Abstract
Azinomycin B is a hybrid polyketide/nonribosomal peptide natural product and possesses antitumor activity by interacting covalently with duplex DNA and inducing interstrand crosslinks. In the biosynthetic study of azinomycin B, a gene (orf1) adjacent to the azinomycin B gene cluster was found to be essential for the survival of the producer, Streptomyces sahachiroi ATCC33158. Sequence analyses revealed that Orf1 belongs to the HTH_42 superfamily of conserved bacterial proteins which are widely distributed in pathogenic and antibiotic-producing bacteria with unknown functions. The protein exhibits a protective effect against azinomycin B when heterologously expressed in azinomycin-sensitive strains. EMSA assays showed its sequence nonspecific binding to DNA and structure-specific binding to azinomycin B-adducted sites, and ChIP assays revealed extensive association of Orf1 with chromatin in vivo. Interestingly, Orf1 not only protects target sites by protein–DNA interaction but is also capable of repairing azinomycin B-mediated DNA cross-linking. It possesses the DNA glycosylase-like activity and specifically repairs DNA damage induced by azinomycin B through removal of both adducted nitrogenous bases in the cross-link. This bifunctional protein massively binds to genomic DNA to reduce drug attack risk as a novel DNA binding protein and triggers the base excision repair system as a novel DNA glycosylase.
INTRODUCTION
Azinomycin B is a promising antitumor natural product isolated from the soil-dwelling bacteria Streptomyces sahachiroi and Streptomyces griseofuscus (1,2). This new structural class of dual alkylating agents is highly effective against tumor cells by interacting covalently with duplex DNA in the major groove and inducing interstrand crosslinks (ICLs) at 5′-GNC or 5′-GNT sequences (3–5). Due to the damage effects of alkylation and cross-linking (Supplementary Figure S1), DNA replication is interrupted and ultimately cell death occurs. The unprecedented azabicyclic ring system and highly active epoxide moiety in azinomycin B are two primary electrophilic functional groups responsible for the ICL process (6). Covalent ICLs arise from nucleophilic opening of the aziridine at C-10 and the epoxide at C-21 by the N-7 position of purine residues located on the two DNA strands (4,7). In addition, the naphthoate moiety, along with other functional groups within the skeleton of the compound, may also serve to influence the efficiency of ICL formation (8–11). Azinomycin B shows a strong propensity to alkylate guanine nucleosides in the major groove of DNA. On the basis of an average G+C content of 70%, there may be a large number of potentially susceptible target sites within the genome of a streptomycete strain. Obviously, efficient cellular self-protection is crucial for azinomycin B producers to protect themselves from the lethal effects of the compound.
Mechanisms of cellular self-protection mainly include target sites modification, drug inactivation, drug binding and export pumps (12). Considering that a single DNA damage is sufficient to cause cell death, multiple modes of resistance determinants are often adopted to ensure efficient self-protection toward a DNA-targeting antibiotic in its producers. For example, two resistance genes, blmA and blmB, were found to protect the bleomycin-producing strain Streptomyces verticillus from DNA damage induced by this cytotoxic agent through drug binding and drug inactivation via acetylation, respectively (13). The important antitumor antibiotic mitomycin C is produced by Streptomyces lavendulae as an inert prodrug that requires enzymatic or chemical reduction to generate a highly effective alkylating agent. Mitomycin self-resistance occurs via three cellular mechanisms: a flavoenzyme (McrA) that oxidizes the active reduced form of mitomycin C (14), a drug binding protein (Mrd) that prevents reductive activation of the prodrug and a membrane-associated protein (Mct) involved in drug efflux (15,16).
It is common that antibiotic biosynthetic genes are clustered with resistance and regulator genes. The azinomycin B biosynthetic gene cluster in S. sahachiroi was reported in 2008. Its biosynthetic pathway, however, is still not completely understood. To date, only the enzymatic cascade leading to the formation of the naphthoate moiety has been unequivocally deciphered (17,18). Since many of the proposed enzymes and reactions are novel, cloning and characterization of correlative genes will contribute to the elucidation of molecular mechanisms of its biosynthesis, self-resistance and regulation. Recently, one locus (aziR) far away from the azinomycin B gene cluster was identified to mediate azinomycin resistance in S. sahachiroi. The aziR gene encodes an aminoglycoside phosphotransferase which normally inactivates antibiotics via phosphorylation, but here it behaves as an azinomycin B binding protein conferring azinomycin resistance (19).
Here we report the identification of another azinomycin resistance locus (orf1) (GenBank accession number ABY83174) adjacent to the azinomycin B gene cluster. Expression of this gene in azinomycin-sensitive strains and deletion of it in a non-azinomycin-producing strain supported that it is, indeed, essential for survival of the azinomycin-producing strain, S. sahachiroi ATCC33158. Whereafter, the action mechanism of this newly discovered resistance determinant was clearly revealed by a series of in vivo and in vitro experiments.
MATERIALS AND METHODS
Strains, plasmids and primers
Strains, plasmids and primer sequences used in this study are summarized in Supplementary Data. Growth and fermentation of S. sahachiroi and isolation of azinomycin B were performed as described previously (20). Streptomyces and Escherichia coli strains were cultured and manipulated as previously reported (21). Restriction and polymerase chain reaction (PCR) enzymes were purchased from Fermentas or Takara. PCR primers were synthesized by GenScript. DNA fragments were cloned generally through amplification using the high PCR fidelity DNA polymerase KOD-plus- (TOBOYO).
Detection of azinomycin resistance
Detection of azinomycin resistance was executed in the orf1 gene deletion mutants and the azinomycin-sensitive strains expressing Orf1 by both disc diffusion test and cross streak method. In the disc diffusion test, filter paper discs spotted with azinomycin B were laid on lawns of the mutant strains, and incubated at 30°C for two days. Resistance levels to azinomycin B were determined by analysis of inhibition zones. In the cross streak method, a single streak of the wild type strain ATCC33158 (positive control), and the non-azinomycin-producing strain ΔaziU3 (negative control), were independently inoculated onto GYM medium and incubated at 30°C for two days, allowing the active organisms to produce and secrete azinomycin B into the medium. The azinomycin-sensitive strains expressing Orf1 were subsequently streaked perpendicularly across growth lines of the control strains, and incubated at 30°C for another two days. Absence of growth adjacent to the wild type strain indicated inhibition of tested cultures.
Electrophoretic mobility shift assay
DNA fragments were acquired by using PCR or annealed with primers listed in Supplementary Table S3. Each DNA fragment was incubated with various amounts of Orf1 protein in 20 μl of a reaction solution (10 mM Tris–HCl, pH 8.0, 10 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol, 10% glycerol and 60 mM KCl) at 30°C for 30 min. The mixture was electrophoresed through a 5% native polyacrylamide gel (PAGE) in 0.5× Tris-borate-EDTA buffer (TBE) and observed after ethidium bromide or SYBR Gold staining. Binding activity of Orf1 to the probe Porf7 labeled with biotin was determined using a chemiluminescent EMSA kit (Beyotime, China) according to the manufacturer's protocol.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) was performed using the EZ-ChIPTM kit (Millipore) according to the manufacturer's instructions. The antisera used in these ChIP assays were raised by immunization of mice with recombinant Orf1. Procedures for chromatin preparation and immunoprecipitation were detailed in Supplementary Methods. Each experiment was performed in duplicate and repeated twice.
