Abstract
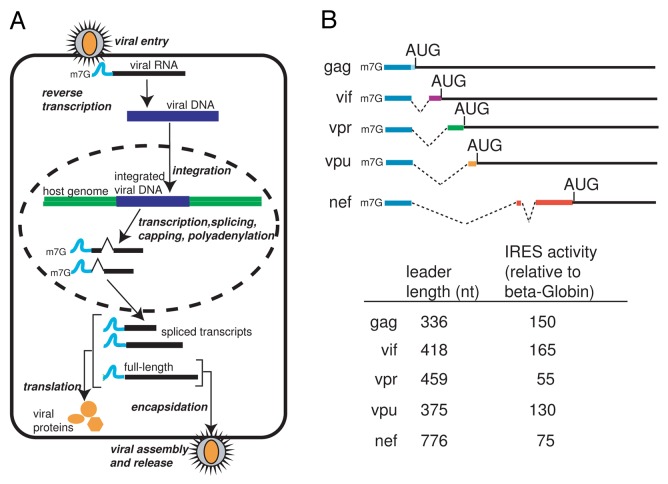
Alternative splicing of the human immunodeficiency virus 1 (HIV-1) RNA transcripts produces mRNAs encoding nine different viral proteins. The leader of each contains a common non-coding exon at the 5' end. Previous studies showed that the leaders from the common exon-containing transcripts gag, nef, vif, vpr and vpu can direct protein synthesis through internal ribosome entry sites (IRESs) with varying efficiencies. Here we explored whether the common exon acts as an IRES element in the context of all the 5' leaders or if each harbors a distinct IRES. We also explored the relationship between the IRESs and initiation codon selection. We find that the common exon adopts a similar conformation in every leader we explored and that the sequence and structure is required for IRES activity. We also find that each leader uses a scanning mechanism for start codon identification. Together, our data point to a model in which the common exon on HIV-1 transcripts acts as the ribosome landing pad, recruiting preinitiation complexes upstream of the initiation codon, followed by scanning to each transcript's initiator AUG.
Keywords: IRES, HIV-1, common exon, translation initiation, viral RNA
Introduction
The 5′ untranslated leader of the HIV-1 genomic RNA contains several highly conserved RNA structural domains that direct diverse important processes in the viral life cycle (Reviewed in 1). One function of this leader is to provide a splice donor site that directs splicing of a subset of over 30 viral transcripts, which provide the templates for viral protein synthesis. Although they are differentially spliced, the leader of each transcript contains a common 289 nucleotide long non-coding exon at the 5′ end with sequence unique to each transcript spliced onto the common exon’s 3′ end (Fig. 1). The synthesis, export, and translation of these mRNAs are highly regulated and critical to viral function. Like other viruses, HIV-1 depends on the host cell translation machinery for viral protein synthesis. Protein synthesis in eukaryotes consists of four phases: initiation, elongation, termination and ribosome recycling. Translation initiation is considered rate-limiting and is a common target for regulating protein synthesis (reviewed in 2). Initiation consists of recruiting and assembling elongation-competent ribosomes at the mRNA start codon and can occur through canonical and non-canonical mechanisms.3 In the case of the HIV-1 gag leader and other viral transcripts, there is evidence that translation can initiate by both a canonical cap-dependent and a non-canonical cap-independent pathway (reviewed in 4).
Figure 1. Overview of the HIV-1 viral life cycle and the mRNAs produced. A. Following entry into the host cell, the viral RNA is reverse-transcribed into a viral cDNA intermediate which is integrated into the host cell genome. The DNA is transcribed and the resulting pre-mRNA is processed into over 30 different RNA species. The resultant viral RNAs are translated into a subset of viral proteins and the full-length RNA is packaged into new virions. B. Diagram of the 5′ leaders from the transcripts encoding for gag, vif, vpr, vpu and nef, displaying splicing patterns, leader length, and IRES activity relative to B-globin 5′ UTR.14 Blue depicts the common 289 nt region present on the 5′ end of all the transcripts.
In the canonical cap- and scanning-dependent translation initiation pathway (reviewed in 5,6), the N7-methylated guanine (m7G) cap at the 5′ end of mRNA (in this case, the HIV-1 gag RNA) serves as a signal for recruitment of the multi-protein complex eukaryotic initiation factor 4F (eIF4F). The heterotrimeric eIF4F complex consists of a cap-binding subunit (eIF4E), an RNA helicase (eIF4A), and a scaffolding protein (eIF4G). In conjunction with eIF4B and/or eIF4H, the complex unwinds local secondary structures and recruits the 43S pre-initiation complex upstream of the major open reading frame (ORF). Start codon identification generally is thought to require 5′ to 3′ scanning of the mRNA by the pre-initiation complex until an appropriate initiation codon is reached.7,8 Base pairing of the initiator tRNA to the AUG start codon, large (60S) ribosome subunit joining, and release of initiation factors create an elongation-competent ribosome in the correct reading frame.
One mechanism of non-canonical translation initiation used by HIV-1 transcripts is driven by an internal ribosome entry site (IRES) RNA. IRESs recruit the translation machinery independently of the 5′ end of the mRNA. IRESs were first identified in picornaviruses, but have since been reported in many other viral RNAs, as well as some cellular mRNAs (reviewed in 9). The mechanism for IRES-directed initiation varies by RNA; in some cases ribosomes are directly recruited to the initiation codon whereas in others the pre-initiation complexes are acquired upstream of the initiation codon and scan to the start site. The factor requirement for IRESs can significantly vary with some requiring a subset of canonical factors to mediate initiation whereas others, such as the cricket paralysis virus IRES, can directly recruit ribosomes (reviewed in 10). For the majority of IRESs, our knowledge of how components of the initiation apparatus are recruited to the mRNA and how start codon recognition is achieved is limited.
IRESs have been identified in HIV-1 RNA sequences from the gag ORF,11 as well as the 5′ leaders of gag, tat, nef, vif, vpr and vpu transcripts.12-14 For most of these IRESs, it is unclear how the ribosome is recruited and positioned on the initiation codon. In this study, we explored the mechanism of internal initiation in the HIV-1 5′ leaders of the gag, nef, vif, vpr and vpu transcripts. We first considered whether the common 289 nt non-coding exon on the 5′ end of these mRNAs was acting as a common IRES element or if the 5′ leaders were acting as unique IRESs. We provide evidence suggesting that the common exon adopts the same conformation in different HIV-1 5′ leaders and this RNA sequence and structure is required for internal initiation in all the leaders, consistent with a role as a common IRES element. In addition, we sought to better understand how this common IRES element recruits the translation machinery and how the ribosome identifies the initiation codon. We show that all HIV-1 5′ leader IRESs require eIF4A for activity and are sensitive to the presence of upstream out-of-frame AUGs, consistent with a ribosome scanning mechanism for initiation codon selection. Together, our data suggest a model in which the common exon on the 5′ end of HIV-1 transcripts acts as the ribosome landing pad, recruiting the translation machinery upstream of the initiation codon followed by 5′ to 3′ ribosome scanning to each transcript’s initiator AUG. Our results demonstrate how a single RNA structural element, grafted onto different transcripts by alternative splicing, can serve to provide a common function to an otherwise diverse family of mRNAs.
