Abstract
mTOR is a protein kinase which integrates a variety of environmental and intracellular stimuli to positively regulate many anabolic processes of the cell, including protein synthesis. It exists within two highly conserved multi-protein complexes known as mTORC1 and 2 mTORC2. Each of these complexes phosphorylates different downstream targets, and play roles in different cellular functions. They also show distinctive sensitivity to the mTOR inhibitor rapamycin. Nevertheless, despite their biochemical and functional differences, recent studies have suggested that the regulation of these complexes is tightly linked to each other. For instance, both mTORC1 and 2 share some common upstream signaling molecules, such as PI3K and tuberous sclerosis complex TSC, which control their activation. Stimulation of the mTOR complexes may also trigger both positive and negative feedback mechanisms, which then in turn either further enhance or suppress their activation. Here, we summarize some recently discovered features relating to the crosstalk between mTORC1 and 2. We then discuss how aberrant mTOR complex crosstalk mechanisms may have an impact on the development of human diseases and drug resistance.
Keywords: mTORC1, mTORC2, SIN1, TSC2, protein synthesis, mRNA translation
Introduction to the mTOR Complexes
mTOR is a master molecular hub which positively controls anabolic, and conversely negatively regulates some catabolic processes. mTOR is present in two biochemically and functionally distinct multi-component complexes termed mTORC1 and mTORC2. Some components are present in both mTORC1 and 2, for instance, mTOR, mLST8,1 DEPTOR,2 Tti1, Tel2,3,4 GRp585 and Rac1.6 Yet each of the mTOR complexes possesses a unique subset of components. RAPTOR7,8 and PRAS409-13 can only be found in mTORC1, whereas RICTOR,14 SIN1,15-17 PROTOR,18 XPLN,19 NBS1,20 IKKα and IKKβ21 are specific to mTORC2 (Fig. 1). Each of these components can either support or suppress the activation of the respective mTOR complex (Table 1). Rapamycin (as a complex with the immunophilin FKBP12) rapidly inhibits some activities of mTORC1, whereas mTORC2 is resistant to short-term rapamycin treatment, although prolonged rapamycin treatment does interfere with mTORC2 in some cell types.22-24 This is thought to be due to the binding of rapamycin/FKBP12 to newly synthesized and unbound mTOR molecules, which prevents mTORC2 assembly.22
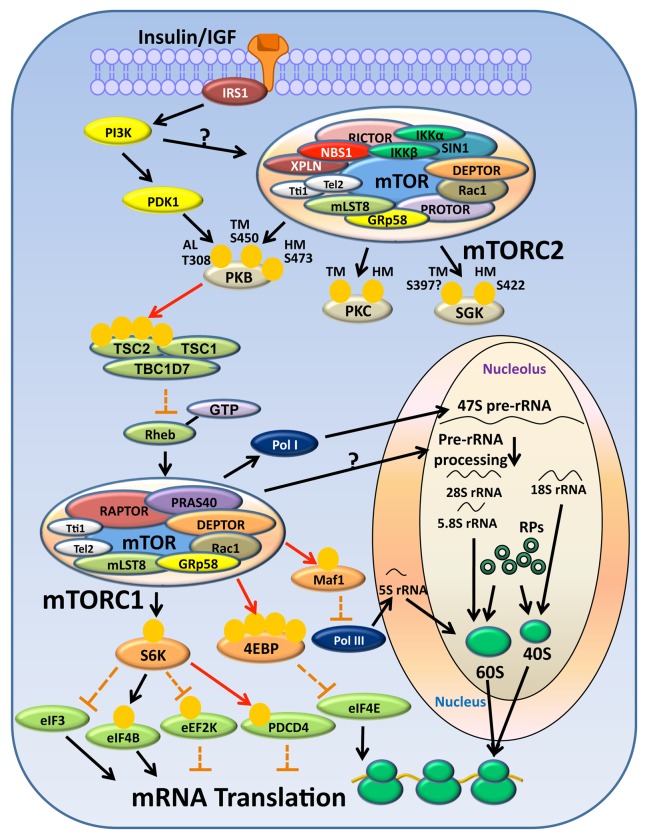
Figure 1. Pathway upstream and downstream of mTOR complexes in response to insulin/IGF stimulation and the regulation of protein synthesis by mTORC1. Insulin/IGF activates mTORC1 through IRS-PI3K-PKB pathway which in turn inhibits the GAP activity of TSC complex toward Rheb, allowing Rheb to activate mTORC1. How PI3K leads to mTORC2 activation remains to be investigated. mTORC2’s downstream targets include PKB, PKC and SGK. mTORC1 may regulate several steps in mRNA translation through its substrates S6K (and its own substrates) and 4E-BP1/2. For instance, eIF3, eIF4B, PDCD4 and eIF4E are implicated in the initiation step of translation, whereas eEF2K is involved in translation elongation. mTORC1 also positively regulates ribosome biogenesis, and this can be mediated through the activation of RNA polymerases and the processing of 47S pre-rRNA. Black arrows: positive effect or activation; red lines: phosphorylation events causing a negative effect or inactivation; discontinuous orange lines: an inhibitory mechanism inactivated in response to upstream stimulation. We indicate all the proteins reported to associate with mTORC1/2, but do not intend to imply all are necessarily associated simultaneously.