DNA cross-linking assay
DNA cross-linking assays were performed as previously described (22–24). DNA was reacted with azinomycin B for DNA cross-linking under typical conditions (25 mM triethanolamine, 1mM EDTA, pH7.2, 37°C for 1 h). Samples were denatured with 0.05 M NaOH for 5 min and then detected by gel electrophoresis. Alternatively, ICL formation of DNA could be achieved by adding the drug directly to the DNA solution for a short incubation. However, this method was not as effective as the typical condition.
DNA protection assay
For drug binding assays, an appropriate amount of azinomycin B and excess Orf1 were first incubated together in the SGDM buffer (20 mM NaH2PO4, pH 7.5, 50 mM NaCl, 5% glycerol, 1 mM dithiothreitol, 10 mM MgCl2) at 30°C for 20 min, linearized pBluescript SK(+) DNA was then supplemented. For site protection assays, substrate DNA and sufficient Orf1 were incubated together at 30°C for 15 min. DNA–protein complexes were then incubated with azinomycin B at increasing concentrations. The above samples were denatured with 0.05 M NaOH for 5 min prior to electrophoresis.
DNA repair assay
A reaction buffer (50 mM NaCl, 10% glycerol) containing excess protein Orf1, 32.8 nM cross-linked linearized pBluescript SK(+) DNA or native DNA was incubated at 30°C for 10 min. The reaction mixtures were either untreated or denatured with alkali and then separated in a 1% agarose gel.
DNA cleavage assay
28 nM supercoiled pOJ260 DNA or 32.8 nM linearized pBluescript SK(+) DNA was treated with azinomycin B at 37°C for 2.5 h. For oligodeoxynucleotides (3.75 μM), the reaction mixture was incubated at 4°C for 20 h. Following extraction of the residual drug with an appropriate amount of chloroform, cross-linked DNA was incubated with Orf1 for 20 min at 30°C. For AP endonuclease cleavage, the Orf1-treated DNA was subject to the reaction with 1 U Endonuclease IV for 30 min at 37°C. All proteins were subsequently removed by digestion with proteinase K (1 mg/ml) for 15 min at 37°C prior to gel detection.
RESULTS
Identification of an essential resistance protein Orf1 in the azinomycin B producer S. sahachiroi
The involvement of specific genes in antibiotic biosynthetic gene clusters is commonly confirmed by gene inactivation. Owing to lack of candidates for self-resistance and regulation within the published azinomycin B biosynthetic gene cluster, the orf1 gene adjacent to its right boundary was investigated in our study. All attempts to disrupt orf1 in the azinomycin B producer S. sahachiroi ATCC33158 failed, whereas deletion of native orf1 was achieved by double-crossover homologous recombination after a second copy of the gene had been introduced into the chromosome at a different location (Supplementary Figure S2). The orf1 gene proved to be essential for the wild type strain. However, it became easy to eliminate orf1 when the azinomycin B biosynthetic pathway was abolished by knockout of the key biosynthetic gene aziU3 (20) (Supplementary Figure S2). Obviously, the essential nature of orf1 is closely bound to production of azinomycin B. Compared to the original strain ΔaziU3, the orf1 deletion mutant strain ΔaziU3Δorf1 exhibited increased sensitivity to azinomycin B (Figure 1A). The difference in azinomycin sensitivity of ΔaziU3 and ΔaziU3Δorf1 supports Orf1 as an azinomycin resistance determinant.
Figure 1.
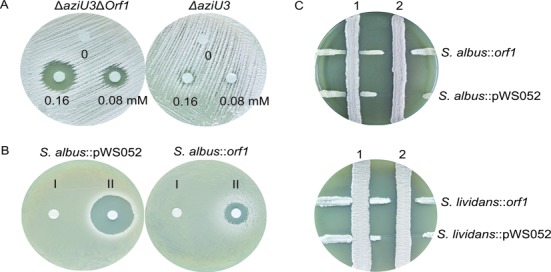
Azinomycin resistance of the orf1 deletion mutant and heterologous expression strains. (A) Azinomycin resistance detection of ΔaziU3 (right) and ΔaziU3Δorf1 (left) with increasing concentrations of azinomycin B. (B) Disc diffusion test assay to detect azinomycin resistance of S. albus::orf1 (right) and S. albus::pWS052 (left). I, 10 μl of methanol (control); II, 10 μl of 0.15 mM azinomycin B dissolved in methanol. (C) Cross streak method assay to detect azinomycin resistance conferred by Orf1 in S. albus (up) and S. lividans TK24 (down). The vertical lines are the azinomycin B producing strain ATCC33158 (2) and the mutant strain ΔaziU3 (1) without production of azinomycin B. The horizontal lines are the tested strains.
To further confirm the function of orf1, heterologous expression of this gene in two sensitive strains, Streptomyces albus and Streptomyces lividans, was pursued. The orf1 gene was cloned into the vector pWS052 under the control of a constitutive promoter PermE*, and introduced into two sensitive strains to give the mutant strains S. albus::orf1 and S. lividans::orf1, respectively. We used two different methods to examine the ability of orf1 to confer resistance against azinomycin B. In cross streak method (Figure 1C), the inhibition distances between the strains expressing Orf1 and the wild type strain ATCC33158 were significantly smaller than those of the negative control strains containing the vector pWS052 alone. No antagonistic effect was found between the test strains and the non-azinomycin-producing strain ΔaziU3. In disc diffusion test (Figure 1B), the strains expressing Orf1, likewise, exhibited apparent increase in resistance to azinomycin B. In these studies, we observed Orf1 consistently conferring azinomycin resistance in both native and sensitive strains.
Bioinformatics analyses indicated that Orf1 is a novel, putative DNA binding protein
BLASTP analysis showed that Orf1 belongs to the DUF1006 family of conserved bacterial proteins with unknown functions. This family has recently been reclassified as the HTH_42 superfamily (PF06224) by the Pfam protein families database. The helix-turn-helix (HTH) is a major structural motif capable of binding DNA and usually found in many regulator proteins (25). The HTH_42 represents a novel DNA-binding domain possibly containing two copies of winged helix-turn-helix motifs. So far, neither precise structure nor definite function for a protein in this family has been published.
In Pfam database, the HTH_42 superfamily consisted of about 1653 unique proteins (derived from 1193 species) with six different domain architectures (Figure 2A). Most of proteins (1582 sequences) contain just one HTH_42 domain, while only 3 proteins have two truncated HTH_42. The remaining 68 sequences including other types of protein domains were proposed to be related to ATP-dependent helicases. The domain architecture of Orf1 belongs to the first group, of which 48 sequences were annotated as putative uncharacterized YcaQ protein. In E. coli strain K12, the YcaQ was identified as one of UVC-toxicity modulating proteins with an unknown role (26). It is worth noting that 31 of these putative YcaQ proteins are from common pathogens in Enterobacteriaceae and 3 come from antibiotic producers. These data coincide with the distribution that HTH_42 proteins are primarily found in species of Salmonella (causes typhoid fever), Escherichia (Gut flora, some strains cause intestinal infection) and Streptomyces (produce antibiotics) (Figure 2B). Statistically, pathogenic bacteria and antibiotic-producing bacteria hold a high percentage in sequences occupancy (37% and 21%, respectively) (Figure 2C). Extremophiles and tolerant bacteria as well as nitrogen-fixing bacteria also share a proportion of HTH_42 proteins. It seems that members of HTH_42 superfamily play a certain role in some bacteria that could escape the unfavorable conditions.
