Abstract
All organisms universally encode, synthesize and utilize proteins that function optimally within a subset of growth conditions. While healthy cells are thought to maintain high translational fidelity within their natural habitats, natural environments can easily fluctuate outside the optimal functional range of genetically encoded proteins. The hyperthermophilic archaeon Aeropyrum pernix (A. pernix) can grow throughout temperature variations ranging from 70 to 100°C, although the specific factors facilitating such adaptability are unknown. Here, we show that A. pernix undergoes constitutive leucine to methionine mistranslation at low growth temperatures. Low-temperature mistranslation is facilitated by the misacylation of tRNALeu with methionine by the methionyl-tRNA synthetase (MetRS). At low growth temperatures, the A. pernix MetRS undergoes a temperature dependent shift in tRNA charging fidelity, allowing the enzyme to conditionally charge tRNALeu with methionine. We demonstrate enhanced low-temperature activity for A. pernix citrate synthase that is synthesized during leucine to methionine mistranslation at low-temperature growth compared to its high-fidelity counterpart synthesized at high-temperature. Our results show that conditional leucine to methionine mistranslation can make protein adjustments capable of improving the low-temperature activity of hyperthermophilic proteins, likely by facilitating the increasing flexibility required for greater protein function at lower physiological temperatures.
INTRODUCTION
Accurate protein synthesis requires exceptional fidelity in the processes that convert the amino acid sequence specified within DNA into proteins. Although errors occurring during DNA and mRNA synthesis have the potential to cause mistranslation, inaccuracies are far more common during protein synthesis process itself (1). Accurate translation not only requires correct tRNA selection by the ribosome, but also the correct ligation of amino acids to cognate tRNAs by aminoacyl-tRNA synthetases (aaRSs) (2). The catalytic sites and editing domains of aaRSs help ensure that only cognate amino acids are used in the aminoacylation reaction in order to prevent tRNA mischarging and subsequent mistranslation (3).
Although mistranslation is generally considered a deleterious occurrence, mistranslation in mammalian cells can be induced by reactive oxygen species and has been shown to be useful for oxidative stress tolerance (4–6). Furthermore, artificial mistranslation in mycobacteria increases antibiotic resistance (7), while artificially constructed mistranslation in Candida albicans can reduce phagocytotic killing by macrophages and increase antifungal resistance (8–10), and artificial mistranslation has also been shown to increase tolerance to oxidative stress in E. coli (11,12). Additionally, mistranslation has been found during varying culture conditions in Saccharomyces cerevisiae (13), Bacillus subtilis (14) and Mycobacterium smegmatis (7,15), although the benefit of such mistranslation remains unknown.
Genetically encoded proteins from hyperthermophilic organisms have extreme rigidity mediated by salt bridges and strong hydrophobic interactions in order to maintain their structure and function at high temperatures (16,17). Although hyperthermophiles are capable of growth far below the optimal functional temperatures of their genetically encoded proteins (18), low temperatures can inhibit thermophilic enzymes by precluding the degree of flexibility they require to function (19). Here, we discover that the hyperthermophilic archaeon, Aeropyrum pernix (A. pernix) globally mistranslates leucine (Leu) to methionine (Met)—a substitution specifically shown to increase protein flexibility (20,21)—during low-temperature growth where the rigidity of genetically encoded proteins could compromise function. Leu-to-Met mistranslation is facilitated by misacylation of tRNALeu with Met by the methionyl-tRNA synthetase (MetRS). We identify mistranslated A. pernix proteins by mass spectrometry, including Leu-to-Met mistranslated citrate synthase. Remarkably, natural A. pernix citrate synthase synthesized during lower translational fidelity at low growth temperature has greater activity at lower temperatures than its counterpart synthesized during high translational fidelity at high growth temperature. Our results demonstrate the utility of natural mistranslation for adjusting the function of proteins for relevant nonoptimal growth conditions encountered in natural environments without the burden of additional genes.
MATERIALS AND METHODS
A. pernix cultivation
A. pernix was obtained from The Leibniz Institute DSMZ (German Collection of Microorganisms and Cell). A. pernix was grown in a medium containing 1 g Difco yeast extract, 5 g tryptone and 1 g Na2S2O3•5H2O per 1 l of synthetic sea water (34 g/l Sigma sea salts). Yeast extract and tryptone were added to the synthetic sea salt solution and autoclaved. Filter sterilized Na2S2O3•5H2O was then added to the basal medium. Growth occurred at 75 or 90°C, 180 rpm in a shaking glycerol bath in an erlenmeyer flask with a 5:1 flask:medium ratio.
Pulse labeling
A total of 50 ml midlog cultures (OD660 ∼0.2) were pelleted for 5 min at 2500 g and resuspended in the initial culture volume in a medium containing 1 mM of all amino acids – Met and 1 g Na2S2O3•5H2O per 1 l of synthetic sea water and growth continued for 1 h. Cells were pelleted for 5 min at 2500 g and resuspended in 300 μl of the medium supernatant and were pulse labeled with 1 μCi/μl 35S-Met for 4 min at the growth temperature before addition of 400 μl of ice cold 0.3 M NaOAc/HOAc, 10 mM EDTA pH 4.8 and placement on ice. Cells were pelleted briefly at 4°C and washed once in 0.3 M NaOAc/HOAc, 10 mM EDTA pH 4.8. Cells were then lysed in 0.3 M NaOAc/HOAc, 10 mM EDTA, 0.5% SDS pH 4.8. RNA was then extracted from lysed cells a total of three times by adding 400 μl acetate saturated phenol chloroform pH 4.8 followed by 1 min of vortexing and 5 min of centrifugation at 17 000 g at 4°C before ethanol precipitation. RNA pellet was suspended in 10 mM NaOAc/HOAc, 1 mM EDTA pH 4.8.
Microarray analysis
tRNA microarray analysis and controls were performed as previously described (22,23). For cross-hybridization control, 100 pmol of unbound eMet and iMet probes were added to the RNA hybridization mix prior to array loading. For thio-modification control, tRNA was deacylated with 0.1 M Tris-HCl pH 9.0 at 37°C for 30 min before loading on the array. A. pernix specific array was created using tRNA sequences obtained from the Genomic tRNA database (24).
