Abstract
DNA methylation differences capture substantial information about the molecular and gene-regulatory states among biological subtypes. Enrichment-based next generation sequencing methods such as MBD-isolated genome sequencing (MiGS) and MeDIP-seq are appealing for studying DNA methylation genome-wide in order to distinguish between biological subtypes. However, current analytic tools do not provide optimal features for analyzing three-group or larger study designs. MethylAction addresses this need by detecting all possible patterns of statistically significant hyper- and hypo- methylation in comparisons involving any number of groups. Crucially, significance is established at the level of differentially methylated regions (DMRs), and bootstrapping determines false discovery rates (FDRs) associated with each pattern. We demonstrate this functionality in a four-group comparison among benign prostate and three clinical subtypes of prostate cancer and show that the bootstrap FDRs are highly useful in selecting the most robust patterns of DMRs. Compared to existing tools that are limited to two-group comparisons, MethylAction detects more DMRs with strong differential methylation measurements confirmed by whole genome bisulfite sequencing and offers a better balance between precision and recall in cross-cohort comparisons. MethylAction is available as an R package at http://jeffbhasin.github.io/methylaction.
INTRODUCTION
Differential DNA methylation distinguishes a broad range of biological subtypes including stages of mammalian embryonic development (1), eusocial insect behavioral types (2), immune cell activation and memory (3,4), human tissue types (5), ages of human blood and brain tissue (6,7) and regions of the human brain (8). There is also considerable utility to detecting differential DNA methylation among disease subtypes, particularly in the case of cancer. DNA methylation differences are well established between tumor and normal tissues (9), and more work remains to define differences across inter- and intra- tumoral heterogeneity of human cancers. Moreover, differential DNA methylation has been detected in many other diseases, including schizophrenia (10), obesity (11), epilepsy (12) and rheumatoid arthritis (13), and may delineate subtypes in these diseases as well.
Next generation sequencing techniques, such as MeDIP-seq (14), MethylCap-seq (15) and MBD-isolated genome sequencing (MiGS/MBD-seq) (16), enrich for methylated DNA fragments from genomic DNA for sequencing library construction and provide mapped read abundances that reveal hyper- and hypo- methylation states. These techniques are advantageous for distinguishing biological and clinical subtypes because they are genome-wide and are substantially more cost effective compared to whole genome bisulfite sequencing (WGBS). This is especially true for experimental designs with larger sample sizes involving two or more groups (17). Moreover, MiGS correlates highly with bisulfite-based sequencing assays (18). The methylome produced by MiGS can be paramount in revealing molecular mechanisms that confer different conditions and disease states because recent work has implicated DNA methylation in modulation of transcription factor binding (19), alternative splicing (20–22), alternative promoters (23) and enhancer function (24) in a genomic-context dependent manner (25,26).
Current computational tools for analyzing enrichment-based sequencing data do not address important requirements for studies focused on biological and disease subtypes. These requirements include support for three or more group comparisons, determination of false discovery rates (FDRs) via bootstrapping and the stratification of differentially methylated regions (DMRs) based on frequency of methylation within groups (Table 1). For example, BayMeth uses a Bayesian model to estimate DNA methylation levels but provides no mechanisms for differential testing (27). While MEDIPS provides two-group statistical testing in non-overlapping windows genome-wide (28), it does not provide region-level P-values or solutions to share information between adjacent windows and to reduce the multiple-testing burden. While designed for ChIP-seq data, diffReps employs a two-stage testing approach that does provide differential region detection. However, the method is restricted to two-group comparisons only, and does not have a means to stratify DMRs by frequency with respect to expected methylation status. In contrast to previous methods, MethylAction provides functionality that specifically fulfills the needs of studies of biological subtypes as it detects all possible hyper- and hypo- methylation patterns of frequent and statistically significant DMRs among any number of experimental groups genome-wide.
Table 1. MethylAction uniquely provides statistically significant differential region detection and bootstrapping FDRs for n-group comparisons. Comparison of features relevant for biological subtype studies for existing data processing tools that could be applied to DNA methylation enrichment sequencing data. The tools compared are MethylAction, MEDIPS (28), diffReps (32) and BayMeth (27).
| MethylAction | MEDIPS | diffReps | BayMeth | |
|---|---|---|---|---|
| Intended for MiGS? | Yes | Yes | ChIP-seq | Yes |
| Two-group differential testing? | Yes (region-level) | Yes (window-level) | Yes (region-level) | No |
| n-group differential testing? | Yes | No | No | No |
| Computes bootstrap FDRs? | Yes | No | No | No |
| Implementation | R package | R package | Perl program | R package |
Here, we describe the functionality of MethylAction and demonstrate its utility in a four-group comparison among benign prostatic tissue and three clinically relevant subgroups of prostate cancer. DMRs were detected for all possible differential methylation patterns between the groups, and bootstrapping narrowed these patterns down to four that had FDRs below 10%. An analysis of the bootstraps (resampling with replacement) revealed that the number of iterations performed was adequate to provide stable estimates of the FDRs. By lowering significance thresholds, FDRs could be reduced even further. We then used a comparison between MeDIP-seq data from skin keratinocytes and skin fibroblasts to compare DMRs detected by MethylAction and those detected by two existing tools that are limited to two-group comparisons (MEDIPS and diffReps). MethylAction found more DMRs than MEDIPS and covered nearly all differential windows found by MEDIPS. Regions unique to diffReps contained low effect sizes between conditions and did not reflect large differences in percent methylation measurements from whole genome bisulfite sequencing (WGBS) available for one of the samples. Additionally, we compared DMRs from prostate cancer and colon cancer data sets detected by all three tools to differential methylation cataloged by The Cancer Genome Atlas (TCGA), and established that MEDIPS tends to high precision, diffReps tends to high recall but low precision, and MethylAction can provide a balance between both precision and recall. By providing region-level statistical analysis, bootstrapping of pattern-level FDRs and stratification by DNA methylation frequency in comparisons not limited to two groups, MethylAction is a valuable tool for the determination of DMRs that distinguish biological subtypes and provides an advance over the capabilities of existing programs.
