Key Points
The combined effects of AID-associated base excision and MMR delay the development of BCL6-driven DLBCL.
UNG single deficiency prevents the development of BCL6-driven DLBCL.
Abstract
Somatic hypermutation and class-switch recombination of the immunoglobulin (Ig) genes occur in germinal center (GC) B cells and are initiated through deamination of cytidine to uracil by activation-induced cytidine deaminase (AID). Resulting uracil-guanine mismatches are processed by uracil DNA glycosylase (UNG)–mediated base-excision repair and MSH2-mediated mismatch repair (MMR) to yield mutations and DNA strand lesions. Although off-target AID activity also contributes to oncogenic point mutations and chromosome translocations associated with GC and post-GC B-cell lymphomas, the role of downstream AID-associated DNA repair pathways in the pathogenesis of lymphoma is unknown. Here, we show that simultaneous deficiency of UNG and MSH2 or MSH2 alone causes genomic instability and a shorter latency to the development of BCL6-driven diffuse large B-cell lymphoma (DLBCL) in a murine model. The additional development of several BCL6-independent malignancies in these mice underscores the critical role of MMR in maintaining general genomic stability. In contrast, absence of UNG alone is highly protective and prevents the development of BCL6-driven DLBCL. We further demonstrate that clonal and nonclonal mutations arise within non-Ig AID target genes in the combined absence of UNG and MSH2 and that DNA strand lesions arise in an UNG-dependent manner but are offset by MSH2. These findings lend insight into a complex interplay whereby potentially deleterious UNG activity and general genomic instability are opposed by the protective influence of MSH2, producing a net protective effect that promotes immune diversification while simultaneously attenuating malignant transformation of GC B cells.
Introduction
Acquired somatic mutations and chromosome translocations are a hallmark of cancer that arise as a pathological result of a DNA repair response to a genotoxic event.1 In contrast, the introduction of nontemplated nucleotides and DNA double-strand breaks (DSBs) is part of the normal developmental program in germinal center (GC) B cells. Somatic hypermutation (SHM) and class-switch recombination (CSR) of the immunoglobulin (Ig) genes promotes antibody diversification in GC B cells, but the nature of these genetic remodeling events makes these cells uniquely vulnerable to malignant transformation.2 Numerous types of B-cell malignancies arise from GC and post-GC B cells including diffuse large B-cell lymphoma (DLBCL), Burkitt lymphoma, follicular lymphoma (FL), lymphoplasmacytic lymphoma, multiple myeloma, and Hodgkin lymphoma.3 SHM and CSR are both initiated by activation-induced cytidine deaminase (AID),4 a GC B-cell enzyme that lacks strict target specificity and is also able to introduce mutations and DSBs into non-Ig genes throughout the genome.5-12 A role for AID in lymphomagenesis is supported by the presence of characteristic somatic mutations within numerous oncogenes associated with human GC and post-GC B-cell malignancies.13-20 In addition, a prominent feature of these cancers is chromosome translocations that arise as a consequence of AID-mediated DSBs within the Ig heavy chain (IgH) class-switch (S) region and a partner oncogene such as BCL6 and cMYC.2,21 Further evidence implicating a direct role for AID in lymphomagenesis stems from several mouse models in which the development and phenotype of B-cell lymphoma is dependent on AID.10,22-24
Despite its DNA modification properties, it is well established that AID does not act alone.25 Base-excision repair (BER) and mismatch repair (MMR) pathways are required to process AID-generated uracil-guanine (U-G) mismatches into mutations and DNA strand lesions.9,26-29 During SHM, removal of uracil by uracil DNA glycosylase (UNG) creates an abasic site that serves as a template for DNA replication, which could result in any nucleotide substitution.30 Alternatively, the abasic site can be excised by apurinic/apyridimic endonuclease (APE) 1 and APE2 and filled in by low-fidelity translesion DNA polymerases.31,32 The MMR heterodimer MutSα (MSH2 and MSH6) can also recognize and facilitate removal of the U-G mismatch with exonuclease 1 (EXO1) activity creating a gap that is filled in by low-fidelity DNA polymerases.33 This permits spreading of mutations to surrounding A/T base pairs. During CSR, DNA single-strand breaks (SSBs) on opposite strands within IgH S regions are created through uracil removal by UNG and APE activity resulting in staggered DSBs if located in close proximity.31 If distantly located, these SSBs provide entry points for MutSα recruitment of EXO1 with consequent strand resection.34 Resulting DSBs are subsequently ligated by canonical nonhomologous and alternative end joining.35 These events are also thought to be responsible for strand lesions that lead to chromosome translocations.2 There are no other known repair pathways involved in the resolution of AID-generated U-G mismatches, and it is unknown how these pathways contribute to malignant transformation of GC B cells. To explore this question, we used a murine model to examine BCL6-driven AID-dependent GC B-cell lymphomagenesis in the absence of UNG (BER) and MSH2 (MMR).
Materials and methods
Mice
All mice were bred onto a C57BL/6 background. IµHABcl6, Ung+/−, and Msh2+/− mice were used to generate IµHABcl6 Ung+/− Msh2+/− and Ung−/− Msh2+/− mice.36-39 Intercrossing of the offspring generated IµHABcl6, IµHABcl6 Ung−/−, IµHABcl6 Msh2−/−, and IµHABcl6 Ung−/− Msh2−/− mice, as well as control Ung−/−, Msh2−/−, and Ung−/− Msh2−/− mice. Genotyping was performed by polymerase chain reaction (PCR) as described previously.26,36 Mice were housed in the Yale Animal Resource Center, and all procedures involving mice were approved by the Yale Institutional Animal Care and Use Committee (Yale IACUC protocol #11403). Statistical analysis of survival was performed by Kaplan-Meier survival and log-rank (Mantel-Cox) tests to assess whether survival differences were significant. The Mann-Whitney U test was used to compare median DLBCL latency.
Flow cytometry, histopathology, and immunohistochemistry
At necropsy, involved tissues were collected for cellular, histologic, and molecular analysis. For analysis of CSR from IgM to IgG1, splenic B cells were activated ex vivo with lipopolysaccharide (20 µg/mL) and interleukin 4 (10 ng/mL) for ∼72 hours. For immunophenotyping, cells were stained with fluorochrome-conjugated antibodies against CD3, B220, IgM, CD95, CD138, and IgG1 (BD Pharmingen). For γH2AX analysis, activated B cells were fixed in 70% ethanol and then incubated with rabbit anti-γH2AX antibody (Abcam 81299) followed by Alexa Fluor 647-conjugated goat-anti-rabbit secondary antibody (Abcam). After washing, cells were incubated with 1 µg/mL of 4,6 diamidino-2-phenylindole to stain DNA. Data were acquired on a FACSCalibur or a Stratedigm S1000 flow cytometer and analyzed with FlowJo software. For histopathology, formalin-fixed paraffin-embedded sections were stained with hematoxylin and eosin and biotinylated peanut agglutinin (PNA) (Vector, B-1075) by standard methods.
Clonality and mutation analysis
Splenic and Peyer’s patch B cells from healthy IµHABcl6 and IµHABcl6 Ung−/− Msh2−/− mice were used as controls. All oligonucleotide primer sequences are listed in supplemental Table 1 (available on the Blood Web site). To assess clonality, the rearranged VH sequence was amplified from genomic DNA using a mixture of forward primers designed to represent most mouse VH gene families and a reverse primer from the JH4 intron as previously described.36 Using this protocol, 4 major bands corresponding to rearranged JH1, JH2, JH3, and JH4 segments can be detected from a normal B-cell population, whereas only 1 major band will arise from clonal malignant B cells.40 Mutation analysis of Ig (JH4 intron, IgH 5′Sµ, and core IgH Sµ) and non-Ig (cMyc, Pim1, RhoH, Cd79a, CD79b, H2afx, Pax5, and Cd83) AID target loci was carried out by PCR amplification using high-fidelity Phusion polymerase (New England Biolabs) followed by sequencing of amplification and cloned products. Sequences were compared with National Center for Biotechnology Information references and with sequences from healthy littermates to identify both clonal and nonclonal mutations. Sequence analysis and statistical comparisons were performed as previously described.9 Clonal and subclonal mutations were only counted once toward the mutation frequency. Deletion frequency was calculated as number of events per cloned sequence. Statistical significance of mutation frequencies was assessed by Pearson’s χ2 test. Fold-enrichment of mutations in AID hotspots (DGYW/WRCH) was calculated as the ratio of observed-to-expected frequency of hotspot C/G to T/A mutations.