Results
Sequences spliced to the common exon do not function as IRESs
We previously demonstrated that the 5′ leaders from the HIV-1 transcripts, gag, nef, vif, vpr and vpu function as IRESs and that in the gag leader, the most important regions for internal initiation are within the common exon.14 As the common exon is found in all the leaders, these observations led us to hypothesize that this exon confers IRES activity to the different HIV-1 leaders. If this hypothesis is correct, then the sequences downstream of the common exon, which we refer to as leader specific sequences (LSS), would not be expected to function as IRESs. To directly test this, we inserted the LSSs of gag, nef, vif, vpr, and vpu into a dual-luciferase reporter vector (Fig. 2A) and measured IRES activity in transfected Jurkat T-cells, a T-cell model permissive for HIV-1 replication. Consistent with our previous report,14 the full-length leaders (common exon + LSS) displayed IRES activity substantially greater (40–160 fold) than the negative control β-globin-containing plasmid (data not shown). When only the LSSs were tested, IRES activity was reduced by 95% compared with the full-length construct (Fig. 2B). Thus, each LSSs in isolation does not appear to function as an IRES. The loss of IRES activity observed in the absence of the common exon indicates that sequences within the common exon are critical for IRES activity in all HIV-1 5′ leaders tested.
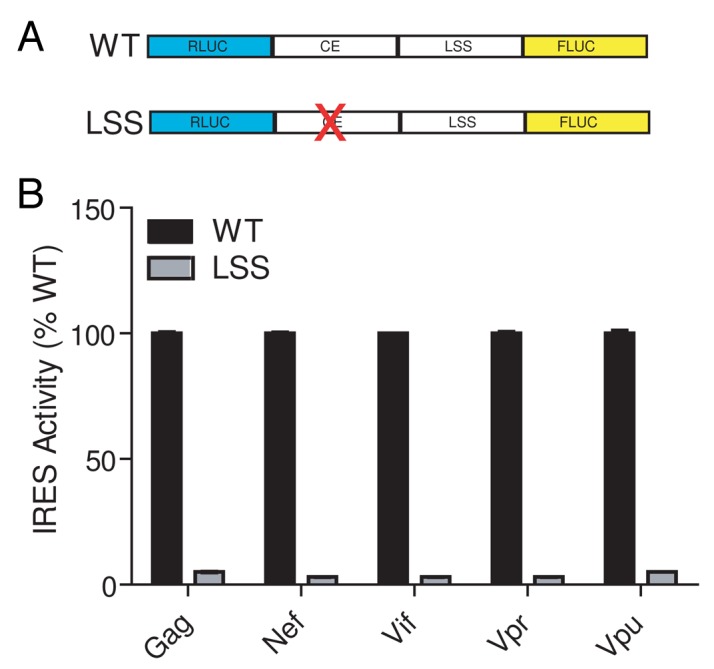
Figure 2. The leader specific sequences do not act as IRESs. A. Diagram of reporter constructs. 5′ leader DNA sequences from the gag, nef, vif, vpr and vpu mRNAs were inserted between two luciferase genes. Transcription from this DNA construct in cells produces a capped and poly-adenylated dicistronic message. Translation of the upstream cistron (renilla luciferase, RLUC) is cap-dependent while translation of the downstream cistron (firefly luciferase, FLUC) will occur if there is an upstream IRES present. IRES activity is defined as the ratio of FLUC to RLUC light production (relative light units, RLUs). WT constructs contain the entire 5′ leader (common exon plus LSS) while the LSS construct has the common exon sequence removed. B. IRES activity was measured from Jurkat T-cells transfected with dual-luciferase DNA constructs containing WT and LSS sequences (as shown in panel A). IRES activity from each WT leader construct is set at 100%.
The common exon adopts a similar conformation in HIV-1 leaders
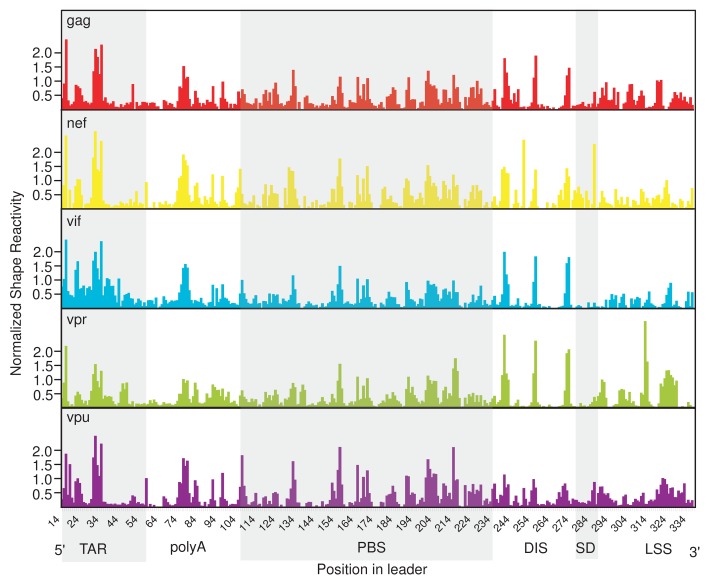
Having demonstrated that the common exon is critical for IRES activity in the HIV-1 leaders tested, we next considered whether the common exon sequence was conferring internal initiation to all of these different mRNAs using a structure that is the same in the context of all of these transcripts. While the secondary structure of the gag RNA has been intensively studied,1,15-20 much less is known about the structures formed by the leaders of other HIV-1 transcripts. Therefore, we interrogated the secondary structure of several full-length HIV-1 leader RNAs with selective 2′ hydroxyl acylation analyzed by primer extension (SHAPE). SHAPE is a chemical probing technique that modifies the 2’ hydroxyl group of flexible RNA nucleotides, such as those found in single-stranded regions, loops and bulges.21 We used this technique to obtain a “fingerprint” of the inherent flexibility of HIV-1 leader RNAs at nucleotide resolution. Within the region of the common exon for which we obtained quantitative SHAPE data (nts 14–289), we observed a similar SHAPE reactivity profile in all leaders tested (Fig. 3B, TAR through SD). Where a peak or valley was observed in the profile of one leader, it was almost always observed in the profile of the others. In contrast, the regions downstream of the common exon (nts 290–336) (Fig. 3B, LSS) had very different SHAPE profiles with no discernible similarities in pattern (a complete data set of SHAPE reactivity for each leader RNA is included in Supplemental Material Table S1). The similar SHAPE profiles of the common exon in different leaders suggests that this RNA region is folding independently of the LSS and is consistent with our data demonstrating that the LSSs do not function as IRESs. To more quantitatively assess the similarities and differences in the SHAPE profiles, we determined how well the SHAPE reactivity of the different RNAs correlated using the Pearson’s product-moment correlation coefficient. We first compared our SHAPE probing of the gag leader to a previously published study16 and found that they were well correlated (R = 0.85). We then compared the reactivity profiles of the common exon in different leaders and found that they were also well correlated (0.70 ≤ R ≤ 0.85, Table 1). In contrast, comparing the SHAPE profile of the LSSs (nts 290–336) to each other revealed no correlation (R ≤ 0, data not shown). Taken together, the well-correlated SHAPE profiles of the common exon in the context of each leader strongly suggest that the common exon is adopting a similar conformation in each.
Figure 3. The common exon forms a similar structure in all HIV-1 transcripts. Quantitated, normalized, and background-corrected modification data from at least two independent SHAPE probing experiments. The relative SHAPE reactivity is on the y-axis and the defined RNA elements of the common exon are shown 5′ to 3′ on the x-axis (gray and white shading). RNA elements include the trans-activating region (TAR), the polyadenylation signal (poly (A)), the primer binding site (PBS), the dimerization initiation site (DIS), the major splice donor site (SD) and the packaging signal (PSI) and the leader specific sequences (289–336 nts).
