Abstract
Progenitor-B cells recombine their immunoglobulin (Ig) loci to create unique antigen receptors. Despite a common recombination machinery, the Ig heavy and Ig light chain loci rearrange in a stepwise manner. We studied pre-pro-B cells and Rag−/− progenitor-B cells to determine whether Ig locus contraction or nuclear positioning is decisive for stepwise rearrangements. We found that both Ig loci were contracted in pro-B and pre-B cells. Igh relocated from the nuclear lamina to central domains only at the pro-B cell stage, whereas, Igκ remained sequestered at the lamina, and only at the pre-B cell stage located to central nuclear domains. Finally, in vitro induced re-positioning of Ig alleles away from the nuclear periphery increased germline transcription of Ig loci in pre-pro-B cells. Thus, Ig locus contraction juxtaposes genomically distant elements to mediate efficient recombination, however, sequential positioning of Ig loci away from the nuclear periphery determines stage-specific accessibility of Ig loci.
INTRODUCTION
During differentiation in bone marrow, each progenitor-B cell assembles a unique immunoglobulin molecule (Ig) through genetic recombination of V, D and J genes in their Ig heavy chain (Igh) and Ig light chain loci (Igκ or Igλ) (1,2). The lymphoid-specific recombination activating gene proteins 1 and 2 (Rag1 and Rag2) are crucial in this process through induction of double strand DNA breaks at recombination signal sequences (RSS) flanking each V, D and J gene (3,4). The V(D)J recombination of Ig is initiated in uncommitted pre-pro-B cells by formation of incomplete Dh–Jh rearrangements followed by rearrangement of Vh to DJh junction in pro-B cells. Subsequently, V to J gene rearrangements in the Ig light chain loci are induced in pre-B cells with expression of functional Ig μ molecules on the cell surface (1,2).
Although the core machinery for V(D)J recombination is identical for the different loci, each locus undergoes recombination in a developmental stage-specific manner. This stage-specific accessibility of the Ig loci for the V(D)J recombination machinery has been extensively addressed and is thought to involve 3 epigenetic processes: DNA and chromatin modifications, nuclear positioning and locus contraction (1,5–6).
It is well-established that non-coding RNA transcription (6,7), and epigenetic modifications of histones and/or DNA (8,9) modulate gene accessibility. Ig genes poised for V(D)J recombination are wrapped around histones that are extensively acetylated and methylated (10,11). Importantly, stage-specific trimethylation of lysine 4 in H3 (H3K4me3) in Ig genes can directly recruit Rag2 (12–14).
During cellular maturation, genes can change their nuclear ‘neighborhoods’ by re-positioning from repressive sites to transcriptionally active compartments, and vice versa (9,15–17). Relocation of genes in the nucleus toward transcription factories occurs through extruding of decondensed chromatin loops into interchromosomal space and intermingling with neighboring chromosome territories (9,15,18). The nuclear periphery represents a repressive region through tethering of chromosomal domains to nuclear lamins, thereby creating lamina associated domains (LADs) (16,17). The nuclear positioning of Ig loci also seems tightly regulated, as they are positioned centrally in committed B-cell progenitors, while in non-B cells they are located at the nuclear periphery (17,19–20).
Committed B-cell progenitors show large-scale Ig locus contraction to provide a diverse antigen receptor repertoire (7,21–27). Initially, Ig locus contraction was thought to only occur at the stage in which the locus undergoes recombination (26,27). However, recent observations indicate that the Igκ locus is contracted in both pro-B and pre-B cells (28,29) with similar levels of long-range interactions (29–31). Since Igκ gene rearrangements rarely occur prior to the pre-B-cell stage (32), the question arises whether Ig locus contraction is decisive for V(D)J recombination.
Here we have examined how Ig locus contraction and nuclear positioning are associated with the stepwise control of V(D)J recombination. We found that nuclear localization rather than Ig locus topology is closely linked with the developmental regulation of Igh and Igκ locus assembly.
MATERIALS AND METHODS
Mice
Rag1−/−, Rag2−/−, Vh81X transgenic, human IgM transgenic, κE1/E2mut and EμR mice were obtained (33–38) and kept on a C57BL/6 background under specific pathogen-free conditions. Mice were euthanized at 5–13 weeks of age. The experiments were performed on pooled cells from 2–4 mice. Experimental procedures were reviewed and approved by the Erasmus University committee of animal experiments.
Cell culture and flow cytometry
CD19+ B cells were enriched from femoral bone marrow suspensions by magnetic separation (Miltenyi Biotec), and cultured for 2–3 weeks in Iscove's Modified Dulbecco's medium containing 10% fetal calf serum, 200 U/ml penicillin, 200 mg/ml streptomycin, 4nM L-glutamine, 50 μM β-mercaptoethanol, and 2 ng/ml of both IL-7 and SCF. E2A−/− hematopoietic progenitors were grown as described previously (39).
Flow cytometric immunophenotyping of bone marrow suspensions and cultured progenitor cells was performed after staining with B220-PerCP-Cy5.5 (RA3–6B2), CD19-APC-Cy7 (1D3), CD43-APC (S7), CD2-PE (RM2–5; all from BD Biosciences) on an LSRII flow cytometer (BD Biosciences) and analyzed with BD FACSDiva Software.
Probe preparation and 3D DNA fluorescence in situ hybridization (FISH)
BAC clones CT7–526A21, RP23–24I12, CT7–34H6 for detecting regions in the murine Igh locus (27), and RP23–234A12, RP24–475M8, RP23–435I4 recognizing regions within murine Igκ locus (all from BACPAC Resources) were used as FISH probes. Probe labeling and DNA FISH were performed as described previously (27,40–41). Images were acquired on a Leica SP5 confocal microscope (Leica Microsystems), followed by deconvolution and analysis with Huygens Professional software (Scientific Volume Imaging) (40,41). Details of the procedures are available in the supplementary Materials and Methods.
Circular chromosome conformation capture with high-throughput sequencing (3C-Seq)
3C libraries were prepared from cultured E2A−/−, Rag1−/− and Rag1−/−Vh81X B-cell progenitors and wild type fetal liver erythroid progenitors (29–30,42), and interactions were studied with 4 Igh and 4 Igκ viewpoints (primers sequences are available in the Supplementary Table S1). More details about the 3C-Seq procedure and data analysis can be found in the supplementary Materials and Methods.
Gene expression profiling
The expression profiles of Rag1−/− pro-B and Rag1−/−Vh81X pre-B cells were previously generated with Affymetrix Mouse Gene 1.0 ST Arrays (29,30), and obtained from the Gene Expression Omnibus (GEO; accession number GSE53896).
Inhibition of histone deacetylase activity
For inhibition of histone deacetylase activity, E2A−/− pre-pro-B cells were incubated at 37°C for 10 h with trichostatin A (TSA, Sigma-Aldrich) or DMSO at 3 ng/ml (Igh) or 10 ng/ml (Igκ) concentration (43,44). Next, 3D DNA FISH was performed with the Alexa-568-labeled CT7–34H6 probe (Ch), Cy5-labeled RP23–435I4 probe (Cκ) and MarinaBlue-labeled nuclear lamina. Germline transcripts from the iEμ and iEκ enhancers were quantified with a TaqMan-based RQ-PCR. More details are available in the supplementary Materials and Methods.