Table 1. List of mTOR complex components and their contribution to substrate recognition or complex activity.
| Complex component | Presence in mTORC1 or 2 | Contribution to substrate recognition or complex activity |
|---|---|---|
| mTOR | Both | Positive to both |
| RAPTOR | 1 | Positive |
| mLST8 | Both | No effect on 1, positive to 2 |
| GRp58 | Both | Positive to 1, no effect on 2 |
| DEPTOR | Both | Negative to both |
| Rac1 | Both | Positive to both |
| PRAS40 | 1 | Negative and/or a substrate? |
| Tti1 | Both | Positive to both |
| Tel2 | Both | Positive to both |
| RICTOR | 2 | Positive |
| SIN1 | 2 | Positive |
| PROTOR | 2 | Positive on SGK1, no effect on others |
| XPLN | 2 | Negative |
| NBS1 | 2 | Positive |
| IKKα | 2 | Positive |
| IKKβ | 2 | Positive |
mTORC1 and protein synthesis
mTORC1 is acutely activated by a variety of anabolic stimuli including growth factors, hormones and nutrients via different signaling pathways, many of which, with the notable exception of amino acid signaling,25 impinge on TSC2. TSC2 serves as a GAP of the small G protein Rheb.26-28 The inhibition of TSC2 upon its phosphorylation, induced by upstream stimuli, favors the accumulation of GTP-bound Rheb, which in turn activates mTORC1 through a yet-to-be-determined mechanism (Fig. 1).26-28 Insulin, for example, inhibits TSC2 function via the PI3K-PKB pathway.29-32 PI3K catalyzes the conversion of PIP2 to PIP3.33 The latter recruits PKB to the plasma membrane, allowing PDK1 to phosphorylate PKB on Thr308 in its activation loop, leading to its activation,34 PKB in turn phosphorylates and inhibits TSC2 (Fig. 1).29-32 Notably, the full activation of PKB also requires its phosphorylation on a hydrophobic site Ser473, which is catalyzed by mTORC2.35 Interestingly, in mouse striatal tissue, another small G protein Rhes, has also been shown to stimulate mTORC1 in its GTP-bound state.36 Nevertheless, unlike Rheb, Rhes also activates mTORC2.36
mTORC1 is tightly linked to several steps of protein synthesis including ribosome biogenesis, translation initiation and elongation. In order to meet the demand for global protein synthesis in response to rapid changes in extracellular nutrient supply, mTOR plays an important role in the control of total ribosome levels within the cell, and this is mediated through the regulation of RNA polymerases and other transcription factors which positively controls the expression of RP or rRNA genes (Fig. 1).37-42 mTORC1 upregulates the transcriptional activity of all three classes of RNA polymerases [Pol I (makes 5S rRNA), Pol II (transcribes RP genes) and Pol III (makes the other 3 major rRNAs: 28S, 18S and 5.8S)] involved in ribosome biogenesis.38 mTORC1 directly phosphorylates Maf1, a repressor of Pol III, and thereby abolishes the inhibitory effect of Maf1 on Poll III transcription (Fig. 1).39,40 mTORC1 also activates TFIIIC, a transcription factor for Pol I and Pol III.39 In yeast, TOR translocates into the nucleus in response to nutrient supply, and is associated with rDNA promoter to regulate the transcription of RP genes and 35S rRNA synthesis.37
The translation of 5′-TOP mRNAs, which encode ribosomal proteins (plus other proteins involved in anabolic processes including some translation factors) is positively regulated by mTORC1.43-45 However, it is still not fully understood how mTORC1 controls 5′-TOP mRNA translation and this remains a key question in the field. In addition, Sonenberg’s group has discovered that the phosphorylation and inhibition of 4E-BPs is responsible for the proliferative effect of mTORC1 signaling,46 and the same group has also recently reported that the translation of many cell cycle-related mRNAs is indeed controlled by mTORC1/4E-BP pathway.47 Of interest, another report from Ruggero’s group has indicated that RICTOR is highly expressed during S phase, and mTORC2 may be a key regulator of the translation of mRNAs encoding proteins that are important in S-phase.48