Table 1. Pearson’s product-moment correlation coefficients for common exon shape reactivities.
| gag | nef | vif | vpr | vpu | |
|---|---|---|---|---|---|
| gag | 1.0 | 0.78 | 0.85 | 0.83 | 0.82 |
| nef | 0.78 | 1.0 | 0.70 | 0.70 | 0.80 |
| vif | 0.85 | 0.70 | 1.0 | 0.79 | 0.74 |
| vpr | 0.83 | 0.70 | 0.79 | 1.0 | 0.70 |
| vpu | 0.82 | 0.80 | 0.74 | 0.70 | 1.0 |
The DIS is used in IRES activity in different HIV-1 leader RNAs
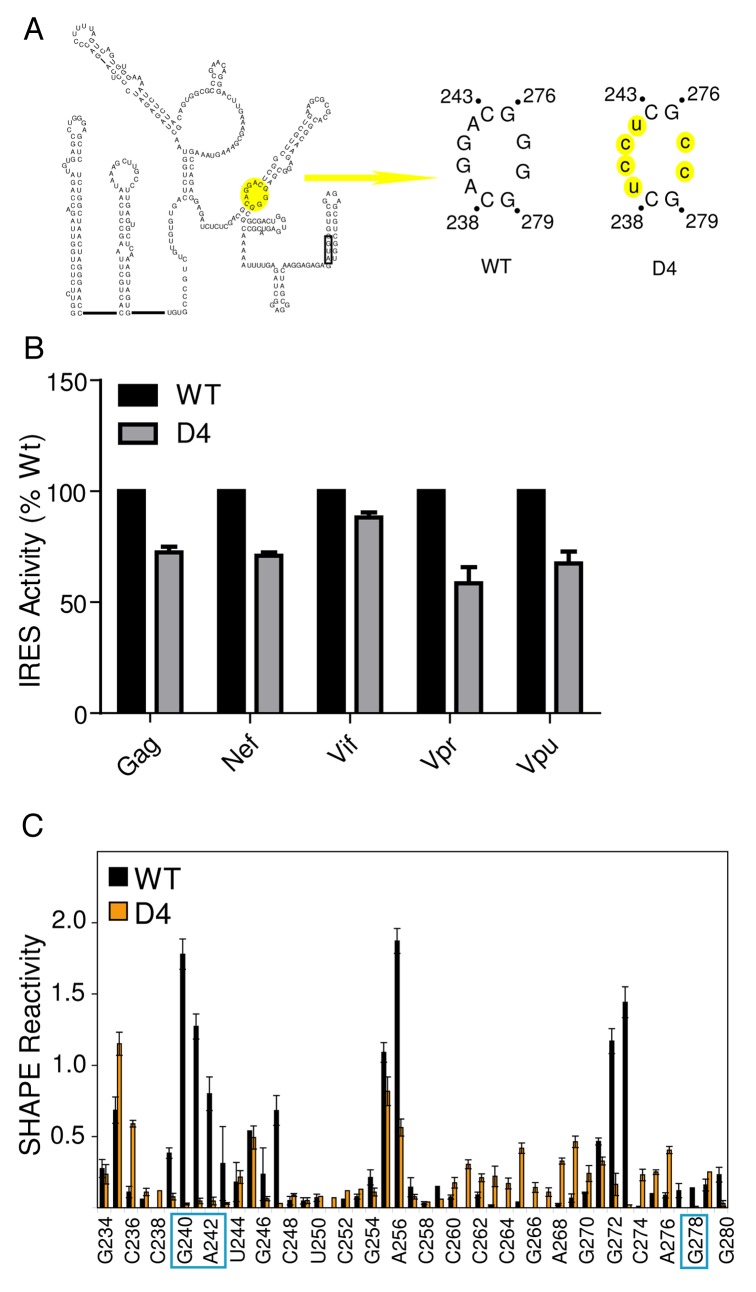
Because the function of an IRES RNA can directly depend on its structure, and having established that the common exon was adopting a similar conformation in HIV-1 leader RNAs, we hypothesized that the different leaders use a similar RNA structure-based mechanism of internal initiation. If true, a mutation made in the common exon that disrupts IRES activity in one HIV-1 leader should also do so in the other leaders. We tested this prediction by inserting the gag IRES inhibitory mutation D414 (Fig. 4A) into the gag, nef, vif, vpr and vpu HIV-1 leader RNAs and measured IRES activity. As in our previous study,14 the D4 mutation in the gag leader inhibited IRES activity relative to the WT counterpart (Fig. 4B). Consistent with our hypothesis, the D4 mutation in the nef, vif, vpr and vpu leaders inhibited IRES activity by 15–40% relative to their WT counterparts. While the D4 mutation did not inhibit IRES activity to the same degree in all leaders, this result suggests that this region of the common exon plays a role in internal initiation in all HIV leaders.
Figure 4. Regions within the DIS are important for IRES activity in HIV-1 transcripts. A. Schematic diagram of one of two proposed secondary structure models of the HIV-1 gag 5′ leader. The AUG hairpin model is presented.16,58 Base-pairing within the DIS domain I is proposed to be identical in each. The D4 mutation is highlighted in yellow in the AUG hairpin. WT denotes the common exon sequence naturally contained in all leader RNAs, and D4 denotes the mutation made in the leader RNAs. Mutated nucleotides are lower case and highlighted in yellow. B. IRES activity measured from Jurkat cells transfected with dual-luciferase DNA constructs containing WT and D4 sequences (as shown in panel A). IRES activity from each WT leader construct is set at 100%. C. Quantitated, normalized, and background-corrected modification data from at least two independent SHAPE probing experiments of the WT gag leader RNA and the D4 mutant RNA. The region surrounding the mutation is presented. The relative SHAPE reactivity is on the y-axis and the sequence is on the x-axis. Black bars denote reactivity of WT RNA, orange bars are reactivity of mutant RNA. The locations of the mutated nucleotides are boxed.
Structural integrity of the DIS is important for IRES function
The D4 region has a high degree of similarity when reactivity profiles of the different leaders are compared (nts 234–282 0.62 < R > 0.96), suggesting that the structural integrity of this region is important for function. To determine if the D4 mutation resulted in structural changes in the IRES RNA, we probed the gag D4 mutant RNA by SHAPE. We observed a significant loss in SHAPE reactivity in the D4 mutant within the DIS (nts 239–242) compared with WT, but no substantial change in the reactivity profile outside of this region (Fig. 4C). This loss in reactivity corresponds to the mutated nucleotides and suggests that they are involved in new base-pairing interactions. While we cannot explicitly identify where these new interactions are occurring, collectively the data indicate that local changes in SHAPE reactivity correlate with a loss in IRES activity and suggest that this activity may be dependent on the DIS element’s structural integrity.