Figure 2.
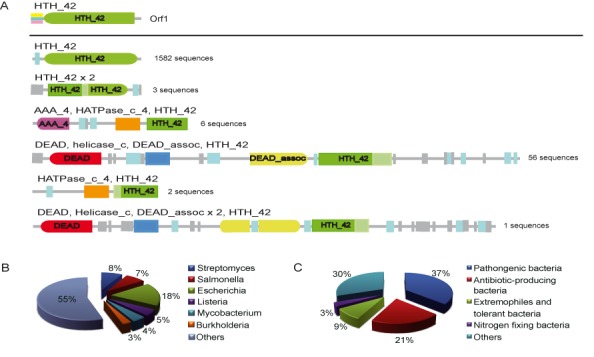
Bioinformatics analyses of Orf1. (A) Typical domain organization of HTH_42 superfamily (PF06224) proteins. The Orf1 protein belongs to the first domain architecture which contains only one HTH_42. (B) The primary species distribution of the PF06224 family is presented in a pie chart. (C) The pie chart displays the percentage of protein sequences from species with specific functional categories.
Additionally, PSI-BLAST (one interation) on the non-redundant protein database identified 17 homologous proteins with concrete descriptions. Besides two winged-helix DNA-binding domain proteins, other proteins were all annotated as prevent-host-death proteins, sharing low sequence identity (average 23%) and similarity (average 39%) with Orf1. These homologous proteins as well as Orf1 are essential for the vitality of their hosts. The comprehensive bioinformatics analyses proposed Orf1 to be a novel putative DNA binding protein only, but no hint of its mechanism in azinomycin B resistance was found.
Orf1 binds sequence nonspecifically to native DNA and structure-specifically to azinomycin B-modified sites
Based on bioinformatics results, Orf1 was initially suspected to be a transcriptional regulator controlling expression of antibiotic resistance genes. Therefore, electrophoretic mobility shift assay (EMSA) experiments were performed to investigate the interaction between Orf1 and potential promoter regions in the azinomycin B gene cluster. Intriguingly, Orf1 was found to retard all DNA fragment migration, including non-promoter regions and sequences from another streptomycete strain used as negative controls (Supplementary Figure S3B), suggesting that Orf1 exhibits little sequence specificity in DNA binding. A representative gel shift pattern for the binding of Orf1 to a 220-bp linearized DNA is shown in Figure 3A. As protein concentration increased, Orf1 produced progressively slower DNA migration, different from the gel retardation pattern of specific interactions. This sequence-independent manner excluded the possibility of Orf1 functioning as a regulator.
Figure 3.
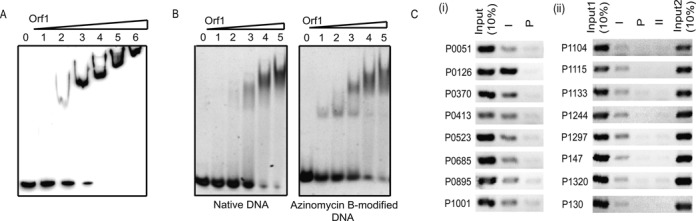
Orf1 is a non-specific DNA binding protein but specifically binds to azinomycin B-modified site. (A) DNA binding activity of Orf1 in vitro by EMSA with a biotinylated 220 bp DNA fragment. Protein concentrations were 0, 0.94, 1.88, 3.75, 7.5, 15.0 and 30 μM, respectively. Orf1-DNA complexes were subjected to electrophoresis in polyacrylamide. (B) EMSA assays for the binding of Orf1 to azinomycin B-modified DNA. A 21 bp oligodeoxynucleotide (1 μM) containing one GCC site (Supplementary Table S3) pre-treated with (right) or without (left) azinomycin B was incubated with various amounts of Orf1 protein (0, 1.12, 2.25, 4.5, 9.0 and 18 μM). The gel analysis of protein–DNA complexes were done following SYBR Gold staining. (C) ChIP assays of Orf1 and nucleic acids interactions in vivo. (i) ChIP assay in S. sahachiroi ATCC33158. ChIP using preimmune (P) or immune (I) sera raised against Orf1. (ii) ChIP assay in S. lividans::orf1 and S. lividans::pWS052. Immunoprecipitation of S. lividans::orf1 with preimmune (P) and immune (I) sera, and immunoprecipitation of S. lividans::pWS052 with immune sera (II) were detected by PCR with multiple pairs of primers. Input 1 and Input 2 were chromatin fractions without immunoprecipitation from two strains, respectively.
To further investigate if Orf1 exhibits some selectivity for 5′-GNT or 5′-GNC sequences which are target sites for azinomycin B induced cross-linking, four oligodeoxynucleotides (ODNs) containing GCC, GCT, GAT and AAT located within AT tracts were used for EMSA experiments. Likewise, Orf1 bound to four ODNs with no obvious difference in affinity (Supplementary Figure S3C). However, a single sharp retardation band of a site-specific protein–DNA complex appeared when the adducted ODN with a single azinomycin B cross-linking site of GCC was incubated with lower concentration of Orf1 (lanes 1–3 in Figure 3B). As protein concentration increased, the amount of the specific protein–DNA complex initially increased (lanes 1 and 2) and then decreased (lanes 3–5) due to the sequence non-specific binding of extra Orf1 to other sites. This phenomenon suggested that the binding mode of Orf1 is biphasic, with higher affinity to azinomycin B-modified sites and lower affinity to native DNA duplex.
ChIP assays revealed extensive association of Orf1 with chromatin in vivo
To test the binding of Orf1 to nucleic acids in physiological environment, chromatin immunoprecipitation (ChIP) assays were carried out. In the ChIP assay, DNA recovered from immune or preimmune precipitates was amplified using primers specific for the genomic regions indicated in Supplementary Tables S4 and S5. For validating non-specific interactions between Orf1 and DNA in vivo, the amplified regions were randomly selected throughout the genome. The Orf1 produced by both the native host and one sensitive host S. lividans whose genome sequence is available, was tested for in vivo DNA binding activity using the purified antibody against Orf1. In addition to the preimmune serum being used as a control, immunoprecipitation obtained from S. lividans::pWS052 cells was used as an additional negative control for ChIP assays in S. lividans::orf1 cells. DNA corresponding to the eight randomly chosen regions in the genome of the native host or the sensitive host was co-immunoprecipitated with Orf1, while significantly reduced levels of DNA were immunoprecipitated in the negative controls (Figure 3C). The results of ChIP assays revealed an extensive association between Orf1 and chromatin in vivo.