MATERIALS AND METHODS
Detection of differential DNA methylation with methylaction
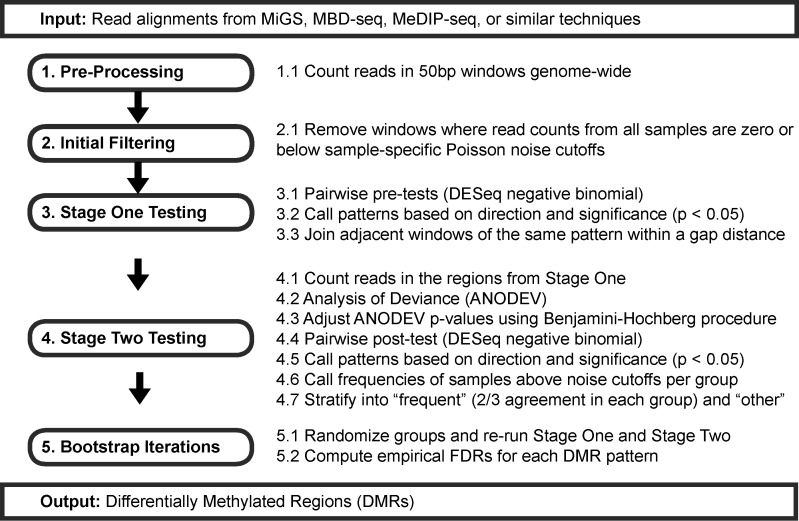
MethylAction is implemented as an R package (http://jeffbhasin.github.io/methylaction) and provides a pre-processing function, a DMR calling function and visualization functions. For pre-processing, MethylAction generates read counts in non-overlapping windows genome-wide. DMR calling involves initial filtering, stage one testing, stage two testing, frequency calling and bootstrapping (Figure 1). Visualization is achieved via export of tracks in BED or BigWig format for the UCSC Genome Browser (29,30), karyograms plotted by ggbio (31) and heatmaps.
Figure 1.
Stages and component steps in MethylAction. Note that the window size, P-value cutoffs, and ‘frequent’ fraction are user-adjustable.
Initial filtering
The genome is divided into equally-sized (50 bp by default) and non-overlapping windows. The number of fragments overlapping each window are then counted to produce a counts matrix. For single-end data this is achieved by extending each sequencing read by a user-provided mean fragment length. For paired-end data, valid mate pairs can be used to establish the fragments used for counting. These read counts are filtered to produce a set of windows deemed to contain signal in at least one sample by removing all windows with either all zero counts or all counts below sample-specific cutoffs. These noise cutoffs are established by generating a histogram of the observed number of windows containing each number of reads (i.e. 1 read, 2 reads, 3 reads, etc.) and comparing the observed number of windows at each read count level to the number expected under a null distribution generated with a Poisson model (16). An FDR is established for each number of reads, and windows with greater than or equal to the lowest level of reads with an FDR of less than 10% (user-adjustable) are considered to contain signal.
Stage one and stage two testing
To fully exploit the genome-wide nature of enrichment-based sequencing data, MethylAction can find differential regions of any length at any location in the assembled and mappable genome. To detect regions of differential methylation across multiple replicated groups, we employed a two-stage testing approach similar to the differential ChIP-seq program diffReps (32) combined with an analysis of deviance (ANODEV). In MethylAction, the first stage performs the negative binomial test from DESeq (33) for each pairwise comparison. Normalization based on library size is also performed using DESeq prior to statistical testing, and these normalized counts are saved for visualization and reporting purposes. The pairwise stage one P-values and the direction of the change are used to detect a pattern, which specifies how the means of all groups relate to each other (i.e. which groups are hyper- or hypo- methylated with respect to each other). Patterns are derived from pairwise comparisons using a decision table (Supplementary Figure S1). By default, a P-value cutoff of 0.05 is used both for the adjusted ANODEV P-values and for the pairwise post-test P-values, and these cutoffs are user-adjustable. Windows with equivalent adjacent patterns, and within a user-specified gap distance (200 bp by default), are then joined to create a set of candidate regions. The coordinates of these regions are then provided to stage two, where reads are re-counted within the region and an ANODEV is performed using DESeq. Because the ANODEV is implemented as a Generalized Linear Model (GLM), adjustment for covariates can be added, enabling adjustment for group variables such as ancestry and for paired subject designs. The ANODEV P-values are adjusted using the Benjamini-Hochberg procedure (34), and pairwise post-tests are performed for all regions with significant P-values (by default, P < 0.05) to confirm their patterns when considered as a contiguous region. The combination of the ANODEV and pairwise testing theoretically allows the method to be used for any number of groups within hardware limitations. In practice, we have tested the method with up to eight groups.
Frequency calling
Finally, DMRs are classified as ‘frequent’ if a user-specified fraction of samples (two thirds by default) within each group have consistent methylation status. DMRs are classified as ‘other’ if the group-wise differences in mean read counts are statistically significant but the DMR either lacks sufficient within-group consistency or does not reflect an expected DMR. Some regions may have statistically significant quantitative differences, but the total read counts across all conditions are either very high or very low, and thus are unlikely to represent changes in DNA methylation. The ‘frequent’ classification was developed to filter these situations and provide a set of DMRs that qualitatively represent an expectation of binary methylation differences. The expectation of methylation used for filtering is determined using the same Poisson cutoff from the initial filtering stage.
Bootstrap or permutation testing
The two-stage approach is advantageous because it can detect regions not specified a priori and controls for error across regions of differences. However, it does not guarantee type I error control across the experiment due to the possible inflation of significance caused by the two rounds of testing (35). We addressed this concern by implementing a rigorous permutation (sampling without replacement) or bootstrapping (sampling with replacement) analysis to empirically quantify the false discovery rates (FDRs) for all patterns of differential methylation detected. This is accomplished by re-running the entire DMR detection procedure through stage one, stage two and frequency calling for randomized sample to group assignments. Empirical FDRs for each pattern are computed by dividing the average number detected in the null cases with a certain pattern by the number of DMRs in the real data with the same pattern. Convenience functions are provided to assist in running, merging and computing FDRs for thousands of permutations across multiple computers or in high performance computing (HPC) environments.
MBD-isolated sequencing (MiGS) data for four-group and two-group comparisons of prostate cancer specimens
MiGS (16) reads were obtained from a study of aggressive prostate cancer deposited in GEO under accession GSE66505 (36). Reads were aligned to hg19 using bowtie2 (37), and reads with MAPQ < 10 were removed. In addition to the benign, low grade and high grade groups provided by this data set, the low grade group was additionally stratified into African American and European American subgroups using the ancestry metadata for the four-group comparison. For the two-group comparison, only the European American samples were used to correspond with the TCGA cohort. In this case, benign samples were compared to the low grade and high grade groups combined together. For both analyses, MethylAction was run using a window size of 50 bp, minimum DMR size of 150 bp, P-value cutoffs of 0.05 for all stages, ancestry as a covariate adjustment, 2/3 as the frequent cutoff, and a join distance of 200 bp.