HIV-1 leader IRESs employ a “land and scan” mechanism for initiation codon identification
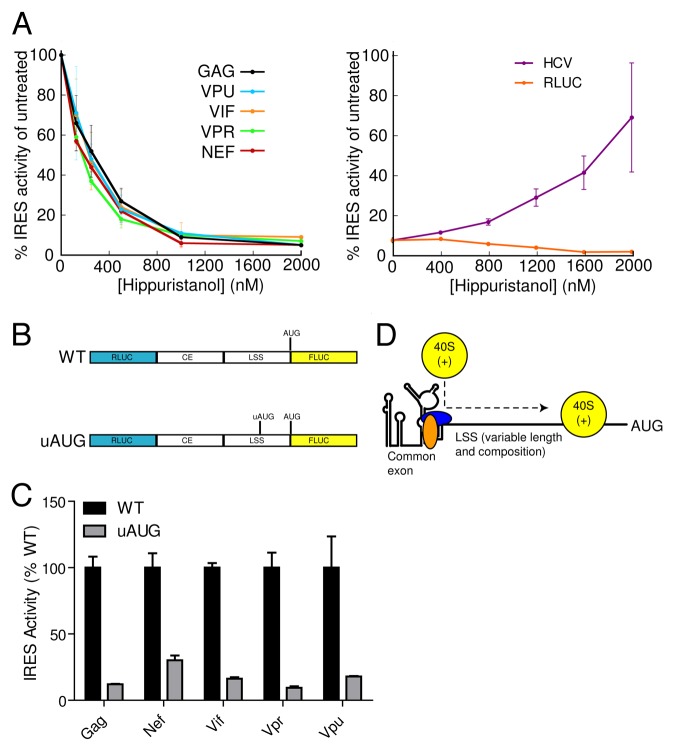
Having identified a common, functionally important IRES structural element in multiple HIV-1 leader RNAs, we next examined the mechanism by which ribosomes identify initiation codons in leaders of different lengths. IRESs position the 40S ribosome at the start codon through either direct recruitment to the initiator codon or via a “land and scan” approach in which the recruited 40S subunit scans to the start codon (Reviewed in 10, 22). The variability in length of the LSSs causes the initiator AUG to be placed as close as 47 nt (gag) and as far as 535 nt (nef) from the 3′ end of the common exon. The disparity in LSS length and sequence suggests that the leaders use a “land and scan” mechanism. To test this, we determined if IRES-harboring mRNAs were dependent on eIF4A for activity. We took advantage of hippuristanol, a small molecule inhibitor of eIF4A, that can be used to distinguish between eIF4A-dependent IRESs (such as poliovirus) and eIF4A-independent IRESs (such as the hepatitis C virus (HCV) IRES).23 Hippuristanol treatment of HeLa cells transfected with HIV-1 leader constructs inhibited IRES activity in a dose-dependent manner (Fig. 5A), with calculated IC50 values ranging from 163 to 296 nM (Table 2). This result is consistent with a previous study demonstrating that translation from the gag 5′UTR is eIF4A-dependent, although in that study capped monocistronic transcripts were used and thus the mode of initiation was likely different.24 As expected, cap-dependent translation from the RLUC reporter was also inhibited by hippuristanol treatment (Fig. 5A). In contrast the control HCV IRES was resistant to and even stimulated by hippuristanol treatment consistent with this IRES not requiring eIF4A for translation initiation (Fig. 5A).
Figure 5.HIV-1 IRES containing transcripts employ a “land and scan” mechanism. A. IRES activity was measured from cells transfected with dual-luciferase DNA constructs containing WT leader sequences and treated with increasing concentrations of the eIF4A inhibitor hippuristanol. B. An uAUG was inserted out of frame from the FLUC AUG in each leader specific sequence. C. IRES activity was measured from Jurkat T-cells transfected with dual-luciferase DNA constructs containing WT and uAUG sequences (as shown in panel B). IRES activity from each WT leader construct is set at 100%. D. Diagram of the “land and scan” model for initiation from different HIV-1 transcripts. In this model, the common exon structure, likely bound by ITAFs (blue and orange ovals), serves as the site for entry of the 40S subunit and associated factors (yellow, labeled 40S(+)). The 40S(+) scans through each LSS to reach the start codon. Different lengths, sequences, structures, bound proteins, etc. in the different LSSs could modulate the efficiency of scanning and thus of translation initiation.
Table 2. IC50 values for hippuristanol inhibition, with the associated error of IC50 values shown in parenthesis.
| Construct | IC50 (nM) |
|---|---|
| gag | 296.2 (114.4) |
| nef | 200.6 (108.8) |
| vif | 250.4 (110.7) |
| vpr | 162.8 (105.4) |
| vpu | 275.4 (114.6) |
The results above demonstrate that HIV-1 leaders require eIF4A for IRES activity and suggest the use of a scanning mechanism, but eIF4A dependence is insufficient to demonstrate a scanning mechanism since eIF4A can function in IRES activity independently of scanning. For example, the encephalomyocarditis virus (EMCV) IRES uses eIF4A to facilitate a conformational change that prepares the IRES for ribosome binding.25 Therefore, we investigated scanning at the RNA level by taking advantage of the sensitivity of a scanning ribosome to AUGs inserted upstream of the canonical initiation codon.26,27 We inserted an AUG start codon (uAUG), in a favorable context ten nucleotides upstream and out of frame with the FLUC AUG in each HIV-1 leader (Fig. 5B). If scanning occurs, the ribosome will recognize the uAUG as the start site and synthesize a frame-shifted nonsense protein instead of the FLUC reporter.28 Insertion of the uAUG strongly inhibited downstream reporter activity in all HIV-1 leaders by ≥ 75% compared with their WT counterparts (Fig. 5C). This result indicates that most ribosomes recognized the uAUG as the start codon rather than the authentic start site and is consistent with ribosome scanning occurring following recruitment of ribosomes to the RNA. The significant, but incomplete, repression of reporter synthesis could arise as a consequence of some scanning ribosomes bypassing the uAUG and initiating at the downstream start codon. Alternatively, a low fraction of ribosomes may be directly transferred from the IRES to the AUG codon. Overall, the data strongly suggest that the majority of internal initiation occurs on the different HIV-1 leaders through a “land and scan” mechanism of internal initiation (Fig. 5D).
Discussion
The presence of a common exon on the 5′ end of HIV-1 transcripts is well documented, but the functional significance of this sequence outside of splicing has not been explored. The fact that different HIV-1 transcripts have IRES activity,14 and all contain the common exon, suggests that the common exon sequence may be involved in internal initiation in all these transcripts. However, considering the diversity in RNA sequence downstream of the common exon in the leaders of these transcripts, the possibility also existed that each leader functioned as an IRES independent of the common exon. Exploring these possibilities, here we show that the common exon is important for IRES activity in different transcripts, that the structure of the exon is the same in all transcripts, and that all of the transcripts likely employ a “land and scan” mechanism of ribosome recruitment (Fig. 5D).
Our finding that the common exon sequence confers IRES activity to these transcripts can be interpreted in different ways. One possibility is that the common exon and each different LSS synergistically function together as an independent IRES element. This could occur if the common exon and each LSS cooperate to fold into a unique IRES RNA structure; removal of the common exon would disrupt formation of this structure and thus eliminate IRES activity. However, our structural probing data are not consistent with this possibility since the results indicate that the common exon adopts a similar conformation in all HIV-1 leaders (Fig. 3). This suggests that the common exon folds independently of the downstream sequences. Thus, we favor the interpretation that the common exon is the primary unit required for internal initiation and that it is solely responsible for ribosome recruitment to the gag, nef, vif, vpr and vpu mRNAs. Further support for this comes from the identification of a region of the common exon, that when mutated, inhibits IRES activity in all HIV-1 leaders (Fig. 4). If each leader depends on its own unique structure and mechanism, it is highly unlikely that the same mutation would inhibit IRES activity in all mRNA species. Thus, the data strongly support the hypothesis that the common exon is functioning as the primary driver of IRES activity in all HIV-1 leaders we tested.