The binding of a large amount of Orf1 within the bacterial chromatin might mask target sites through protein–DNA interactions to prevent azinomycin B-induced damage. If Orf1 serves to protect chromatin against azinomycin B, it should be in abundance. Therefore, we measured cellular Orf1 levels at different stages in the life cycle of ATCC33158 by quantitative immunoblotting (Supplementary Figure S6A). In western blot analysis, expression of Orf1 was undetectable or only found in trace amounts in the early growth phase, but rose sharply after 24 h and reached a peak at 36 h. Subsequently, levels dipped slightly from 48 h to 60 h and came back up at 72 h (Supplementary Figure S6B). It is reasonable that the initial and then increasing production of Orf1 apparently preceded azinomycin B production to serve as a protector during drug production (Supplementary Figure S6C). The Orf1 protein has also been detected in mutant strains that do not accumulate azinomycin B (data not shown), which implies the orf1 gene expression is not directly induced by azinomycin B. The Orf1 (40.8 kDa) constituted ≈0.18% of the total protein levels (Supplementary Table S6). If we translated this into molecules per cell, however, there were about 106 molecules of Orf1/cell, approximately on par with the number of target sites. These findings support the plausibility of Orf1 conferring azinomycin B resistance by preventing antibiotic occupancy of the accessible target sites on chromatin in vivo.
In vitro DNA protection assays demonstrated Orf1 prevents DNA from azinomycin-mediated cross-linking via direct target site protection
For further confirmation, we established a series of in vitro DNA protection assays based on the cross-linking effect of the drug. The DNA interstrand cross-linking activities of azinomycin B were initially determined using the linearized plasmid pBluescript SK(+) in an agarose gel assay (Figure 4A). The linearized plasmid DNA was incubated with the drug or a control solvent prior to alkali denaturation and gel electrophoresis. ICLs induced by azinomycin B covalently link the two strands of the DNA double helix together, and hinder separation of the two strands by denaturation. Thus, native DNA exhibits a different mobility in agarose gels than cross-linked DNA after alkali treatment. DNA cross-linking assays were successfully performed under typical conditions as described previously (22–24). With increasing concentrations of azinomycin B, the amount of cross-linked double stranded DNA able to withstand alkali denaturation gradually increased (Figure 4A). In the DNA protection assay, the Orf1 protein was first bound to DNA, and then the protein–DNA complex was incubated with azinomycin B. Sufficient amount of protein was used in the test to observe protective effect. Protein–DNA complexes resisted cross-linking so that DNA was completely denatured into single strands by subsequent alkali treatment (lanes 5–8 in Figure 4B). Conversely, the amount of cross-linked double stranded DNA in the control group (without Orf1) gradually raised as azinomycin B concentrations increased (lanes 1–4 in Figure 4B). These results indicated that Orf1 protects DNA against azinomycin B mediated cross-linking.
Figure 4.

Characterization of the DNA glycosylase activity of Orf1 in vitro. (A) Detection of azinomycin B induced DNA cross-linking using alkali sedimentation. The linearized pBluescript SK(+) DNA was treated with 0, 25, 50, 75 and 125 μM of azinomycin B for 1 h prior to alkali denaturation and gel electrophoresis. The control of ssDNA (lane 1) was generated by alkali denaturation of the dsDNA control (lane 6). (B) Site protection effect of Orf1. The linearized pBluescript SK(+) DNA was incubated with (indicated as pr-DNA) or without (indicated as DNA ) Orf1 for 15 min and then subjected to cross-linking reaction at increasing concentrations of azinomycin B (0, 50, 100 and 150 μM). (C) Repair of DNA interstrand cross-links by Orf1. Orf1 protein was added after DNA cross-linking reaction and recovery of ssDNA was observed. (D) Cross-linking effects of the plasmid pOJ260 DNA. The supercoiled DNA pOJ260 (scDNA) containing tiny amounts of open circular DNA (ocDNA) was treated with 0, 50, 100, 200 and 400 μM of azinomycin B for 1 h at 37°C and then extracted with chloroform to eliminate the remaining drug prior to gel electrophoresis. (E) The circular DNA with different levels of cross-linking (prepared as described above) was incubated with Orf1 (30 μM) for 20 min at 30°C. DNA-bound Orf1 proteins were removed by digestion with proteinase K (1 mg/ml) for 15 min at 37°C before gel detection. (F) Repair of the highly cross-linked circular DNA by Orf1. The ICL DNA was incubated with Orf1 (63.6 μM) for 20 min at 30°C and treated with proteinase K prior to gel detection. The native circular DNA was set as a DNA control (lanes 4–6) and BSA was set as a protein control (lanes 2 and 5). (I) Repair of the highly cross-linked linear pBluescript SK(+) DNA by Orf1. (G) and (H) Products of Endo IV cleavage reaction with the Orf1 treated crosslinked circular (G) and linear (H) DNA.
In order to detect the possibility of drug inactivation and sequestration by Orf1, we conducted another DNA protection experiment where the order of addition of the reagents was changed, but no concentrations where altered. The Orf1 was first incubated with a moderate amount of azinomycin B and then DNA was added. As shown in Supplementary Figure S7A, DNA was not protected from cross-linking regardless if previously sufficient amounts of Orf1 were present in the reaction, which suggested that pre-incubation with Orf1 could not affect the cross-linking activity of azinomycin B. An ultrafiltration process was also applied to detect the interaction between Orf1 and azinomycin B (Supplementary Figure S5). All filtrate separated from the Orf1/azinomycin B pre-incubation reaction using a 10 k ultrafiltration device displayed prominent and equivalent cross-linking regardless of the Orf1 protein concentration used (Supplementary Figure S5A). In contrast, cross-linked DNA decreased as the amount of drug was reduced (Supplementary Figure S5B). Taken together, it is evident that Orf1 neither inactivates nor binds azinomycin B in vitro, hence protective effect in Figure 4B was achieved because of its DNA binding ability.
Orf1 initiates the repair of azinomycin B induced DNA damage by BER
Orf1 binds to not only native DNA but also cross-linked DNA in the experiments (Figure 3B and Supplementary Figure S4A). The specific binding activity of Orf1 with cross-linked DNA has prompted us to detect its capacity to repair DNA damage through setting up a DNA repair assay system. Linearized plasmid pBluescript SK(+) DNA was first cross-linked by the drug at 37°C for 1 h, and then incubated with excess Orf1 at 30°C for 10 min before alkali denaturation and agarose gel analysis. As shown in Figure 4C, cross-linking was reversed by Orf1, such that DNA was denatured into single strands (lane 7), whereas the negative control without Orf1 (lane 6) maintained the double-stranded structure under alkali denaturation. The reverse cross-linking of DNA implied that Orf1 made an incision in at least one DNA strand near the ICL sites. Three possible mechanisms of ICL DNA repaired by Orf1 were schematically presented in Figure 6B. Besides direct removal of alkyl adducts, alkylation lesions are usually repaired by excision of damaged DNA regions through base excision repair (BER) or nucleotide excision repair (NER). As relatively low-molecular weight DNA was not observed during alkaline sedimentation, incision was more likely to occur between azinomycin B and the adducted nucleoside, instead of at phosphodiester backbone.
Figure 6.