Statistics
Statistical significance was calculated with the non-parametric Mann-Whitney U test or the χ2 test. The Mann-Whitney U test was used to analyze statistical significance between two groups of data following unknown (not normal) distribution, whereas χ2 test was used to compare the distribution of two data sets of two groups. All statistical tests were performed using GraphPad Prism version 5.0. P values <0.05 were considered as significant.
RESULTS
Igh locus contraction in pro-B and pre-B cells
To study the role of 3D organization of the Igh locus in the stepwise Igh and Igκ gene rearrangement processes, we employed 3D DNA FISH in uncommitted pre-pro-B cells, and in committed pro-B cells and pre-B cells. Each of these subsets was obtained from specific mouse models to enable studies on the Igh and Igκ loci in their germline configuration. Uncommitted pre-pro-B cells (B220+CD19-CD43+; Figure 1A) were cultured from E2A−/− mice and did not contain complete Vh to DJh, nor incomplete Dh to Jh rearrangements (39). Pro-B cells were derived from Rag1 or Rag2-deficient mice and expressed CD19 and CD43 in absence of Ig gene rearrangements (35). Pre-B cells were derived from transgenic mice expressing a functional murine (Vh81X) or human Igh μ chain on a Rag-deficient background or from Rag-deficient pro-B cells transduced with human Igh μ chain. These cells express a pre-BCR, while their endogenous Ig loci are preserved in germline configuration and remain non-functional (34,36).
Figure 1.
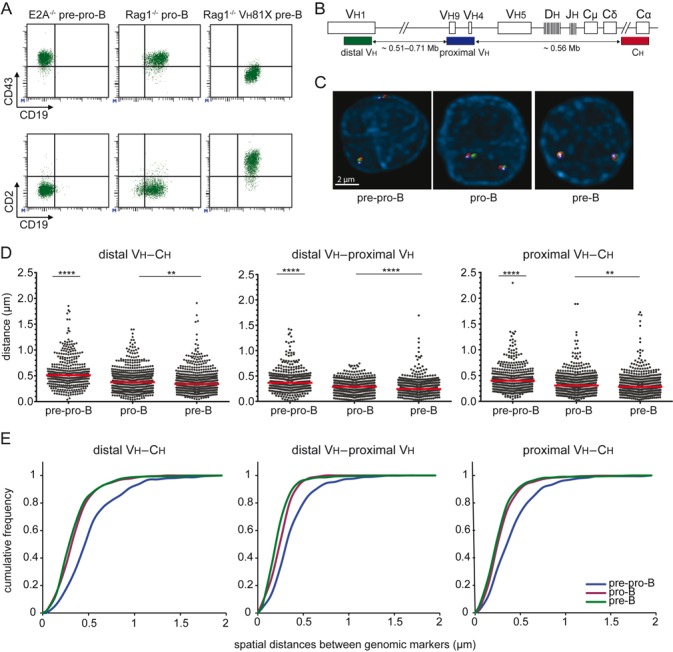
The Igh locus is contracted in pro-B and pre-B cells. (A) Flow cytometric analysis of cultured precursor-B cells from E2A−/−, Rag1−/− and Rag1−/− Vh81X mice confirmed their differentiation block at the pre-pro-B, pro-B and pre-B cells stage, respectively (45). (B) Schematic representation of the murine Igh locus. Bacterial artificial chromosome clones used as 3D FISH probes are indicated (27). The distal Vh probe was conjugated with Alexa488, the proximal Vh probe with Cy5 and the Ch probe with Alexa568. The indicated distance separating each of the three probes and their positions within the Igh locus were determined from the Ensembl mouse genome database. (C) Representative FISH images of Igh loci in the different populations. (D) Scatter plots show the spatial distances separating the FISH probes with red lines representing median distances. 2–4 mice were used for each condition and at least 200 alleles were analyzed per population. The non-parametric Mann-Whitney test was used to calculate significance levels: **, P < .01; ****, P < .0001. (E) Cumulative frequency plots showing the distribution of spatial distances between the distal Vh, proximal Vh and Ch probes in the 3 B-cell subsets.
To determine Igh locus contraction during B-cell development (45), we measured spatial distances between 3 BAC probes hybridized to distal Vh, proximal Vh and Ch regions of the Igh locus (Figure 1B–C). Spatial distances between each of these three regions were significantly shorter in pro-B cells than in pre-pro-B cells, in line with previous findings (25–27). More interestingly, we found that the spatial distances measured in pre-B cells were also significantly shorter than in pre-pro-B cells, and were similar to the spatial distances in pro-B cells (Figure 1D–E). The contraction was not affected by the choice of model, because the distances were similar between Rag1−/− and Rag2−/− progenitor-B cells, and between Vh81X and hIgM transgenics (Supplementary Figure S1A–B). Furthermore, transduction of hIgM into cultured Rag2−/− pro-B cells also did not affect contraction (Supplementary Figure S1C). The Igh locus was also contracted in freshly isolated pro-B cells, excluding effects of culture in the spatial organization of Igh (Supplementary Figure S1A).
Thus, in line with previous observations, the Igh locus is contracted in pro-B cells, but, contrary to previous observations (26), it was not decontracted in pre-B cells. The 3D chromatin organization might therefore facilitate locus accessibility for recombination, however, it does not seem decisive for closure of Igh in pro-B cells.
Igκ locus contraction in pro-B and pre-B cells is independent of the intronic κ enhancer (iEκ)
We next evaluated in our model system with germline Ig genes whether the Igκ locus was contracted in pre-B cells only (26), or already in pro-B cells (28,29). 3D DNA FISH was performed with 3 BAC probes detecting distal Vκ, proximal Vκ and Cκ regions (Figure 2A–B). The spatial distances between the Igκ probes were similar between pro-B cells and pre-B cells as previously published (42), and significantly shorter than in pre-pro-B cells (Figure 2C–D). Igκ contraction was similar between Rag1−/−, Rag2−/−, Vh81X, hIgM transgenic and hIgM transduced progenitor-B cells, and freshly isolated pro-B cells, excluding effects of genetic background or cell culture (Supplementary Figure S1D–1E).
Figure 2.
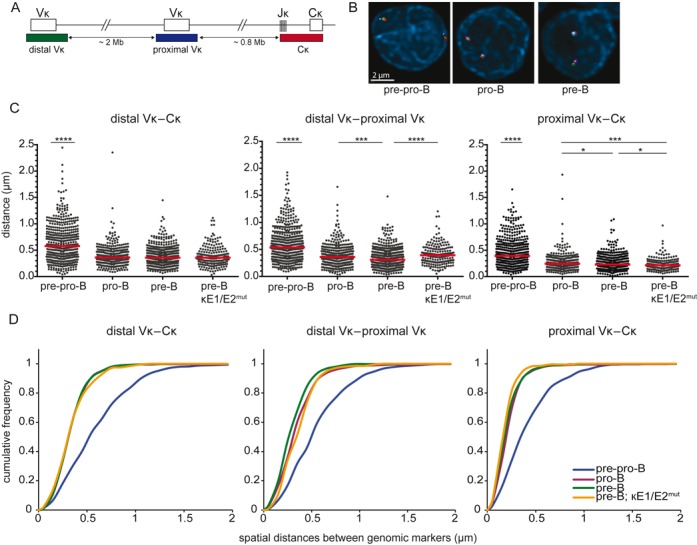
Igκ locus contraction in pro-B and pre-B cells is independent of iEκ. (A) Schematic representation of the murine Igκ locus including the bacterial artificial chromosome clones used as 3D FISH probes: distal Vκ – Alexa488, proximal Vκ – Cy5, Cκ – Alexa568. The indicated distances separating each of the 3 probes and their positions within the Igκ locus were determined from the Ensembl mouse genome database. (B) Representative images of the Igκ locus in the 3 B-cell subsets. (C) Scatter plots show the distances between distal Vκ, proximal Vκ, and Cκ regions in cultured E2A−/− pre-pro-B, Rag1−/− pro-B, Rag1−/− Igμ pre-B and Rag2−/− κE1/E2mut pre-B cells. For each condition 2–3 mice were used and at least 200 alleles were analyzed per population. Red horizontal lines represent median distances. The non-parametric Mann-Whitney test was used to calculate significance levels: *, P < .05; ***, P < .001; ****, P < .0001. (D) Cumulative frequency plots showing the distribution of spatial distances between the distal Vκ, proximal Vκ and Cκ region probes in the 4 progenitor-B cell subsets.