Gene expression profiling
Total RNA from tumors was purified using the RNeasy kit (Qiagen). Complementary RNA labeling, hybridization to Illumina MouseWG-6 v2.0 BeadChips, and scanning were performed according to the manufacturer’s instructions (Illumina). Analysis was performed using the lumi and limma in Bioconductor R package. The expression data reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database under series accession number GSE48304.
Spectral karyotyping
Metaphase slides were prepared from lymphoma cells grown in culture according to standard cytogenetic procedures. Mouse SkyPaint probe (Applied Spectral Imaging [ASI]) was added to each slide according to standard ASI protocol. Image acquisition was performed with a COOL-1300 SpectraCube camera (ASI) mounted on an Olympus BX43 microscope using a SKY optical filter (ASI). For each sample, a minimum of 20 metaphases was analyzed for chromosome abnormalities using the HiSKY v6.0 software (ASI).
Results
BCL6-driven lymphomas can arise in the combined absence of BER and MMR
BCL6 is considered the master regulator of the GC response where its physiological role in B cells is to establish a molecular environment that permits the accumulation of mutations through transcriptional repression of DNA damage response, cell cycle arrest, and B-cell maturation.41 BCL6 is often constitutively expressed in human DLBCL, typically as a result of a chromosome translocation with an Ig locus. Similarly, through deregulated expression of BCL6, IµHABcl6 mice spontaneously develop a clonal GC-derived lymphoma that emulates human DLBCL.36 In these mice, enforced B-cell-specific expression of BCL6 is achieved through the insertion of a full-length hemagglutinin (HA)-tagged murine Bcl6 coding sequence downstream of the IgH Iµ promoter. In the absence of AID, tumor incidence in these mice is markedly reduced and phenotype is restricted to marginal zone lymphoproliferations, supporting the notion that AID is required for GC-derived lymphomagenesis.24 In nonmalignant B cells that are deficient in both UNG and MSH2, U-G mismatches are not recognized and are simply replicated, revealing the footprint of AID by yielding C/G to T/A transitions.9,26,28,29 Thus, to investigate the role of AID-associated BER and MMR in the pathogenesis of GC lymphoma, we crossed IµHABcl6 mice onto a background deficient in both UNG (Ung−/−)39 and MSH2 (Msh2−/−).38
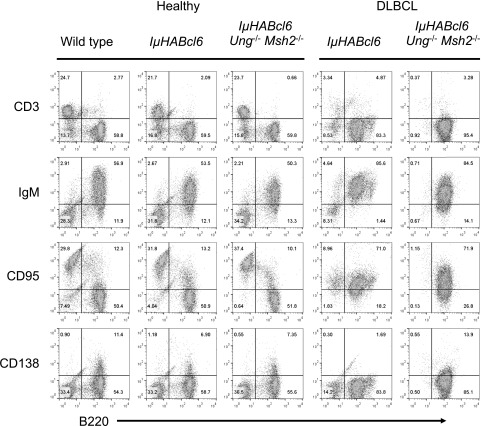
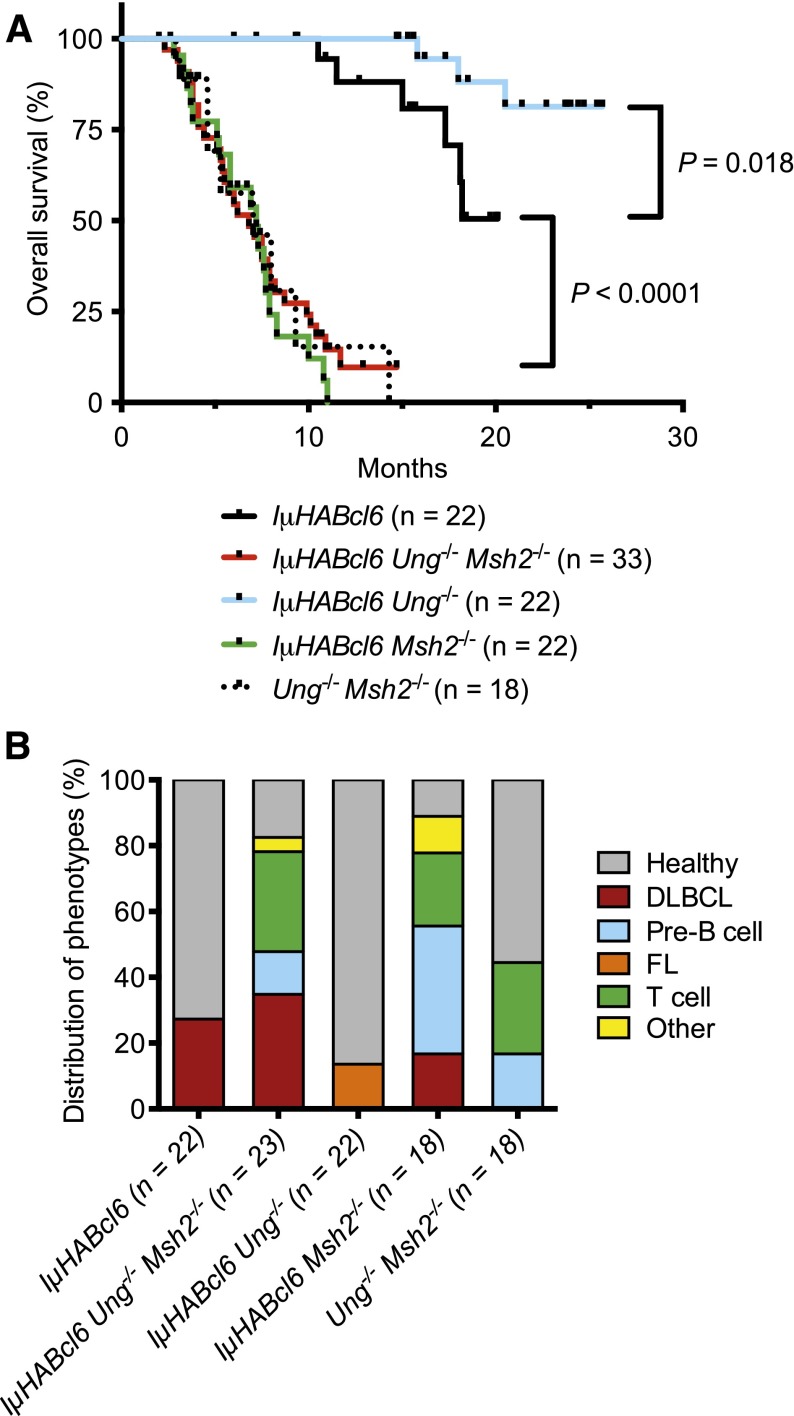
Young (<3 months) healthy IµHABcl6 and IµHABcl6 Ung−/− Msh2−/− mice displayed similar GC architecture (supplemental Figure 1) and a normal distribution of B and T cells (Figure 1). Consistent with previous studies,24,36 6 of 22 (27.3%) IµHABcl6 mice became sick starting at ∼12 months of age. However, 29 of 33 (87.9%) IµHABcl6 Ung−/− Msh2−/− mice became sick rapidly with an onset as early as 3 months and a median survival of 6.8 months (Figure 2A). Sick mice in both groups were found to be moribund with enlarged spleens and variable nodal and thymic involvement. All IµHABcl6 tumors analyzed were derived from mature B220+ IgM+ CD138− B cells (Figure 1). Of 19 IµHABcl6 Ung−/− Msh2−/− tumors available for characterization, immunophenotyping demonstrated 8 mature B220+ IgM+ CD138− B-cell lymphomas (Figure 1), 3 pre-B-cell lymphomas, 7 T-cell lymphomas, and 1 histiocytic sarcoma (Figure 2B). UNG-deficient mice have been shown to develop an FL, whereas MSH2-deficient mice develop numerous malignancies but predominantly pre-B-cell and T-cell lymphomas.37,38,42,43 As a central component of MMR, MSH2 provides global protection against genomic instability.44,45 Indeed, among 18 Ung−/− Msh2−/− control mice, median survival was 7 months with 3 pre-B-cell lymphomas and 5 T-cell lymphomas but no mature B-cell lymphomas (Figure 2). Thus, the development of pre-B-cell and T-cell malignancies in IµHABcl6 Ung−/− Msh2−/− mice is independent of BCL6 and is likely a reflection of MSH2 deficiency. Moreover, these data demonstrate that mature BCL6-driven B-cell lymphomas can arise independent of AID-associated BER and MMR.