Proposed action modes of Orf1. (A) Three strategies might be adopted by Orf1 to resist azinomycin B. (a) Drug binding. Orf1 binds azinomycin B and sequesters it from target DNA. (b) Site protection. Binding of Orf1 occupies target sites and protects them from the drug. (c) DNA repair. Orf1 binds interstrand cross-linked DNA and repairs DNA damage. (B) Schematic diagrams of three proposed mechanisms for ICL DNA repair by Orf1: (1) Orf1 makes an incision at phosphodiester backbones of azinomycin B-adducted DNA; (2) Orf1 binds to azinomycin B modified double-stranded DNA region and consequently releases azinomycin B from the adducted base without incision of DNA; (3) Orf1 cleaves the N-glycoside bond of the damaged base to liberate an abasic site. In the diagram, ‘P’ indicates phosphate group and ‘a’ indicates abasic site. (C) The BER-like repair mechanism initiated by Orf1.
To find out the pattern of incision, we initially used the supercoiled plasmid pOJ260 (scDNA) containing tiny amounts of open circular DNA (ocDNA) as a substrate in ICL DNA cleavage assays. Intercalative binding of azinomycin B to duplex DNA unwinds the double helix and relaxes the tight conformation, resulting in a slower DNA migration through a gel (27,28). As seen in Figure 4D, the rate of DNA migration was inversely proportional to the level of cross-linking. The supercoiled DNA as well as the ocDNA was progressively relaxed by increased azinomycin B and migrated in agarose gel at continuously decreasing rates. When Orf1 was supplemented, most of cross-linked DNA was repaired and exhibited the same mobility as ocDNA except a little bit left in lane 5 due to insufficiency of the repair enzyme (Figure 4E). The residual highly cross-linked DNA was also effectively converted into the similar conformation of ocDNA after incubation with a raised concentration of Orf1 (Figure 4F, lane 3), indicating that Orf1 specifically repaired the azinomycin B modified DNA. The reverse of cross-linking was protein concentration-dependent, which produced progressively faster migrating products as the Orf1 concentrations increased (Supplementary Figure S7B). It is apparent that Orf1 removed azinomycin B from the cross-linked DNA by cleavage and thus led to the rewinding of DNA. The same rewinding effects were clearly observed in the cleavage assays of the highly cross-linked linear DNA (Figure 4I and Supplementary Figure S7D). Notably, the original conformation of DNA was not completely restored by Orf1 (Supplementary Figure S7B and S7D). Although cross-linking was reversed, transformation activity of cross-linked pOJ260 DNA was not restored at all after treatment with Orf1 (data not shown). Moreover, the prolonged treatment of ICL DNA with higher concentration of Orf1 caused breaks in DNA strands (Supplementary Figure S7C, lane 5). The uncompacted conformation and easy degradation of repaired DNA suggested the cleavage sites of Orf1 were located within the DNA strands, which excluded the direct reversal and supported the BER mechanism proposed in Figure 6C.
The first step of BER is catalyzed by DNA glycosylases that remove the damaged nitrogenous base to create an apurinic or apyrimidnic site (AP site). AP endonucleases specifically recognize the abasic DNA and cleave the phosphosdiester bond exclusively at the AP sites (29). Thus, we used Endonuclease IV (Endo IV), a kind of AP endonucleases to detect if depurination occurred during the DNA repair process. As seen in Figure 4G and H, Endo IV cleavage occurred massively at phosphodiester backbone of the Orf1-treated supercoiled and linear ICL DNA (lane 6), thus leading to complete DNA degradation into a smear of smaller fragments, while there was only a slight degradation in the reactions with the Orf1-untreated ICL DNA due to spontaneous depurination of N7-alkylated products (lane 5). Depurination phenomena were not detected in the control reactions with native circular and linear DNA (lane 4), although Orf1 could bind to it. This result indicated that numerous AP sites were produced by Orf1 in DNA strands of the azinomycin-DNA adducts.
Orf1 removes two adducted bases in the cross-link as a novel DNA glycosylase
To determine whether Orf1 hydrolyzes the glycosidic bonds of both purines that are cross-linked, 21-mer DNA duplex with one GCC site (Supplementary Table S3) was treated with azinomycin B and subjected to the cleavage reaction of Orf1 and Endo IV. As previously reported in literature (9,24,30), this triplet readily reacted with azinomycin B, giving rise to two kinds of lower-mobility products in a 20% denaturing polyacrylamide gel. One was monoalkylated intermediate which ran close to the substrate DNA, the other was cross-linked DNA at much lower mobility (lane 1 in Figure 5A). With the addition of Orf1, two kinds of modified DNA disappeared, and instead the intensity of native DNA band greatly enhanced (lane 2), which indicated that the monoalkylation and the cross-link had been successfully reversed by Orf1. The succedent treatment with Endo IV liberated smaller fragments with sizes that corresponded to two predicted split AP products (both are 9-/11-nt DNA duplexes designated as M1 and M2), and led to a drop-off in the intensity of 21-mer DNA band (lane 6). In contrast, Endo IV did not incise the modified DNA without Orf1 pretreatment (lane 5). A limited amount of smaller fragments also appeared in the absence of Endo IV (lane 2), which implied that an alkali-induced strand scission at the abasic sites happened during denaturing PAGE (also seen in Supplementary Figure S8A). To avoid the unexpected DNA strand breaks induced by alkali and observe the formation of double-stranded break (DSB) in the cross-link repair, the same DNA samples were analyzed using native PAGE again. Figure 5B showed that apparent short DNA band of expected size appeared only after sequential treatment with Orf1 and Endo IV (lane 6). For further confirmation, another 21-mer DNA duplex with different arm lengths around G-G cross-link was used in the cleavage assay. As expected, two distinct DNA duplexes (a 10-/12-nt and an 8-/10-nt) were generated (lane 6 in Supplementary Figure S8B). The results of denaturing and native PAGE analyses clearly demonstrated that Orf1 incised two adducted G bases in both strands of the cross-linked ODN. Orf1 is supposed to be a new kind of DNA glycosylases without any similarity to known DNA repair proteins, which proved to specifically recognize azinomycin B-mediated DNA crosslinks and remove the adducted bases to repair DNA damage by inducing a BER-like pathway.
Figure 5.
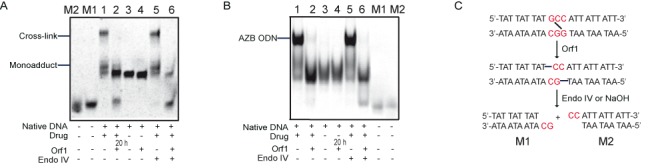
Removal of monoalkylated and cross-linked bases by Orf1 in vitro. Products of Orf1 and Endo IV cleavage reactions with the cross-linked/monoalkylated oligodeoxynucleotide (AZB ODN) were respectively detected by denaturing PAGE (A) and native PAGE (B). M1 and M2 (control DNA) were synthesized according to the predicted cleaved products in (C).