The intron enhancers in Igh (iEμ) and Igκ (iEκ) are known to contribute to stage-specific Ig gene rearrangements. Mice carrying homozygous mutations in E-box motifs in the iEκ element (κE1/E2mut mice) show reduced efficiencies of Igκ rearrangements (38), while Igκ alleles with targeted replacement of iEκ by iEμ (EμR mice) already rearrange at the pro-B-cell stage (37). To study whether Igκ locus contraction can take place in the absence of a functional iEκ, we crossed κE1/E2mut mice on the Rag2−/− Igμ background, and performed 3D DNA FISH with the three Igκ BAC probes. The spatial distances measured between Igκ BAC probes in κE1/E2mut pre-B cells were similar to pre-B cells with a wildtype iEκ and were significantly shorter than in pre-pro-B cells (Figure 2C–D). Additional analysis of κE1/E2mut pro-B and EμR pro-B cells also revealed full contraction of these Igκ loci (Supplementary Figure S2). These results suggest that contraction of the Igκ locus in pro-B and pre-B cells does not critically depend on a functional iEκ. Thus, the Igκ locus undergoes locus contraction in pro-B cells and remained contracted in pre-B cells, independent of iEκ functionality.
Long-range interactions within the Igh and Igκ loci
To further study the nature of the contracted Igh and Igκ loci in pro-B and pre-B cells, we analyzed interactions within these Ig loci using Chromosome Conformation Capture and sequencing (3C-Seq). Viewpoints in iEμ and iEκ were selected and their long-range interactions were studied in non-lymphoid erythroid progenitors, E2A−/− pre-pro-B, Rag1−/− pro-B and Rag1−/− Vh81X pre-B cells.
The bulk of interactions for iEμ were found with sites on other chromosomes or other regions on chromosome 12 outside of Igh (Figure 3A). Still many interactions were confined to the 3.5 Mb Igh locus (Figure 3B), creating its own discrete topologically associated domain (TAD) (46). This confinement of interactions within the Igh locus was also found with 3 additional viewpoints located in the distal Vh, proximal Vh and 3′RR regions (Supplementary Figure S3A–B). Still, pro-B and pre-B cells contained clearly more interactions between iEμ and Vh regions than pre-pro-B cells and erythroid progenitors (Figure 3A–B). Moreover, pro-B cells carried specific interactions between iEμ and the distal Vh region (Figure 3B), whereas in pre-B cells these were mainly found between iEμ and proximal Vh genes (Figure 3B). To study whether these interactions preferentially involved Vh genes, we determined the frequencies of interactions with fragments that contained a Vh gene (+Vh gene) or not (–Vh gene; Figure 3C). Of the 869 BglII fragments within the Vh region, only a minority contained one or more of the 110 Vh genes. B-lineage cells showed more interactions between iEμ and the entire Vh region than erythroid progenitors. Erythroid progenitors displayed low amounts of interactions to the Vh region (Figure 3C). These numbers were significantly higher in pre-pro-B-cells and the highest numbers were found in pro-B and pre-B cells. Moreover, the committed pro-B and pre-B cells showed relatively more interactions with +Vh fragments than uncommitted pre-pro-B cells. Thus, committed B-cell precursors showed frequent interactions between iEμ and Vh genes, with distal Vh interactions being more frequent in pro-B than in pre-B cells.
Figure 3.
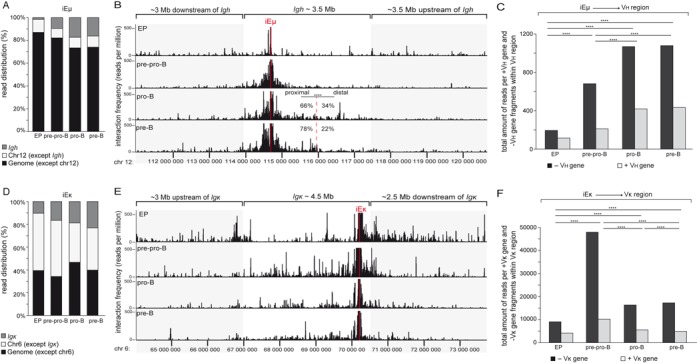
Long-range interactions within the Igh and Igκ loci. (A) Bar graphs showing the relative distribution of 3C-Seq reads in the Igh locus, the rest of chromosome 12 (chr12 except Igh) and rest of the genome (genome except chr12). Read distributions across the genome were analyzed for iEμ enhancer viewpoint in erythroid progenitors (EP), cultured E2A−/− pre-pro-B, Rag1−/− pro-B and Rag1−/− Vh81X pre-B cells. Indicated cell fractions were obtained from 4 mice. Data represent average of two biological replicates. (B) 3C-Seq long-range interactions of the iEμ viewpoint along ∼10 Mb range of chromosome 12 were plotted as reads per million for erythroid progenitors (EP), E2A−/− pre-pro-B, Rag1−/− pro-B and Rag1−/− Vh81X pre-B cells. The frequency of reads within proximal Vh and distal Vh regions are presented for Rag1−/− pro-B and Rag1−/− Vh81X pre-B cells as% of total interactions within the entire Vh region. The χ2 test was used to calculate significance level. ****, P < .0001. (C) Bar graphs showing total amount of 3C-Seq interactions within the Vh region of Igh analyzed for BglII fragments that contain Vh genes (+Vh gene) or not (–Vh gene). Bars represent sum of interactions. The χ2 test was used to calculate significance levels between +Vh gene and -Vh gene fragments of two cell types. ****, P < .0001. (D) Bar graphs showing the relative distribution of 3C-Seq reads in the Igκ locus, the rest of chromosome 6 (chr6 except Igκ) and rest of the genome (genome except chr6). Read distributions across the genome were analyzed for iEκ enhancer viewpoint in EP, cultured E2A−/− pre-pro-B, Rag1−/− pro-B and Rag1−/− Vh81X pre-B cells. Indicated cell fractions were obtained from 4 mice. Data represent average of two biological replicates. (E) 3C-Seq long-range interactions of the iEκ enhancer viewpoint along ∼10 Mb range of chromosome 6 were plotted as reads per million for EP, E2A−/− pre-pro-B, Rag1−/− pro-B and Rag1−/− Vh81X pre-B cells. (F) Bar graphs showing total amount of 3C-Seq interactions within the Vκ region of Igκ analyzed for BglII fragments that contain Vκ genes (+Vκ gene) or not (–Vκ gene). Bars represent sum of interactions. The χ2 test was used to calculate significance levels between +Vκ gene and −Vκ gene fragments of two cell types. ****, P < .0001.