Expression and purification of A. pernix proteins
Recombinant MetRS and citrate synthases were expressed from a pET28a expression plasmid containing the A. pernix gene under IPTG control. Constructs were transformed into BL21 E. coli and cells were grown at 37°C at 200 rpm in terrific broth with 50 μg/ml kanamycin. IPTG (1 mM) was added at OD600 of 0.8 and expression continued for 4 h before harvest. Cells were resuspended in 125 mM NaCl, 25 mM Tris-HCl pH 7.5, 20 mM imidazole before lysis and removal of cell debris by centrifugation. Enzymes were purified via elution from a Ni-NTA column using 75 mM, 200 mM and 500 mM imidazole fractions. Eluted fractions were analyzed via SDS-PAGE and the purest fractions were dialyzed overnight in 125 mM NaCl, 25 mM Tris-HCl pH 7.5. Dialyzed samples were concentrated with centrifugal filters.
Purification of A. pernix tRNAs
Large midlog A. pernix cultures were pelleted by centrifugation and lysed in 0.3 M NaOAc/HOAc, 10 mM EDTA, 0.5% SDS pH 4.8. RNA was extracted by adding an equal volume of acetate saturated phenol chloroform pH 4.8 followed by 1 min of vortexing and 5 min of centrifugation at 17 000 g at 4°C before ethanol precipitation of aqueous phase. tRNA was purified by running RNA pellets on an 8% denaturing polyacrylamide gel and cutting type I and type II tRNA bands identified through UV shadowing. tRNA was eluted in 200 mM KCl/50 mM K-acetate, ethanol precipitated, resuspended in water and refolded prior to the charging reaction.
In vitro aminoacylation reactions
Reactions were performed in 20 μl containing 1 μM MetRS, 10 μg total A. pernix tRNA, and 2.5 μCi/μl 35S-Met, 2 mM ATP, and 1mM DTT in 50 mM HEPES-KOH pH 7.5, 100 mM potassium glutamate and 10 mM magnesium acetate. Reactions were run for 10 min at 75 or 90°C and were stopped by adding 45 μl 0.3 M NaOAc/AcOH, 10 mM EDTA pH 4.8 and an equal volume of acetate saturated phenol/chloroform. Mixture was centrifuged for 5 min at 17 000 g and 4°C before removal of aqueous layer and ethanol precipitation of the RNA and resuspension in 10 mM NaOAc/AcOH, 1 mM EDTA pH 4.8.
Mass spectrometry
Total A. pernix protein sample were prepared by pelleting a midlog A. pernix culture grown at 75°C and washing pellet in 50 mM ammonium bicarbonate (NH4HCO3), 0.5 M NaCl prior to cell lysis in 50 mM ammonium bicarbonate (NH4HCO3) and removal of cell debris by centrifugation. Sample was digested with 1 μg of trypsin at 37°C overnight, acidified and dried before injection into LTQ-Orbitrap Velos Pro (ThermoFisher Scientific, Waltham, MA, USA) coupled with a nanoLC Ultra (Eksigent, Dublin, USA). The protein digests were first loaded onto a trap column and peptide separation was carried out on a C18 column. The Orbitrap mass analyzer was operated in positive ionization mode using collision induced dissociation (CID) to fragment the HPLC separated peptides. All MS/MS samples were analyzed using Protein Prospector (http://prospector.ucsf.edu/). Protein Prospector was set up to search an A. pernix protein library containing protein sequences for all annotated A. pernix genes. Oxidation of methionine, carbamidomethyl of cysteine, N-terminal acetylation and Met loss, and leucine to methionine substitutions were specified as variable modifications. Since the sample was derived from crude lysate, the data set was also searched for spectra with potassium and sodium adducts.
Citrate synthase assays
A. pernix lysates were created by pelleting 75 or 90°C midlog cultures and resuspending the pellets in 125 mM NaCl, 25 mM Tris-HCl pH 7.5. Cell debris was pelleted and supernatants were removed and protein concentration was measure via Bradford assay and was standardized to 200 μg/ml. Spectrophotometric citrate synthase assays were performed in 80 μl reactions containing 500 μM oxaloacetate, 100 μM DTNB, 25 μM Acetyl-CoA, 5 mM triethanolamine-HCl pH 8.0, 100 mM Tris-HCl pH 8.0 as previously described (25). Lysate (5 μl) was added to preheated reaction components (60, 80 or 100°C) and aliquots were removed and placed on ice to stop the reaction. Absorbance at 412 nm was measured for the aliquots and reactions slopes were calculated and converted to citrate synthase enzyme units.
RESULTS
A. pernix misacylates Met to tRNALeu at low growth temperatures
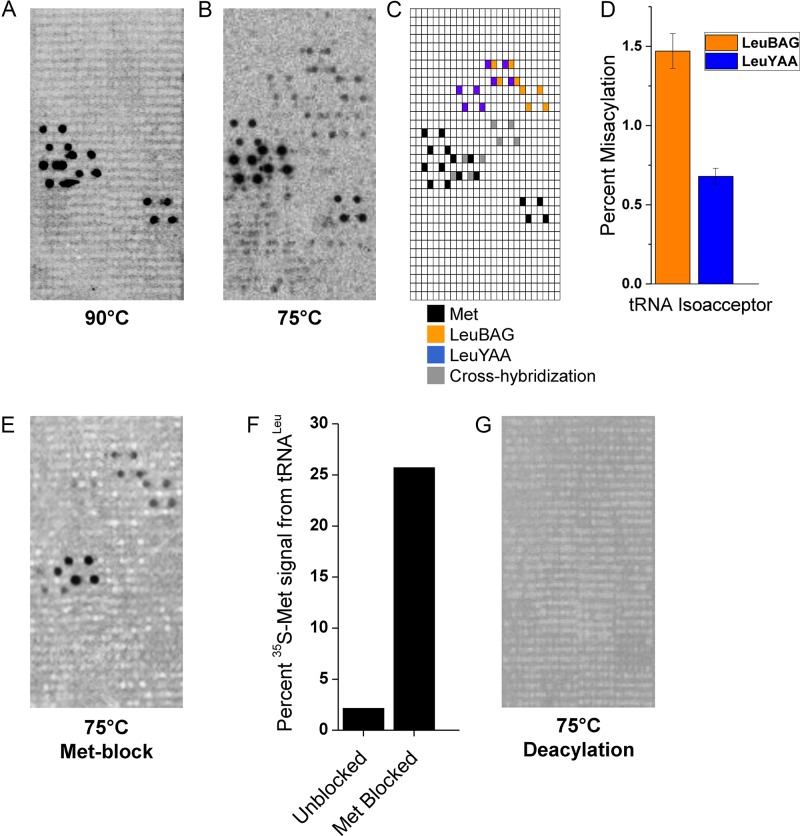
To determine whether mistranslation could be employed during physiologically relevant nonoptimal growth conditions in archaea, we assayed for tRNA misacylation with methionine in the hyperthermophilic archaeon Aeropyrum pernix. A. pernix grows optimally at 90°C, yet it is capable of growth between 70–100°C (26) and we took advantage of this large growth range to determine whether mistranslation played a role in this high level of adaptability. We examined mistranslation occurring at the level of tRNA mischarging using previously established 35S-methionine (Met) pulse labeling in combination with tRNA microarray analysis (5,13). The fidelity of tRNA charging reactions for 35S-Met pulse-labeled samples can be determined by using tRNA microarrays to separate tRNA isoacceptors and subsequent phosphorimaging can determine which tRNAs were aminoacylated with 35S-Met. Using custom designed tRNA microarrays that contain complementary DNA probes for all A. pernix tRNAs, we assayed Met charging fidelity at the optimal 90°C growth temperature and a substantially lower 75°C growth temperature. Remarkably, tRNALeu are specifically misacylated at 75°C, but not at 90°C (Figure 1A–C). Normalizing the total Met signals of the cognate tRNAMets to 100%, we found that ∼1.5% of Met signal is present at the LeuBAG (B = C,G,U) probe that covers the tRNALeuUAG, tRNALeuCAG, tRNALeuGAG isoacceptors and ∼0.6% of Met signal is present at the LeuYAA (Y = C,U) probe that covers the tRNALeuUAA, tRNALeuCAA isoacceptors (Figure 1D). In total, about 2% of total Met is charged to tRNALeu during growth at 75°C. This total misacylation fraction is comparable to those in the mildly stressed mammalian cells (5).