MAP-seq data for two-group comparisons of colon cancer specimens
Enrichment sequencing reads generated by MAP-seq (38,39) were obtained for paired normal colon and colon cancer samples (40) from GEO accession GSE21442. MethylAction was run using the same settings as for the prostate MiGS data.
MeDIP-seq and WGBS data for two-group comparison of skin cells
MeDIP-seq (14) read alignments for penis foreskin fibroblast primary cells and penis foreskin keratinocyte primary cells were obtained from the Roadmap Epigenomics Project (41). The data consist of samples from three individual donors with each cell type collected from each donor for a total of six MeDIP-seq experiments. Aligned reads in BED format were obtained from GEO accessions GSM707022, GSM941726, GSM958180, GSM707021, GSM941725, GSM958182 and converted to BAM format using bedtools bamtobed (42). The read depths for two donors were greater than those of the third and these samples were downsampled to match the mean of the depth of the two samples from the donor with the lowest depth (42 994 232 reads) using samtools (43). This was to minimize normalization differences potentially confounding DMR comparison between tools. Data were pre-processed using a fragment size of 266 bp, and MethylAction was run using a window size of 50 bp, minimum DMR size of 250 bp, P-value cutoffs of 0.05 for all stages, donor ID as a covariate adjustment, 2/3 as the frequent cutoff and a join distance of 200 bp. Whole genome bisulfite sequencing (WGBS) data were available for both cell types from a single donor only (skin03), and processed percent methylation measurements were obtained from GEO accessions GSM1127120 and GSM1127056 in WIG format.
Two-group DMR detection using MEDIPS and diffReps
Data were pre-processed for MEDIPS (28) using the MEDIPS.createSet() function with the options: extend = 266, shift = 0, window_size = 50 and uniq = FALSE. Differential windows were called for MEDIPS using the MEDIPS.meth() function with the options: p.adjust = fdr, diff.method = edgeR, MeDIP = FALSE, CNV = FALSE and minRowSum = 20. Significant windows were selected using the MEDIPS.selectSig() function with the options: p.value = 0.05 and adj = TRUE. The diffReps (32) Perl script was run using the options: –meth nb, –frag 266, –window 200, –pval 0.05, and differential regions were loaded from the saved report text file into R for analysis. For the colon cancer and prostate cancer data sets, fragment size of 120 bp was used.
Computational performance comparison
Run time and peak RAM usage was measured while running MethylAction, MEDIPS and diffReps using Syrupy (https://github.com/jeetsukumaran/Syrupy). All runs were performed using individual (one node per run) Linux nodes in a cluster environment with 20 CPU cores (Intel Xeon E5–2680v2) and 64GB of RAM available on each. Disk access was provided by a Lustre distributed filesystem. Each run was replicated four times. For MethylAction, 6 cores were used for preprocessing, and 10 were used for the call to methylaction(). For diffReps, –nproc was set to 19.
DMR comparison among MethylAction, MEDIPS and diffReps
While MethylAction and diffReps output regions of statistical significance, MEDIPS produces statistical significance at the window level only. To compare the programs in a two-group setting, we joined all contiguous genomic regions covered by one or more significant output range from any program into a set of consensus DMRs using the reduce() function from the GenomicRanges R package (44). These regions were filtered to those with length 250 bp or greater for the skin data and 150 bp or greater for the prostate and colon cancer data. Then, the number of consensus regions covered by at least one significant output range from each data set were counted and compared to produce Venn diagrams using the VennDiagram R package (45). All heatmaps were plotted using the code from the maHeatmap() function provided by MethylAction. For the skin data, reported percent methylation values for all CpG sites falling within a DMR's coordinates (specified by chromosome, start, end) were averaged. The differences between these averages between fibroblasts and keratinocytes were then computed and distributions plotted for each direction of DMR. For visualization of effect sizes for shared and unique regions to each tool, reads were re-counted in these regions using the getCounts() function from MethylAction.
Cross-cohort and platform precision and recall analysis
Differential DNA methylation in prostate (PRAD) and colon (COAD) cancer present in The Cancer Genome Atlas (TCGA) was considered as a ‘gold standard’ for this comparison. Raw data from the Illumina HumanMethylation 450K microarray were obtained from TCGA (http://tcga-data.nci.nih.gov/). Probes known to be cross-hybridizing (46) or overlapping with a common SNP (minor allele frequency > 1%) were excluded. For the prostate cancer data, only those listed as race ‘white’ (the majority of samples) were used to be more comparable to the European American samples from our own data with respect to genetic background. Normalization was performed using the preprocessFunnorm() function available in the minfi R package (47). Beta-values and M-values were computed for each CpG site (48) and differential methylation was tested using the moderated t-statistics on the M-values with limma (49). A CpG was considered differentially methylated if there was a delta beta of ≥ 0.1 between the tumor and normal means and a Benjamini-Hochberg adjusted P-value < 0.05 reported by limma.
Precision and recall were computed with respect to the array sites, as each site can only overlap with a single DMR, whereas DMRs can overlap with multiple array sites. Each array site was classified as hypermethylated, hypomethylated, or no change based on direction and the significance criteria stated above. For the enrichment sequencing DMRs, the absence of DMR was considered as the no change class. A 3 × 3 confusion matrix tabulating how each classification compares between data sets was constructed for each DMR set (produced by MethylAction, MEDIPS, or diffReps) versus the TCGA-derived classifications. Precision was computed as the fraction of ‘true’ (contained in TCGA) hypermethylation classifications out of all hypermethylation classifications produced by the given tool. Recall was computed as the fraction of ‘true’ (contained in TCGA) hypermethylation classifications out of all hypermethylation classified by the TCGA data.