DISCUSSION
Although orf1 is the second azinomycin resistance gene found in S. sahachiroi, gene inactivation experiments reported here have demonstrated that it is essential for azinomycin production. In contrast to the other resistance protein AziR which acts by binding to the drug, the Orf1 may act as a new DNA glycosylase to confer azinomycin resistance. To better elucidate the mode of action of Orf1, probable interactions among drug (azinomycin B), target (DNA) and resistance protein (Orf1) were intensively analyzed and are illustrated in Figure 6A. Orf1 may confer the azinomycin resistance through drug binding (a), site protection (b) and/or DNA repair (c). Based on its DNA binding ability, the site protection strategy via protein–DNA interaction was first put forward. It may sound inconceivable for Orf1 to protect millions of potential target sites in a Streptomyces genome. The number of target sites susceptible to azinomycin B attack is, in fact, much lower than expected, due to the high density of chromosome structure and binding of nucleoid-associated proteins (31). Moreover, the aziE which is located within the azinomycin B gene cluster and encodes a putative export protein, was found to be employed in the drug efflux system (Wang Shan & Jing He, unpublished). Drug sequestration associated with efficient efflux systems which plays an important role in bacterial self-protection, maintains a low concentration of intracellular drug (32). The relatively fewer target sites, plus low drug concentrations in bacterial cells, are propitious conditions for Orf1 to implement efficacious site protection. This supposition was further supported by in vivo ChIP and in vitro DNA protection assays. Neither drug binding nor drug inactivation was identified by in vitro experiments, which both ruled out direct Orf1/drug interactions. Our studies in vitro subsequently discovered the DNA repair capability of Orf1, which is the most intriguing result of our observation.
To our knowledge, three mechanisms exist to repair DNA alkylating/cross-linking damages (Figure 6B). Among them, NER is the most common mechanism that could remove ICL lesions induced by a variety of antitumor antibiotics such as anthramycin and mitomycin C (33,34) by using the UvrABC endonuclease enzyme complex in E. coli cells. The excision of the adducted nucleotide is performed with the UvrA recognizing the helix-distorting lesion, the UvrB unwinding the damaged region, and the UvrC cleaving the phosphodiester bonds near the lesion (35,36). An additional matter of interest is that UvrA and UvrB proteins are capable of repairing the helix-stabilizing anthramycin-N2 guanine DNA adducts in an alternative way: UvrA coupled with UvrB strongly binds DNA and releases DNA-bound anthramycin without incision of DNA (37). A few homologs of UvrA proteins are found to confer self-resistance toward DNA-targeting drugs in antibiotics-producing bacteria. For example, UvrA-like proteins derived from chromomycin A3 (38) and the enediyne antitumor antibiotic C-1027 (39) biosynthetic gene clusters have been reported to participate in self-resistance of the corresponding drugs. The daunorubicin resistance protein (DrrC) in Streptomyces peucetius was speculated to bind within DNA regions intercalated by daunorubicin and release pre-bound drug to prevent DNA damage, due to its sequence homology and similar DNA binding properties with UvrA. The drrC, as well as orf1, is vital to self-resistance and antibiotic production. Moreover, as previously reported, inactivation of drrC was only accomplished in non-antibiotic-producing conditions. The DNA binding activity of DrrC is mediated by ATP and enhanced by daunorubicin (DNR) (40,41), while interaction of Orf1 and DNA is independent of ATP and unaffected by azinomycin B (Figure 4C and Supplementary Figure S4A).
Pyrimidine dimers and many of alkylation lesions are commonly repaired by direct reversal as the second repair mechanism in Figure 6B, but which is rarely seen in the removal of DNA ICL and drug self-resistance (42). The last but particularly important mechanism is BER, which repairs small, non-helix distorting base lesions like oxidation, alkylation and deamination (43). Recently, YtKR2, a DNA glycosylase homologous protein, was first reported to confer self-resistance toward a DNA-targeting antibiotic by triggering the BER system (44), which removes N3-YTM-alkylated adenine from monoadduct induced by yatakemycin (YTM). Shortly afterward, our study also found out the Orf1 protein, involved in self-resistance to azinomycin B, possesses DNA glycosylase-like activity. BER is normally initiated by DNA glycosylases and further processed by AP endonucleases. Monofunctional glycosylases only remove damaged base by cleaving the N-glycoside bond to liberate an AP site, whereas bifunctional glycosylases possess an extra AP lyase activity that hydrolyzes the phosphodiester bond 3′ to the AP site. Orf1 is supposed to be a novel type of monofunctional glycosylases because it only catalyzed base excision in ICL DNA cleavage assay in vitro. Orf1 specifically recognizes interstrand azinomycin B-DNA crosslinks and removes modified bases located on the two DNA strands, forming apurinic (AP) sites. The ubiquitous AP endonucleases in cells cleave the phosphodiester bond near the AP site to generate a 3′ hydroxyl group adjacent to 5′ deoxyribosephosphate (dRP). DNA polymerase then removes the damaged region using its 5′ to 3′ exonuclease activity and resynthesizes the missing part using the complementary strand as a template. Considering that BER is usually implicated in the elimination of mono-functional alkylation, we speculate that removal of monoadducts prior to crosslink formation seems to be more rational within the cell.
The Orf1 protein minimizes the amount of azinomycin B bound to genomic DNA and repairs incidental alkylation to ensure efficient protection. As a protein has the typical domain architecture of HTH_42, Orf1 is the first member of this superfamily to have a defined biological function in bacterial self-resistance identified. Contrary to most regulatory proteins with HTH motifs, we have shown Orf1 binds to DNA irrespective of any particular sequence. This kind of DNA binding pattern is commonly observed in nucleoid-associated proteins (NAPs) including histone-like DNA-binding proteins (HLPs) and high-mobility group proteins (HMGs) (45–47). The E. coli HU protein, a well-documented NAP, binds linear DNA fragments with low affinity and this binding was only detectable in vitro under low salt conditions (48,49). High concentration of NaCl (up to 150 mM), however, had no effect on the formation and mobility of the Orf1-DNA complex (Supplementary Figure S4B). Moreover, HU protein was found to bind specifically to DNA recombination and repair intermediates with high affinity although it binds to DNA duplex without sequence specificity. This structure-specific manner also occurred in the binding of Orf1 to the azinomycin B modified DNA. NAPs are usually considered to be involved in all DNA-dependent functions, including replication, repair, recombination and gene regulation (50). A novel bifunctional NAP identified in Streptomyces has recently been shown to modulate DNA conformation and transcription during development and stress (47). There were no obvious difference in phenotypic characteristics of colony morphology, spore formation and growth rate between the non-azinomycin-producing strain ΔaziU3 and the orf1 gene deletion mutant strain ΔaziU3Δorf1 observed. Thus Orf1 may not be involved in primary metabolism, but specific to the secondary metabolic pathway of azinomycin B.
Taken altogether, our present study revealed a novel DNA glycosylase, which could confer resistance in azinomycin B biosynthesis via distinct DNA binding ability and DNA repair capability. Intensive biochemical and structural biology studies of Orf1 will not only elucidate its enzymatic characteristics but also provide insight into the unique structure-function relationship of these newly found HTH_42 superfamily proteins.