Figure 1.
Mistranslation in A. pernix occurs during low-temperature growth. (A) tRNA microarray showing high-fidelity tRNA charging with Met during growth at 90°C. (B) tRNA microarray showing tRNALeu misacylation with Met during growth at 75°C. (C) Array map showing locations of relevant tRNA probes. (D) Quantification of tRNALeu misacylation from panel B as a percent of tRNAMet signal. (E) tRNA microarray excluding the possibility of Met-tRNA cross-hybridization to Leu probes through addition of free Met probes in hybridization mixture. (F) Quantification of Met blocked panel E and unblocked panel B showing the intensity of tRNALeu signal relative to tRNAMet signal. (G) tRNA microarray excluding the possibility of signal emanating from tRNA thiomodifications by chemical deacylation of charged tRNA prior to array hybridization.
We ensured that 35S-Met signals emanating from tRNALeu probes were not due to tRNAMet cross-hybridization by adding excess free iMet and eMet array probes to the hybridization mixture and observing no change in the tRNALeu signal (Figure 1E). Complete removal of tRNAMet signal was difficult due to its overabundance, so we quantified the signals from the Met blocked array and found that tRNAMet signals are reduced approximately 10-fold, while tRNALeu signals remain unchanged (Figure 1F). Additionally, we excluded the possibility of signal from 35S-thio tRNA nucleotide modifications by chemical deacylation of the amino acids ligated to tRNAs prior to array hybridization (Figure 1G).
A. pernix methionyl-tRNA synthetase (MetRS) specifically misacylates tRNALeu at lower temperatures in vitro
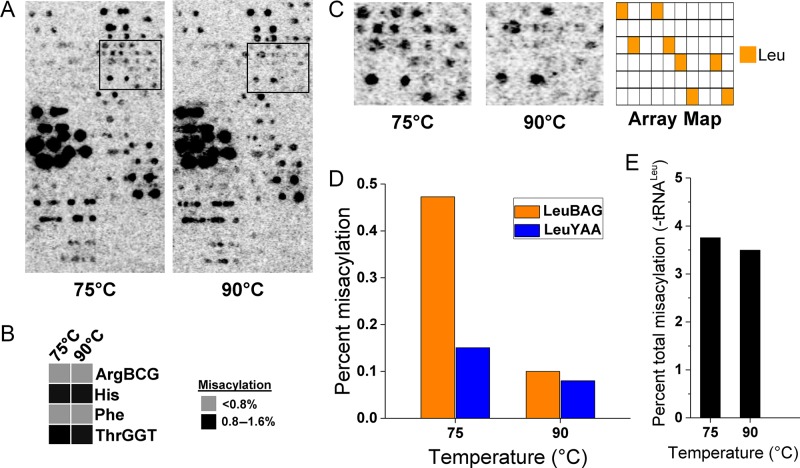
To elucidate the biochemical process responsible for the temperature dependent mischarging of tRNALeu at lower temperature, we characterized the charging fidelity of purified, recombinant A. pernix MetRS using purified total A. pernix tRNA. The recombinant MetRS enzyme misacylates several non-Met tRNAs in vitro at both 75°C and 90°C (Figure 2A and B). However, misacylation of tRNALeu only occurs appreciably during in vitro charging at 75°C, but not 90°C (Figure 2C). We quantified the array signals from tRNALeu (Figure 2D) and found that about 5-fold more Met is charged to LeuBAG tRNAs at 75°C compared to 90°C. Additionally, about 2-fold more Met is charged to LeuYAA tRNAs at 75°C compared to 90°C. Although the A. pernix MetRS significantly mischarges tRNAArgBCG, tRNAHis, tRNAPhe and tRNAThrGGT species in vitro, but not in vivo, this mischarging phenomenon has been observed in vitro by MetRSs from other species (13,27), indicating that additional controls for mischarging are present in vivo. Using Clustal alignment, we were unable to determine any tRNAMet similarities or common identity elements (28) in these mischarged tRNA species that were not present in other non-Met tRNAs not mischarged by MetRS in vitro. Nonetheless, we quantified the total signal from these other non-Met tRNA species and found that they are misacylated identically at 75°C and 90°C (Figure 2E). Taken together, these data suggest that temperature dependent MetRS fidelity plays an important role for the conditional mischarging of tRNALeu at lower temperature in vivo.
Figure 2.
A. pernix methionine tRNA synthetase misacylates tRNALeu at lower temperatures. (A) tRNA microarrays showing tRNAs charged with Met by the recombinant A. pernix MetRS in vitro at 75°C and 90°C. (B) Quantification of in vitro misacylation to individual nonMet-tRNAs not observed in vivo as a percent of tRNAMet signal from panel A. (C) Zoomed array section from panel A showing probes for tRNALeu. (D) Quantification of in vitro tRNALeu misacylation as a percent of tRNAMet signal. All tRNALeu are misacylated approximately 4-fold more at 75°C than at 90°C in vitro. (E) Quantification of total in vitro non-Met tRNA misacylation—excluding tRNALeu—as a percent of tRNAMet signal at 75°C and 90°C.