Similar to Igh, the iEκ viewpoint showed many interactions with sites on other chromosomes or other regions on chromosome 6 outside Igκ (Figure 3D and Supplementary Figure S3C–D). Interactions within Igκ increased with more mature subsets and were most frequent in pre-B cells (Figure 3D). Long-range interactions within Igκ were found in pre-pro-B, pro-B and pre-B cells. In contrast to pre-pro-B and pro-B cells, these interactions were more dispersed in pre-B cells, showing fewer hotspots with high interaction frequencies (Figure 3E–F) Taken together, both Igh and Igκ loci were associated with locus contraction in committed B-lineage cells indicating that locus contraction as such does not correlate with the developmental regulation of Ig locus assembly.
Nuclear positioning is associated with stage-specific induction of Ig gene rearrangements
To study whether the nuclear positioning of Ig loci orchestrates the stepwise Ig gene rearrangements, we analyzed spatial distances between the Igh and Igκ loci and the nuclear lamina using 3D DNA FISH. The Igh locus was preferentially positioned at the nuclear periphery in uncommitted E2A−/− pre-pro-B cells, and became more centrally positioned in Rag1−/− pro-B cells (Figure 4A). Importantly, the Igh alleles were positioned back to the nuclear lamina compartments in Rag1−/− Igμ pre-B cells. On the other hand, Igκ was located at the nuclear periphery in both pre-pro-B and pro-B cells, and only in pre-B cells Igκ was located in the center (Figure 4B). The re-positioning involved both Ig loci in each cell as shown by independent analysis of the most lamina-proximal and lamina-distal loci (Supplementary Figure S4A–B). The stage-specific positioning did not appear to involve the iEκ: the Igκ allele with the iEμR knock-in was also located at the nuclear lamina in pro-B cells, and the Igκ alleles with κE1/E2mut were not fully retained at the nuclear periphery in pre-B cells (Supplementary Figures S5 and S6). Thus, the Ig locus positioning in the nucleus, rather than the large-scale contractions, is directly associated with the sequential rearrangement of the Ig loci.
Figure 4.
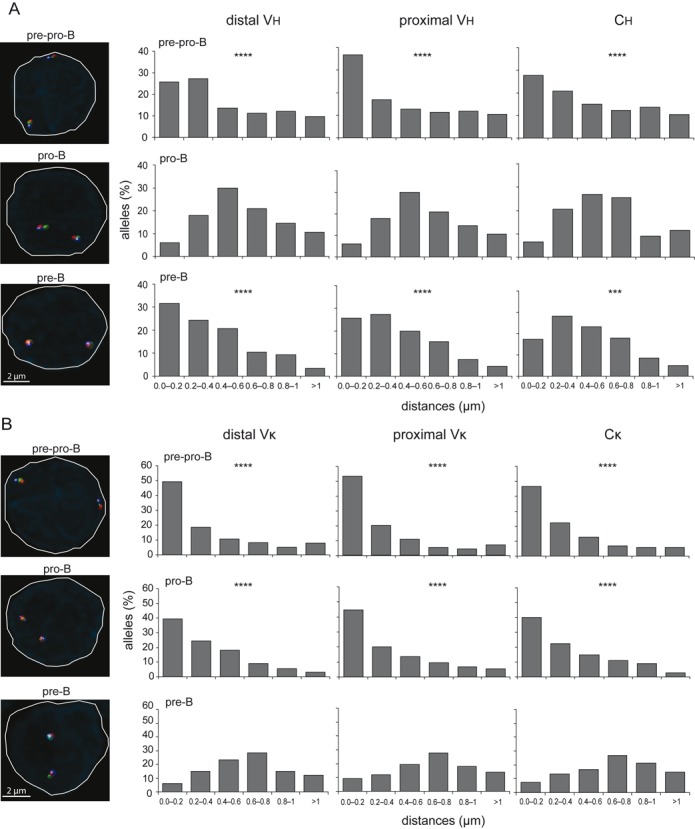
Stage-specific positioning of Ig loci away from the nuclear lamina. (A) Bar graphs showing the frequencies of Igh alleles positioned within a distance range from the nuclear lamina in cultured E2A−/− pre-pro-B, Rag1−/− pro-B and Rag1−/− hIgM pre-B cells. (B) Bar graphs depicting frequencies of Igκ alleles positioned within a distance range from the nuclear lamina in cultured E2A−/− pre-pro-B, Rag1−/− pro-B and Rag1−/−hIgM pre-B cells. In A and B, X-axis represents the distance ranges from the nuclear lamina and Y-axis represents the% of alleles positioned with a certain distance range. Distances were measured with 3D DNA FISH of >100 alleles, obtained from 2 mice. Statistical significance was calculated with the χ2 test between Rag1−/− pro-B and E2A−/− pre-pro-B and Rag1−/−hIgM pre-B cells in (A) or between Rag1−/− hIgM pre-B and E2A−/− pre-pro-B and Rag1−/− pro-B cells in (B) based on the sum of alleles located <0.4 μm from the lamina and the sum of alleles located >0.4 μm of the lamina. Significant changes in distribution of distances were calculated with the χ2 test; ***, P < .001; ****, P < .0001. Representative microscope images on the left indicate differential nuclear positioning of Ig loci in the three precursor-B-cell populations.
Positioning away from the nuclear lamina increases germline Ig loci transcription
To study whether the nuclear re-positioning was related to the changes in transcriptional activity, we investigated the nuclear localization of Igh and Igκ in E2A−/− pre-pro-B cells treated with histone deacetylase inhibitor trichostatin A (TSA). TSA was previously shown to modulate gene localization at the nuclear periphery by disturbing the interactions of LAD-derived sequences with the nuclear lamina (43,44). As a read out for locus accessibility, germline transcription from the Ig loci was assessed. From previously generated microarray studies (29,30) and with RQ-PCR, we analyzed Igh and Igκ germline transcripts. In line with established concepts, we found high expression of Igh germline transcripts in Rag1−/− pro-B cells, while Igκ germline transcripts were high in Rag1−/−Vh81X pre-B cells (Figure 5A–C and Supplementary Table S2).
Figure 5.