Figure 1.
IµHABcl6 and IµHABcl6 Ung−/−Msh2−/− mice have similar lymphocyte development and develop similar lymphomas. Representative flow cytometric analysis of splenocytes from healthy (n = 3 for each genotype) and sick (n = 6 IµHABcl6 and n = 19 IµHABcl6 Ung−/− Msh2−/−) mice. Cells were stained with T-cell (CD3), mature B-cell (B220, IgM), GC B-cell (CD95), and plasma cell (CD138) markers as indicated. For healthy spleens, only the lymphocyte gate is shown. For diseased spleens, cells were ungated.
Figure 2.
Absence of UNG and/or MSH2 influences lymphomagenesis in IµHABcl6 mice. (A) Kaplan-Meier overall survival curves for IµHABcl6 mice with indicated genotypes. Median survival for IµHABcl6 and IµHABcl6 Ung−/− mice was not reached. P values were calculated using log-rank (Mantel-Cox) tests. (B) Distribution of tumor types among the different genotypes (“other” indicates histiocytic sarcoma and squamous cell carcinoma).
BER and MMR regulate the latency of BCL6-driven lymphomagenesis
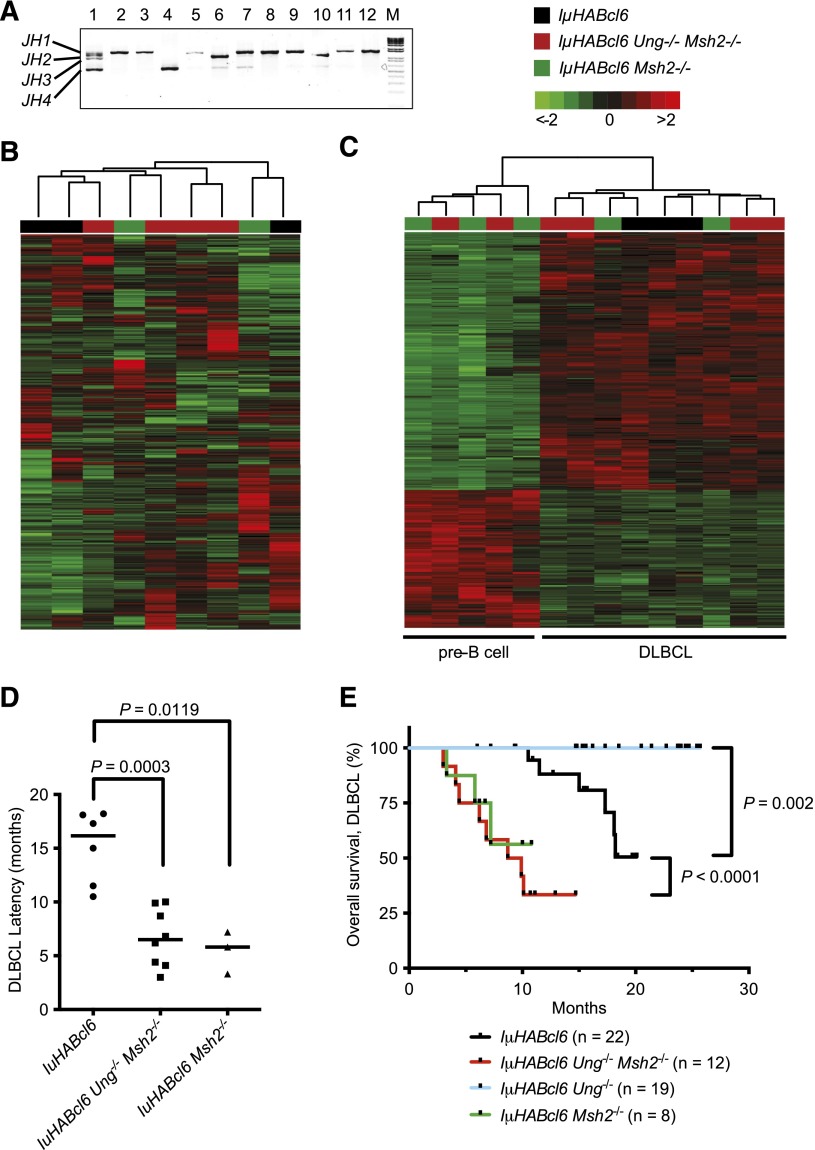
Focused examination of the mature B-cell lymphomas from both genotypic groups showed clonal IgH gene rearrangements (Figure 3A), expression of GC markers (Fas/CD95, PNA) (Figures 1 and 4), and disruption of lymphoid architecture with infiltration by large lymphoid cells consistent with GC-derived DLBCL (Figure 4). Analysis of gene expression profiles of representative tumors did not reveal any consistent differences between DLBCLs from each genotype (Figure 3B) and clearly distinguished the DLBCLs from pre-B-cell lymphomas (Figure 3C; supplemental Table 2). Although the background effect of MSH2 deficiency on the development of other malignancies precludes an accurate comparison of the true incidence of DLBCL between genotypes, the median time to development of disease was 2.5-fold shorter in IµHABcl6 Ung−/− Msh2−/− mice compared with IµHABcl6 mice (Figure 3D). This led to a significantly shortened overall survival for mice with DLBCL in the IµHABcl6 Ung−/− Msh2−/− group (Figure 3E). Thus, BER and MMR pathways exert a combined protective effect by increasing the latency period to BCL6-driven lymphomagenesis.
Figure 3.
IµHABcl6 Ung−/−Msh2−/− and IµHABcl6 Msh2−/− mice die of early onset DLBCL. (A) Clonality analysis of mature B-cell lymphomas was carried out on all B-cell tumors using Ig variable region amplification. Four major PCR fragments corresponding to respective JH1, JH2, JH3, and JH4 arrangement are labeled on the left. Multiple bands are present in a polyclonal population of healthy B cells (lane 1), and a single or dominant band indicates a clonal population. Representative specimens are shown. Lanes 2 to 4: DLBCLs from IµHABcl6 mice. Lanes 5 to 7: FLs from IµHABcl6 Ung−/− mice. Lanes 8, 9: DLBCLs from IµHABcl6 Msh2−/− mice. Lanes 10 to 12: DLBCLs from IµHABcl6 Ung−/− Msh2−/− mice. M, 1-kb DNA marker. (B) Unsupervised hierarchal clustering of gene expression profiles obtained from Illumina Mouse WG-6 v2.0 microarrays of representative B220+ IgM+ CD138− IµHABcl6 (n = 3), IµHABcl6 Ung−/− Msh2−/− (n = 4), and IµHABcl6 Msh2−/− (n = 2) mature GC B-cell lymphomas (color coded). Rows represent different gene probes, and columns denote individual samples. The scale indicates relative changes in gene expression normalized by the standard deviation (−2 to 2 in log2 units; 0 represents the mean expression level of a given gene across samples). (C) Supervised hierarchical clustering of gene expression data from representative IµHABcl6 (n = 3), IµHABcl6 Ung−/− Msh2−/− (n = 6), and IµHABcl6 Msh2−/− (n = 5) B-cell lymphomas. The pre-B-cell lymphomas and DLBCLs (defined by immunophenotyping and histology) are noted on the bottom and have distinct gene expression profiles (supplemental Table 2). (D) Median time to development of DLBCL in IµHABcl6 mice (squares) was 16.2 months compared with 6.5 months in the IµHABcl6 Ung−/− Msh2−/− mice (circles) and 5.8 months in IµHABcl6 Msh2−/− mice (triangles). Bars denote the median. P values were calculated using the 1-tailed Mann-Whitney U test. (E) Kaplan-Meier overall survival curves for IµHABcl6 mice with indicated genotypes that developed DLBCLs. P values were calculated using the log-rank (Mantel-Cox) test.