Additional evidence supporting the hypothesis that the common exon is functioning as a common IRES element in different HIV-1 transcripts comes from our mechanistic analysis of initiation codon identification. Specifically, the HIV-1 leaders we tested all require eIF4A for activity and are all sensitive to uAUGs, indicative of a mechanism in which the ribosome scans through the LSS to reach the start codon. Given the diversity in IRES mechanisms for ribosome recruitment and start codon identification, these results suggest a unified mechanism for IRES activity from different HIV-1 leaders. A mechanism that includes scanning for start codon identification may also explain why IRES activity is variable among HIV-1 common exon-containing leaders. If ribosomes are recruited upstream of the LSS, then downstream sequences could modulate the efficiency by which ribosomes scan to the initiator AUG. This modulation might depend on LSSs length, secondary structure, context of the initiator AUG, and/or protein-mRNA interactions. Individually or in combination, these features could impact the efficiency of formation of an elongation-competent ribosome at the AUG and subsequent translation efficiency. Alternatively, the LSSs may effect initial ribosome recruitment to the leaders.
The identification of an IRES inhibitory mutation in the lower loop of the DIS provides further insight into the mechanism of internal initiation from the common exon IRES. In the context of the gag leader, introduction of this mutation results in a significant and local change in SHAPE reactivity (Fig. 4C). Specifically, we observed a loss in reactivity within the lower loop of the DIS where the mutation is introduced. Within the common exon, no other significant changes in SHAPE reactivity were observed, suggesting that the mutation results in a local structural change, and thus IRES activity from the common exon likely depends on the integrity of this lower loop structure. Because this mutation results in a local change in structure and only partially inhibits IRES activity, we hypothesize that this loop structure contributes to IRES activity by binding an IRES trans-acting factor (ITAF). ITAFs are proteins that are not part of the canonical initiation factor set, but that bind to IRESs and facilitate their function.29-31 This hypothesis, while not fully tested, is supported by studies in which the addition of HeLa lysate altered the chemical probing pattern in this region of the leader, implying that proteins bind to and alter the RNA structure.32 Precedent for an ITAF requirement has been established with the gag 5′ leader, and proteins such as hnRNP A1,33 DDX3, eIF5A and hRIP34 have all been identified as putative HIV ITAFs. It is possible that one of these proteins, or a yet to be identified ITAF, binds the lower loop of the DIS and facilitates IRES activity. The D4 mutation also provides insight as to why the related HIV-2 5′ leader does not contain an IRES.35-38 Differences in the sequence and structure of the D4 region of the DIS elements in HIV-1 and HIV-2 could potentially change the ability to bind putative ITAFs.
While our results are consistent with a common, unified mechanism for internal initiation from common exon-containing transcripts, there may be some differences. A recent study found that in the context of HIV-1 and poliovirus infection, vpr protein synthesis is abolished, while gag protein synthesis is maintained.39 If these are both being translated by IRES elements (as we propose), then the reported differences may be highlighting functional variation due to events after ribosome recruitment.39 Because poliovirus infection results in cleavage of eIF4G and PABP40-43 and if the ribosomes recruited to the vpr mRNA template have a greater dependency on eIF4G and/or PABP for scanning or subunit joining (compared with those recruited to the gag template), then translation of vpr mRNA would be more affected. Alternatively, the poliovirus mRNA templates may be titrating canonical factors or ITAFs for which the vpr 5′ UTR has a greater dependency. While we did not specifically examine the dependence of the HIV-1 IRESs on eIF4G and PABP, our findings indicate that the factor requirements of these IRESs converge: all require the common exon, eIF4A, and a “land-and-scan” mechanism for initiation codon identification.
The HIV-1 translation initiation literature contains reports of at least five different mechanisms for initiation of protein synthesis. These include the canonical cap-dependent mechanism,44,45 three different cap-dependent but eIF4E-independent mechanisms,46-49 and two different IRES-dependent mechanisms.11,12 These apparently conflicting reports may actually be providing insight into how the virus has evolved to use a number of different strategies to maximize its ability to initiate under a variety of cellular conditions.38,50 HIV-1 uses diverse nuclear export pathways and splicing strategies, thus it may not be surprising that it has evolved to use more than one mode of ribosome recruitment. Given that viral infection poses many stresses on the cell which result in global inhibition of cap-dependent translation initiation, including cleavage of eIF4GI and PABP,51-53 the ability of multiple HIV-1 transcripts to initiate both cap-dependently and internally, and to do so in a regulated and flexible manner, may confer an advantage to the virus. By extension, a similar strategy could be used by cellular mRNAs as a means to regulate translation of families of mRNAs generated by alternative splicing or with related, but not identical, leaders.
Materials and Methods
Plasmid construction
IRES activity was studied using the dual-luciferase reporter vector pRL.54 The 5′ UTRs of HIV-1 gag, nef, vif, vpr and vpu were cloned between the RLUC and FLUC sequences of the pRL vector as described previously.14 For clarity, we generically refer to these constructs as “WT leader-pRL.” The dual-luciferase reporter vector pRL containing the β-Globin 5′ leader insert was a gift from the Krushel lab and the HCV IRES-containing reporter plasmid has been described previously.55 LSS constructs were generated by PCR amplification of the LSS from the respective WT leader-pRL plasmid, adding a 5′ EcoRI and 3′ NcoI site, and subcloned back into the EcoRI and NcoI sites of pRL. The D4 and uAUG mutations were generated by PCR amplification of the respective WT leader-pRL plasmid, using construct-specific, mutation-encoding primers to produce two overlapping fragments of the 5′ leader harboring the desired sequence changes. The fragments were annealed, extended, and amplified by a second PCR with common primers and subcloned back into the EcoRI and NcoI sites of pRL. We generically refer to these constructs as D4-prL or uAUG-prL. Primers used in this study are listed in Supplementary Material Table S2.
Plasmids harboring the templates for in vitro transcription of RNA were generated by PCR amplification of WT leader sequences from the respective WT leader-pRL plasmid or the gag D4 mutation from the D4-prL construct. The 5′ primer added an EcoRI site followed by a T7 promoter and a hammerhead ribozyme and the 3′ primer added a hepatitis delta ribozyme and a BamHI site. The insert was cloned into the EcoR1 and BamHI sites of a pUC19 vector. We generically refer to the WT constructs as “WT-pUC19” and the gag D4 construct as “D4-pUC19.” All plasmids used in this study were verified by sequencing.
Cell Culture, Transfections, and Hippuristanol treatments
Jurkat cells (ATCC #TIB-152) were maintained in RPMI 1640 supplemented with 10% FBS at 37 °C in a 5% CO2 atmosphere. Cells were transfected with Lipofectamine LTX and Plus reagents (Invitrogen) at a density of 100,000 cells per well in a 24 well dish. Cells were transfected with 1 µg plasmid DNA, 1 µL Plus reagent and 1 µL Lipofectamine LTX, according to the manufacturer’s protocol. Cells were transfected in standard media containing 4ug/mL Concanavalin A (Sigma). Twenty-four hours post-transfection, cells were harvested with 100 µL 1X Passive Lysis Buffer (Promega). Samples were either stored at -20 °C, or assayed directly for luciferase activity.