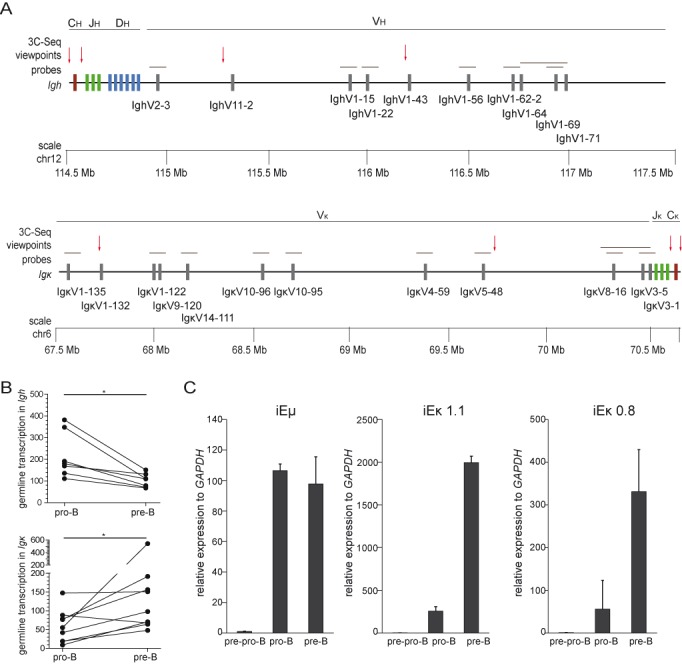
Developmentally regulated germline transcription of Ig loci. (A) Schematic representation of Igh (top panel) and Igκ (bottom panel) loci with indicated positions of certain V (grey rectangles), D (blue rectangles), J (green rectangles) and C genes (purple rectangles). In addition, the positions of microarray probes (grey horizontal lines) for detection of germline transcripts, and 3C-Seq viewpoints (red arrows) are depicted. (B) Graphs represent germline transcripts in Igh and Igκ paired between Rag1−/− pro-B and Rag1−/− Vh81X pre-B cells. Data were derived from previously generated expression profiles (Supplementary Table S2 (29,30), and significance was calculated with the Mann-Whitney U test; *, P < 0.05. (C) Germline transcript levels of iEμ and iEκ as assessed in E2A−/− pre-pro-B, Rag1−/− pro-B and Rag1−/− Vh81X pre-B with RQ-PCR in 2 independent cultures. Bars represent average with SD. Gene expression levels were normalized to the levels of GAPDH, and the values in E2A−/− pre-pro-B cells were set to 1.
Following 10 h incubation of E2A−/− pre-pro-B cells with TSA, spatial distances between the Ch and Cκ probes, and the nuclear lamina were measured by 3D DNA FISH. Indeed, incubation with TSA resulted in positioning of both Igh and Igκ away from the nuclear lamina in these uncommitted early precursor cells (Figure 6A–C). This movement was associated directly with increased expression of germline transcription of both the Igh and Igκ loci (Figure 6D–E).
Figure 6.
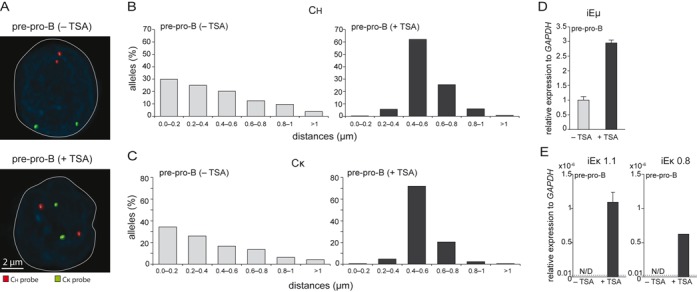
Positioning away from the nuclear lamina increases germline transcription of Igh and Igκ. (A) Representative microscope images indicate differential nuclear positioning of Igh (red) and Igκ (green) loci in E2A−/− pre-pro-B-cells cultured with (+) or without (−) TSA. Bar graphs showing the frequencies of (B) Ch regions and (C) Cκ regions positioned within a distance range from the nuclear lamina in E2A−/− pre-pro-B cultured with (+) or without (−) TSA. >100 alleles were acquired for each condition. Germline transcript levels of (D) iEμ and (E) iEκ as assessed with RQ-PCR in 2 independent cultures. Bars represent average with SD. ND, not detectable. Dashed lines indicate the detection limit at cycle-threshold 40.
Taken together, these data indicate that large-scale contraction of Ig loci facilitates equal utilization of V genes during Ig gene rearrangements. However, it is the spatial positioning in the nucleus that orchestrates the Ig loci accessibility for V(D)J recombination machinery and thereby directs the sequential nature of Ig gene rearrangements in precursor-B cells (model in Figure 7).
Figure 7.
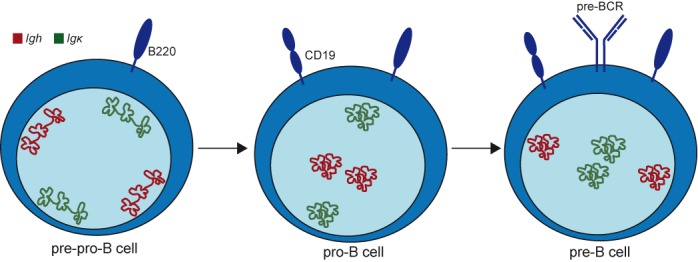
A model for nuclear positioning of Igh and Igκ loci during B-cell development. Nuclei in pre-pro-B, pro-B and pre-B cells are indicated with schematic drawing of differentially positioned Igh (red) and Igκ (green) loci. Both Ig loci were already contracted in the earliest committed pro-B-cell stage and retained this configuration in pre-B cells. In contrast, positioning away from the nuclear lamina was tightly regulated for Igh in pro-B cells and for Igκ in pre-B cells. Thus, nuclear localization rather than Ig locus topology is closely linked with the developmental regulation of Igh and Igκ locus assembly.
DISCUSSION
We studied locus contraction, long-range chromatin interactions and nuclear positioning of the germline Igh and Igκ alleles in three consecutive progenitor-B-cell subsets with 3D DNA FISH and 3C-Seq. This integrated approach enabled us to identify that both Ig loci were already contracted in the earliest committed pro-B-cell stage and retained this configuration in pre-B cells. In contrast, positioning away from the nuclear lamina was tightly regulated for Igh in pro-B cells and for Igκ in pre-B cells, and correlated with germline transcription. Thus, nuclear localization rather than Ig locus topology is closely linked with the developmental regulation of Igh and Igκ locus assembly.
In line with previous observations, we observed contraction of Igh in pro-B cells as compared to pre-pro-B cells (25–27). However, contrasting a previous study using wild type precursor-B-cells (26), our Rag-deficient cells kept the Igh locus contracted in pre-B cells. The absence of functional Rag in our system creates an artificial situation, because no DNA breaks are induced that could affect gene expression programs in the progenitor-B cells (47). Still, the immunophenotypes of our Rag-deficient pro-B and pre-B cells were normal, as was the stage-specific upregulation of Igh and Igκ germline transcripts. Rag-proficient pre-B cells will carry rearranged Ig genes with unknown spans of excised DNA. Moreover, FISH signals could be derived from excision products. Therefore, a model with Igh and Igκ in germline configuration is best-suited to study their 3D structural organization.
Similar to Igh, we observed that Igκ was already contracted in pro-B cells (28,29). This contraction, as well as abundant intra-locus interactions have been described before (7,21–23,29–31,48). The germline Ig loci on our studies retained cis-regulatory elements that potentially drive locus contraction. These include the intergenic control region 1 (IGCR1) downstream of the Vh region (22,23), and the contracting element for recombination (Cer) located within the Vκ-Jκ intervening region (49). These elements likely function similarly as the newly described regulatory elements between V and DJ region of the TCRβ locus to mediate locus topology (50,51). Thus, by choosing Rag-deficient models we did not study a heterogeneous pool of cells with and without deletion of these intergenic elements. The main limitation is the retained presence of IGCR1 in Igh that might affect Igh locus contraction resulting in differences between rearranged and non-rearranged alleles.