RESULTS
MethylAction detects subtype-specific DMRs in a four-group comparison of prostate cancer specimens
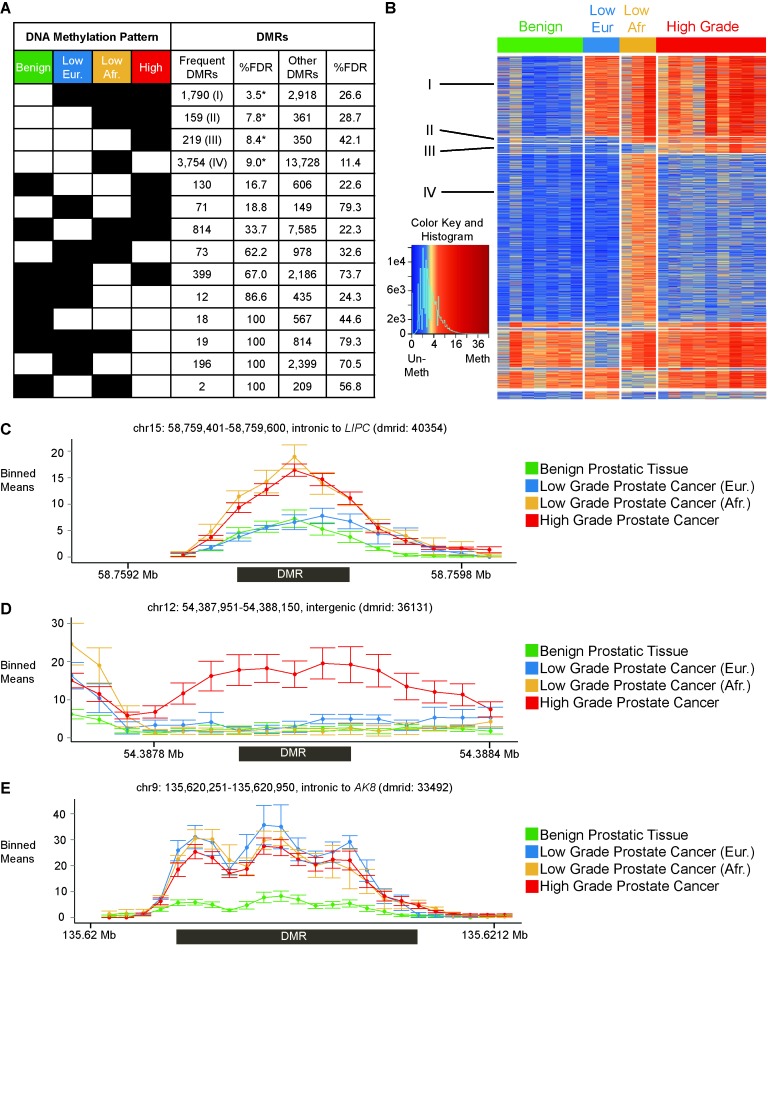
To illustrate the use of MethylAction in investigating biological subtypes, we performed a four-group analysis among benign prostatic tissue and three distinct and clinically relevant subsets of prostate cancer. Histopathologically low grade tumors from African Americans have increased risk of disease recurrence when compared to low grade tumors from European Americans (50). Thus, we used MethylAction to detect regions where DNA methylation is present in both low grade cancers from African Americans and high grade cancers but is absent in low grade samples from European Americans or benign prostatic tissue (Supplementary Table S1). MethylAction found 159 ‘frequent’ DMRs where hypermethlyation is unique to these two groups, and analysis of 2500 bootstraps calculated an FDR of 7.8% (Figure 2A). A notable aspect of MethylAction is that all possible patterns of hyper- and hypo- methylation among the groups are detected with FDRs computed for each pattern (Supplementary Figure S1). This includes both ‘frequent’ DMRs that meet consistency criteria (see Materials and Methods) and any statistical differences in read counts (‘other’ DMRs). The FDR estimates enable prioritization of patterns least likely to occur by chance in the data set. In this case, other interesting patterns from the ‘frequent’ subset with FDRs below 10% include hypermethylation shared by all three cancer groups (1790 DMRs, 3.5% FDR), hypermethylation unique to high grade disease (219 DMRs, 8.4% FDR) and hypermethylation unique to low grade tumors from African Americans (3754 DMRs, 9.0% FDR). The sample-level sequencing data for all DMRs can be plotted on a heatmap, which visualizes the results of the pattern and frequency calling performed by MethylAction (Figure 2B). Example plots of mean read levels among the four groups at loci of interest demonstrate the performance of the method in capturing DMRs that are both specific to certain groups with known clinical distinctions from the others (Figure 2C and D) and shared among all disease subgroups (Figure 2E). Additionally, DMRs can be spatially visualized by creating a karyogram (Supplementary Figure S2).
Figure 2.
MethylAction detects differentially methylated regions (DMRs) that distinguish among benign prostatic tissue and three clinically relevant subgroups of prostate cancer. (A) Number of DMRs detected for all possible patterns of hyper- (black squares) and hypomethylation (white squares). The table is sorted by false discovery rates (FDRs) that are the result of 2500 bootstraps. Patterns with FDR < 10% are indicated with an asterisk. ‘Frequent’ DMRs require the methylation status of two thirds or more of the samples in a group to agree. (B) Heatmap of read count distributions for all ‘frequent’ DMRs detected, ordered by pattern as in (A). Patterns with FDR < 10% are indicated with numerals corresponding to those indicated in (A). Columns represent samples, and rows represent DMRs. Normalized read counts have been divided by the window size and square root-transformed for visualization purposes. (C) Example hypermethylation DMR that is shared between high grade and African low grade. The x-axis represents genomic coordinates, and the y-axis represents normalized read counts. The read counts are plotted as the mean ± standard error for 50 bp non-overlapping windows. The region of the DMR called by MethylAction is indicated by the box under the x-axis. (D) Example hypermethylation DMR that is specific to high grade. (E) Example hypermethylation DMR that is shared by European low grade, African low grade and high grade.
Bootstrapping determines empirical false discovery rates (FDRs) for each pattern of differential methylation among four groups
A unique aspect of MethylAction is built-in support for permutation (sampling without replacement) or bootstrapping (sampling with replacement) of the entire DMR calling procedure (Table 1). To establish confidence in our FDR estimates, we re-sampled the 2500 iterations into smaller subsets and demonstrated that 2500 iterations were more than sufficient to produce stable estimates of FDRs for the ‘frequent’ DMR subset (Supplementary Figure S3A). These estimates of FDRs can be used to justify lower P-value cutoffs, and we found that some ‘frequent’ patterns with FDRs greater than 10% can be filtered at lower ANODEV P-values to produce subsets of DMRs with lower FDRs if desired (Supplementary Figure S3B). Thus, sufficient bootstrapping to attain stable estimates of FDRs is essential in comparisons involving more than two groups, as the FDRs aid in prioritization of the most robust patterns and can be used to select more significant subsets of DMRs if FDRs are unacceptably high at the default ANODEV P-value cutoff of 0.05.