Translation initiation is generally considered the rate-limiting phase of protein synthesis and especially as the step at which mRNA-specific control is exerted. It relies on a subset of highly organized protein-protein interactions between the eIFs, which serve to recruit the mRNA to the 43S preinitiation complex (comprised of 40S ribosome subunit, eIF2-GTP-methionyl(Met)-tRNAi ternary complex, eIF3 and the eIF2 specific GAP eIF5). mRNA recruitment is mediated by the eIF4F (comprising the mRNA helicase eIF4A, eIF4E and the scaffold protein eIF4G) group of factors (reviewed in49,50). The formation of eIF4F is prevented by the binding of 4E-BPs to eIF4E, and this inhibitory interaction is abolished upon 4E-BP phosphorylation by mTORC1 (Fig. 1).51-56 Inhibition of mTORC1 strongly impairs the translation of mRNAs containing a 5′-TOP sequences, and recent studies have indicated that this is largely dependent on the effect of mTORC1 on 4E-BPs.43 Another well-known mTORC1 downstream target is S6K (two main isoforms, S6K1/2), which are phosphorylated and activated by mTORC1.57 Active S6Ks can phosphorylate and activate eIF4B,58 an RNA-binding protein that enhances eIF4A’s helicase activity; or phosphorylate and inactivate PDCD4, by promoting its degradation via binding to the ubiquitin ligase SCFβTRCP, and thereby releasing eIF4A from the inhibitory association with PDCD4.59 On the other hand, inactive S6K1 binds to eIF3, a scaffold protein which provides the platform for the association of mRNA and the 43S preinitiation complex, and the inhibitory binding of S6K1 to eIF3 can be prevented upon S6K1 activation by mTORC1 (Fig. 1).60,61
eEF2 mediates the translocation of the ribosome along the mRNA by one codon. The activity of eEF2 is inhibited upon phosphorylation at Thr56 by the highly specific Ca2+/calmodulin-dependent kinase, eEF2K.62 eEF2K also undergoes rapamycin-sensitive inhibitory phosphorylation at Ser78, Ser359 and Ser366.63-65 The phosphorylation of Ser359 and Ser366 (the latter catalyzed by S6Ks) severely impairs eEF2K kinase activity while the phosphorylation of Ser78 prevents the recruitment of calmodulin to eEF2K and therefore also inhibits eEF2K (Fig. 1).63-65
The regulation and functions of mTORC2
In comparison to mTORC1, the mechanism by which mTORC2 is regulated is far less well understood. Nonetheless, mTORC2 activity and signaling to PKB can also be acutely stimulated by insulin via a PI3K-dependent manner (Fig. 1),66-68 so further study is essential to elucidate the mechanism by which PI3K activates mTORC2. mTORC2 is responsible for the phosphorylation and full activation of several AGC kinases, including PKB,35 SGK1,69 cPKCs (including α, βI, βII and γ) and ε, an nPKC (Fig. 1).14,70,71 During synthesis, these AGC kinases are folded and stabilized upon phosphorylation of their TM by mTORC2, and for full activation, mTORC2 phosphorylates a further site within the HM.70,71 For example, mTORC2 associates with polysomes and phosphorylates the nascent PKB polypeptide at Thr450 (TM), which allows PKB to escape from being ubiquitinated.72 Upon stimulation of the PI3K pathway, PKB translocates to the plasma membrane and is phosphorylated by PDK1 at Thr308 (AL)34 and on Ser473 (HM) by mTORC2 (Fig. 1).35