The consistent Ig locus contraction is potentially mediated by the ubiquitously expressed CTCF and YY1, which mediate the global DNA folding (21,31,52–54), and the B-cell specific transcription factors E2A, EBF1, Pax5, Ikaros and PU.1, which likely further facilitate contraction (24,31,53,55–57). This is supported by observations in CTCF−/− pro-B cells where the Igh locus was more compacted than in E2A−/− pre-pro-B cells and only slightly decontracted as compared to wild type pro-B cells (21). Because these factors are highly expressed in both pro-B and pre-B cells (Supplementary Table S3 and (29)), it is well-possible that they function similarly in both developmental stages to maintain the contracted conformation of the chromatin fiber. Importantly, our refined model for Ig locus organization supports a role for Ig locus contraction in efficient V(D)J recombination, but excludes a role in developmental regulation of Ig gene rearrangements.
We found that the E2A-binding sites in iEκ were unimportant for Igκ locus contraction, despite their role in Igκ gene rearrangements (37,38). Previously, deletion of the iEμ and 3′RR enhancers in Igh were found to be redundant for contraction or long-range interactions in Igh (48,58). This could imply that the enhancers in Ig loci are not involved in 3D structural organization, and mediate efficient V(D)J recombination through different means. Multiple DNA-binding proteins have been implicated in Ig locus contraction and they can bind to regulatory elements disseminated along the loci, such as PAIR elements within the Vh region (53). CTCF binding to the Cer region within Igκ mediated the locus contraction (49). Likely CTCF binding to IGCR1 also established the Igh folding. Thus, the enhancers could be involved in contraction of wild type alleles, but the disruption of a single transcription factor binding site would only have a minor impact on large-scale chromatin contraction.
Although the decontracted Igκ locus in E2A−/− pre-pro-B cells is not yet a target for V(D)J recombination (31), we found a large number of intra-locus interactions. Further analysis showed that these mostly involved fragments that did not contain Vκ genes. Thus, the extent of spatial decontraction in pre-pro-B cells still allowed physical contacts between chromatin domains within the Ig loci, and in fact enabled freedom for random interactions. Rather, spatial contraction in pro-B and pre-B cells restricted these interactions with a preference for DNA regions containing V genes, specifically for Igκ following pre-BCR signaling (29).
Stage-specific accessibility of Igh and Igκ, as measured by increased germline transcription, was associated with positioning of the loci away from the nuclear periphery specifically at the stage of increased germline transcription. Movement of Igh away from the nuclear lamina in pro-B cells has been observed before (19), but we are the first to show re-location towards the nuclear lamina in pre-B cells. Unexpectedly, our finding of Igκ positioning at the nuclear lamina in pro-B cells contradicts previous observations (19). This is potentially due to the fact that in these studies 2D FISH was performed and the nuclear lamina were not stained. Goldmit et al. found that of the centrally-located Igκ alleles in pre-B cells, one was silenced by association with heterochromatin, forming the basis of allelic exclusion (59). In their studies, Igκ alleles in pro-B cells hardly associated with heterochromatin, most likely because they are associated with the nuclear lamina as observed here. Thus, in pro-B cells Igκ is inaccessible at the nuclear lamina, and this is a different process from heterochromatic recruitment necessary for allelic exclusion.
The nuclear lamina were previously shown to act as a repository for transcriptionally inactive chromatin domains (16,17), and repositioning of genes can occur at specific stages of cellular differentiation (9,15–16,18). Increased acetylation of Ig loci due to histone deacetylase inhibitor TSA resulted in movement of Ig loci toward the nuclear interior in E2A−/− pre-pro-B cells and increase of germline Ig transcription. This suggests that locus mobility and gene transcription stay in relation and are controlled by histone modifications. Recently, histone modifications were shown to direct V(D)J recombination (10). Specifically, Rag2 is recruited through binding to H3K4me3 histone marks (13,14). These histone modifications were detected within the Igh locus in pro-B cells, and in Igκ in the pre-B cell stage (12). It is conceivable that nuclear positioning towards the nuclear center enables the induction of H3K4me3 histone modifications. Alternatively, H3K4me3 histone modifications poise the Ig loci to migrate away from the nuclear lamina. A potential signal guiding histone modifications could be induced by IL-7. Recently, Mandal et al. showed that IL-7 signaling inhibited Igκ rearrangements in pro-B cells by recruiting STAT5 heterodimers to iEκ, which further induced Ezh2 methyltransferase and thereby silenced the Igκ locus through H3K27me3 modifications (60). These modifications could keep Igκ positioned at the nuclear lamina, while subsequent attenuation of IL-7R signaling by Ikaros would then induce re-positioning of Igκ and enable induction of gene rearrangements (61,62).
It still remains puzzling, however, which mechanisms control allelic exclusion of the second non-rearranged Igh locus in pre-B cells. We here used a model system with an Ig heavy chain transgene to study the locus organization and positioning of Igh alleles in the germline. As a result, all of our observations in pre-B cells concerned the relocation of non-functional Igh alleles towards the nuclear periphery and the down regulation of germline transcripts (Figure 4B, Supplementary Figure S4A–B). Thus, repositioning of Igh toward nuclear lamina and away from transcription factories could be a potential mechanism to decrease the locus accessibility and mediate allelic exclusion. However, the question remains whether a functionally rearranged Igh locus with a genomically proximal Vh promotor and iEμ will also move towards the nuclear periphery or remain transcriptionally active in the nuclear center (63). Addressing this issue would require a more complex model system with a functional Igh rearrangement on one allele and sufficient genomic markers to distinguish it from the other germline allele with DNA probes.
Taken together, our studies confirm that murine Igh and Igκ loci undergo contraction prior to gene rearrangement to allow juxtaposing of genomically distant gene segments in order to create equal opportunities for recombination. However, we here show that nuclear positioning is associated with germline transcription and the developmental regulation of Igh and Igk locus assembly. These combined new insights are important for future studies on the factors that regulate stepwise control of Ig gene rearrangements and indicate that these are likely to be controlled via nuclear positioning of Ig loci.
ACCESSION NUMBERS
The high-throughput sequencing data sets have been submitted to the Sequence Read Archive (SRA), accession number: SRP055900.
Supplementary Material
Acknowledgments
We thank Gert van Cappellen and Gert-Jan Kremers (Optical Imaging Center, Erasmus MC) for support with confocal microscopy, Ralph Stadhouders (Cell Biology, Erasmus MC) for his help with 3C-Seq experiments and data analysis, Rutger Brouwer (Biomics, Erasmus MC) and Harmen van der Werken (Cell Biology, Erasmus MC) for their computational contribution, and Y. Xu for providing the κE1/E2mut and EμR mice.