In vivo Leu-to-Met mistranslated proteins
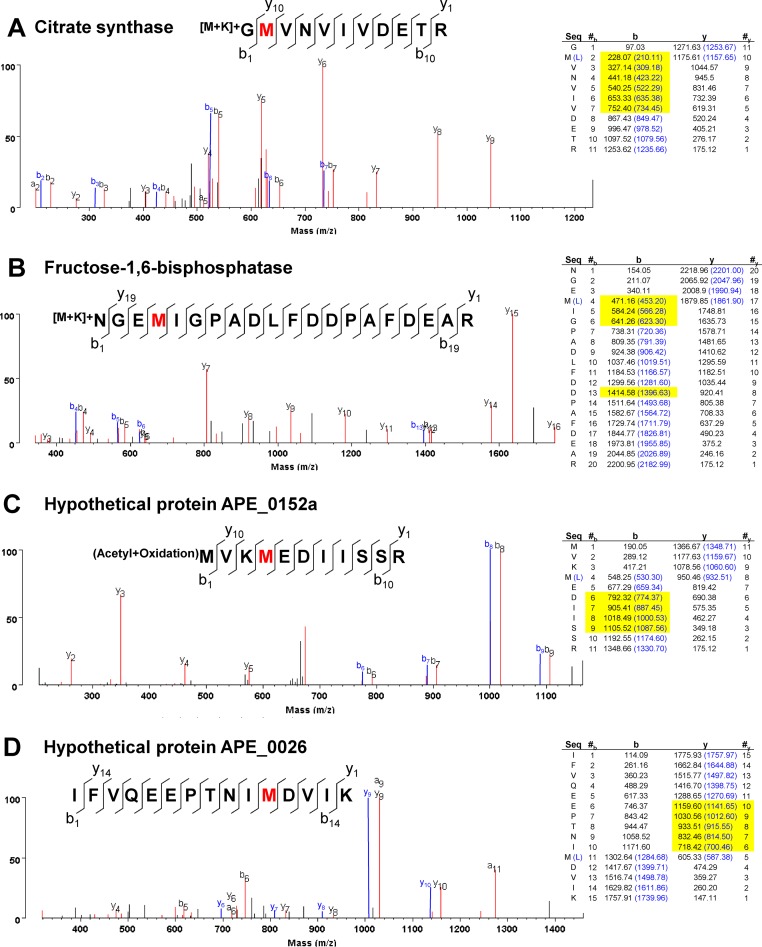
We next verified that mischarged tRNALeu are indeed used in translation via mass spectrometry analysis on whole cell A. pernix lysate growing at 75°C to identify in vivo Leu-to-Met substitutions. We found several Leu-to-Met events within A. pernix cells growing at 75°C within different proteins (Figure 3). Met (MW 149.2) substitution for Leu (MW 131.2) is demonstrated in the 18Da mass shift in the relevant peaks for the mistranslated peptides in the MS/MS spectra. For example, the genomic sequence of the citrate synthase peptide shown in Figure 3A is GLVNVIVDETR. Upon L-to-M substitution, all identifiable b-ions at and C-terminal to the Leu position show an 18Da mass shift (b2–b7). Because of their low abundance, mistranslated peptides are difficult to detect via mass spectroscopy, and quantitation is nearly impossible due to the limitations in proteome coverage. We were able to find 4 high-confidence Leu-to-Met substitutions from 715 spectra that matched to 312 A. pernix proteins. This detection ratio of 0.56% (4/715) is similar to the amount of Met-substituted peptides found in yeast among the proteins in glycolysis and fermentation (0.66%, (13)).
Figure 3.
Mistranslated A. pernix proteins. Total protein mass spectrometry spectra with corresponding ion fragmentation tables from A. pernix grown at 75°C showing Leu-to-Met substitutions in red for citrate synthase (A), fructose-1,6-bisphosphate (B), hypothetical protein APE0152a (C) and hypothetical proteinAPE_0026 (D). Blue peaks in the spectra and blue masses in the ion fragmentation tables show the theoretical data for the genome-encoded, wild-type peptide sequence. The 18Da mass shift associated with Leu-to-Met substitution is illustrated in the differences between the corresponding experimental and theoretical peaks. The identifiable mass shifted peaks are highlighted in yellow.
Of the Leu-to-Met substitutions identified in vivo, we focused on Leu-to-Met substitution in citrate synthase (Figure 3A) to investigate the effects of low-temperature mistranslation on enzymatic activity since this enzyme has already been extensively used as a model for the structural basis of thermostability in hyperthermophiles (29–31). Interestingly, mesophilic proteomes have greater Met content than hyperthermophilic proteomes (32). Similarly, there is greater Met content in the mesophilic citrate synthases compared to hyperthermophilic citrate synthases (29), which substantiates the rationale for Leu-to-Met mistranslation during low-temperature growth in A. pernix.