Supplementary Material
Acknowledgments
We thank YuHui Sun (Wuhan University) for providing the plasmid pWHU77.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [30800020, 30970059 and 31270136]; New Century Excellent Talents grant from the Ministry of Education of China [NECT-08-0779]; Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry (SRF for ROCS, SEM) [to J. H.]; Fundamental Research Funds for the Central Universities [2009PY006]. Funding for open access charge: The National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1.Nagaoka K., Matsumoto M., Oono J., Yokoi K., Ishizeki S., Nakashima T. Azinomycins A and B, new antitumor antibiotics. I. Producing organism, fermentation, isolation, and characterization. J. Antibiot. 1986;39:1527–1532. doi: 10.7164/antibiotics.39.1527. [DOI] [PubMed] [Google Scholar]
- 2.Yokoi K., Nagaoka K., Nakashima T. Azinomycins A and B, new antitumor antibiotics. II. Chemical structures. Chem. Pharm. Bull. 1986;34:4554–4561. doi: 10.1248/cpb.34.4554. [DOI] [PubMed] [Google Scholar]
- 3.Hodgkinson T.J., Shipman M. Chemical synthesis and mode of action of the azinomycins. Tetrahedron. 2001;57:4467–4488. [Google Scholar]
- 4.Armstrong R.W., Salvati M.E., Nguyen M. Novel interstrand cross-links induced by the antitumor antibiotic carzinophilin/azinomycin B. J. Am. Chem. Soc. 1992;114:3144–3145. [Google Scholar]
- 5.Kelly G.T., Liu C., Smith R. III, Coleman R.S., Watanabe C.M. Cellular effects induced by the antitumor agent azinomycin B. Chem. Biol. 2006;13:485–492. doi: 10.1016/j.chembiol.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 6.Hartley J.A., Hazrati A., Kelland L.R., Khanim R., Shipman M., Suzenet F., Walker L.F. A synthetic azinomycin analogue with demonstrated DNA cross‐linking activity: insights into the mechanism of action of this class of antitumor agent. Angew. Chem. Int. Ed. 2000;39:3467–3470. [PubMed] [Google Scholar]
- 7.Fujiwara T., Saito I., Sugiyama H. Highly efficient DNA interstrand crosslinking induced by an antitumor antibiotic, carzinophilin. Tetrahedron Lett. 1999;40:315–318. [Google Scholar]
- 8.Coleman R.S. Issues of orthogonality and stability: synthesis of the densely functionalized heterocyclic ring system of the antitumor agents azinomycins A and B. Synlett. 1998;141:1031–1039. [Google Scholar]
- 9.Coleman R.S., Perez R.J., Burk C.H., Navarro A. Studies on the mechanism of action of azinomycin B: definition of regioselectivity and sequence selectivity of DNA cross-link formation and clarification of the role of the naphthoate. J. Am. Chem. Soc. 2002;124:13008–13017. doi: 10.1021/ja025563k. [DOI] [PubMed] [Google Scholar]
- 10.Coleman R.S., Burk C.H., Navarro A., Brueggemeier R.W., Diaz-Cruz E.S. Role of the azinomycin naphthoate and central amide in sequence-dependent DNA alkylation and cytotoxicity of epoxide-bearing substructures. Org. Lett. 2002;4:3545–3548. doi: 10.1021/ol0267275. [DOI] [PubMed] [Google Scholar]
- 11.Landreau C.A., LePla R.C., Shipman M., Slawin A.M., Hartley J.A. Delineating noncovalent interactions between the azinomycins and double-stranded DNA: Importance of the naphthalene substitution pattern on interstrand cross-linking efficiency. Org. Lett. 2004;6:3505–3507. doi: 10.1021/ol048653y. [DOI] [PubMed] [Google Scholar]
- 12.Cundliffe E. How antibiotic-producing organisms avoid suicide. Ann. Rev. Microbiol. 1989;43:207–233. doi: 10.1146/annurev.mi.43.100189.001231. [DOI] [PubMed] [Google Scholar]
- 13.Masanori S., Thompson C.J., Takanori K., Katsuyuki S., Rolf D., Raimundo V., Julian D. Characterisation by molecular cloning of two genes from Streptomyces verticillus encoding resistance to bleomycin. Gene. 1994;151:11–16. doi: 10.1016/0378-1119(94)90626-2. [DOI] [PubMed] [Google Scholar]
- 14.August P.R., Flickinger M.C., Sherman D.H. Cloning and analysis of a locus (mcr) involved in mitomycin C resistance in Streptomyces lavendulae. J. Bacteriol. 1994;176:4448–4454. doi: 10.1128/jb.176.14.4448-4454.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheldon P.J., Johnson D.A., August P.R., Liu H.W., Sherman D.H. Characterization of a mitomycin-binding drug resistance mechanism from the producing organism. Streptomyces lavendulae. J. Bacteriol. 1997;179:1796–1804. doi: 10.1128/jb.179.5.1796-1804.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheldon P.J., Mao Y., He M., Sherman D.H. Mitomycin resistance in Streptomyces lavendulae includes a novel drug-binding-protein-dependent export system. J. Bacteriol. 1999;181:2507–2512. doi: 10.1128/jb.181.8.2507-2512.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Q., He Q., Ding W., Tang M., Kang Q., Yu Y., Deng W., Zhang Q., Fang J., Tang G. Characterization of the azinomycin B biosynthetic gene cluster revealing a different iterative type I polyketide synthase for naphthoate biosynthesis. Chem. Biol. 2008;15:693–705. doi: 10.1016/j.chembiol.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 18.Ding W., Deng W., Tang M., Zhang Q., Tang G., Bi Y., Liu W. Biosynthesis of 3-methoxy-5-methyl naphthoic acid and its incorporation into the antitumor antibiotic azinomycin B. Mol. Biosyst. 2010;6:1071–1081. doi: 10.1039/b926358f. [DOI] [PubMed] [Google Scholar]
- 19.Foulke-Abel J., Kelly G.T., Zhang H., Watanabe C.M. Characterization of AziR, a resistance protein of the DNA cross-linking agent azinomycin B. Mol. Biosyst. 2011;7:2563–2570. doi: 10.1039/c1mb05136a. [DOI] [PubMed] [Google Scholar]
- 20.Wang S., Zhao R., Liu K., Zhu M., Li A., He J. Essential role of an unknown gene aziU3 in the production of antitumor antibiotic azinomycin B verified by utilizing optimized genetic manipulation systems for Streptomyces sahachiroi. FEMS Microbiol. Lett. 2012;337:147–154. doi: 10.1111/1574-6968.12020. [DOI] [PubMed] [Google Scholar]
- 21.Kieser T., Bibb M.J., Buttner M.J., Chater K.F., Hopwood D.A. Norwich: The John Innes Foundation; 2000. [Google Scholar]
- 22.Ponti M., Forrow S.M., Souhami R.L., D'lncalci M., Hartley J.A. Measurement of the sequence specificity of covalent DNA modification by antineoplastic agents using Taq DNA polymerase. Nucleic Acids Res. 1991;19:2929–2933. doi: 10.1093/nar/19.11.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casely-Hayford M.A., Pors K., James C.H., Patterson L.H., Hartley J.A., Searcey M. Design and synthesis of a DNA-crosslinking azinomycin analogue. Org. Biomol. Chem. 2005;3:3585–3589. doi: 10.1039/b508908e. [DOI] [PubMed] [Google Scholar]