MethylAction detects more DMRs consistent with expectations of differential methylation when compared with existing tools
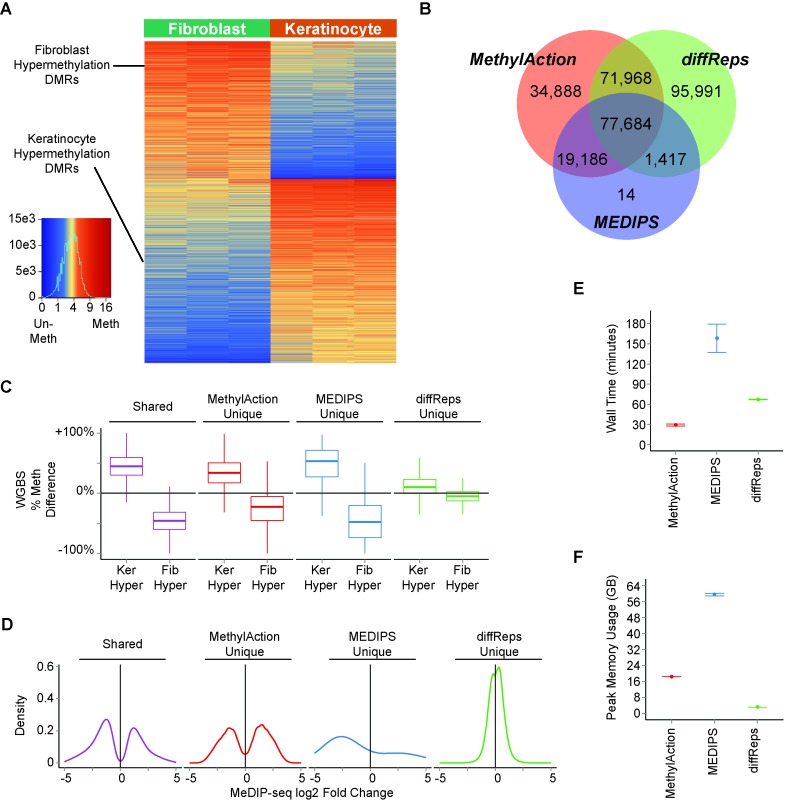
Because the existing tools, diffReps and MEDIPS, are limited to two-group analyses, we selected a two-group comparison of MeDIP-seq data between skin fibroblasts and skin keratinocytes (41) to compare the differential methylation results generated by these three tools. This comparison highlights the use of DMR detection to compare between developmental lineages, which are also known to have opposing gene expression signatures with relevance for understanding the process of wound healing (51). MethylAction detected 100 239 DMRs hypermethylated in keratinocytes and 104 031 DMRs hypermethylated in fibroblasts. The ‘frequent’ subset of 28 179 keratinocyte hypermethylation DMRs and 21 001 fibroblast hypermethylation DMRs are visualized in Figure 3A. Note that bootstrapping was not performed due to the paired nature of the comparison, wherein each type of cell was obtained from the same set of donors.
Figure 3.
Comparison among DMRs detected by MethylAction, MEDIPS and diffReps between MeDIP-seq data for skin fibroblasts and skin keratinocytes. (A) Heatmap of read count distributions for all ‘frequent’ DMRs detected by MethylAction. Columns represent samples, and rows represent DMRs. Normalized read counts have been divided by the number of windows in the DMR and square root-transformed for visualization purposes. (B) Venn diagram of the number of outputted differential regions unique to each or in common among all of the three tools. The DMR sets from all three analysis results were reduced into a set of consensus regions to enable the comparison. Both the ‘frequent’ and the ‘other’ DMRs from MethylAction were used. (C) Boxplots showing the distribution of the difference in percent methylation as measured by whole genome bisulfite sequencing (WGBS) in one of the skin donors for shared DMRs and DMRs unique to each tool. Differences were computed as % methylation in keratinoctyes minus % methylation in fibroblasts. (D) Distributions of log2 fold changes in MeDIP-seq reads between fibroblasts and keratinocytes for consensus regions shared by all three tools or unique to each tool. (E) Comparison of total time elapsed (wall time) for a complete run of each tool. Values shown are the mean±SEM of four separate program executions. (F) Peak RAM usage (the maximum RAM usage over the course of program execution when sampled in 1 second increments) for each tool shown as mean±SEM of four separate program executions.
We first compared the genomic ranges of all DMRs output by MethylAction (both the ‘frequent’ and ‘other’ DMRs) to the differential regions produced by the other tools. MethylAction DMRs did not greatly differ in width or CpG density distributions between the ‘frequent’ and ‘other’ groups (Supplementary Figure S4). Regions from all three tools were reduced into consensus regions, and the comparison revealed 77 684 regions common among all three tools (Figure 3B). Nearly all regions detected by MEDIPS were also detected by the other two tools, as only 14 regions were unique to the MEDIPS set. There were a large number of regions both unique to diffReps (95 991) and MethylAction (34 888). This raises the question of how many of these regions unique to each tool represent actual differences in DNA methylation and how strong the differences are in these regions with respect to fold changes.
Because simply detecting more DMRs does not indicate a DMR set is more reflective of true differential methylation, we compared percent methylation values from whole genome bisulfite sequencing (WGBS) data available for one of the three skin cell donors among the unique regions reported by each tool (Figure 3C). The regions shared between all three tools had high differences in percent methylation: a median of 45% more in keratinocytes versus fibroblasts for keratinocyte hypermethylation DMRs and a median of 46% less in keratinocytes versus fibroblasts for fibroblast hypermethylation DMRs. While the large number of regions unique to MethylAction and the much smaller number unique to MEDIPS had median differences of 18% or more, the regions unique to diffReps had very low percent differences (9% and –4%) with distributions closely overlapping no change. This analysis indicates that the DMRs unique to MethylAction represent validated changes in DNA methylation that are not detected by the other two tools. When considering the fold changes between sequencing reads for the two groups, the fold changes for the DMRs unique to MethylAction are comparable to those for the DMRs shared by all tools (Figure 3D). In contrast, the diffReps regions show very small effect sizes, consistent with these regions not representing differential methylation in the WGBS sample. The performance of MethylAction is not at the expense of computational timing, as MethylAction had the shortest runtime of all three tools on our hardware (Figure 3E). While MethylAction had a higher peak RAM usage than diffReps, it used considerably less RAM than our run of MEDIPS (Figure 3F). Thus, MethylAction detected a set of DMRs that was more comprehensive than MEDIPS, show methylation changes when compared to WGBS data, and have fold changes in the MeDIP-seq data comparable to the regions shared by all three tools.