HeLa cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS at 37 °C in a 5% CO2 atmosphere. Twenty-four hours prior to transfection, cells were plated at 60% confluency in a 24 well dish. HeLa cells were transfected with 250 ng DNA, 0.25 µL Plus reagent and 0.5 µL Lipofectamine LTX, according to the manufacturer’s protocol (Invitrogen). Twenty-four hours post-transfection, cells were harvested as described for Jurkat cells.
For experiments assessing IRES sensitivity to hippuristanol, HeLa cells were transfected (using Lipofectamine 2000) with expression plasmids encoding dicistronic mRNAs harboring the different 5′ UTRs as intercistronic region. Three hours after transfection, the media was replaced, and the indicated concentrations of hippuristanol added for 6 h. Cells were harvested in passive lysis buffer and luciferase activity measured using the dual-luciferase assay system (Promega).
Luciferase assays
Lysates were analyzed for luciferase activity using the dual-luciferase assay reporter system (Promega). Lysates were analyzed in a 96 well plate format on a Glomax plate reader. Aliquots of 20–50 µL lysate were analyzed with 100 µL LAR II reagent and 100 µL 1 x Stop N Glo reagent (Promega). Results are reported as the average measurement obtained from at least three independent transfections, with each construct transfected in triplicate and the results normalized to the negative control B-globin plasmid.
In vitro transcription and RNA purification
Template DNA used for in vitro transcription of WT or the gag D4 leader RNAs was amplified from the respective WT-pUC19 or D4-pUC19 plasmid with M13-Forward and a reverse primer containing the sequence 5′-CCAGCGAGGAGGCTGGGACCATGC-3′ (“internal delta primer”) that sits just 3′ of the leader sequence in the hepatitis delta ribozyme. The resulting PCR product contains a T7 promoter followed by a 5′ hammerhead ribozyme, the template of interest, and a partial 3′ hepatitis delta ribozyme. The PCR reaction was digested with DpnI to remove any contaminating plasmid DNA and the resultant PCR templates were used in a 100 µL in vitro transcription reaction using the MegaScript T7 Kit (Ambion), according to the manufacturer’s protocol. The template DNA was digested with DNase I and the remaining RNA was ethanol precipitated. The RNA pellet was resuspended in 1 mL of water and an additional 1 mL of triethylammonium acetate (TEAA) buffer (pH 7.0) was added. The solution was passed through a 0.22 µM syringe filter, heated and separated by reverse phase high performance liquid chromatography (RP-HPLC) on a Varian PLRP-S1000A HPLC column (150 × 7.5 mm, 8 μm) equipped with a precolumn (4.6 mm × 20 mm) at 75 °C on an Agilent 1260 infinity HPLC system. The RNA was passed through a gradient of buffer A (100 mM TEEA, pH 7.0) and buffer B (100 mM TEAA, 50% v/v Acetonitrile) and the chromatograms were monitored at 260 nm and the peak corresponding to the full-length RNA was collected. The gradient conditions and elution times varied by RNA. Nef: 98% buffer A for six minutes, 78% for 64 min, 71.6% for seven minutes. Gag, Vif, Vpr, and Vpu: 90% buffer A for two minutes, 82% for 48 min, 70% for 15 min. Collected RNA samples were concentrated using a centrifugal filter with an Amicon Ultra 50,000 Dalton molecular weight cutoff (Millipore). Purity was assessed by PAGE and concentration was determined spectrophotometrically.
Selective 2′ hydroxyl acylation analyzed by primer extension (SHAPE)
SHAPE was performed essentially as previously described.56 In brief, 20 pmoles RNA in 24 ul of RNase-free water was heated at 85 °C for 1 min, and cooled at RT for 5 min. A total of 12 ul of RNA folding buffer (222mM HEPES pH 8.0, 20mM MgCl2, 333mM NaCl) was added and incubated at 37 °C for 10 min. The reaction was divided equally between two tubes. One tube received 2 ul of NMIA prepared in DMSO; the other received 2 µL of neat DMSO. The final NMIA concentration varied based on the RNA and ranged from 1.25–5 mM. Reactions were incubated at 37 °C for 35 min and desalted using Micro Bio-Spin 30 Tris Chromatography Columns (BioRad).
Reverse transcription (RT) was performed with 9 µL of desalted RNA and 3 µL of 0.3 µM FAM labeled primer (for the nef RNA, the primer sequence was 5′-ctattccttcgggcctgtcg-3′, for the remaining RNAs, the primer sequence was the internal delta primer). RT reactions were heated in a thermocycler at 65 °C for 5 min followed by 42 °C for 20 min and placed on ice. Six µll Superscript III enzyme mix (a 4:1:1 mixture of 5x first strand buffer (Invitrogen), 100 mM DTT, and a solution that is 10 mM in each dNTP) was added. The reactions were returned to the thermocycler and heated at 52 °C for 1 min. Each reaction received 1 µL of Superscript III (Invitrogen) and the reaction proceeded at 52 °C for 20 min, 60 °C for 5 min and a 4 °C hold. Sequencing reactions were performed essentially as described above, using 2 pmoles RNA, a Hex or Fam labeled primer, and an enzyme mix with 1 µL of ddNTP in place of the corresponding dNTP. One modification reaction or DMSO reaction was combined with two sequencing reactions and desalted using G25 sephadex (BioRad). The desalted cDNAs were added to HiDi Formamide (ABI) and resolved by capillary electrophoresis on an ABI 3500 genetic analyzer. The resultant ABIF files were analyzed using QuSHAPE software57 following the default parameters. The alignment of the ladders was inspected and corrected manually when necessary. Each RNA was analyzed in two or more independent experiments and the normalized SHAPE reactivities were averaged. The averages and standard deviation reactivities for the full leader sequences are provided in the Supplemental Material Table S1.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank current and former Kieft Lab members for thoughtful discussions and technical assistance, David Costantino for reading of this manuscript, and Nahum Sonenberg (McGill University), Anne Willis (Medical Research Council, UK) and Leslie Krushel (MD Anderson Cancer Center) for plasmids. This work was supported by the National Institutes of Health (GM081346 and GM097333 to JSK, T32 AI052066 to TDMP) and the Canadian Institutes of Health Research (MOP-106530 to JP). TDMP was an American Heart Association Predoctoral Fellow (10PRE260143). JSK is an Early Career Scientist of the Howard Hughes Medical Institute.
Author Contributions
TDMP and JW conducted the bulk of the experiments and analysis, with the exception of the hippuristanol l-inhibition experiments, which were conducted by RC in the laboratory of JP. TDMP, JW, and JSK designed the study and the experiments, and wrote the manuscript with input from all authors.