- 24.LePla R.C., Landreau C.A., Shipman M., Hartley J.A., Jones G.D. Azinomycin inspired bisepoxides: influence of linker structure on in vitro cytotoxicity and DNA interstrand cross-linking. Bioorg. Med. Chem. Lett. 2005;15:2861–2864. doi: 10.1016/j.bmcl.2005.03.091. [DOI] [PubMed] [Google Scholar]
- 25.Brennan R.G., Matthews B.W. The helix-turn-helix DNA binding motif. J. biol. Chem. 1989;264:1903–1906. [PubMed] [Google Scholar]
- 26.Rooney J.P., Patil A., Joseph F., Endres L., Begley U., Zappala M.R., Cunningham R.P., Begley T.J. Cross-species Functionome analysis identifies proteins associated with DNA repair, translation and aerobic respiration as conserved modulators of UV-toxicity. Genomics. 2011;97:133–147. doi: 10.1016/j.ygeno.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zang H., Gates K.S. DNA binding and alkylation by the ‘left half’ of azinomycin B. Biochemistry. 2000;39:14968–14975. doi: 10.1021/bi001998d. [DOI] [PubMed] [Google Scholar]
- 28.Casely-Hayford M.A., Pors K., Patterson L.H., Gerner C., Neidle S., Searcey M. Truncated azinomycin analogues intercalate into DNA. Bioorg. Med. Chem. Lett. 2005;15:653–656. doi: 10.1016/j.bmcl.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 29.Cabrera G., Barria C., Fernandez C., Sepulveda S., Valenzuela L., Kemmerling U., Galanti N. DNA repair BER pathway inhibition increases cell death caused by oxidative DNA damage in Trypanosoma cruzi. J. Cell Biochem. 2011;112:2189–2199. doi: 10.1002/jcb.23138. [DOI] [PubMed] [Google Scholar]
- 30.LePla R.C., Landreau C.A., Shipman M., Jones G.D. On the origin of the DNA sequence selectivity of the azinomycins. Org. Biomol. Chem. 2005;3:1174–1175. doi: 10.1039/b502188j. [DOI] [PubMed] [Google Scholar]
- 31.Dronkert M.L., Kanaar R. Repair of DNA interstrand cross-links. Mutat. Res. 2001;486:217–247. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- 32.Cundliffe E., Demain A.L. Avoidance of suicide in antibiotic-producing microbes. J. Ind. Microbiol. Biotechnol. 2010;37:643–672. doi: 10.1007/s10295-010-0721-x. [DOI] [PubMed] [Google Scholar]
- 33.Walter R.B., Pierce J., Case R., Tang M.S. Recognition of the DNA helix stabilizing anthramycin-N2 guanine adduct by UVRABC nuclease. J. Mol. Biol. 1988;203:939–947. doi: 10.1016/0022-2836(88)90119-2. [DOI] [PubMed] [Google Scholar]
- 34.Weng M.W., Zheng Y., Jasti V.P., Champeil E., Tomasz M., Wang Y., Basu A.K., Tang M.S. Repair of mitomycin C mono-and interstrand cross-linked DNA adducts by UvrABC: a new model. Nucleic Acids Res. 2010;38:6976–6984. doi: 10.1093/nar/gkq576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Truglio J.J., Croteau D.L., Van Houten B., Kisker C. Prokaryotic nucleotide excision repair: the UvrABC system. Chem. Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 36.Wang H., Lu M., Tang M.S., Van Houten B., Ross J.B.A., Weinfeld M., Le X.C. DNA wrapping is required for DNA damage recognition in the Escherichia coli DNA nucleotide excision repair pathway. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12849–12854. doi: 10.1073/pnas.0902281106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang M.S., Nazimiec M.E., Doisy R.P., Pierce J.R., Hurley L.H., Alderete B.E. Repair of helix-stabilizing anthramycin-N2 guanine DNA adducts by UVRA and UVRB proteins. J. Mol. Biol. 1991;220:855–866. doi: 10.1016/0022-2836(91)90358-d. [DOI] [PubMed] [Google Scholar]
- 38.Menéndez N., Braña A.F., Salas J.A., Méndez C. Involvement of a chromomycin ABC transporter system in secretion of a deacetylated precursor during chromomycin biosynthesis. Microbiology. 2007;153:3061–3070. doi: 10.1099/mic.0.2007/007922-0. [DOI] [PubMed] [Google Scholar]
- 39.Galm U., Hager M.H., Van Lanen S.G., Ju J., Thorson J.S., Shen B. Antitumor antibiotics: bleomycin, enediynes, and mitomycin. Chem. Rev. 2005;105:739–758. doi: 10.1021/cr030117g. [DOI] [PubMed] [Google Scholar]
- 40.Lomovskaya N., Hong S.K., Kim S.U., Fonstein L., Furuya K., Hutchinson R.C. The Streptomyces peucetius drrC gene encodes a UvrA-like protein involved in daunorubicin resistance and production. J. Bacteriol. 1996;178:3238–3245. doi: 10.1128/jb.178.11.3238-3245.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Furuya K., Richard Hutchinson C. The DrrC protein of Streptomyces peucetius, a UvrA-like protein, is a DNA-binding protein whose gene is induced by daunorubicin. FEMS Microbiol. Lett. 1998;168:243–249. doi: 10.1111/j.1574-6968.1998.tb13280.x. [DOI] [PubMed] [Google Scholar]
- 42.Mishina Y., Duguid E.M., He C. Direct reversal of DNA alkylation damage. Chem. Rev. 2006;106:215–232. doi: 10.1021/cr0404702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fromme J.C., Verdine G.L. Base excision repair. Adv. Protein Chem. 2004;69:1–41. doi: 10.1016/S0065-3233(04)69001-2. [DOI] [PubMed] [Google Scholar]
- 44.Xu H., Huang W., He Q.L., Zhao Z.X., Zhang F., Wang R., Kang J., Tang G.L. Self-resistance to an antitumor antibiotic: a DNA glycosylase triggers the base-excision repair system in yatakemycin biosynthesis. Angew. Chem. Int. Ed. 2012;51:10532–10536. doi: 10.1002/anie.201204109. [DOI] [PubMed] [Google Scholar]
- 45.Saito K., Kikuchi T., Yoshida M. The mechanism of sequence non-specific DNA binding of HMG1/2-box B in HMG1 with DNA. Protein Eng. 1999;12:235–242. doi: 10.1093/protein/12.3.235. [DOI] [PubMed] [Google Scholar]
- 46.Liu D., Yumoto H., Murakami K., Hirota K., Ono T., Nagamune H., Kayama S., Matsuo T., Miyake Y. The essentiality and involvement of Streptococcus intermedius histone-like DNA-binding protein in bacterial viability and normal growth. Mol. Microbiol. 2008;68:1268–1282. doi: 10.1111/j.1365-2958.2008.06232.x. [DOI] [PubMed] [Google Scholar]
- 47.Aldridge M., Facey P., Francis L., Bayliss S., Del Sol R., Dyson P. A novel bifunctional histone protein in Streptomyces: a candidate for structural coupling between DNA conformation and transcription during development and stress. Nucleic Acids Res. 2013;41:4813–4824. doi: 10.1093/nar/gkt180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kamashev D., Rouviere-Yaniv J. The histone-like protein HU binds specifically to DNA recombination and repair intermediates. EMBO J. 2000;19:6527–6535. doi: 10.1093/emboj/19.23.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balandina A., Kamashev D., Rouviere-Yaniv J. The Bacterial Histone-like Protein HU Specifically Recognizes Similar Structures in All Nucleic Acids DNA, RNA, AND THEIR HYBRIDS. J. Biol. Chem. 2002;277:27622–27628. doi: 10.1074/jbc.M201978200. [DOI] [PubMed] [Google Scholar]
- 50.Grove A. Functional evolution of bacterial histone-like HU proteins. Curr. Issues Mol. Biol. 2011;13:1–12. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