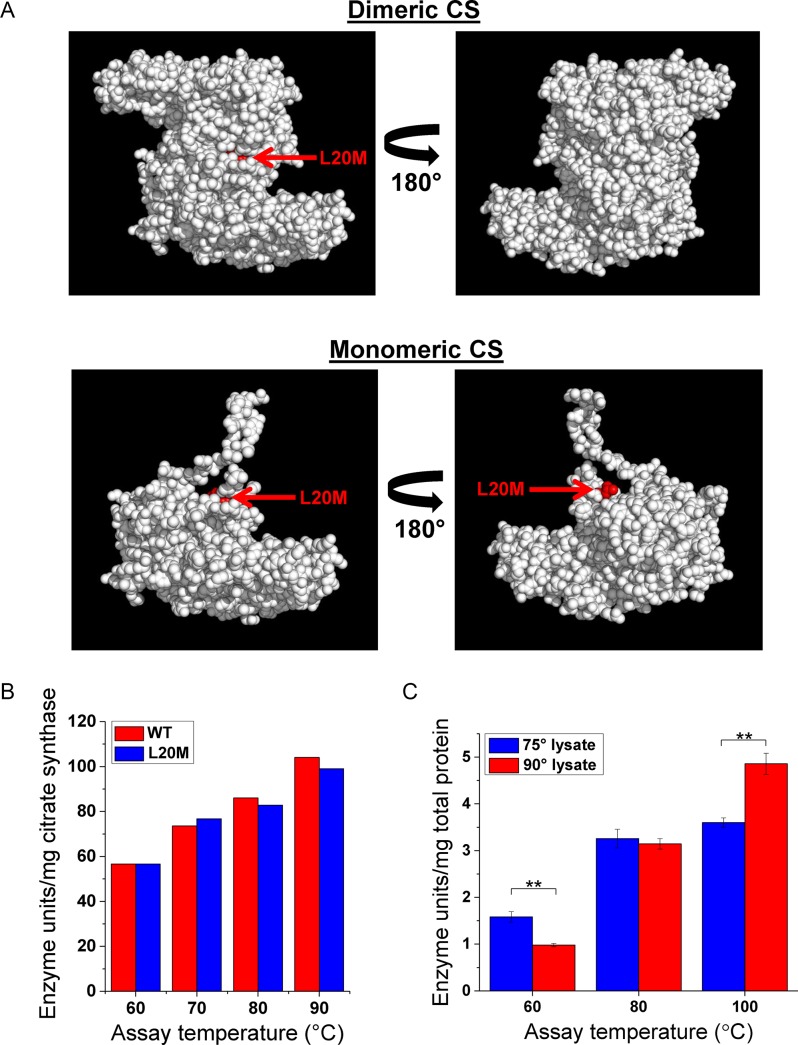
The in vivo citrate synthase Met substitution we detected via mass spectrometry resided at Leu residue 20. To determine approximately where the detected Leu20-to-Met residue resides within citrate synthase, we analyzed the citrate synthase crystal structure from the related crenarchaeon Sulfolobus solfataricus. Citrate synthase from S. solfataricus (and all crystalized citrate synthases from archaea) are homodimers. We mapped the Met substitution identified in A. pernix to the dimerization interface (Figure 4A). This residue becomes almost completely buried once the enzyme dimerizes and may therefore increase flexibility between the citrate synthase subunits by altering subunit contacts through a change from a branched (Leu) to a straight (Met) hydrophobic side chain. Interestingly, mutations at the dimerization interface in an archaeal hyperthermophilic citrate synthase have previously been shown to significantly destabilize the enzyme as well as reduce its temperature optimum for function (33,34).
Figure 4.
Met mistranslation of citrate synthase. (A) Structure of citrate synthase from the related organism Sulfolobus solfataricus showing the particular Leu-to-Met substitution identified by mass spectrometry in red in both the monomeric and dimeric forms. (B) Specific citrate synthase activity of the purified recombinant WT and single L20M mutant citrate synthase across relevant temperatures. (C) Total citrate synthase activity from A. pernix lysates obtained from growth at 75°C or 90°C and assayed at low, moderate and high temperatures (**P < 0.01, Student t-test).
Citrate synthase synthesized during Leu-to-Met mistranslation has greater low-temperature activity
We next tested whether temperature dependent mistranslation could indeed adjust the protein activity in a temperature dependent manner. We first assayed the enzymatic activity of the purified, recombinant A. pernix wild-type and L20M mutant citrate synthases using in vitro assays, and did not find appreciable differences in their enzymatic activities across various physiologically relevant temperatures (Figure 4B). This assay using recombinant proteins, however, is unlikely to reflect the mistranslated citrate synthase enzymes present in A. pernix cells. First, the natural enzyme is likely a mixture of homo and heterodimers of the wild-type and the L20M mutant, whereas the recombinant enzymes are only homodimers. Second and more importantly, the A. pernix citrate synthase has 33 leucine residues and the in vivo Leu-to-Met mistranslated enzymes are likely a diverse mixture of mistranslated protein variants, each containing a Leu-to-Met substitution at distinct locations. Only one variant was detected in our mass spectrometry analysis due to the inherently low coverage of mistranslated peptides.
For these reasons, we performed the same in vitro citrate synthase assay on A. pernix cell lysates obtained from both 75°C and 90°C growth, representing a high and a low mistranslation condition, respectively, in order to determine the citrate synthase activities of the naturally mistranslated enzyme and how this activity is affected by temperature. To establish that there were no significant differences in citrate synthase abundance between the lysates, we ensured the citrate synthase activity was equivalent at 80°C in our assay, which represents a moderate temperature for activity where Leu-to-Met mistranslation should neither be markedly beneficial nor deleterious (Figure 4C). However, when assayed at 60°C, the citrate synthase activity from the 75°C mistranslated lysate is 1.5-fold higher than the 90°C high-fidelity lysate (Figure 4C). Conversely, when assayed at 100°C, the high-fidelity lysate has 1.35-fold higher activity. Targeted mutations in recombinants proteins that compromise protein activity or stability at higher temperatures to create more flexibility and increase activity at lower temperatures have been reported for several other thermophilic enzymes (35–39). The lower citrate synthase activity of the high-fidelity protein at 60°C or the lower activity of the low-fidelity protein at 100°C can be either due to decreased enzyme activities or reduced level of correctly folded proteins or a combination of both. In any case, the net result of activity or folding change is that the mistranslated citrate synthase has a higher activity at lower temperatures. The distinct property of the Leu-to-Met mistranslated citrate synthase demonstrates that mistranslated enzymes can naturally have greater activity at lower physiological temperatures than their genetically encoded counterparts.
DISCUSSION
All organisms have a repertoire of proteins encoded within the genome that help facilitate adaptation to varying conditions. Some organisms also contain multiple genes for the same differentially adapted protein. Although useful, maintaining multiple genes for differential adaptation can be costly and require transcriptional programming to respond to environmental conditions. The alternative, Leu-to-Met mistranslational strategy of altering the genetically encoded proteins described here offers another way for cells to optimize their protein activity to current conditions without the burden of added genes (6). In addition, adaptation via tRNA misacylation can be activated much more quickly because the MetRS can simply respond to temperature in the case of A. pernix and charged tRNAs can turn over in a matter of seconds in cells (40).
Naturally inducible mistranslation via tRNA misacylation with methionine was first described in mammalian cells and was shown to benefit cells under oxidative stress (4,5). Our work here extends a variation of this adaptive mechanism to the archaeal domain and provides another function for Met mistranslation. The Leu-to-Met mistranslation in response to lower temperatures makes a very specific alteration to the hyperthermophilic proteins encoded within the A. pernix genome. High thermostability in proteins is mostly conferred through increased electrostatic and hydrophobic interactions, but the alterations in thermostable proteins that facilitate the rigidity required to maintain protein structure at high temperatures (32) can prevent the requisite flexibility for function at lower temperatures (19). A Met-to-Leu exchange would be capable of disrupting the robust stabilizing interactions within thermophilic proteins, since the Met side chain has a similar volume as Leu, but it is more flexible and has more degrees of freedom. An obvious disadvantage of inducible mistranslation for any amino acid substitution is the potential of generating protein mutants that can form harmful aggregates. This disadvantage may be minimized through the specific use of mistranslation with Met, which has a side chain that is both hydrophobic and polar and therefore can be utilized in both the interior and exterior of proteins.