REFERENCES
- 1.Berkhout B.. Structure and function of the human immunodeficiency virus leader RNA. Prog Nucleic Acid Res Mol Biol 1996; 54:1 - 34; http://dx.doi.org/ 10.1016/S0079-6603(08)60359-1; PMID: 8768071 [DOI] [PubMed] [Google Scholar]
- 2.Sonenberg N, Hinnebusch AG.. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 2009; 136:731 - 45; http://dx.doi.org/ 10.1016/j.cell.2009.01.042; PMID: 19239892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson RJ.. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem Soc Trans 2005; 33:1231 - 41; http://dx.doi.org/ 10.1042/BST20051231; PMID: 16246087 [DOI] [PubMed] [Google Scholar]
- 4.Balvay L, Lopez Lastra M, Sargueil B, Darlix JL, Ohlmann T.. Translational control of retroviruses. Nat Rev Microbiol 2007; 5:128 - 40; http://dx.doi.org/ 10.1038/nrmicro1599; PMID: 17224922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinnebusch AG, Lorsch JR.. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol 2012; 4:4; http://dx.doi.org/ 10.1101/cshperspect.a011544; PMID: 22815232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aitken CE, Lorsch JR.. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol 2012; 19:568 - 76; http://dx.doi.org/ 10.1038/nsmb.2303; PMID: 22664984 [DOI] [PubMed] [Google Scholar]
- 7.Kozak M.. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol 1991; 115:887 - 903; http://dx.doi.org/ 10.1083/jcb.115.4.887; PMID: 1955461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozak M.. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 1986; 44:283 - 92; http://dx.doi.org/ 10.1016/0092-8674(86)90762-2; PMID: 3943125 [DOI] [PubMed] [Google Scholar]
- 9.Jackson RJ.. The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb Perspect Biol 2013; 5:5; http://dx.doi.org/ 10.1101/cshperspect.a011569; PMID: 23378589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plank TD, Kieft JS.. The structures of nonprotein-coding RNAs that drive internal ribosome entry site function. Wiley Interdiscip Rev RNA 2012; 3:195 - 212; http://dx.doi.org/ 10.1002/wrna.1105; PMID: 22215521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buck CB, Shen X, Egan MA, Pierson TC, Walker CM, Siliciano RF.. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J Virol 2001; 75:181 - 91; http://dx.doi.org/ 10.1128/JVI.75.1.181-191.2001; PMID: 11119587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brasey A, Lopez-Lastra M, Ohlmann T, Beerens N, Berkhout B, Darlix JL, Sonenberg N.. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J Virol 2003; 77:3939 - 49; http://dx.doi.org/ 10.1128/JVI.77.7.3939-3949.2003; PMID: 12634354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charnay N, Ivanyi-Nagy R, Soto-Rifo R, Ohlmann T, López-Lastra M, Darlix JL.. Mechanism of HIV-1 Tat RNA translation and its activation by the Tat protein. Retrovirology 2009; 6:74; http://dx.doi.org/ 10.1186/1742-4690-6-74; PMID: 19671151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plank TD, Whitehurst JT, Kieft JS.. Cell type specificity and structural determinants of IRES activity from the 5′ leaders of different HIV-1 transcripts. Nucleic Acids Res 2013; 41:6698 - 714; http://dx.doi.org/ 10.1093/nar/gkt358; PMID: 23661682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW Jr., Swanstrom R, Burch CL, Weeks KM.. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 2009; 460:711 - 6; http://dx.doi.org/ 10.1038/nature08237; PMID: 19661910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM.. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol 2008; 6:e96; http://dx.doi.org/ 10.1371/journal.pbio.0060096; PMID: 18447581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baudin F, Marquet R, Isel C, Darlix JL, Ehresmann B, Ehresmann C.. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J Mol Biol 1993; 229:382 - 97; http://dx.doi.org/ 10.1006/jmbi.1993.1041; PMID: 8429553 [DOI] [PubMed] [Google Scholar]
- 18.Berkhout B, Ooms M, Beerens N, Huthoff H, Southern E, Verhoef K.. In vitro evidence that the untranslated leader of the HIV-1 genome is an RNA checkpoint that regulates multiple functions through conformational changes. J Biol Chem 2002; 277:19967 - 75; http://dx.doi.org/ 10.1074/jbc.M200950200; PMID: 11896057 [DOI] [PubMed] [Google Scholar]
- 19.Damgaard CK, Andersen ES, Knudsen B, Gorodkin J, Kjems J.. RNA interactions in the 5′ region of the HIV-1 genome. J Mol Biol 2004; 336:369 - 79; http://dx.doi.org/ 10.1016/j.jmb.2003.12.010; PMID: 14757051 [DOI] [PubMed] [Google Scholar]
- 20.Huthoff H, Berkhout B.. Two alternating structures of the HIV-1 leader RNA. RNA 2001; 7:143 - 57; http://dx.doi.org/ 10.1017/S1355838201001881; PMID: 11214176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM.. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc 2005; 127:4223 - 31; http://dx.doi.org/ 10.1021/ja043822v; PMID: 15783204 [DOI] [PubMed] [Google Scholar]
- 22.Filbin ME, Kieft JS.. Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol 2009; 19:267 - 76; http://dx.doi.org/ 10.1016/j.sbi.2009.03.005; PMID: 19362464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, Pelletier J.. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol 2006; 2:213 - 20; http://dx.doi.org/ 10.1038/nchembio776; PMID: 16532013 [DOI] [PubMed] [Google Scholar]
- 24.de Breyne S, Chamond N, Décimo D, Trabaud MA, André P, Sargueil B, Ohlmann T.. In vitro studies reveal that different modes of initiation on HIV-1 mRNA have different levels of requirement for eukaryotic initiation factor 4F. FEBS J 2012; 279:3098 - 111; http://dx.doi.org/ 10.1111/j.1742-4658.2012.08689.x; PMID: 22759308 [DOI] [PubMed] [Google Scholar]
- 25.Kolupaeva VG, Lomakin IB, Pestova TV, Hellen CU.. Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosomal entry site. Mol Cell Biol 2003; 23:687 - 98; http://dx.doi.org/ 10.1128/MCB.23.2.687-698.2003; PMID: 12509466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cigan AM, Feng L, Donahue TF.. tRNAi(met) functions in directing the scanning ribosome to the start site of translation. Science 1988; 242:93 - 7; http://dx.doi.org/ 10.1126/science.3051379; PMID: 3051379 [DOI] [PubMed] [Google Scholar]
- 27.Pestova TV, Borukhov SI, Hellen CU.. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 1998; 394:854 - 9; http://dx.doi.org/ 10.1038/29703; PMID: 9732867 [DOI] [PubMed] [Google Scholar]
- 28.Kozak M.. Selection of initiation sites by eucaryotic ribosomes: effect of inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Res 1984; 12:3873 - 93; http://dx.doi.org/ 10.1093/nar/12.9.3873; PMID: 6328442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komar AA, Hatzoglou M.. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle 2011; 10:229 - 40; http://dx.doi.org/ 10.4161/cc.10.2.14472; PMID: 21220943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis SM, Holcik M.. For IRES trans-acting factors, it is all about location. Oncogene 2008; 27:1033 - 5; http://dx.doi.org/ 10.1038/sj.onc.1210777; PMID: 17767196 [DOI] [PubMed] [Google Scholar]
- 31.Stoneley M, Willis AE.. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene 2004; 23:3200 - 7; http://dx.doi.org/ 10.1038/sj.onc.1207551; PMID: 15094769 [DOI] [PubMed] [Google Scholar]