MethylAction DMRs provide an improved balance between cross-cohort and cross-platform precision and recall in comparison to existing tools
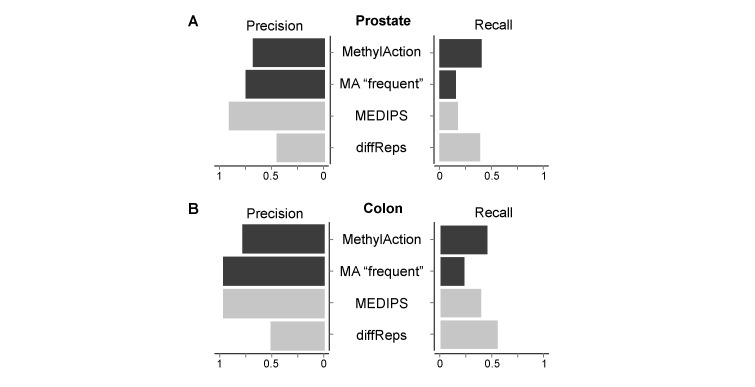
To compare the accuracy of each tool in a controlled setting, we performed a cross-cohort and cross-platform comparison between two enrichment-sequencing cohorts and methylation microarray data from The Cancer Genome Atlas (TCGA). Using all three tools, DMRs were detected and compared for both a prostate cancer and colon cancer cohort (Supplementary Figure S5). Differential methylation from the TCGA cohort was considered as the gold standard, and precision (fraction of differential regions called by the tool that are differential in TCGA) and recall (fraction of regions that are differential in TCGA that are called by the tool) of cancer hypermethylation classification was calculated for MethylAction, MEDIPS and diffReps. For the prostate data set (Figure 4A), MEDIPS had the highest precision (0.92), followed by ‘frequent’ MethylAction DMRs (0.76), and all MethylAction DMRs (0.69). By comparison, diffReps had much lower precision (0.46), yet had the second highest recall (0.4). While the set of all MethylAction DMRs achieved a recall comparable to diffReps (0.41), the MEDIPS DMRs had much lower recall (0.18). The ‘frequent’ MethylAction DMRs were similar in performance, though with somewhat less precision and recall (0.16) than MEDIPS. In colon cancer, MethylAction finds 28 278 DMRs not detected by MEDIPS while having only a slight reduction in precision and achieving a recall closer to that of diffReps (Figure 4B and Supplementary Figure S5D). Therefore, in a cross-cohort and platform comparison, MethylAction is capable of achieving a better balance between precision, recall and total number of DMRs than existing tools.
Figure 4.
Precision and recall calculations for hypermethylation detection for each tool when comparing DMRs from enrichment sequencing cohorts to data from The Cancer Genome Atlas (TCGA). (A) Precision (fraction of ‘true’ hypermethylation events out of all hypermethylation called by the tool, where ‘true’ indicates agreement with TCGA) and recall (fraction of ‘true’ hypermethylation events out of all hypermethylation events called in the TCGA data) fractions for each tool between a prostate cancer MiGS cohort and the TCGA PRAD cohort. (B) Precision and recall fractions for each tool between a colon cancer MAP-seq cohort (40) and the TCGA COAD cohort.
DISCUSSION
MethylAction is a valuable tool for identifying DMRs that can distinguish biological subtypes, including clinically relevant disease subtypes. This is accomplished by providing analyses lacking in existing tools, namely, support for any number of groups, bootstrap FDRs and stratification by methylation frequency within groups. In a four-group comparison among benign prostate and three clinical subtypes, MethylAction detected all possible patterns of DMRs between the groups. Bootstrap FDRs were essential for narrowing the patterns down to those that are the most robust and establishing confidence in the DMR detection. Comparisons involving more than two groups are of increasing interest due to the need to discover possible molecular drivers of disease subtypes for precision medicine. Exceeding the two-group limit can also be powerful for studies of normal physiology and development, where multiple cell types from the same tissue can be compared in order to elucidate possible epigenetic regulators of cell fate.
We demonstrated the ability of MethylAction to detect more DMRs that more likely represent biologically relevant differential DNA methylation in both cancer biology and developmental contexts and across multiple different enrichment sequencing protocols for data produced by different laboratories. However, these comparisons to existing tools are restricted to be two-group comparisons by the fact that MEDIPS and diffReps are limited to this study design. However, there are interesting biological questions to explore in each example involving subgroups that are not tractable with the existing tools. For example, MethylAction can analyze tumors containing different somatic mutations or analyze melanocyte data alongside the fibroblasts and keratinocytes in a single analysis and statistical framework. Because of the regional nature of the DMRs, simply running these tools for all pair-wise comparisons and reducing them is non-trivial, and would also increase the multiple testing burden in the absence of an ANOVA or ANODEV-style approach such as that taken by MethylAction.
Our head-to-head comparisons demonstrated the importance of MethylAction's region-based testing approach. MEDIPS only tests for differential reads within windows, which creates a large burden of multiple testing that reduces power and likely explains the smaller number of DMRs detected by this tool. Moreover, MEDIPS does not support consideration of the paired design of the skin and colon cohorts, which can cause a loss of power. MethylAction still performs the generalized linear model (GLM)-based ANODEV in the two-group cases, which allows for covariate adjustments. While diffReps uses a two-stage testing approach (without performing an ANODEV) to attain significance across differential regions, it is designed for ChIP-seq data and does not have faculties for stratifying the results based on expectations of differential DNA methylation. While diffReps finds a large number of regions that are not reported by MethylAction, these regions have very low fold changes in MeDIP-seq reads between samples. Our analysis of WGBS data for both cell types from one of the donors indicates that these only rarely represent true changes in DNA methylation status, whereas the regions unique to MethylAction represent more robust differences. By implementing a pre-filtering approach based on Poisson thresholds and performing two stages of hierarchical testing, MethylAction takes a data-driven approach to reduce the multiple testing burden. While this may inflate type I error rates (35), we quantify this effect by providing permutations and bootstraps to compute empirical FDRs, which is feasible for the large prostate cancer cohort that can be divided into more than two groups. Then, by post-filtering to groups that are the most consistent and assigning the ‘frequent’ classification, MethylAction defines a subset of DMRs with lower FDRs. Compared to diffReps and MEDIPS, MethylAction outputs a set of DMRs that more closely match expectations of qualitative differential DNA methylation, and this expectation is supported by comparison to WGBS data.