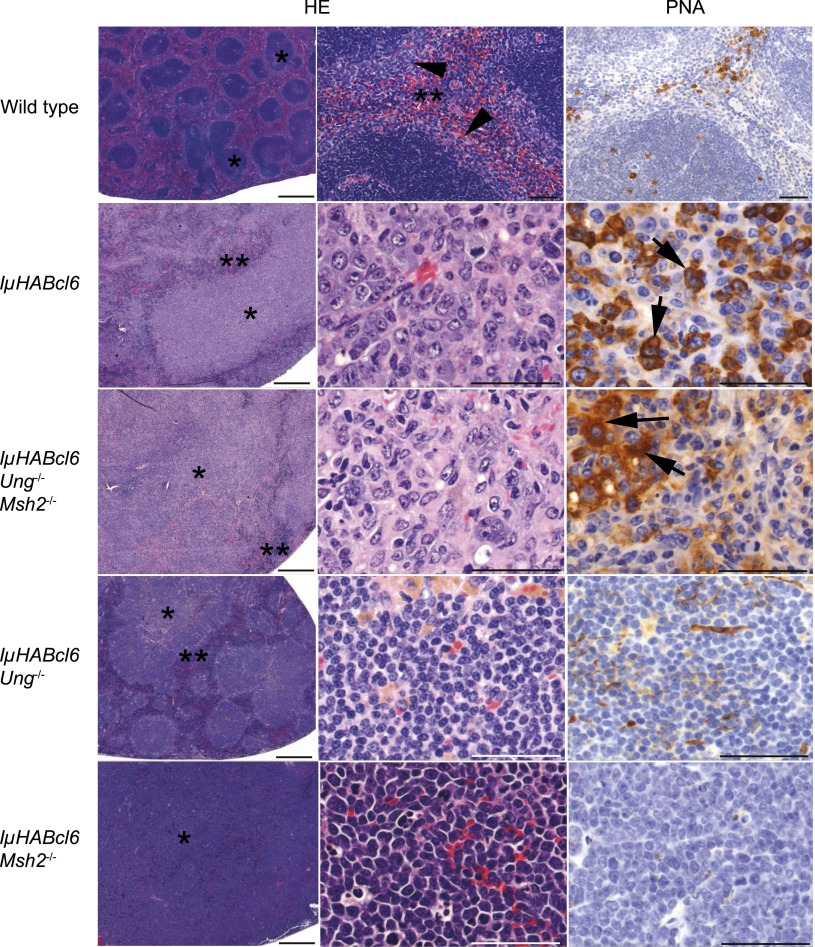
Figure 4.
Histologic analysis of B-cell tumor types. Representative hematoxylin and eosin (HE) and PNA-stained sections of mouse spleen from wild-type and IµHABcl6 mice with lymphoma from indicated genotypes. Wild-type mouse spleen is composed of white pulp with follicles (*) and a distinct marginal zone (arrowheads) separated by distinct areas of red pulp (**) with scattered PNA+ cells within follicles and the red pulp. In contrast, mice with lymphoma have marked expansion of the white pulp (*) with little or no visible red pulp (**). Neoplastic lymphocytes from IµHABcl6 and IµHABcl6 Ung−/− Msh2−/− mice are large and pleomorphic with abundant cytoplasm and are frequently PNA+ (arrows), consistent with DLBCL. Lymphomas from IµHABcl6 Ung−/− mice have expanded follicles composed of small monotypic lymphocytes that are predominantly PNA−, consistent with FL. Pre-B-cell lymphomas, shown here from IµHABcl6 Msh2−/− mice, are composed of medium-sized monotypic PNA− lymphocytes. Images were obtained on a Zeiss Axio Imager A1 microscope. Short scale bars represent 500 µM, and long scale bars represent 50 µM.
Absence of UNG prevents development of BCL6-driven DLBCL
To investigate the contribution of individual BER and MMR pathways to the pathogenesis of GC lymphoma, we generated IµHABcl6 Ung−/− and IµHABcl6 Msh2−/− single-deficient mice. The majority of IµHABcl6 Ung−/− mice remained healthy beyond 20 months with only 3 of 22 (13.6%) mice becoming sick starting at ∼16 months (P = .018; Figure 2). These mice were found to have clonal splenic lymphomas composed of mature B220+ IgM+ CD138− B cells (data not shown). Similar to the low penetrant FLs that arise after a prolonged latency in the absence of UNG,37 these lymphomas had expanded follicles with a population of small lymphocytes that were negative or only weakly positive for PNA (Figure 4). Thus, in the setting of deregulated BCL6, loss of UNG confers a significant survival advantage by preventing GC B cells from evolving into DLBCL. Conversely, this also suggests that UNG might play an active role in facilitating events that lead to GC B-cell transformation.
Absence of MSH2 promotes development of BCL6-driven DLBCL
In contrast to IµHABcl6 Ung−/− mice, 20 of 22 (90.9%) IµHABcl6 Msh2−/− mice rapidly died of malignancy starting at ∼3 months and had a median survival of 6 months (Figure 2A). Of 16 tumors available for analysis, there were 3 DLBCLs, 7 pre-B-cell lymphomas (Figure 4), 4 T-cell lymphomas, 1 histiocytic sarcoma, and 1 squamous cell carcinoma (Figure 2B). Similar to the IµHABcl6 Ung−/− Msh2−/− mice, the development of non-GC B-cell malignancies in the IµHABcl6 Msh2−/− mice is likely a reflection of MSH2 deficiency that occurs independent of BCL6. Analysis of gene expression profiles clustered the IµHABcl6 Msh2−/− DLBCLs with DLBCLs from IµHABcl6 and IµHABcl6 Ung−/− Msh2−/− mice (Figure 3B-C). In addition, independent analysis of latency and overall survival specifically from DLBCL indicated a trend that was similar to that of IµHABcl6 Ung−/− Msh2−/− mice (Figure 3D-E). Although the technical challenge of also deleting AID from our model limited our ability to fully assess its contribution to UNG and MSH2-dependent lymphomagenesis, the development of other malignancies from cell types that do not express AID exposes a critical role for MSH2 that extends beyond immune diversification. Altogether, these results demonstrate that IµHABcl6, IµHABcl6 Ung−/− Msh2−/−, and IµHABcl6 Msh2−/− mice develop the same GC B-cell lymphoma but have the most favorable survival when MSH2 is present. This indicates that in contrast to UNG-mediated BER, MMR provides a protective advantage against BCL6-driven DLBCL.
BER and MMR protect against widespread cytidine deamination
The impact of UNG and MSH2 single deficiency on the development of BCL6-driven DLBCL highlights the presence of opposing pathways to lymphomagenesis. However, the rapid development of DLBCL in the absence of both UNG and MSH2 suggests the existence of a complex and unbalanced interplay between BER and MER whereby the tumor-promoting potential of MSH2 deficiency is dominant over the protective effect of UNG deficiency. To gain insight into these mutagenic mechanisms, we sequenced AID target loci in B-cell tumors from selected genotypes.