- 32.Vallejos M, Deforges J, Plank TD, Letelier A, Ramdohr P, Abraham CG, Valiente-Echeverría F, Kieft JS, Sargueil B, López-Lastra M.. Activity of the human immunodeficiency virus type 1 cell cycle-dependent internal ribosomal entry site is modulated by IRES trans-acting factors. Nucleic Acids Res 2011; 39:6186 - 200; http://dx.doi.org/ 10.1093/nar/gkr189; PMID: 21482538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monette A, Ajamian L, López-Lastra M, Mouland AJ.. Human immunodeficiency virus type 1 (HIV-1) induces the cytoplasmic retention of heterogeneous nuclear ribonucleoprotein A1 by disrupting nuclear import: implications for HIV-1 gene expression. J Biol Chem 2009; 284:31350 - 62; http://dx.doi.org/ 10.1074/jbc.M109.048736; PMID: 19737937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Henao-Mejia J, Liu H, Zhao Y, He JJ.. Translational regulation of HIV-1 replication by HIV-1 Rev cellular cofactors Sam68, eIF5A, hRIP, and DDX3. J Neuroimmune Pharmacol 2011; 6:308 - 21; http://dx.doi.org/ 10.1007/s11481-011-9265-8; PMID: 21360055 [DOI] [PubMed] [Google Scholar]
- 35.de Breyne S, Soto-Rifo R, López-Lastra M, Ohlmann T.. Translation initiation is driven by different mechanisms on the HIV-1 and HIV-2 genomic RNAs. Virus Res 2013; 171:366 - 81; http://dx.doi.org/ 10.1016/j.virusres.2012.10.006; PMID: 23079111 [DOI] [PubMed] [Google Scholar]
- 36.Chatterjee P, Garzino-Demo A, Swinney P, Arya SK.. Human immunodeficiency virus type 2 multiply spliced transcripts. AIDS Res Hum Retroviruses 1993; 9:331 - 5; http://dx.doi.org/ 10.1089/aid.1993.9.331; PMID: 8512748 [DOI] [PubMed] [Google Scholar]
- 37.Strong CL, Lanchy JM, Dieng-Sarr A, Kanki PJ, Lodmell JS.. A 5’UTR-spliced mRNA isoform is specialized for enhanced HIV-2 gag translation. J Mol Biol 2009; 391:426 - 37; http://dx.doi.org/ 10.1016/j.jmb.2009.06.046; PMID: 19559029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ricci EP, Soto Rifo R, Herbreteau CH, Decimo D, Ohlmann T.. Lentiviral RNAs can use different mechanisms for translation initiation. Biochem Soc Trans 2008; 36:690 - 3; http://dx.doi.org/ 10.1042/BST0360690; PMID: 18631141 [DOI] [PubMed] [Google Scholar]
- 39.Monette A, Valiente-Echeverría F, Rivero M, Cohen EA, Lopez-Lastra M, Mouland AJ.. Dual mechanisms of translation initiation of the full-length HIV-1 mRNA contribute to gag synthesis. PLoS One 2013; 8:e68108; http://dx.doi.org/ 10.1371/journal.pone.0068108; PMID: 23861855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson SR, Sarnow P.. Regulation of host cell translation by viruses and effects on cell function. Curr Opin Microbiol 2000; 3:366 - 70; http://dx.doi.org/ 10.1016/S1369-5274(00)00106-5; PMID: 10972496 [DOI] [PubMed] [Google Scholar]
- 41.Gradi A, Svitkin YV, Imataka H, Sonenberg N.. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci U S A 1998; 95:11089 - 94; http://dx.doi.org/ 10.1073/pnas.95.19.11089; PMID: 9736694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kerekatte V, Keiper BD, Badorff C, Cai A, Knowlton KU, Rhoads RE.. Cleavage of Poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff?. J Virol 1999; 73:709 - 17; PMID: 9847377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joachims M, Van Breugel PC, Lloyd RE.. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J Virol 1999; 73:718 - 27; PMID: 9847378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miele G, Mouland A, Harrison GP, Cohen E, Lever AM.. The human immunodeficiency virus type 1 5′ packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J Virol 1996; 70:944 - 51; PMID: 8551634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berkhout B, Arts K, Abbink TE.. Ribosomal scanning on the 5′-untranslated region of the human immunodeficiency virus RNA genome. Nucleic Acids Res 2011; 39:5232 - 44; http://dx.doi.org/ 10.1093/nar/gkr113; PMID: 21393254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai MC, Wang SW, Cheng L, Tarn WY, Tsai SJ, Sun HS.. Human DDX3 interacts with the HIV-1 Tat protein to facilitate viral mRNA translation. PLoS One 2013; 8:e68665; http://dx.doi.org/ 10.1371/journal.pone.0068665; PMID: 23840900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soto-Rifo R, Rubilar PS, Ohlmann T.. The DEAD-box helicase DDX3 substitutes for the cap-binding protein eIF4E to promote compartmentalized translation initiation of the HIV-1 genomic RNA. Nucleic Acids Res 2013; 41:6286 - 99; http://dx.doi.org/ 10.1093/nar/gkt306; PMID: 23630313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soto-Rifo R, Rubilar PS, Limousin T, de Breyne S, Décimo D, Ohlmann T.. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J 2012; 31:3745 - 56; http://dx.doi.org/ 10.1038/emboj.2012.220; PMID: 22872150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sharma A, Yilmaz A, Marsh K, Cochrane A, Boris-Lawrie K.. Thriving under stress: selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity. PLoS Pathog 2012; 8:e1002612; http://dx.doi.org/ 10.1371/journal.ppat.1002612; PMID: 22457629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chamond N, Locker N, Sargueil B.. The different pathways of HIV genomic RNA translation. Biochem Soc Trans 2010; 38:1548 - 52; http://dx.doi.org/ 10.1042/BST0381548; PMID: 21118124 [DOI] [PubMed] [Google Scholar]
- 51.Jowett JB, Planelles V, Poon B, Shah NP, Chen ML, Chen IS.. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol 1995; 69:6304 - 13; PMID: 7666531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Castelló A, Franco D, Moral-López P, Berlanga JJ, Alvarez E, Wimmer E, Carrasco L.. HIV- 1 protease inhibits Cap- and poly(A)-dependent translation upon eIF4GI and PABP cleavage. PLoS One 2009; 4:e7997; http://dx.doi.org/ 10.1371/journal.pone.0007997; PMID: 19956697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deshmane SL, Mukerjee R, Fan S, Del Valle L, Michiels C, Sweet T, Rom I, Khalili K, Rappaport J, Amini S, et al.. Activation of the oxidative stress pathway by HIV-1 Vpr leads to induction of hypoxia-inducible factor 1alpha expression. J Biol Chem 2009; 284:11364 - 73; http://dx.doi.org/ 10.1074/jbc.M809266200; PMID: 19204000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE.. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene 1998; 16:423 - 8; http://dx.doi.org/ 10.1038/sj.onc.1201763; PMID: 9467968 [DOI] [PubMed] [Google Scholar]
- 55.Filbin ME, Kieft JS.. HCV IRES domain IIb affects the configuration of coding RNA in the 40S subunit’s decoding groove. RNA 2011; 17:1258 - 73; http://dx.doi.org/ 10.1261/rna.2594011; PMID: 21606179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McGinnis JL, Duncan CD, Weeks KM.. High-throughput SHAPE and hydroxyl radical analysis of RNA structure and ribonucleoprotein assembly. Methods Enzymol 2009; 468:67 - 89; http://dx.doi.org/ 10.1016/S0076-6879(09)68004-6; PMID: 20946765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karabiber F, McGinnis JL, Favorov OV, Weeks KM.. QuShape: rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA 2013; 19:63 - 73; http://dx.doi.org/ 10.1261/rna.036327.112; PMID: 23188808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, et al.. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science 2011; 334:242 - 5; http://dx.doi.org/ 10.1126/science.1210460; PMID: 21998393 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.