Leu-to-Met substitutions have already been experimentally shown to decrease protein stability without drastically altering enzymatic activity (20). In fact, numerous single Leu-to-Met substitutions in T4 lysozyme were sufficient to destabilize the protein, and multiple Leu-to-Met substitutions resulted in even greater loss in protein stability (20,21). Furthermore, while rigid hyperthermophilic proteins have reduced Met content (32), extremely flexible psychrophilic proteins—including psychrophilic citrate synthases (41,42)—frequently have greater Met content than their mesophilic counterparts (42,43), which corroborates the role of Met for naturally facilitating protein flexibility (42,44). Moreover, there are numerous examples of destabilizing mutations in thermophilic proteins which can increase catalysis at lower temperatures by compromising stability at higher temperatures (35–39). Therefore, not only are Leu-to-Met substitutions an experimentally validated method to reduce protein stability, but protein destabilization is a validated method to increase enzymatic activity at lower temperatures in thermophilic proteins. Although these biochemical principles have been demonstrated in vitro with recombinant proteins, here we show that A. pernix performs this process naturally to harness the potentially improved activity from protein destabilization when growing at lower temperatures.
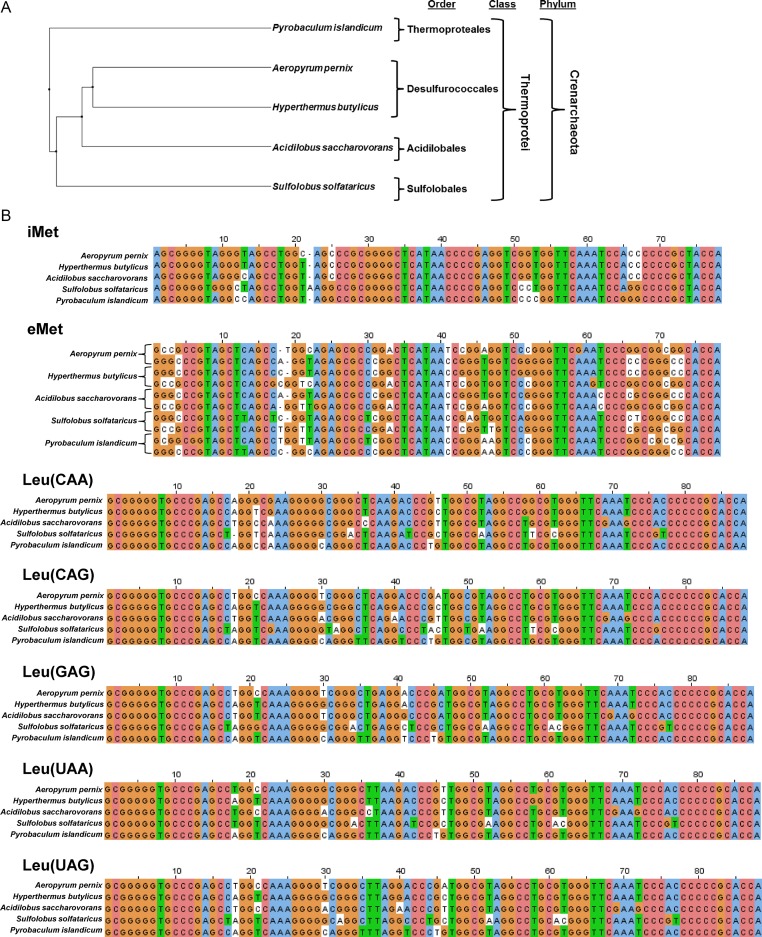
Although it is advantageous for the MetRS to perform conditional tRNALeu acceptance at lower temperatures, it is surprising that the decrease in fidelity is specific for the type II tRNALeu, which contains an extra variable stem-loop (45,46). Previous studies on bacterial MetRSs have shown that the anticodon sequence CAU is the most important identity element for tRNAMet (27,47,48). Consistent with this basis for tRNAMet recognition, tRNALeu could be recognized by the A. pernix MetRS on the basis of anticodon sequences that include CAG and CAA which differ by only one nucleotide from the anticodon of tRNAMet. Whether other identity elements independent of the anticodon sequences also mediate or contribute to tRNALeu recognition by MetRS remains to be determined. Other organisms in the phylum Crenarchaeota which belong to different taxonomic orders have remarkable tRNAMet and tRNALeu sequence similarities with A. pernix (Figure 5). Due to the high level of tRNALeu sequence conservation, the veritable tRNALeu identity elements (28,49) that facilitate their recognition by the A. pernix MetRS may also be present in tRNALeu from other Crenarchaeota, perhaps allowing other hyperthermophiles to perform Leu-to-Met mistranslation as well.
Figure 5.
tRNAMet and tRNALeu conservation in Crenarchaeota. (A) Phylogenetic tree showing the relationship between Crenarchaeota species analyzed for tRNA sequence conservation. (B) tRNAMet and tRNALeu alignments for A. pernix and other Crenarchaeota from panel A.
In summary, we show that A. pernix can optimize proteins to low growth temperature without the burden of additional genes through Leu-to-Met mistranslation. This process is mediated by global Leu-to-Met substitutions in the proteome, which can destabilize the rigid, genetically encoded proteins in order to adopt a more flexible, mesophilic-like proteome. This response seems to be mediated by the temperature-dependent fidelity of the A. pernix MetRS. We show that the citrate synthase from A. pernix synthesized during Leu-to-Met mistranslation is indeed more active at low temperatures, but is compromised at high temperatures likely due to its destabilization. Global Leu-to-Met mistranslation in hyperthermophiles could allow entire proteomes to achieve greater function at lower temperatures, where increased activity is a greater benefit than extreme thermostability.
Acknowledgments
The authors are grateful to Dr Xiaoyun Wang for help with mass spectrometry data analysis and Dr Sophie Alvarez for mass spectrometry sample processing. We also thank Dr Elizabeth Wiltrout for help designing and creating the custom A. pernix microarray and Sarah Pan for performing some enzyme assays.
FUNDING
NIH MCB Training [T32 GM007183 to M.S.] and the NIH Director's Pioneer Award [DP1GM105386 to T.P.]. Funding for open access charge: NIH [DP1GM105386].