MethylAction is also able to achieve a favorable balance between precision and recall when DMRs are compared to a gold standard. Here, we used differentially methylated CpGs detected in methylation microarray data from TCGA for both prostate cancer and colon cancer. In the case of prostate cancer, MEDIPS had very high precision but low recall, whereas diffReps had higher recall at the expense of precision. MethylAction, on the other hand, had slightly higher recall than diffReps while maintaining a precision of 0.69. In the case of colon cancer, MethylAction was more comparable to MEDIPS, as MEDIPS had a higher recall in this example. The ‘frequent’ stratification of MethylAction was also able to provide an increase of 0.18 in precision, highlighting the usefulness of considering both subsets for different data sets. It is important to note that the majority of DMRs detected by MethylAction from enrichment sequencing were not even assayed by the array design, as only 9071 out of 24 788 total DMRs (36.6%) overlapped with one or more site on the array. This underscores the benefit of the genome-wide data set produced by enrichment sequencing which can enable the detection of DNA methylation changes at distal regulatory elements. The use of TCGA data as a gold standard here is an arbitrary choice, and it should be noted that lack of perfect recall or precision is not necessarily a methodological defect of the DMR detection software, and can represent biological and technical differences between the different cohorts and platforms. By comparing the tools head-to-head, we establish that for the subset of DMRs that do overlap with the TCGA sites, MethylAction can perform as well as or better than the existing tools. This implies that MethylAction likely also has comparable performance for the additional regions not covered by the arrays and in cohorts involving multiple groups that the other tools are not designed to process.
As the importance of epigenetics in understanding the molecular differences between biological conditions continues to grow, cost-effective detection of DMRs will be essential. DMRs can serve not only as biomarkers but can also reveal functional differences between conditions that change gene regulation via a multitude of mechanisms (52). Recent advances in epigenetic editing have demonstrated the use of TAL effectors in both methylating (53) and de-methylating (54) specific loci in the genome and have unlocked opportunities for targeted functional testing of DMRs. Thus, the combination of MeDIP-seq/MiGS and DMR detection among biological subtypes using MethylAction provides a substantial solution for research seeking molecular determinants of clinical phenotypes. Such studies will lead to advances both in precision medicine and our understanding of the function of DNA methylation in regulating genes and conferring phenotypes.
Supplementary Material
Acknowledgments
The authors acknowledge the Roadmap Epigenomics Consortium and the TCGA Research Network for generating data used in this report.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Cancer Institute of the National Institutes of Health [R01CA154356 to A.T., F31CA195887 to J.M.B]. J.M.B. is a predoctoral student in the Molecular Medicine PhD Program of the Cleveland Clinic Lerner College of Medicine of Case Western Reserve University, funded in part by the ‘Med into Grad’ initiative of the Howard Hughes Medical Institute (HHMI). Funding for open access charge: National Institutes of Health [R01CA154356].
Conflict of interest statement. None declared.
REFERENCES
- 1.Smith Z.D., Chan M.M., Mikkelsen T.S., Gu H., Gnirke A., Regev A., Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyko F., Foret S., Kucharski R., Wolf S., Falckenhayn C., Maleszka R. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 2010;8:e1000506. doi: 10.1371/journal.pbio.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komori H.K., Hart T., LaMere S.A., Chew P.V., Salomon D.R. Defining CD4 T cell memory by the epigenetic landscape of CpG DNA methylation. J. Immunol. 2015;194:1565–1579. doi: 10.4049/jimmunol.1401162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohkura N., Hamaguchi M., Morikawa H., Sugimura K., Tanaka A., Ito Y., Osaki M., Tanaka Y., Yamashita R., Nakano N., et al. T Cell Receptor Stimulation-Induced Epigenetic Changes and Foxp3 Expression Are Independent and Complementary Events Required for Treg Cell Development. Immunity. 2012;37:785–799. doi: 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 5.Lokk K., Modhukur V., Rajashekar B., Martens K., Magi R., Kolde R., Koltsina M., Nilsson T.K., Vilo J., Salumets A., et al. DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome Biol. 2014;15:r54. doi: 10.1186/gb-2014-15-4-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spiers H., Hannon E., Schalkwyk L.C., Smith R., Wong C.C.Y., O'Donovan M.C., Bray N.J., Mill J. Methylomic trajectories across human fetal brain development. Genome Res. 2015;25:338–352. doi: 10.1101/gr.180273.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ladd-Acosta C., Pevsner J., Sabunciyan S., Yolken R.H., Webster M.J., Dinkins T., Callinan P.A., Fan J.-B., Potash J.B., Feinberg A.P. DNA Methylation Signatures within the Human Brain. Am. J. Hum. Genet. 2007;81:1304–1315. doi: 10.1086/524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varley K.E., Gertz J., Bowling K.M., Parker S.L., Reddy T.E., Pauli-Behn F., Cross M.K., Williams B.A., Stamatoyannopoulos J.A., Crawford G.E., et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wockner L.F., Noble E.P., Lawford B.R., Young R.M., Morris C.P., Whitehall V.L.J., Voisey J. Genome-wide DNA methylation analysis of human brain tissue from schizophrenia patients. Transl. Psychiatry. 2014;4:e339. doi: 10.1038/tp.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benton M.C., Johnstone A., Eccles D., Harmon B., Hayes M.T., Lea R.A., Griffiths L., Hoffman E.P., Stubbs R.S., Macartney-Coxson D. An analysis of DNA methylation in human adipose tissue reveals differential modification of obesity genes before and after gastric bypass and weight loss. Genome Biol. 2015;16:8. doi: 10.1186/s13059-014-0569-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller-Delaney S.F.C., Das S., Sano T., Jimenez-Mateos E.M., Bryan K., Buckley P.G., Stallings R.L., Henshall D.C. Differential DNA methylation patterns define status epilepticus and epileptic tolerance. J. Neurosci. 2012;32:1577–1588. doi: 10.1523/JNEUROSCI.5180-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y., Aryee M.J., Padyukov L., Fallin M.D., Hesselberg E., Runarsson A., Reinius L., Acevedo N., Taub M., Ronninger M., et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol. 2013;31:142–147. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacinto F.V., Ballestar E., Esteller M. Methyl-DNA immunoprecipitation (MeDIP): hunting down the DNA methylome. Biotechniques. 2008;44:35–43. doi: 10.2144/000112708. [DOI] [PubMed] [Google Scholar]