mTORC2 was originally identified as a positive regulator of actin cytoskeletal organization and polarization,14,73-75 it also regulates cell growth,76,77 proliferation,76,78 cell survival19,24,79 and differentiation19,80-82 in certain cell types. In contrast to mTORC1, the role of mTORC2 in mRNA translation is far from clear. It has been shown that the effect of mTOR on 5′-TOP mRNA translation is mediated through mTORC1 but not mTORC2.43 However, mTORC2 may also control mRNA translation in other cell types which have not been tested before. Interestingly, in some cells,24,83 but not all,84 the stimulation of the mTORC2-PKB pathway leads to phosphorylation and inhibition of GSK3β. Active GSK3β phosphorylates eIF2Bε (the catalytic subunit of eIF2B) and inhibit eIF2B’s activity.85 eIF2B is the GEF that generates active GTP-bound eIF2. eIF2-GTP, in turn, binds to the initiator Met-tRNAi and thereby promotes 43S preinitiation complex formation and hence translation initiation. Therefore, it is plausible that the suppression of GSK3β by mTORC2-PKB activation releases eIF2Bε from the inhibitory phosphorylation by GSK3β, which may result in increased rates of translation initiation. However, this remains to be tested.
“On/Off” Switches for the mTOR Complexes
Although mTORC1 and 2 share a number of components in common, the effects of them on each of the complexes may differ (Table 1). For instance, mLST8 is essential for the integrity and activation of mTORC2, yet its deletion has no effect on mTORC1.84 In contrast, GRp58 is required for the assembly and stimulation of mTORC1, but it is dispensable for mTORC2.5 Nevertheless, some mTORC components do exert similar effects on both mTOR complexes, such as Rac1, deletion of which in MEFs and lymphocytes leads to a reduction in cell size.6 Rac1 is a positive regulator of both mTORC1 and mTORC2.6 Tel2, an essential protein for mammalian embryonic development and cell growth, positively regulates the maturation and stability of mTOR,3 and it has been demonstrated that Tel2 and its associated protein Tti1 are crucial for the assembly of both mTOR complexes.4
DEPTOR
DEPTOR is a negative regulator of both mTORCs,2 and is expressed at low levels in most cancer cells, coincident with enhanced activities of mTORC1 and 2 and improved resistance to nutrient deprivation-induced apoptosis, yet striking exceptions occur in many multiple myelomas where DEPTOR is often overexpressed.2 Although the activation of mTORC1 is suppressed in those cells, mTORC2 is “re-activated” as a result of the alleviation of the negative feedback mechanism exerted by S6K, downstream of mTORC1, toward PI3K. Therefore survival rates of these tumor cells are also improved.2 As a positive feedback mechanism, the phosphorylation of DEPTOR (by S6K1, RSK or CK1α) in response to extracellular stimuli results in its ubiquitination and degradation, which releases mTOR from inhibition by DEPTOR.86
PKB and the TSC complex
As mentioned before, phosphorylation of PKB by mTORC2 at Ser473 (HM) is required for the full activation of PKB against only some substrates,35,84 and although PKB lies upstream of mTORC1, this phosphorylation event is not required for the activation of mTORC1, because knocking-down RICTOR84 or SIN115 (mTORC2 essential component) in MEFs had no effect on the phosphorylation of mTORC1 upstream regulator TSC2 or mTORC1 downstream targets S6K1 and 4E-BPs, and thus mTORC2 cannot be considered as an upstream regulator of mTORC1. In contrast, the phosphorylation of PKB at Thr308 (AL) by PDK-1 is crucial for insulin signaling to mTORC1.34 Nevertheless, PKB Ser473 phosphorylation is essential for the regulation of FoxO, transcription factors which are excluded from the nucleus and thus inactivated upon mTORC2-PKB stimulation.84 Therefore, phosphorylation of Ser473 has differential effects on different PKB substrates. However, in primary, non-transformed, non-immortalized, diploid human fibroblasts, mTORC2 does lie upstream of mTORC1, because knocking-down RICTOR in those cells abolishes the phosphorylation of S6K1.76
