1 palms (highlighted in orange color) were field planted. P–pisifera palm and T–tenera palm. (c) The boxplots represent median values, percentile 25–75 and outliers for the oil-to-dry mesocarp trait (O/DM) of the three progeny testing populations (Tenera), derived from the pedigrees shown in a and b. (d) The genotype composition for each progeny testing population. Red and blue indicates G/A as negative genotype and G/G as positive genotype for O/DM, respectively.
In summary, we report the most comprehensive use of high density SNP genotyping in oil palm to date, the use of a GWAS approach to identify SNP variants associated with differences in the key oil-to-dry mesocarp yield trait, and confirmation of their action in an independent cross. Based on these results, we have implemented a MAS programme to breed new parental lines for commercial oil palm hybrid production. The reported study also lays the foundations for a genomic selection model for oil palm and will act as a model for other perennial tree crops.
Methods
Sampling and DNA preparation
For re-sequencing, we sampled 132 palms belonging to 59 origins (Supplementary Table 1) maintained at the Sime Darby Plantation R&D Centre in Malaysia. The sampling was then extended to the GWAS discovery populations derived from Deli x AVROS (1,459 palms) and Nigerian x AVROS (586 palms).The sample selection was based on progeny/BPRO of relevance to the breeding programme, followed by phenotypic data availability. After a field census, we selected 5–15 palms from each progeny. For validation, we identified a nested-mating designed trial of Deli x AVROS that was generated previously to determine the combining ability of different dura palms to a common Pisifera-12 (Fig. 4a,b). Three progeny testing populations (46 tenera) with their parents (3 Deli dura, 1 AVROS pisifera and 1 AVROS tenera) were sampled. Total genomic DNA was isolated from young leaf tissue (frond 0) using the DNAeasy Plant Mini Kit (Qiagen).
Whole-genome re-sequencing and genotyping
The 132 samples were pooled based on an equal molar concentration of DNA from each sample to form the sequencing DNA pool. A library was prepared for re-sequencing using Illumina HiSeq 2000 to generate 100-bp pair-end reads to give a 35x genome coverage from 924,271,650 raw reads. The pair-end reads were trimmed, filtered and aligned to the published oil palm genome10 using BWA Mapper33 with default parameters. A total of 7,755,949 putative SNPs were then called and filtered using SAMtools34, with parameters of the minimal mapping quality score of the SNP being 25, minimal depth 3x, and minimal SNP distance from a gap of 2bp. We removed 1,085,204 SNPs that were generated from E. oleifera and 746,092 SNPs based on coverage (minimal 17 or maximal 53), genotype quality with minimal score of 8 and minor allele frequency (MAF < 0.05). The other filtering step removed 5,330,765 SNPs based on the technical requirements of Illumina, including the removal of pairs of SNPs with distance less than 60bp apart, ambiguous nucleotides, indels, non-biallelic and A/T or C/G types. We identified 593,888 quality SNPs. According to linkage disequilibrium with a r2 cut-off set at 0.3, 200,000 SNPs with an average density of one SNP per 11Kb were submitted to Illumina for design score calculation using Illumina’s Assay Design Tool for Infinium. The Infinium array, termed as OP200K was used to assay the GWAS discovery populations (~250 ng DNA/sample). The overnight amplified DNA samples were then fragmented by a controlled enzymatic process that did not require gel electrophoresis. The re-suspended DNA samples were hybridized to the BeadChips after an overnight incubation in the capillary flow-through chamber. The allele specific hybridizations were fluorescently labeled and detected by an Illumina BeadArray Reader. The raw reads were then analyzed using the GenomeStudio Data Analysis software for automated genotyping calling and quality control. To generate the genotypic dataset for GWAS, only the SNPs that had minor allele frequency (MAF) >0.01 and >90% call rate were accepted. The missing genotype of accepted SNPs was subsequently imputed based on the mean of each marker35.
Genetic stratification and population analyses
A Neighbor-Joining (NJ) tree was used to infer the genetic stratification of the GWAS discovery populations. A Hamming’s pairwise distance matrix for all SNP sites was calculated to plot the NJ tree. The genome-wide LD decay rates in the Deli x AVROS and Nigerian x AVROS were important to anticipate the requirements for the suitable mapping resolution of the SNP array for GWAS. The rate is defined as the chromosomal distance at which the average pairwise correlation coefficient (r2) dropped to the half of its maximum value. In this study, we calculated the pairwise r2 for all SNPs in a 1-Kb window and averaged across the whole genome based on the composite method in the R package SNPRelate36.