Conflict of interest statement. None declared.
REFERENCES
- 1.Reynolds N.M., Lazazzera B.A., Ibba M. Cellular mechanisms that control mistranslation. Nat. Rev. Microbiol. 2010;8:849–856. doi: 10.1038/nrmicro2472. [DOI] [PubMed] [Google Scholar]
- 2.Dale T., Uhlenbeck O.C. Amino acid specificity in translation. Trends Biochem. Sci. 2005;30:659–665. doi: 10.1016/j.tibs.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Ibba M., Soll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 4.Lee J.Y., Kim D.G., Kim B.G., Yang W.S., Hong J., Kang T., Oh Y.S., Kim K.R., Han B.W., Hwang B.J., et al. Promiscuous methionyl-tRNA synthetase mediates adaptive mistranslation to protect cells against oxidative stress. J. Cell Sci. 2014;127:4234–4245. doi: 10.1242/jcs.152470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Netzer N., Goodenbour J.M., David A., Dittmar K.A., Jones R.B., Schneider J.R., Boone D., Eves E.M., Rosner M.R., Gibbs J.S., et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature. 2009;462:522–526. doi: 10.1038/nature08576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan T. Adaptive translation as a mechanism of stress response and adaptation. Annu. Rev. Genet. 2013;47:121–137. doi: 10.1146/annurev-genet-111212-133522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Javid B., Sorrentino F., Toosky M., Zheng W., Pinkham J.T., Jain N., Pan M., Deighan P., Rubin E.J. Mycobacterial mistranslation is necessary and sufficient for rifampicin phenotypic resistance. Proc. Natl. Acad. Sci. U.S.A. 2014;111:1132–1137. doi: 10.1073/pnas.1317580111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miranda I., Silva-Dias A., Rocha R., Teixeira-Santos R., Coelho C., Goncalves T., Santos M.A., Pina-Vaz C., Solis N.V., Filler S.G., et al. Candida albicans CUG mistranslation is a mechanism to create cell surface variation. mBio. 2013;4:1–9. doi: 10.1128/mBio.00285-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miranda I., Rocha R., Santos M.C., Mateus D.D., Moura G.R., Carreto L., Santos M.A. A genetic code alteration is a phenotype diversity generator in the human pathogen Candida albicans. PloS One. 2007;2:e996. doi: 10.1371/journal.pone.0000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bezerra A.R., Simoes J., Lee W., Rung J., Weil T., Gut I.G., Gut M., Bayes M., Rizzetto L., Cavalieri D., et al. Reversion of a fungal genetic code alteration links proteome instability with genomic and phenotypic diversification. Proc. Natl. Acad. Sci. U.S.A. 2013;110:11079–11084. doi: 10.1073/pnas.1302094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan Y., Wu J., Ung M.H., De Lay N., Cheng C., Ling J. Protein mistranslation protects bacteria against oxidative stress. Nucleic Acids Res. 2015;43:1740–1748. doi: 10.1093/nar/gku1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling J., Soll D. Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4028–4033. doi: 10.1073/pnas.1000315107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiltrout E., Goodenbour J.M., Frechin M., Pan T. Misacylation of tRNA with methionine in Saccharomyces cerevisiae. Nucleic Acids Res. 2012;40:10494–10506. doi: 10.1093/nar/gks805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyerovich M., Mamou G., Ben-Yehuda S. Visualizing high error levels during gene expression in living bacterial cells. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11543–11548. doi: 10.1073/pnas.0912989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leng T., Pan M., Xu X., Javid B. Translational misreading in Mycobacterium smegmatis increases in stationary phase. Tuberculosis (Edinb) 2015;95:678–681. doi: 10.1016/j.tube.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 16.Vogt G., Woell S., Argos P. Protein thermal stability, hydrogen bonds, and ion pairs. J. Mol. Biol. 1997;269:631–643. doi: 10.1006/jmbi.1997.1042. [DOI] [PubMed] [Google Scholar]
- 17.Elcock A.H. The stability of salt bridges at high temperatures: implications for hyperthermophilic proteins. J. Mol. Biol. 1998;284:489–502. doi: 10.1006/jmbi.1998.2159. [DOI] [PubMed] [Google Scholar]
- 18.Vieille C., Zeikus G.J. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 2001;65:1–43. doi: 10.1128/MMBR.65.1.1-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vihinen M. Relationship of protein flexibility to thermostability. Protein Eng. 1987;1:477–480. doi: 10.1093/protein/1.6.477. [DOI] [PubMed] [Google Scholar]
- 20.Gassner N.C., Baase W.A., Matthews B.W. A test of the ‘jigsaw puzzle’ model for protein folding by multiple methionine substitutions within the core of T4 lysozyme. Proc. Natl. Acad. Sci. U.S.A. 1996;93:12155–12158. doi: 10.1073/pnas.93.22.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gassner N.C., Baase W.A., Mooers B.H., Busam R.D., Weaver L.H., Lindstrom J.D., Quillin M.L., Matthews B.W. Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability. Biophys. Chem. 2003;100:325–340. doi: 10.1016/s0301-4622(02)00290-9. [DOI] [PubMed] [Google Scholar]
- 22.Dittmar K.A., Sorensen M.A., Elf J., Ehrenberg M., Pan T. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 2005;6:151–157. doi: 10.1038/sj.embor.7400341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zaborske J.M., Narasimhan J., Jiang L., Wek S.A., Dittmar K.A., Freimoser F., Pan T., Wek R.C. Genome-wide analysis of tRNA charging and activation of the eIF2 kinase Gcn2p. J. Biol. Chem. 2009;284:25254–25267. doi: 10.1074/jbc.M109.000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan P.P., Lowe T.M. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reisch A.S., Elpeleg O. Biochemical assays for mitochondrial activity: assays of TCA cycle enzymes and PDHc. Methods Cell Biol. 2007;80:199–222. doi: 10.1016/S0091-679X(06)80010-5. [DOI] [PubMed] [Google Scholar]