Intriguingly, although genetic ablation of the mTORC1 upstream negative regulator TSC2 (TSC2−/−) in MEFs greatly enhances mTORC1 activity, the stimulation of mTORC2 and hence PKB Ser473 phosphorylation are abolished in these cells.87 This effect is independent of the GAP activity of TSC2 toward Rheb, or negative feedback mechanisms (see below) that inactivate PI3K.87 The TSC1/2 complex can also associate with mTORC2.87 Recently, a third subunit of the TSC1/2 complex called TBC1D7 was identified.88 TBC1D7 is required for the GAP activity of TSC1/2 and is therefore inhibitory to mTORC1 activation. shRNA knockdown of TBC1D7 in HeLa cells also causes a reduction in insulin-induced PKB Ser473 phosphorylation, implying that as well as TSC2, TBC1D7 is also a positive regulator of mTORC2.88 In contrast, mTORC2 activity is reportedly unaffected in TSC2−/− ELT3 cells, implying that the positive effect of TSC2 on mTORC2 activity may cell type-dependent.89 Notably, knocking-down of RICTOR by siRNA in TSC2−/− ELT3 cells impair cell proliferation and survival, and this was apparently caused by a reduction in RhoA activity, because constitutively active RhoA could restore the decrease in DNA synthesis caused by RICTOR knock-down in these cells.89
FoxO
On the other hand, FoxO, which is regulated by the mTORC2-PKB pathway, also plays an important role in maintaining the homeostatic balance between mTORC1 and 2. Under energy stress conditions, activated FoxO1 can promote the expression of Sestrin3,90 a member of the sestrin family of PA26-related proteins which regulate cell growth and survival.91 In turn, Sestrin3 suppresses the activation of mTORC1 in a TSC2-dependent manner.90 In contrast, active FoxO1 also induces the expression of RICTOR at both mRNA and protein levels, which promotes mTORC2 complex formation and PKB stimulation.90
Negative Feedback Interplay between mTORCs
IRS
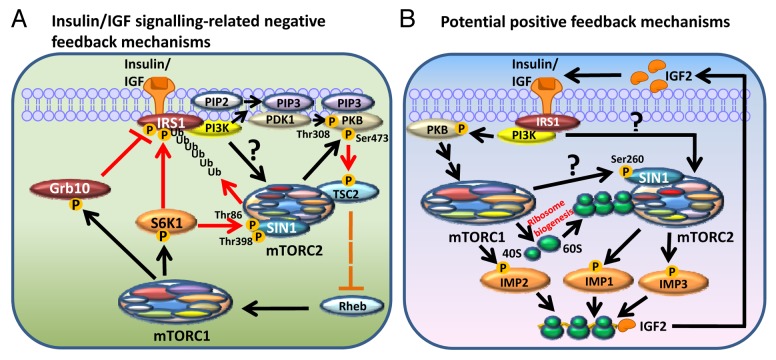
Knocking-down RAPTOR, an mTORC1 essential component, enhances the activation of mTORC2 by insulin, because mTORC1 inhibition alleviates mTORC2 from the suppression by several negative feedback loops (Fig. 2A). In the first, prolonged mTORC1 activation leads to a decrease in the transcription of IRS1, which is required for insulin or IGF signaling to activate PI3K (reviewed in92). Chronic mTORC1 activation also causes the inhibitory phosphorylation of IRS1 which disrupts its association with InsR or IGF1R. This was first found to be mediated by S6K1 (Fig. 2A).93 Since then, many additional IRS1 phosphorylation sites have been discovered which negatively affect IRS1 function. Besides mTORC1 itself and S6K1, several other protein kinases are also able to phosphorylate these sites, some of which are mTORC2 downstream targets like PKB and GSK3; others also include mTORC1 upstream regulators such as ERK and AMPK (reviewed in92). In a negative feedback mechanism, the inhibition of IRS1 upon prolonged mTORC1 stimulation diminishes the activation of PI3K, which normally promotes the activation of both mTORC1 and mTORC2 (Fig. 2A).66-68