Phenotypic data compilation and genome wide association study (GWAS)
Oil-to-dry-mesocarp (O/DM) is a direct measurement of crude palm oil (CPO) extracted from dry mesocarp tissue using a solvent. The individual palms were phenotyped to produce a reliable mean O/DM value for analysis as per standard industry practice37 with modifications38. The O/DM difference between Deli x AVROS and Nigerian x AVROS groups was tested for significance by a Student-t test. Subsequently, association analysis was conducted on 1,459 Deli x AVROS and 586 Nigerian x AVROS, respectively, based on a simple linear model in the R package GenABEL39, and a compressed MLM with P3D analysis29 in the rrBLUP35 programme. The total number of common SNPs in both groups was 55,054 SNPs with MAF >0.01. We accounted for the genetic sub-structure resulting from cryptic relatedness by including a kinship matrix40 as a random effect in the compressed MLM method. The whole-genome significance cut-offs were fixed at p ≤ 10−4, and ≤10−7, based on a Bonferroni correction method. The Quantile-Quantile plots and Manhattan plots were then constructed using the R package qqman41. We also evaluated the inflated false-positive signals for both methods according to the genomic inflation factor (GIF) estimated in R package GenABEL39.
SNP effect and statistical analyses
The common association signals in both groups based on p ≤ 10−4 were selected to measure the SNP effects on O/DM phenotype and further validated in a Deli x AVROS breeding trial separately. One-way ANOVA tests with multiple comparisons were performed to test the SNP genotypes against the O/DM trait variation. The same approach was applied to determine the SNP effects of the combination of significant SNPs based on the number of positive alleles carried by individual palms. All the statistical analyses were implemented in Minitab 1442. The genotype composition in pie charts were also incorporated into a boxplot of the SNP effects on O/DM variation across progeny testing populations to evaluate the combining abilities between dura palms and the common pisifera. The profiled SNP effects were subsequently used to implement MAS on newly propagated dura and pisifera seedlings at the pre-nursery stage and followed by field planting.
Data availability
All the SNPs used in the OP200K genotyping array have been deposited in dbSNP under the handle of SDTC_BB with NCBI submitted SNP (ss) accession numbers of 181006940–1810592638.
Additional Information
How to cite this article: Teh, C.-K. et al. Genome-wide association study identifies three key loci for high mesocarp oil content in perennial crop oil palm. Sci. Rep. 6, 19075; doi: 10.1038/srep19075 (2016).
Supplementary Material
Acknowledgments
We would like to acknowledge the contribution of the collaborating breeders from Oil Sime Darby Palm Breeding Unit and molecular breeders from Sime Darby Molecular Breeding Unit for sampling of oil palm materials. We also thank Sime Darby Bioinformatics Unit for providing analytical supports. The SNP genotyping service was done by DNA Landmarks Inc., Canada. The study was conducted in Sime Darby Plantation R&D Centre, which is fully supported by a grant from Sime Darby Plantation Division, Malaysia.
Footnotes
This study is fully supported by grants from Sime Darby Plantation Division, Malaysia. Two authors i.e. F.C. (National University of Singapore, Singapore) and S.M. (University of Nottingham, UK) have received consultancy fees under the same grants. The rest of the authors as the employees to Sime Darby Plantation Division declare that they have no relevant conflicts of interest.
Author Contributions C.T., F.C., D.A. and H.K. conceived and designed the study; A.O., Q.K. and C.T. performed data analysis and interpretations; M.M. performed breeding trial and trait recording. S.A. prepared and supplied plant materials and DNA samples. C.T., S.M., F.C., A.O. and Q.K. drafted the manuscript. All authors read and approved the final manuscript.
References
- Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007). [DOI] [PMC free article] [PubMed]
- Huang X. et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42, 961–967 (2010). [DOI] [PubMed] [Google Scholar]
- Mather K. A. et al. The Extent of Linkage Disequilibrium in Rice (Oryza sativa L.). Genetics 177, 2223–2232 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally K. L. et al. Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proceedings of the National Academy of Sciences 106, 12273–12278 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G. et al. A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat Genet 45, 957–961 (2013). [DOI] [PubMed] [Google Scholar]
- Li H. et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet 45, 43–50 (2013). [DOI] [PubMed] [Google Scholar]
- Atwell S. et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465, 627–631 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappa E. P. et al. Impacts of Population Structure and Analytical Models in Genome-Wide Association Studies of Complex Traits in Forest Trees: A Case Study in Eucalyptus globulus. PLoS ONE 8, e81267 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama K. et al. Demonstration of Genome-Wide Association Studies for Identifying Markers for Wood Property and Male Strobili Traits in Cryptomeria japonica. PLoS ONE 8, e79866 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. et al. Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds. Nature 500, 335–339 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley C. W. S. In The Oil Palm. Ch.2, 1–36 (Longman, London, 1967). [Google Scholar]
- Mayes S., Jack P. L., Corley R. H. V. & Marshall D. F. Construction of a RFLP genetic linkage map for oil palm (Elaeis guineensis Jacq.). Genome 40, 116–122 (1997). [DOI] [PubMed] [Google Scholar]
- Rance K. A., Mayes S., Price Z., Jack P. L. & Corley R. H. V. Quantitative trait loci for yield components in oil palm (Elaeis guineensis Jacq.). Theoretical and Applied Genetics 103, 1302–1310 (2001). [Google Scholar]
- Singh R. et al. Mapping quantitative trait loci (QTLs) for fatty acid composition in an interspecific cross of oil palm. BMC Plant Biology 9, 114–114 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billotte N. et al. QTL detection by multi-parent linkage mapping in oil palm (Elaeis guineensis Jacq.). Theoretical and Applied Genetics 120, 1673–1687 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting N.-C. et al. High density SNP and SSR-based genetic maps of two independent oil palm hybrids. BMC Genomics 15, 309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. et al. A consensus linkage map of oil palm and a major QTL for stem height. Scientific Reports 5, 8232 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamin K. A. A hundred and fifty years of oil palm in Indonesia: from the Bogor Botanical Garden to the industry.. in International Oil Palm Conference ‘Commodity of the Past, Today and the Future’, 3-23 (1998).