- 15.Brinkman A.B., Simmer F., Ma K., Kaan A., Zhu J., Stunnenberg H.G. Whole-genome DNA methylation profiling using MethylCap-seq. Methods. 2010;52:232–236. doi: 10.1016/j.ymeth.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 16.Serre D., Lee B.H., Ting A.H. MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 2010;38:391–399. doi: 10.1093/nar/gkp992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aberg K.A., McClay J.L., Nerella S., Xie L.Y., Clark S.L., Hudson A.D., Bukszár J., Adkins D., Consortium S.S., Hultman C.M., et al. MBD-seq as a cost-effective approach for methylome-wide association studies: demonstration in 1500 case–control samples. Epigenomics. 2012;4:605–621. doi: 10.2217/epi.12.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bock C., Tomazou E.M., Brinkman A.B., Müller F., Simmer F., Gu H., Jäger N., Gnirke A., Stunnenberg H.G., Meissner A. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat. Biotechnol. 2010;28:1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee D.-S., Shin J.-Y., Tonge P.D., Puri M.C., Lee S., Park H., Lee W.-C., Hussein S.M.I., Bleazard T., Yun J.-Y., et al. An epigenomic roadmap to induced pluripotency reveals DNA methylation as a reprogramming modulator. Nat. Commun. 2014;5:5619. doi: 10.1038/ncomms6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yearim A., Gelfman S., Shayevitch R., Melcer S., Glaich O., Mallm J.-P., Nissim-Rafinia M., Cohen A.-H.S., Rippe K., Meshorer E., et al. HP1 is involved in regulating the global impact of DNA methylation on alternative splicing. Cell Rep. 2015;10:1122–1134. doi: 10.1016/j.celrep.2015.01.038. [DOI] [PubMed] [Google Scholar]
- 21.Maunakea A.K., Chepelev I., Cui K., Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013;23:1256–1269. doi: 10.1038/cr.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shukla S., Kavak E., Gregory M., Imashimizu M., Shutinoski B., Kashlev M., Oberdoerffer P., Sandberg R., Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maunakea A.K., Nagarajan R.P., Bilenky M., Ballinger T.J., D'Souza C., Fouse S.D., Johnson B.E., Hong C., Nielsen C., Zhao Y., et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hon G.C., Rajagopal N., Shen Y., McCleary D.F., Yue F., Dang M.D., Ren B. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat. Genet. 2013;45:1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 26.Lay F.D., Liu Y., Kelly T.K., Witt H., Farnham P.J., Jones P.A., Berman B.P. The role of DNA methylation in directing the functional organization of the cancer epigenome. Genome Res. 2015;25:467–477. doi: 10.1101/gr.183368.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riebler A., Menigatti M., Song J.Z., Statham A.L., Stirzaker C., Mahmud N., Mein C.A., Clark S.J., Robinson M.D. BayMeth: improved DNA methylation quantification for affinity capture sequencing data using a flexible Bayesian approach. Genome Biol. 2014;15:R35. doi: 10.1186/gb-2014-15-2-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lienhard M., Grimm C., Morkel M., Herwig R., Chavez L. MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinforma. Oxf. Engl. 2014;30:284–286. doi: 10.1093/bioinformatics/btt650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The Human Genome Browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kent W.J., Zweig A.S., Barber G., Hinrichs A.S., Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin T., Cook D., Lawrence M. ggbio: an R package for extending the grammar of graphics for genomic data. Genome Biol. 2012;13:R77. doi: 10.1186/gb-2012-13-8-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen L., Shao N.-Y., Liu X., Maze I., Feng J., Nestler E.J. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PloS One. 2013;8:e65598. doi: 10.1371/journal.pone.0065598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- 35.Lun A.T.L., Smyth G.K. De novo detection of differentially bound regions for ChIP-seq data using peaks and windows: controlling error rates correctly. Nucleic Acids Res. 2014;42:e95. doi: 10.1093/nar/gku351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhasin J.M., Lee B.H., Matkin L., Taylor M.G., Hu B., Xu Y., Magi-Galluzzi C., Klein E.A., Ting A.H. Methylome-wide sequencing detects DNA hypermethylation distinguishing indolent from aggressive prostate cancer. Cell Rep. 2015;15:S2211–S1247. doi: 10.1016/j.celrep.2015.10.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skene P.J., Illingworth R.S., Webb S., Kerr A.R.W., James K.D., Turner D.J., Andrews R., Bird A.P. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol. Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Illingworth R., Kerr A., DeSousa D., Jørgensen H., Ellis P., Stalker J., Jackson D., Clee C., Plumb R., Rogers J., et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Illingworth R.S., Gruenewald-Schneider U., Webb S., Kerr A.R.W., James K.D., Turner D.J., Smith C., Harrison D.J., Andrews R., Bird A.P. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 2010;6:e1001134. doi: 10.1371/journal.pgen.1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roadmap Epigenomics Consortium. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen H., Boutros P.C. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 2011;12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y., Lemire M., Choufani S., Butcher D.T., Grafodatskaya D., Zanke B.W., Gallinger S., Hudson T.J., Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics Off. J. DNA Methylation Soc. 2013;8:203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fortin J.-P., Labbe A., Lemire M., Zanke B.W., Hudson T.J., Fertig E.J., Greenwood C.M., Hansen K.D. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014;15:503. doi: 10.1186/s13059-014-0503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du P., Zhang X., Huang C.-C., Jafari N., Kibbe W.A., Hou L., Lin S.M. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamoah K., Deville C., Vapiwala N., Spangler E., Zeigler-Johnson C.M., Malkowicz B., Lee D.I., Kattan M., Dicker A.P., Rebbeck T.R. African American men with low-grade prostate cancer have increased disease recurrence after prostatectomy compared with Caucasian men. Urol. Oncol. 2015;33:e15. doi: 10.1016/j.urolonc.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marionnet C., Pierrard C., Vioux-Chagnoleau C., Sok J., Asselineau D., Bernerd F. Interactions between fibroblasts and keratinocytes in morphogenesis of dermal epidermal junction in a model of reconstructed skin. J. Invest. Dermatol. 2006;126:971–979. doi: 10.1038/sj.jid.5700230. [DOI] [PubMed] [Google Scholar]
- 52.Schübeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 53.Bernstein D.L., Le Lay J.E., Ruano E.G., Kaestner K.H. TALE-mediated epigenetic suppression of CDKN2A increases replication in human fibroblasts. J. Clin. Invest. 2015;125:1998–2006. doi: 10.1172/JCI77321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maeder M.L., Angstman J.F., Richardson M.E., Linder S.J., Cascio V.M., Tsai S.Q., Ho Q.H., Sander J.D., Reyon D., Bernstein B.E., et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat. Biotechnol. 2013;31:1137–1142. doi: 10.1038/nbt.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.