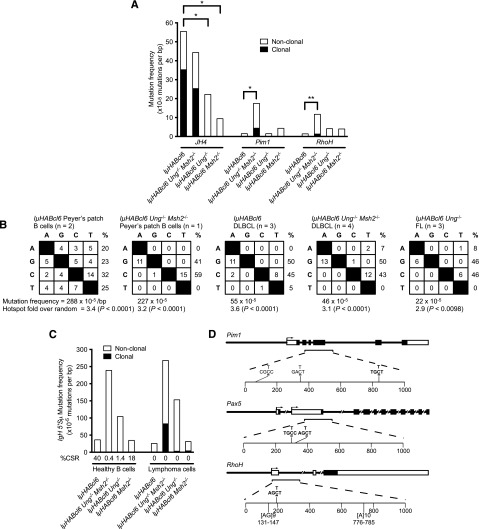
Analysis of the IgH JH4 intronic region from IµHABcl6 and IµHABcl6 Ung−/− Msh2−/− DLBCLs revealed similar frequencies of both clonal and nonclonal mutations, whereas only nonclonal mutations were detected in IµHABcl6 Ung−/− FLs and IµHABcl6 Msh2−/− DLBCLs (Figure 5A). As expected, the majority of IgH JH4 intronic mutations in healthy IµHABcl6 Ung−/−Msh2−/− GC B cells and IµHABcl6 Ung−/−Msh2−/− DLBCLs were C/G to T/A transitions (Figure 5B). However, in contrast to healthy IµHABcl6 GC B cells, the majority of mutations in the remaining lymphomas were also C/G to T/A transitions (Figure 5B). Overall mutation frequencies within the IgH JH4 intronic region were 4- to 24-fold lower in tumors compared with healthy IµHABcl6 GC B cells (Figure 5B), yet analysis of the IgH 5′Sµ region did not demonstrate any significant differences in mutation frequencies between ex vivo activated B cells from healthy mice and mature B-cell lymphomas (Figure 5C). Nonetheless, much like many human lymphomas,46 these tumors remained IgM+ and do not undergo CSR, suggesting an underlying defect in downstream processing of AID-generated U-G mismatches (Figure 5C). Additional sequencing of 8 non-Ig AID target genes (cMyc, Pim1, RhoH, Cd79a, CD79b, H2afx, Pax5, and Cd83)9 in 12 B-cell tumors (7 DLBCL, 3 FL, and 2 pre-B cell) revealed 6 unique clonal mutations within Pim1 (3 mutations), Pax5 (2 mutations), and RhoH (1 mutation) (Figure 5A,D). These mutations were found only in DLBCLs from IµHABcl6 Ung−/− Msh2−/− mice and were all C/G to T/A transitions. Further analysis of Pim1 and RhoH also revealed nonclonal mutation frequencies in IµHABcl6 Ung−/− Msh2−/− DLBCLs that were significantly higher than in the other lymphomas (Figure 5A). Collectively, the presence of clonal and nonclonal C/G to T/A transitions indicates that ongoing AID activity targets Ig and non-Ig loci in these lymphomas. In addition, although the IgH JH4 intron appears to be targeted less frequently by AID in these lymphomas, the higher frequency of clonal and nonclonal mutations in non-Ig genes in the absence of UNG and MSH2 supports the notion that the net effect of BER and MMR might restrict lymphomagenesis by preventing widespread accumulation of C/G to T/A transition mutations.
Figure 5.
Ig and non-Ig gene mutation patterns in IµHABcl6 GC B-cell lymphomas. (A) Sequence analysis of AID target genes was performed on genomic DNA from mature B-cell tumors that developed in IµHABcl6 (n = 3), IµHABcl6 Ung−/− Msh2−/− (n = 4), IµHABcl6 Ung−/− (n = 3), and IµHABcl6 Msh2−/− (n = 2) mice. With exception of the IµHABcl6 Ung−/− FLs, all tumors in this graph are DLBCLs. A total of ∼632 kb of sequencing data were obtained. No mutations were detected in any pre-B-cell lymphomas, consistent with their AID-independent stage of development (not shown). Mutation frequencies for JH4, Pim1, and RhoH are shown as indicated. Bars denote mutation frequency on the y-axis with black and white fill indicating clonal and nonclonal mutations, respectively. Statistical significance was assessed by Pearson’s χ2 test. NS, not significant; *P < .05; **P < .01. (B) The pattern of nucleotide substitutions detected in the IgH JH4 regions from mature B-cell tumors with the indicated genotypes was compared with that of Peyer’s patch GC B cells from healthy 4-month-old IµHABcl6 and IµHABcl6 Ung−/− Msh2−/− mice. Nucleotides in the left column are mutated to the nucleotides in the top row. If the same nucleotide substitution occurred at the same site in multiple clones, it was counted only once. Percentage of mutations occurring at a specific nucleotide base is calculated in the last column. Total mutation frequency and hotspot fold over random (calculated as the ratio of observed-to-expected frequency of hotspot C/G to T/A mutations) is below each box. (C) Analysis of CSR and subsequent mutational analysis of Ig 5′Sµ region was performed on ex vivo activated healthy B cells (white bars) and mature B-cell lymphomas (black bars) from the indicated genotypes. Numbers on the x-axis denote the percentage of cells that underwent CSR from IgM to IgG1 (determined by flow cytometry). (D) Clonal mutation plots of Pim1, Pax5, and RhoH. Genomic loci are shown with untranslated (open boxes) and coding (filled boxes) regions. An ∼1-kb region (brackets and dashed lines) downstream of the major transcriptional start site (arrows) was sequenced. Analysis of ∼96 kb of total sequencing data from 12 B-cell tumors (7 DLBCL, 3 FL, and 2 pre-B cell) revealed 6 unique clonal mutations within Pim1, Pax5, and RhoH. All mutations were found in DLBCLs from IµHABcl6 Ung−/− Msh2−/− mice. Mutations (underlined) and surrounding nucleotides are shown. Bolded sequences indicate an AID hotspot motif. The [AG]9 and [A]10 microsatellite repeats within the RhoH locus are indicated.
Absence of MMR reveals UNG-dependent and independent genomic instability
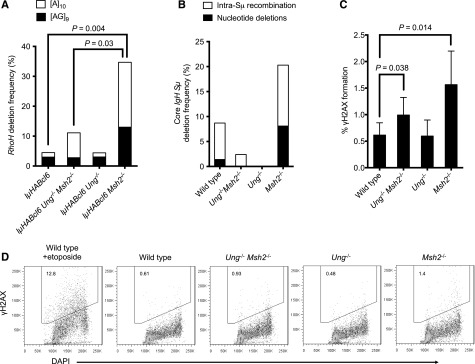
During our non-Ig gene mutational analysis, we also identified numerous deletions within RhoH. This locus contains 2 microsatellite repeats within the 1-kb region downstream of the transcriptional start site: 1 [AG]9 dinucleotide repeat and 1 [A]10 mononucleotide repeat (Figure 5D). Mono- and dinucleotide deletions within these microsatellite repeats were present in DLBCLs from all genotypes but were most prominent in the IµHABcl6 Msh2−/− DLBCLs (Figure 6A). Deletion frequency in the IµHABcl6 Ung−/− Msh2−/− DLBCLs was lower than the IµHABcl6 Msh2−/− DLBCLs and further decreased in the MSH2-sufficient genotypes. This indicates that a subset of these deletions is dependent on the presence of UNG. In addition to their roles in immune diversification, MSH2 is required for maintaining stability of microsatellite repeat sequences, and UNG regulates genomic uracil load incorporated during DNA replication.45,47 In the absence of UNG, aberrant A/U base pairs within microsatellite repeats would simply be replicated. However, in the absence of MSH2, UNG-generated abasic sites can be processed into strand lesions with an imbalance in BER leading to microsatellite instability.47,48 A similar analysis of healthy ex vivo activated B cells also demonstrated a high frequency of UNG-dependent deletion and intra-Sµ recombination events involving both AID hotspot motifs and repetitive DNA sequences within the core IgH Sµ region (Figure 6B and supplemental Figure 2). Altogether, these findings are consistent with the presence of both UNG-dependent and independent mechanisms for initiating genomic instability in GC B cells that are normally offset by the actions of MSH2.
Figure 6.
Deletions within RhoH and DNA DSBs are increased in a UNG-dependent manner in the absence of MSH2. (A) Sequence analysis of the RhoH locus identified mono- and dinucleotide deletions within [A]10 (white bar) and [AG]9 (black bar) microsatellite repeats. A total of 30 deletions were identified in 254 sequences representing ∼450 kb of sequencing data. Deletion frequency is represented as a percentage of the number of cloned sequences that contained a unique deletion. Statistical significance was calculated using Pearson’s χ2 test. (B) Sequence analysis of the core IgH Sµ region identified unique intra-Sµ recombination events (white bar) and deletions (black bar) within AID hotspot motifs and repetitive sequences. A total of 25 events were identified in 228 sequences representing ∼168 kb of sequencing data. (C) Splenic B cells were obtained from wild-type mice and healthy mice deficient in UNG, MSH2, and both. Cells were activated ex vivo with lipopolysaccharide and interleukin 4 for 48 hours followed by measurement of γH2AX formation by flow cytometry. The graph depicts the mean and standard deviation of % γH2AX+ cells from 4 to 6 different mice of each genotype. Statistical significance was calculated by 2-tailed Student t test. (D) Representative flow cytometric analysis of γH2AX formation (y-axis) from each genotype. DAPI staining for DNA content is shown on the x-axis. Wild-type B cells treated with 20 μM etoposide for 15 minutes at the end of activation were used as a positive control for γH2AX formation. DAPI, 4,6 diamidino-2-phenylindole.