- 26.Sako Y., Nomura N., Uchida A., Ishida Y., Morii H., Koga Y., Hoaki T., Maruyama T. Aeropyrum pernix gen. nov., sp. nov., a novel aerobic hyperthermophilic archaeon growing at temperatures up to 100 degrees C. Int. J. Syst. Bacteriol. 1996;46:1070–1077. doi: 10.1099/00207713-46-4-1070. [DOI] [PubMed] [Google Scholar]
- 27.Jones T.E., Alexander R.W., Pan T. Misacylation of specific nonmethionyl tRNAs by a bacterial methionyl-tRNA synthetase. Proc. Natl. Acad. Sci. U.S.A. 2011;108:6933–6938. doi: 10.1073/pnas.1019033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giege R., Sissler M., Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Danson M.J., Hough D.W. Structure, function and stability of enzymes from the Archaea. Trends Microbiol. 1998;6:307–314. doi: 10.1016/s0966-842x(98)01316-x. [DOI] [PubMed] [Google Scholar]
- 30.Wells S.A., Crennell S.J., Danson M.J. Structures of mesophilic and extremophilic citrate synthases reveal rigidity and flexibility for function. Proteins. 2014;82:2657–2670. doi: 10.1002/prot.24630. [DOI] [PubMed] [Google Scholar]
- 31.Bell G.S., Russell R.J., Connaris H., Hough D.W., Danson M.J., Taylor G.L. Stepwise adaptations of citrate synthase to survival at life's extremes. From psychrophile to hyperthermophile. Eur. J. Biochem./FEBS. 2002;269:6250–6260. doi: 10.1046/j.1432-1033.2002.03344.x. [DOI] [PubMed] [Google Scholar]
- 32.Szilagyi A., Zavodszky P. Structural differences between mesophilic, moderately thermophilic and extremely thermophilic protein subunits: results of a comprehensive survey. Structure. 2000;8:493–504. doi: 10.1016/s0969-2126(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 33.Arnott M.A., Michael R.A., Thompson C.R., Hough D.W., Danson M.J. Thermostability and thermoactivity of citrate synthases from the thermophilic and hyperthermophilic archaea, Thermoplasma acidophilum and Pyrococcus furiosus. J. Mol. Biol. 2000;304:657–668. doi: 10.1006/jmbi.2000.4240. [DOI] [PubMed] [Google Scholar]
- 34.Moore V., Kanu A., Byron O., Campbell G., Danson M.J., Hough D.W., Crennell S.J. Contribution of inter-subunit interactions to the thermostability of Pyrococcus furiosus citrate synthase. Extremophiles. 2011;15:327–336. doi: 10.1007/s00792-011-0363-6. [DOI] [PubMed] [Google Scholar]
- 35.Theriot C.M., Semcer R.L., Shah S.S., Grunden A.M. Improving the catalytic activity of hyperthermophilic Pyrococcus horikoshii prolidase for detoxification of organophosphorus nerve agents over a broad range of temperatures. Archaea. 2011;2011:565127. doi: 10.1155/2011/565127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki T., Yasugi M., Arisaka F., Yamagishi A., Oshima T. Adaptation of a thermophilic enzyme, 3-isopropylmalate dehydrogenase, to low temperatures. Protein Eng. 2001;14:85–91. doi: 10.1093/protein/14.2.85. [DOI] [PubMed] [Google Scholar]
- 37.Merz A., Yee M.C., Szadkowski H., Pappenberger G., Crameri A., Stemmer W.P., Yanofsky C., Kirschner K. Improving the catalytic activity of a thermophilic enzyme at low temperatures. Biochemistry. 2000;39:880–889. doi: 10.1021/bi992333i. [DOI] [PubMed] [Google Scholar]
- 38.Roovers M., Sanchez R., Legrain C., Glansdorff N. Experimental evolution of enzyme temperature activity profile: selection in vivo and characterization of low-temperature-adapted mutants of Pyrococcus furiosus ornithine carbamoyltransferase. J. Bacteriol. 2001;183:1101–1105. doi: 10.1128/JB.183.3.1101-1105.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lebbink J.H., Kaper T., Bron P., van der Oost J., de Vos W.M. Improving low-temperature catalysis in the hyperthermostable Pyrococcus furiosus beta-glucosidase CelB by directed evolution. Biochemistry. 2000;39:3656–3665. doi: 10.1021/bi991483q. [DOI] [PubMed] [Google Scholar]
- 40.Jakubowski H., Goldman E. Quantities of individual aminoacyl-tRNA families and their turnover in Escherichia coli. J. Bacteriol. 1984;158:769–776. doi: 10.1128/jb.158.3.769-776.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell R.J., Gerike U., Danson M.J., Hough D.W., Taylor G.L. Structural adaptations of the cold-active citrate synthase from an Antarctic bacterium. Structure. 1998;6:351–361. doi: 10.1016/s0969-2126(98)00037-9. [DOI] [PubMed] [Google Scholar]
- 42.Siddiqui K.S., Cavicchioli R. Cold-adapted enzymes. Annu. Rev. Biochem. 2006;75:403–433. doi: 10.1146/annurev.biochem.75.103004.142723. [DOI] [PubMed] [Google Scholar]
- 43.Thomas T., Cavicchioli R. Archaeal cold-adapted proteins: structural and evolutionary analysis of the elongation factor 2 proteins from psychrophilic, mesophilic and thermophilic methanogens. FEBS Lett. 1998;439:281–286. doi: 10.1016/s0014-5793(98)01375-1. [DOI] [PubMed] [Google Scholar]
- 44.Marshall C.J. Cold-adapted enzymes. Trends Biotechnol. 1997;15:359–364. doi: 10.1016/S0167-7799(97)01086-X. [DOI] [PubMed] [Google Scholar]
- 45.Brennan T., Sundaralingam M. Structlre of transfer RNA molecules containing the long variable loop. Nucleic Acids Res. 1976;3:3235–3250. doi: 10.1093/nar/3.11.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palencia A., Crepin T., Vu M.T., Lincecum T.L. Jr, Martinis S.A., Cusack S. Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase. Nat. Struct. Mol. Biol. 2012;19:677–684. doi: 10.1038/nsmb.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schulman L.H., Pelka H. Anticodon switching changes the identity of methionine and valine transfer RNAs. Science. 1988;242:765–768. doi: 10.1126/science.3055296. [DOI] [PubMed] [Google Scholar]
- 48.Nakanishi K., Ogiso Y., Nakama T., Fukai S., Nureki O. Structural basis for anticodon recognition by methionyl-tRNA synthetase. Nat. Struct. Mol. Biol. 2005;12:931–932. doi: 10.1038/nsmb988. [DOI] [PubMed] [Google Scholar]
- 49.Tocchini-Valentini G., Saks M.E., Abelson J. tRNA leucine identity and recognition sets. J. Mol. Biol. 2000;298:779–793. doi: 10.1006/jmbi.2000.3694. [DOI] [PubMed] [Google Scholar]