Footnotes
Present address: Menno C. van Zelm, Department of Immunology and Pathology, Central Clinical School, Monash University, Melbourne, Victoria, Australia.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This study was performed in the department of Immunology as part of the Molecular Medicine Postgraduate School of the Erasmus MC, and was supported by a Veni grant from The Netherlands Organization for Health Research and Development ZonMW [916.116.090 to M.C.v.Z.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Bossen C., Mansson R., Murre C. Chromatin Topology and the Regulation of Antigen Receptor Assembly. Annu. Rev. Immunol. 2012;30:337–356. doi: 10.1146/annurev-immunol-020711-075003. [DOI] [PubMed] [Google Scholar]
- 2.Jung D., Giallourakis C., Mostoslavsky R., Alt F.W. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 3.Curry J.D., Geier J.K., Schlissel M.S. Single-strand recombination signal sequence nicks in vivo: evidence for a capture model of synapsis. Nat. Immunol. 2005;6:1272–1279. doi: 10.1038/ni1270. [DOI] [PubMed] [Google Scholar]
- 4.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 5.Cobb R.M., Oestreich K.J., Osipovich O.A., Oltz E.M. Accessibility control of V(D)J recombination. Adv. Immunol. 2006;91:45–109. doi: 10.1016/S0065-2776(06)91002-5. [DOI] [PubMed] [Google Scholar]
- 6.Yancopoulos G.D., Alt F.W. Developmentally Controlled and Tissue-Specific Expression of Unrearranged Vh Gene Segments. Cell. 1985;40:271–281. [PubMed] [Google Scholar]
- 7.Verma-Gaur J., Torkamani A., Schaffer L., Head S.R., Schork N.J., Feeney A.J. Noncoding transcription within the Igh distal V-H region at PAIR elements affects the 3D structure of the Igh locus in pro-B cells. Proc. Natl. Acad. Sci. U.S.A. 2012;109:17004–17009. doi: 10.1073/pnas.1208398109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilbert N., Boyle S., Fiegler H., Woodfine K., Carter N.P., Bickmore W.A. Chromatin architecture of the human genome: Gene-rich domains are enriched in open chromatin fibers. Cell. 2004;118:555–566. doi: 10.1016/j.cell.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 9.Schneider R., Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Gene Dev. 2007;21:3027–3043. doi: 10.1101/gad.1604607. [DOI] [PubMed] [Google Scholar]
- 10.Xu C.R., Feeney A.J. The Epigenetic Profile of Ig Genes Is Dynamically Regulated during B Cell Differentiation and Is Modulated by Pre-B Cell Receptor Signaling. J. Immunol. 2009;182:1362–1369. doi: 10.4049/jimmunol.182.3.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi N.M., Loguercio S., Verma-Gaur J., Degner S.C., Torkamani A., Su A.I., Oltz E.M., Artyomov M., Feeney A.J. Deep Sequencing of the Murine Igh Repertoire Reveals Complex Regulation of Nonrandom V Gene Rearrangement Frequencies. J. Immunol. 2013;191:2393–2402. doi: 10.4049/jimmunol.1301279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji Y., Resch W., Corbett E., Yamane A., Casellas R., Schatz D.G. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y., Subrahmanyam R., Chakraborty T., Sen R., Desiderio S. A plant homeodomain in Rag-2 that binds hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matthews A.G.W., Kuo A.J., Ramon-Maiques S., Han S.M., Champagne K.S., Ivanov D., Gallardo M., Carney D., Cheung P., Ciccone D.N., et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:U1106–U1118. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osborne C.S., Chakalova L., Brown K.E., Carter D., Horton A., Debrand E., Goyenechea B., Mitchell J.A., Lopes S., Reik W., et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 16.Peric-Hupkes D., Meuleman W., Pagie L., Bruggeman S.W., Solovei I., Brugman W., Graf S., Flicek P., Kerkhoven R.M., van Lohuizen M., et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy K.L., Zullo J.M., Bertolino E., Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 18.Simonis M., Klous P., Splinter E., Moshkin Y., Willemsen R., de Wit E., van Steensel B., de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat. Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 19.Kosak S.T., Skok J.A., Medina K.L., Riblet R., Le Beau M.M., Fisher A.G., Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 20.Skok J.A., Brown K.E., Azuara V., Caparros M.L., Baxter J., Takacs K., Dillon N., Gray D., Perry R.P., Merkenschlager M., et al. Nonequivalent nuclear location of immunoglobulin alleles in B lymphocytes. Nat. Immunol. 2001;2:848–854. doi: 10.1038/ni0901-848. [DOI] [PubMed] [Google Scholar]
- 21.Degner S.C., Verma-Gaur J., Wong T.P., Bossen C., Iverson G.M., Torkamani A., Vettermann C., Lin Y.C., Ju Z., Schulz D., et al. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc. Natl. Acad. Sci. U.S.A. 2011;108:9566–9571. doi: 10.1073/pnas.1019391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo C., Gerasimova T., Hao H., Ivanova I., Chakraborty T., Selimyan R., Oltz E.M., Sen R. Two forms of loops generate the chromatin conformation of the immunoglobulin heavy-chain gene locus. Cell. 2011;147:332–343. doi: 10.1016/j.cell.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo C., Yoon H.S., Franklin A., Jain S., Ebert A., Cheng H.L., Hansen E., Despo O., Bossen C., Vettermann C., et al. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011;477:424–430. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuxa M., Skok J., Souabni A., Salvagiotto G., Roldan E., Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes. Dev. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jhunjhunwala S., van Zelm M.C., Peak M.M., Cutchin S., Riblet R., van Dongen J.J., Grosveld F.G., Knoch T.A., Murre C. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roldan E., Fuxa M., Chong W., Martinez D., Novatchkova M., Busslinger M., Skok J.A. Locus ‘decontraction’ and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nat. Immunol. 2005;6:31–41. doi: 10.1038/ni1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sayegh C.E., Jhunjhunwala S., Riblet R., Murre C. Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes Dev. 2005;19:322–327. doi: 10.1101/gad.1254305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fitzsimmons S.P., Bernstein R.M., Max E.E., Skok J.A., Shapiro M.A. Dynamic changes in accessibility, nuclear positioning, recombination, and transcription at the Ig kappa locus. J. Immunol. 2007;179:5264–5273. doi: 10.4049/jimmunol.179.8.5264. [DOI] [PubMed] [Google Scholar]
- 29.Stadhouders R., de Bruijn M.J., Rother M.B., Yuvaraj S., Ribeiro de Almeida C., Kolovos P., Van Zelm M.C., van Ijcken W., Grosveld F., Soler E., et al. Pre-B Cell Receptor Signaling Induces Immunoglobulin kappa Locus Accessibility by Functional Redistribution of Enhancer-Mediated Chromatin Interactions. PLoS Biol. 2014;12:e1001791. doi: 10.1371/journal.pbio.1001791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ribeiro de Almeida C., Stadhouders R., de Bruijn M.J., Bergen I.M., Thongjuea S., Lenhard B., van Ijcken W., Grosveld F., Galjart N., Soler E., et al. The DNA-binding protein CTCF limits proximal Vkappa recombination and restricts kappa enhancer interactions to the immunoglobulin kappa light chain locus. Immunity. 2011;35:501–513. doi: 10.1016/j.immuni.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 31.Lin Y.C., Benner C., Mansson R., Heinz S., Miyazaki K., Miyazaki M., Chandra V., Bossen C., Glass C.K., Murre C. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat. Immunol. 2012;13:1196–1204. doi: 10.1038/ni.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novobrantseva T.I., Martin V.M., Pelanda R., Muller W., Rajewsky K., Ehlich A. Rearrangement and expression of immunoglobulin light chain genes can precede heavy chain expression during normal B cell development in mice (vol 189, pg 75, 1999) J. Exp. Med. 1999;189:1361–1361. doi: 10.1084/jem.189.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hao Z.Y., Rajewsky K. Homeostasis of peripheral B cells in the absence of B cell influx from the bone marrow. J. Exp. Med. 2001;194:1151–1163. doi: 10.1084/jem.194.8.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin F., Chen X., Kearney J.F. Development of VH81X transgene-bearing B cells in fetus and adult: sites for expansion and deletion in conventional and CD5/B1 cells. Int. Immunol. 1997;9:493–505. doi: 10.1093/intimm/9.4.493. [DOI] [PubMed] [Google Scholar]