As part of the DNA damage signaling response, histone H2AX is phosphorylated to γH2AX and forms foci at sites of DSBs and SSBs.49 To further investigate the opposing roles of BER and MMR in regulating genomic stability in B cells, we measured γH2AX formation in healthy ex vivo activated B cells from mice deficient in UNG, MSH2, and both. Although no obvious differences in γH2AX formation could be detected by immunoblotting (not shown), analysis of single cells by flow cytometry demonstrated a small but consistent and statistically significant increase in γH2AX+ B cells from Msh2−/− mice compared with B cells from wild-type and Ung−/− mice (Figure 6C-D and supplemental Table 3). In addition, γH2AX formation was significantly higher in Ung−/− Msh2−/− B cells than wild-type B cells but lower than that seen in Msh2−/− B cells. This provides additional evidence of interplay between BER and MER whereby DNA strand lesions can arise in activated B cells in an UNG-dependent manner but are offset by the actions of MSH2.
Consistent with previous findings, spectral karyotyping of IµHABcl6 DLBCLs demonstrated complex chromosome abnormalities.36 We also identified numerous clonal and subclonal chromosome abnormalities in IµHABcl6 Ung−/− Msh2−/− DLBCLs and IµHABcl6 Ung−/− FLs (Table 1 and supplemental Figure 2). Although there were no recurrent translocations or deletions, chromosome 15 (location of cMyc) was involved in an aneuploidy in 7 of 9 karyotypes analyzed. Thus, genomic instability can develop in GC B-cell lymphomas through mechanisms independent of AID-associated BER and MMR pathways. A specific cause for this is not clear but it is likely multifactorial, with our findings supporting previous reports implicating a role for microsatellite instability when MMR is impaired, along with the added stress to DNA damage response mechanisms in the setting of an imbalance in genomic uracil homeostasis.44,50-52
Table 1.
Composite karyotypes from representative tumor samples
| Mouse ID | Genotype | Pathology | JH4 intron | Spectral karyotype |
|---|---|---|---|---|
| 340 | IµHABcl6 | DLBCL | M | 40∼43 < 2n>,XX,+8[11],+8x2[6],Der(10)T(10A4;13?A)T(13?B;17D)[3],+15[19],Der(17) T(10A4;17D)[3][cp21] / 84∼88 < 4n>,XXXX,+8x2∼6[4],+15x2[4][cp4] |
| 379 | IµHABcl6 | DLBCL | M | 42∼44 < 2n>,X,-X[14],Del(X?A6)[5],Dp(5)[14],Der(10)[3],+13[7],+13x2[2],+15[17],+15x2[2],+16[12],+16x2[2],+19 [19][cp19] / 84∼89 < 4n>,XX or XXX,-X[2],-Xx2[4],Dp(5)x2[6],+15x2[3],+15x3[3],+16x2[5],+16x4[1],+19x2[6][cp6] |
| 438 | IµHABcl6 | DLBCL | UM | 40∼42 < 2n>,XY,Del(2G)[3],Der(4)T(4E2;13A3)[3],+13[1],+15[3],+16[4],+17[3][cp24] / 84 < 4n>,XXYY,Der(4)T(4E2;13A3)x2,+13,+15,+16,+16[cp1] |
| 167 | IµHABcl6 Ung−/−Msh2−/− | DLBCL | M | 40∼41,XX,T(2C2;17A2)[21],+15[2],+19[10][cp30] |
| 375 | IµHABcl6 Ung−/−Msh2−/− | DLBCL | M | 40,XY,Dp(1E-F)[2][cp30] |
| 414 | IµHABcl6 Ung−/−Msh2−/− | DLBCL | M | 40,XX[5] / 40∼42,XX,T(8E1;13A3)[2],Der(10)?Del(10D)Dp(10A-C)[3],Der(10)Dp(10A-C)T(10C;16C2)[22],+15[5],+Der(15)T(10D;15D1)[21][cp34] / 79∼83 < 4n>,XXXX,Der(10)?Del(10D)Dp(10A-C)x2[1],Der(10)Dp(10A-C)T(10C;16C2)[1],+15x2[2],+Der(15)T(10D;15D1)x2[1][cp6] |
| 491 | IµHABcl6 Ung−/−Msh2−/− | DLBCL | M | 40,XX,+Rb(XA1.7A1)[30][cp30] |
| 448 | IµHABcl6 Ung−/− | FL | M | 41 < 2n>,X,-X[30],+15[30],+17[30][cp30] |
| 496 | IµHABcl6 Ung−/− | FL | M | 40∼43 < 2n>,XY,+Y[2],+2[6],+5[2],Del(6B3-E1)[24],+15[20],+17[11],+mar[3][cp30] |
[ ], cell number analyzed; cp, composite; Del, deletion; Der, derivative chromosome; Der(N)T, nonreciprocal translocation; Dp, duplication; M, mutated; mar, marker chromosome; Rb, Robertsonian translocation; T, reciprocal translocation; UM, unmutated.
Discussion
Immune diversification in GC B cells occurs in 2 phases: AID-mediated cytidine deamination during phase 1 followed by BER and MMR of resulting U-G mismatches during phase 2. Although this adaptation facilitates SHM and CSR of the Ig genes, AID binds to ∼12 000 sites across the genome, and off-target events are associated with GC and post-GC B-cell malignancies.2,12 Although evidence suggests that AID may initiate the process of lymphomagenesis,10,22-24 we demonstrate here that downstream BER and MMR pathways ultimately regulate the malignant fate and timing of GC B-cell transformation. We propose that lymphoma may arise as a result of events that take place in both phases of immune diversification. When phase 1 is left unchecked, as occurs in the absence of both UNG and MSH2, widespread production of U-G mismatches is associated with the accumulation of somatic C/G to T/A transition mutations within non-Ig genes and a significantly shorter latency to the development of BCL6-driven DLBCL. Moreover, a critical role for MSH2 in general genomic maintenance is made evident by the additional development of other malignancies independent of BCL6. Thus, although the combined actions of UNG and MSH2 prevent the accumulation of potentially lymphomagenic C/G to T/A transition mutations, these findings also suggest that MMR imparts an additional and important layer of genomic protection to GC B cells.
During phase 2 of immune diversification, UNG targets areas of high uracil density to create the majority of AID-initiated DNA breaks, with MSH2 acting downstream of UNG either independently or as a backup mechanism for resecting distantly separated U-G mismatches into strand lesions.53 Absence of MSH2 or MSH6 causes ∼50% to 80% reductions in CSR, whereas absence of UNG causes >95% reductions in CSR in mice and humans.26-29,54,55 In addition, UNG deficiency in mice impairs formation of translocations between IgH and cMyc and reduces the incidence of post-GC plasmacytomas in Bcl-xL transgenic mice.56,57 Consistent with the notion that absence of UNG might limit the formation of lymphomagenic DNA lesions, our findings demonstrate that UNG deficiency prevents the development of BCL6-driven DLBCL. This phenotype resembles the protective effect of AID deficiency in IµHABcl6 mice,24 indicating that under physiological conditions, downstream UNG-mediated processing of U-G mismatches is an essential step in AID-initiated lymphomagenesis.
In addition to forming MutSα with MSH6, MSH2 heterodimerizes with MSH3 to form MutSβ. Both MMR complexes maintain genomic stability by repairing small insertion and deletion loops generated within microsatellite repeats during DNA replication,45 but the ability of MutSα to repair nucleotide mismatches imparts a distinct role in antibody diversification. Despite this mutagenic role and consistent with the reduced capacity of MSH2 to facilitate the formation of AID-initiated strand lesions in the absence of UNG, the lack of DLBCL development in IµHABcl6 Ung−/− mice illustrates that MMR alone cannot promote BCL6-driven lymphomagenesis. Conversely, in the absence of MSH2, GC B cells exhibit extreme microsatellite instability,52 with our findings demonstrating increased formation of UNG-dependent γH2AX foci in activated B cells prior to the development of lymphoma and an associated rise in microsatellite instability in MSH2-deficient DLBCLs. Although we cannot determine the extent of AID dependence, this is consistent with a dual protective role for MSH2 in GC B cells in the maintenance of microsatellite stability and the restriction of UNG-dependent lesions.