Figure 2. Negative and possible positive feedback mechanisms of the mTOR complexes. (A) Negative feedback loops in insulin/IGF signaling. This includes (i) the stabilization of Grb10 by mTORC1; (ii) the inhibition of IRS1 by S6K1, or ubiquitination and degradation of IRS1 induced by mTORC2; and (iii) SIN1 phosphorylation by S6K1. (B) Potential positive feedback mechanisms of the mTOR complexes. This includes upregulation of IGF2 mRNA translation via the phosphorylation and stimulation of IMPs; mTORC1 increases the synthesis of ribosomes which promote mTORC2 activity; and the phosphorylation of SIN1 at Ser260 by mTOR, which is required for mTORC2 complex assembly and activation. Black arrows: upregulation or phosphorylation event leads to activation; red arrows: downregulation or phosphorylation event leading to inactivation; red “T” lines: inhibition of the phosphorylation event which leads to inactivation; discontinuous orange lines: an inhibitory mechanism released in response to upstream stimulation.
Interestingly, it has also recently been found that mTORC2 can also negatively regulate IRS1 by inducing the phosphorylation (at Ser86) and stabilization of Fbw8, which in turn promotes IRS1 degradation (Fig. 2A).94 Fbw8 is a substrate recognition subunit of the Cullin7 E3 ubiquitin ligase complex which promotes the ubiquitination and degradation of IRS1 (reviewed in95). Activation of mTORC2 downregulates the levels of IRS1 via Fbw8.94 There is an increase in IRS1 levels in MEF cells lacking the mTORC2 essential component SIN1 (SIN1−/−). This enhances serum-mediated S6K1 phosphorylation on Thr389, indicative of increased mTORC1 activity,94 and provides an indirect IRS1-mediated feedback mechanism whereby mTORC2 stimulation negatively regulates the activity of mTORC1.
Grb10
Another negative feedback loop involves a recently identified mTORC1 substrate Grb10 (growth factor bound-receptor protein 10).96,97 Grb10 is a negative regulator of the insulin/IGF signaling pathway because it inhibits the autophosphorylation of InsR (Tyr1150/Tyr1151)/IGF1R (Tyr1135/Tyr1136), tyrosine phosphorylation of IRS1/2, and the recruitment of PI3K to IRS1 (Fig. 3).96,97 Several phosphorylation sites in Grb10 have been found to be phosphorylated by mTORC1 in vitro or sensitive to treatment of cells with mTOR inhibitors. The phosphorylation of Grb10 stabilizes it, which in turn destabilizes IRS1 (Fig. 2A).96,97 The discovery of Grb10 as an mTORC1 substrate and IRS1 as a S6K1 substrate are of great importance to understanding type 2 diabetes mellitus, as abnormal insulin signaling is a major feature of insulin resistance. It is believed that nutrient overload, as in the case of obese individuals, leads to over-stimulation of mTORC1 and subsequent downregulation of insulin signaling pathways, although it is not fully clear what mechanisms underlie this effect. On the other hand, long-term inhibition of mTORCs by rapamycin also leads to the development of insulin resistance and impaired glucose homeostasis, and this is primarily due to the inhibition of mTORC2.98
Phosphorylation of RICTOR on Thr1135
Several groups have identified Thr1135 as a phosphorylation site in RICTOR, an essential component of mTORC2.99-102 Thr1135 phosphorylation is modulated by growth factor and nutrient availability. Interestingly, S6K1 is the kinase responsible for phosphorylating this site, which promotes the sequestration of RICTOR by 14–3-3 proteins.99,101,102 This may provide a negative feedback loop between the two mTOR complexes. Indeed, PKB phosphorylation at Ser473 is increased upon insulin stimulation in RICTOR-null (RICTOR−/−) MEFs expressing RICTOR T1135A (non-phosphorylatable threonine to alanine mutant), in comparison to when wild-type RICTOR is expressed in those cells, whereas the phosphorylation of other mTORC2 downstream targets, such as PKCs and SGK1, remains unaffected.99,101 Paradoxically, RICTOR Thr1135 phosphorylation does not affect the integrity of mTORC2 or its kinase activity.99-102 One explanation for this is that RICTOR Thr1135 phosphorylation reduces the affinity of mTORC2 for PKB, but not PKCs or SGK1, independently of mTOR kinase activity. Moreover, this phosphorylation event is unaffected in SIN1−/− MEFs, implying that the phosphorylation of RICTOR at Thr1135 does not require mTORC2.100 Therefore, the significance of RICTOR Thr1135 phosphorylation is far from understood and it may indeed play roles in mTORC2-independent RICTOR functions.100 Furthermore, it was also shown by Gao and colleagues103 that RICTOR Thr1135 phosphorylation has a negative effect on its association with cullin-1 and Rbx1 to form a functional E3 ubiquitin ligase, which promotes the ubiquitination of SGK1. Therefore, RICTOR Thr1135 phosphorylation stabilizes SGK1 by impairing SGK1 degradation, which provides a rational for the accumulation of SGK1 in several human cancers. The same group has also found that RICTOR Thr1135 cannot only be phosphorylated by S6K1, but also by ectopically expressed PKB and SGK1 in 293T cells.103
A regulatory input via SIN1
Because RICTOR Thr1135 phosphorylation does not affect mTORC2 kinase activity,99-102 and RICTOR and SIN1 are essential components of mTORC2,14-17,73 it is possible that mTORC1 activation triggers a negative feedback loop via SIN1. Indeed, Liu et al. have recently been discovered that upon stimulation, S6K1 can phosphorylate SIN1 at Thr86 and Thr398, which will lead to the dissociation of mTORC2 complex and inactivation (Fig. 2A).104 In comparison to the negative feedback loops mentioned above, this mechanism does not apply only to insulin/IGF signaling, but also to other growth factors like EGF and PDGF.104 Upon EGF stimulation, HeLa cells expressing the non-phosphorylatable SIN1 T86A or T398A mutant display sustained PKB phosphorylation on Ser473 and Thr308, whereas EGF treatment only leads to transient PKB phosphorylation in SIN1−/− MEFs expressing wild-type SIN1.104 Paradoxically, an earlier study performed by Humphrey et al. has demonstrated that PKB rather than S6K1, is the SIN1 Thr86 kinase.105 In their study, the phosphorylation of SIN1 on Thr86 had no effect on mTORC2 complex assembly and, instead of suppressing mTORC2 activation as reported by Liu et al., it actually enhanced the activity of mTORC2.105 However, Liu et al.’s study was performed mainly in HeLa cells,104 an epithelial cancer cell line, whereas Humphrey et al. primarily used adipocytes.105 Indeed, in agreement with Humphrey et al., Liu et al. found that both PKB and S6K1 can phosphorylate SIN1 at Thr86 in adipocytes.104 Nevertheless, it is difficult to reconcile the functional differences regarding SIN1 Thr86 phosphorylation between these two studies – stimulatory to mTORC2 according to Humphrey et al.,105 but inhibitory to mTORC2 (and perhaps also mTORC1) in the work of Liu et al.104 Interestingly, some ovarian cancer patients carry SIN1 mutations [R81T (arginine to threonine) and S84L (serine to leucine)] close to Thr86.106 These mutations impair the phosphorylation of SIN1 at Thr86, which disrupts the S6K1-SIN1 negative feedback loop, resulting in sustained PKB phosphorylation at Ser473 and elevated oncogenic/pro-survival signaling.104
Implications of negative feedback loops for cancer therapies