- 35.Mombaerts P., Iacomini J., Johnson R.S., Herrup K., Tonegawa S., Papaioannou V.E. Rag-1-Deficient Mice Have No Mature Lymphocytes-B and Lymphocytes-T. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 36.Nussenzweig M.C., Shaw A.C., Sinn E., Danner D.B., Holmes K.L., Morse H.C. 3rd, Leder P. Allelic exclusion in transgenic mice that express the membrane form of immunoglobulin mu. Science. 1987;236:816–819. doi: 10.1126/science.3107126. [DOI] [PubMed] [Google Scholar]
- 37.Inlay M.A., Lin T.X., Gao H.H., Xu Y. Critical roles of the immunoglobulin intronic enhancers in maintaining the sequential rearrangement of IgH and Igk loci. J. Exp. Med. 2006;203:1721–1732. doi: 10.1084/jem.20052310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inlay M.A., Tian H., Lin T.X., Xu Y. Important roles for E protein binding sites within the immunoglobulin kappa chain intronic enhancer in activating V(kappa)J(kappa) rearrangement. J. Exp. Med. 2004;200:1205–1211. doi: 10.1084/jem.20041135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ikawa T., Kawamoto H., Wright L.Y., Murre C. Long-term cultured E2A-deficient hematopoietic progenitor cells are pluripotent. Immunity. 2004;20:349–360. doi: 10.1016/s1074-7613(04)00049-4. [DOI] [PubMed] [Google Scholar]
- 40.Jensen K., Rother M.B., Brusletto B.S., Olstad O.K., Aass H.C.D., van Zelm M.C., Kierulf P., Gautvik K.M. Increased ID2 Levels in Adult Precursor B Cells as Compared with Children Is Associated with Impaired Ig Locus Contraction and Decreased Bone Marrow Output. J. Immunol. 2013;191:1210–1219. doi: 10.4049/jimmunol.1203462. [DOI] [PubMed] [Google Scholar]
- 41.Nodland S.E., Berkowska M.A., Bajer A.A., Shah N., de Ridder D., van Dongen J.J., LeBien T.W., van Zelm M.C. IL-7R expression and IL-7 signaling confer a distinct phenotype on developing human B-lineage cells. Blood. 2011;118:2116–2127. doi: 10.1182/blood-2010-08-302513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stadhouders R., Kolovos P., Brouwer R., Zuin J., van den Heuvel A., Kockx C., Palstra R.J., Wendt K.S., Grosveld F., van Ijcken W., et al. Multiplexed chromosome conformation capture sequencing for rapid genome-scale high-resolution detection of long-range chromatin interactions. Nat. Protoc. 2013;8:509–524. doi: 10.1038/nprot.2013.018. [DOI] [PubMed] [Google Scholar]
- 43.Zink D., Amaral M.D., Englmann A., Lang S., Clarke L.A., Rudolph C., Alt F., Luther K., Braz C., Sadoni N., et al. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J. Cell. Biol. 2004;166:815–825. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zullo J.M., Demarco I.A., Pique-Regi R., Gaffney D.J., Epstein C.B., Spooner C.J., Luperchio T.R., Bernstein B.E., Pritchard J.K., Reddy K.L., et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–1487. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
- 45.Hardy R.R., Hayakawa K. B cell development pathways. Annu. Rev. Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 46.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bredemeyer A.L., Helmink B.A., Innes C.L., Calderon B., McGinnis L.M., Mahowald G.K., Gapud E.J., Walker L.M., Collins J.B., Weaver B.K., et al. DNA double-strand breaks activate a multi-functional genetic program in developing lymphocytes. Nature. 2008;456:819–823. doi: 10.1038/nature07392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Medvedovic J., Ebert A., Tagoh H., Tamir I.M., Schwickert T.A., Novatchkova M., Sun Q., Huis In ‘t Veld P.J., Guo C., Yoon H.S., et al. Flexible long-range loops in the VH gene region of the Igh locus facilitate the generation of a diverse antibody repertoire. Immunity. 2013;39:229–244. doi: 10.1016/j.immuni.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiang Y., Park S.K., Garrard W.T. Vkappa gene repertoire and locus contraction are specified by critical DNase I hypersensitive sites within the Vkappa-Jkappa intervening region. J. Immunol. 2013;190:1819–1826. doi: 10.4049/jimmunol.1203127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Majumder K., Koues O.I., Chan E.A., Kyle K.E., Horowitz J.E., Yang-Iott K., Bassing C.H., Taniuchi I., Krangel M.S., Oltz E.M. Lineage-specific compaction of Tcrb requires a chromatin barrier to protect the function of a long-range tethering element. J. Exp. Med. 2015;212:107–120. doi: 10.1084/jem.20141479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murre C. Ensuring an equal playing field for antigen receptor loci variable regions. J. Exp. Med. 2015;212:2. doi: 10.1084/jem.2121insight1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Degner S.C., Wong T.P., Jankevicius G., Feeney A.J. Cutting edge: developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J. Immunol. 2009;182:44–48. doi: 10.4049/jimmunol.182.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ebert A., McManus S., Tagoh H., Medvedovic J., Salvagiotto G., Novatchkova M., Tamir I., Sommer A., Jaritz M., Busslinger M. The distal V(H) gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity. 2011;34:175–187. doi: 10.1016/j.immuni.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 54.Liu H., Schmidt-Supprian M., Shi Y., Hobeika E., Barteneva N., Jumaa H., Pelanda R., Reth M., Skok J., Rajewsky K. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 2007;21:1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bain G., Maandag E.C., Izon D.J., Amsen D., Kruisbeek A.M., Weintraub B.C., Krop I., Schlissel M.S., Feeney A.J., van Roon M., et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 56.Lin H., Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 57.Reynaud D., Demarco I.A., Reddy K.L., Schjerven H., Bertolino E., Chen Z.S., Smale S.T., Winandy S., Singh H. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat. Immunol. 2008;9:927–936. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perlot T., Alt F.W., Bassing C.H., Suh H., Pinaud E. Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14362–14367. doi: 10.1073/pnas.0507090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goldmit M., Ji Y., Skok J., Roldan E., Jung S., Cedar H., Bergman Y. Epigenetic ontogeny of the Igk locus during B cell development. Nat. Immunol. 2005;6:198–203. doi: 10.1038/ni1154. [DOI] [PubMed] [Google Scholar]
- 60.Mandal M., Powers S.E., Maienschein-Cline M., Bartom E.T., Hamel K.M., Kee B.L., Dinner A.R., Clark M.R. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat. Immunol. 2011;12:1212–1220. doi: 10.1038/ni.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heizmann B., Kastner P., Chan S. Ikaros is absolutely required for pre-B cell differentiation by attenuating IL-7 signals. J. Exp. Med. 2013;210:2823–2832. doi: 10.1084/jem.20131735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schwickert T.A., Tagoh H., Gultekin S., Dakic A., Axelsson E., Minnich M., Ebert A., Werner B., Roth M., Cimmino L., et al. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat. Immunol. 2014;15:283–293. doi: 10.1038/ni.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Holwerda S.J., van de Werken H.J., Ribeiro de Almeida C., Bergen I.M., de Bruijn M.J., Verstegen M.J., Simonis M., Splinter E., Wijchers P.J., Hendriks R.W., et al. Allelic exclusion of the immunoglobulin heavy chain locus is independent of its nuclear localization in mature B cells. Nucleic Acids Res. 2013;41:6905–6916. doi: 10.1093/nar/gkt491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