It is unknown how AID-associated BER and MMR pathways compete for processing of U-G mismatches or what factors determine whether repair will occur in a high- or low-fidelity manner. Nonetheless, our findings support a model whereby the actions of UNG and MSH2 contribute to SHM and CSR of the Ig genes while simultaneously attenuating malignant transformation by limiting the accompanying genome-wide AID-generated uracil load and maintaining genomic integrity. This combined effort is composed of a complex interplay between BER and MMR, which individually manifest as opposing roles with UNG promoting the formation of potentially deleterious intermediates including abasic sites, SSBs, DSBs, and deletions while MSH2 restricts the formation of UNG-dependent lesions outside the Ig loci and provides an additional layer of general protection from genomic instability. The end result is a net protective effect that delays lymphomagenesis in the setting of an initial genetic hit (in this case, deregulated expression of BCL6), but any breakdown in this balance of repair might subsequently permit the accumulation of lymphomagenic mutations. Consistent with this model, it is not surprising that germ line and somatic mutations of genes involved in MMR (including MSH2, MSH6, and EXO1) have been found in up to 30% of human DLBCL, whereas mutations of BER genes occur with much less frequency.58,59 Additional features of the IµHABcl6 DLBCL model that resemble human disease include lack of CSR and a predominance of mutations at C/G base pairs. Functional impairment of CSR in human activated B-cell DLBCL is strongly associated with a predisposition to chromosome translocations and is thought to be because of acquired defects in DNA repair.46 Similarly, C/G mutation bias within Ig and non-Ig AID target loci in human DLBCL and lymphoma cell lines is a common finding and is consistent with impairment of MMR.13 Altogether, this suggests that disruption of the balance of AID-associated BER and MMR may be a recurrent event in GC B-cell malignancy that provides a selective advantage and contributes to the pathogenesis and heterogeneity of these diseases.
Acknowledgments
The authors thank J. Koeman and the Cytogenetics Core at the Van Andel Research Institute for cytogenetic analysis, the Keck Biotechnology Facility at Yale University for DNA sequencing and microarray hybridization, Michael Schadt at Yale Mouse Research Pathology for histology, Amos Brooks at Yale Research Histology Laboratory for immunohistochemistry, and Riccardo Dalla-Favera at Columbia University for providing the IµHABcl6 mice. The authors also thank David Schatz and Velizar Shivarov for thoughtful discussions and critical review of the manuscript.
This work was supported by the National Institutes of Health, National Cancer Institute (NCI K08CA140718) (M.P.S.), a Scholar Award from the American Society of Hematology (M.P.S.), and the Concern Foundation (M.P.S.).
Footnotes
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE48304).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: X.G. performed the experiments, analyzed data, and cowrote the manuscript; C.J.B. reviewed the histology and immunohistochemistry; Z.L. analyzed and interpreted gene expression profiling data; M.P.S. coordinated the study, designed and performed experiments, analyzed data, and wrote the manuscript; and all authors reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Matthew P. Strout, Yale Cancer Center, Section of Hematology, Yale University School of Medicine, 300 George St, New Haven, CT 06511; e-mail: matthew.strout@yale.edu.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Robbiani DF, Nussenzweig MC. Chromosome translocation, B cell lymphoma, and activation-induced cytidine deaminase. Annu Rev Pathol. 2013;8:79–103. doi: 10.1146/annurev-pathol-020712-164004. [DOI] [PubMed] [Google Scholar]
- 3.Shaffer AL, III, Young RM, Staudt LM. Pathogenesis of human B cell lymphomas. Annu Rev Immunol. 2012;30:565–610. doi: 10.1146/annurev-immunol-020711-075027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102(5):553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 5.Chiarle R, Zhang Y, Frock RL, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147(1):107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hasham MG, Donghia NM, Coffey E, et al. Widespread genomic breaks generated by activation-induced cytidine deaminase are prevented by homologous recombination. Nat Immunol. 2010;11(9):820–826. doi: 10.1038/ni.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato L, Begum NA, Burroughs AM, et al. Nonimmunoglobulin target loci of activation-induced cytidine deaminase (AID) share unique features with immunoglobulin genes. Proc Natl Acad Sci USA. 2012;109(7):2479–2484. doi: 10.1073/pnas.1120791109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein IA, Resch W, Jankovic M, et al. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell. 2011;147(1):95–106. doi: 10.1016/j.cell.2011.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu M, Duke JL, Richter DJ, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451(7180):841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 10.Robbiani DF, Bunting S, Feldhahn N, et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol Cell. 2009;36(4):631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staszewski O, Baker RE, Ucher AJ, Martier R, Stavnezer J, Guikema JE. Activation-induced cytidine deaminase induces reproducible DNA breaks at many non-Ig Loci in activated B cells. Mol Cell. 2011;41(2):232–242. doi: 10.1016/j.molcel.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamane A, Resch W, Kuo N, et al. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat Immunol. 2011;12(1):62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasqualucci L, Neumeister P, Goossens T, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412(6844):341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 14.Reiniger L, Bödör C, Bognár A, et al. Richter’s and prolymphocytic transformation of chronic lymphocytic leukemia are associated with high mRNA expression of activation-induced cytidine deaminase and aberrant somatic hypermutation. Leukemia. 2006;20(6):1089–1095. doi: 10.1038/sj.leu.2404183. [DOI] [PubMed] [Google Scholar]
- 15.Bödör C, Bognár A, Reiniger L, et al. Aberrant somatic hypermutation and expression of activation-induced cytidine deaminase mRNA in mediastinal large B-cell lymphoma. Br J Haematol. 2005;129(3):373–376. doi: 10.1111/j.1365-2141.2005.05454.x. [DOI] [PubMed] [Google Scholar]
- 16.Deutsch AJ, Aigelsreiter A, Staber PB, et al. MALT lymphoma and extranodal diffuse large B-cell lymphoma are targeted by aberrant somatic hypermutation. Blood. 2007;109(8):3500–3504. doi: 10.1182/blood-2006-06-030494. [DOI] [PubMed] [Google Scholar]
- 17.Hardianti MS, Tatsumi E, Syampurnawati M, et al. Activation-induced cytidine deaminase expression in follicular lymphoma: association between AID expression and ongoing mutation in FL. Leukemia. 2004;18(4):826–831. doi: 10.1038/sj.leu.2403323. [DOI] [PubMed] [Google Scholar]
- 18.Mottok A, Renné C, Seifert M, et al. Inactivating SOCS1 mutations are caused by aberrant somatic hypermutation and restricted to a subset of B-cell lymphoma entities. Blood. 2009;114(20):4503–4506. doi: 10.1182/blood-2009-06-225839. [DOI] [PubMed] [Google Scholar]