- Rosenquist E. A. The genetic base of oil palm breeding populations. In The International Workshop on Oil Palm Germplasm and Ultilization, 27–56 (1986). [Google Scholar]
- Kushairi A. & Rajanaidu N. In Advances in oil palm research, Vol. 1 (eds. Basiron Y., Jalani B. S. & Chan K. W.) 39–98 (Malaysian Palm Oil Board, Kuala Lumpur, 2000). [Google Scholar]
- Beirnaert A. & Vanderweyen R. Contribution à l’étude génétique et biométrique des variétiés d’Elaeis guineensis Jacq. In Publ. Inst. Nat. Etude Agron. Congo Belge. Ser. Sci. Vol. 27, 1–101 (1941). [Google Scholar]
- Hardon J. J., Corley R. H. V. & Lee C. H. Breeding and selecting the oil palm, 63–81 (Academic Press, London, 1987). [Google Scholar]
- Corley R. H. V. & Tinker P. B. In The Oil Palm, Ch. 5, 133–187 (Blackwell, 2003). [Google Scholar]
- Lim K. Y. Opening address. In The National Seminar on Opportunities for Maximizing Production through Better OER and Offshore Investment in Oil Palm 13–14 (Palm Oil Research Institute of Malaysia, Kuala Lumpur, 1998). [Google Scholar]
- Astle W. & Balding D. J. Population Structure and Cryptic Relatedness in Genetic Association Studies. Statist. Sci. 24, 451–471 (2009). [Google Scholar]
- Devlin B. & Roeder K. Genomic Control for Association Studies. Biometrics 55, 997–1004 (1999). [DOI] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M. & Donnelly P. Inference of population structure using multilocus genotype data. Genetics 155, 945- 959 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis G. R., Cardon L. R. & Cookson W. O. C. A General Test of Association for Quantitative Traits in Nuclear Families. American Journal of Human Genetics 66, 279–292 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. et al. Mixed linear model approach adapted for genome-wide association studies. Nat Genet 42, 355–360 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeennor S. & Volkaert H. Mapping of quantitative trait loci (QTLs) for oil yield using SSRs and gene-based markers in African oil palm (Elaeis guineensis Jacq.). Tree Genetics & Genomes 10, 1–14 (2014). [Google Scholar]
- Bourgis F. et al. Comparative transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proceedings of the National Academy of Sciences 108, 12527–12532 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh H. F. et al. Differential Metabolite Profiles during Fruit Development in High-Yielding Oil Palm Mesocarp. PLoS ONE 8, e61344 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. & Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endelman J.B. Ridge Regression and Other Kernels for Genomic Selection with R Package rrBLUP. Plant Gen. 4, 250–255 (2011). [Google Scholar]
- Zheng X. et al. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–3328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaak G., Sparnaaij L. D. & Menendez T. In Breeding and inheritance in the oil palm (Elaeis guineensis Jacq.) Part II, Vol. 4, 146–155 (J.W. Afr. Ins. Oil Palm Res., 1963). [Google Scholar]
- Rao V. et al. A critical reexamination of the method of bunch analysis in oil palm breeding. Palm Oil Research Institute Malaysia Occ Paper 9, 1–28 (1983). [Google Scholar]
- Aulchenko Y. S., Ripke S., Isaacs A. & van Duijn C. M. GenABEL: an R library for genome-wide association analysis. Bioinformatics 23, 1294–1296 (2007). [DOI] [PubMed] [Google Scholar]
- VanRaden P. M. Efficient Methods to Compute Genomic Predictions. Journal of Dairy Science 91, 4414–4423 (2008). [DOI] [PubMed] [Google Scholar]
- Turner S. D. qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots, doi: http://dx.doi.org/10.1101/005165 (2014).
- Du Feu C. MINITAB 14. Teaching Statistics 27, 30–32 (2005). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the SNPs used in the OP200K genotyping array have been deposited in dbSNP under the handle of SDTC_BB with NCBI submitted SNP (ss) accession numbers of 181006940–1810592638.