- 19.Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280(5370):1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 20.Jiang Y, Soong TD, Wang L, Melnick AM, Elemento O. Genome-wide detection of genes targeted by non-Ig somatic hypermutation in lymphoma. PLoS One. 2012;7(7):e40332. doi: 10.1371/journal.pone.0040332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu Z, Tsai AG, Akasaka T, et al. BCL6 breaks occur at different AID sequence motifs in Ig-BCL6 and non-Ig-BCL6 rearrangements. Blood. 2013;121(22):4551–4554. doi: 10.1182/blood-2012-10-464958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotani A, Kakazu N, Tsuruyama T, et al. Activation-induced cytidine deaminase (AID) promotes B cell lymphomagenesis in Emu-cmyc transgenic mice. Proc Natl Acad Sci USA. 2007;104(5):1616–1620. doi: 10.1073/pnas.0610732104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovalchuk AL, duBois W, Mushinski E, et al. AID-deficient Bcl-xL transgenic mice develop delayed atypical plasma cell tumors with unusual Ig/Myc chromosomal rearrangements. J Exp Med. 2007;204(12):2989–3001. doi: 10.1084/jem.20070882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pasqualucci L, Bhagat G, Jankovic M, et al. AID is required for germinal center-derived lymphomagenesis. Nat Genet. 2008;40(1):108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- 25.Stavnezer J. Complex regulation and function of activation-induced cytidine deaminase. Trends Immunol. 2011;32(5):194–201. doi: 10.1016/j.it.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rada C, Di Noia JM, Neuberger MS. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell. 2004;16(2):163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 27.Schrader CE, Edelmann W, Kucherlapati R, Stavnezer J. Reduced isotype switching in splenic B cells from mice deficient in mismatch repair enzymes. J Exp Med. 1999;190(3):323–330. doi: 10.1084/jem.190.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen HM, Tanaka A, Bozek G, Nicolae D, Storb U. Somatic hypermutation and class switch recombination in Msh6(-/-)Ung(-/-) double-knockout mice. J Immunol. 2006;177(8):5386–5392. doi: 10.4049/jimmunol.177.8.5386. [DOI] [PubMed] [Google Scholar]
- 29.Xue K, Rada C, Neuberger MS. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2-/- ung-/- mice. J Exp Med. 2006;203(9):2085–2094. doi: 10.1084/jem.20061067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maul RW, Saribasak H, Martomo SA, et al. Uracil residues dependent on the deaminase AID in immunoglobulin gene variable and switch regions. Nat Immunol. 2011;12(1):70–76. doi: 10.1038/ni.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stavnezer J, Linehan EK, Thompson MR, et al. Differential expression of APE1 and APE2 in germinal centers promotes error-prone repair and A:T mutations during somatic hypermutation. Proc Natl Acad Sci USA. 2014;111(25):9217–9222. doi: 10.1073/pnas.1405590111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weill JC, Reynaud CA. DNA polymerases in adaptive immunity. Nat Rev Immunol. 2008;8(4):302–312. doi: 10.1038/nri2281. [DOI] [PubMed] [Google Scholar]
- 33.Bardwell PD, Woo CJ, Wei K, et al. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat Immunol. 2004;5(2):224–229. doi: 10.1038/ni1031. [DOI] [PubMed] [Google Scholar]
- 34.Schrader CE, Guikema JE, Linehan EK, Selsing E, Stavnezer J. Activation-induced cytidine deaminase-dependent DNA breaks in class switch recombination occur during G1 phase of the cell cycle and depend upon mismatch repair. J Immunol. 2007;179(9):6064–6071. doi: 10.4049/jimmunol.179.9.6064. [DOI] [PubMed] [Google Scholar]
- 35.Cortizas EM, Zahn A, Hajjar ME, Patenaude AM, Di Noia JM, Verdun RE. Alternative end-joining and classical nonhomologous end-joining pathways repair different types of double-strand breaks during class-switch recombination. J Immunol. 2013;191(11):5751–5763. doi: 10.4049/jimmunol.1301300. [DOI] [PubMed] [Google Scholar]
- 36.Cattoretti G, Pasqualucci L, Ballon G, et al. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7(5):445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 37.Nilsen H, Stamp G, Andersen S, et al. Gene-targeted mice lacking the Ung uracil-DNA glycosylase develop B-cell lymphomas. Oncogene. 2003;22(35):5381–5386. doi: 10.1038/sj.onc.1206860. [DOI] [PubMed] [Google Scholar]
- 38.Reitmair AH, Schmits R, Ewel A, et al. MSH2 deficient mice are viable and susceptible to lymphoid tumours. Nat Genet. 1995;11(1):64–70. doi: 10.1038/ng0995-64. [DOI] [PubMed] [Google Scholar]
- 39.Nilsen H, Rosewell I, Robins P, et al. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol Cell. 2000;5(6):1059–1065. doi: 10.1016/s1097-2765(00)80271-3. [DOI] [PubMed] [Google Scholar]
- 40.Delbos F, De Smet A, Faili A, Aoufouchi S, Weill JC, Reynaud CA. Contribution of DNA polymerase eta to immunoglobulin gene hypermutation in the mouse. J Exp Med. 2005;201(8):1191–1196. doi: 10.1084/jem.20050292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Basso K, Dalla-Favera R. BCL6: master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv Immunol. 2010;105:193–210. doi: 10.1016/S0065-2776(10)05007-8. [DOI] [PubMed] [Google Scholar]
- 42.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82(2):321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 43.Campbell MR, Nation PN, Andrew SE. A lack of DNA mismatch repair on an athymic murine background predisposes to hematologic malignancy. Cancer Res. 2005;65(7):2626–2635. doi: 10.1158/0008-5472.CAN-04-3158. [DOI] [PubMed] [Google Scholar]
- 44.Hegan DC, Narayanan L, Jirik FR, Edelmann W, Liskay RM, Glazer PM. Differing patterns of genetic instability in mice deficient in the mismatch repair genes Pms2, Mlh1, Msh2, Msh3 and Msh6. Carcinogenesis. 2006;27(12):2402–2408. doi: 10.1093/carcin/bgl079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bak ST, Sakellariou D, Pena-Diaz J. The dual nature of mismatch repair as antimutator and mutator: for better or for worse. Front Genet. 2014;5:287. doi: 10.3389/fgene.2014.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lenz G, Nagel I, Siebert R, et al. Aberrant immunoglobulin class switch recombination and switch translocations in activated B cell-like diffuse large B cell lymphoma. J Exp Med. 2007;204(3):633–643. doi: 10.1084/jem.20062041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krokan HE, Sætrom P, Aas PA, Pettersen HS, Kavli B, Slupphaug G. Error-free versus mutagenic processing of genomic uracil--relevance to cancer. DNA Repair (Amst) 2014;19:38–47. doi: 10.1016/j.dnarep.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 48.Hofseth LJ, Khan MA, Ambrose M, et al. The adaptive imbalance in base excision-repair enzymes generates microsatellite instability in chronic inflammation. J Clin Invest. 2003;112(12):1887–1894. doi: 10.1172/JCI19757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reina-San-Martin B, Difilippantonio S, Hanitsch L, Masilamani RF, Nussenzweig A, Nussenzweig MC. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J Exp Med. 2003;197(12):1767–1778. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shalhout S, Haddad D, Sosin A, et al. Genomic uracil homeostasis during normal B cell maturation and loss of this balance during B cell cancer development. Mol Cell Biol. 2014;34(21):4019–4032. doi: 10.1128/MCB.00589-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pettersen HS, Galashevskaya A, Doseth B, et al. AID expression in B-cell lymphomas causes accumulation of genomic uracil and a distinct AID mutational signature. DNA Repair (Amst) 2015;25:60–71. doi: 10.1016/j.dnarep.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 52.Frey S, Bertocci B, Delbos F, Quint L, Weill JC, Reynaud CA. Mismatch repair deficiency interferes with the accumulation of mutations in chronically stimulated B cells and not with the hypermutation process. Immunity. 1998;9(1):127–134. doi: 10.1016/s1074-7613(00)80594-4. [DOI] [PubMed] [Google Scholar]
- 53.Dingler FA, Kemmerich K, Neuberger MS, Rada C. Uracil excision by endogenous SMUG1 glycosylase promotes efficient Ig class switching and impacts on A:T substitutions during somatic mutation. Eur J Immunol. 2014;44(7):1925–1935. doi: 10.1002/eji.201444482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol. 2002;12(20):1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 55.Imai K, Slupphaug G, Lee WI, et al. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat Immunol. 2003;4(10):1023–1028. doi: 10.1038/ni974. [DOI] [PubMed] [Google Scholar]
- 56.Ramiro AR, Jankovic M, Callen E, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440(7080):105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kovalchuk AL, Ansarah-Sobrinho C, Hakim O, et al. Mouse model of endemic Burkitt translocations reveals the long-range boundaries of Ig-mediated oncogene deregulation. Proc Natl Acad Sci USA. 2012;109(27):10972–10977. doi: 10.1073/pnas.1200106109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Miranda NF, Peng R, Georgiou K, et al. DNA repair genes are selectively mutated in diffuse large B cell lymphomas. J Exp Med. 2013;210(9):1729–1742. doi: 10.1084/jem.20122842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Couronné L, Ruminy P, Waultier-Rascalou A, et al. Mutation mismatch repair gene deletions in diffuse large B-cell lymphoma. Leuk Lymphoma. 2013;54(5):1079–1086. doi: 10.3109/10428194.2012.739687. [DOI] [PubMed] [Google Scholar]