The discovery of these negative feedback mechanisms indeed greatly contributes to a better understanding of cancer development and cancer drug resistance. For instance, a well-known secondary effect related to rapamycin analogs is the alleviation of several negative feedback loops, including those mentioned above, leading to sustained and enhanced stimulation of PI3K, which may then result in the over-activation of mTORC2 and PKB, hence promoting cancer cell survival. mTOR inhibition also triggers feedback activation of RTKs.107 For instance, the tyrosine phosphorylation and trans-activation of the EGFR in response to rapamycin treatment may activate other prosurvival kinases like ERK and RSK.108 Similarly, second generation mTOR inhibitors which simultaneously suppress the activity of mTORC2 as well as mTORC1 may also trigger growth factor receptor (VEGFR, IGF1R and EGFR) tyrosine phosphorylation and activation, and this is mediated by an increase in the activity of FoxO upon mTORC2-PKB inhibition, because transient knock-down of FoxO by siRNA can block the phosphorylation events induced by the mTOR inhibitors.109 However, it remains to be elucidated how activation of FoxO, a transcriptional regulator, leads to phosphorylation of growth factor receptors.
Possible Positive Feedback Crosstalk of the mTOR Complexes
IMPs
In the last section, we listed examples whereby, upon mTORC1 activation, negative feedback loops start to operate to shut down mTORC2 (and also mTORC1) upstream signals. However, the stimulation of mTOR complexes may also triggers some positive feedback mechanisms which consequently further enhance their activation. One such possible mechanism involves IMPs and the translational regulation of IGF2 (Fig. 2B).110,111 It has been demonstrated that active mTORC1 phosphorylates IMP2 at Ser162 and Ser164,110 while IMP1 and IMP3 are phosphorylated by mTORC2 at Ser181 or Ser183, respectively.111 All these phosphorylation events are stimulatory to IMPs, promoting the binding of IMPs to the IGF2 mRNA and enhancing its translation by cap-independent internal ribosomal entry.110,111 Presumably, increased levels of IGF2 will further enhance the activation of mTORC1 and 2, although this is yet to be confirmed.
Ribosome biogenesis
It has been shown that active mTORC2 associates with ribosomes and that this binding can be promoted by insulin signaling and PI3K stimulation.68 Importantly, transient knock-down of mNIP7, which is required for the maturation of rRNA and therefore ribosome production,112 or genes encoding RPs such as L7, L26 and S16, by siRNA in HeLa cells, severely impairs mTORC2 activation in response to insulin whereas the stimulation of mTORC1 remains unaffected.68 Therefore, it is plausible that the upregulation of ribosome biogenesis by mTORC1 may actually promote mTORC2 activation (Fig. 2B). However, this also requires experimental confirmation.
SIN1
We have mentioned that in HeLa cells, upon growth factor stimulation, the essential mTORC2 component SIN1 appears to undergo inhibitory phosphorylation on Thr86 and Thr398, leading to the dissociation of mTORC2 and hence inactivation,104 although according Humphrey et al., in adipocytes, SIN1 Thr86 positively regulates mTORC2 activity.104,105 Another SIN1 phosphorylation site found by Sarbassov’s group, Ser260, which is sensitive to glucose starvation and ATP depletion, positively regulates mTORC2 assembly (Fig. 2B).113 Phosphorylation of SIN1 at Ser260 phosphorylation is catalyzed by mTOR,113 although it remains unclear which mTOR complex (or perhaps both) is (are) responsible for its phosphorylation. Notably, tumors induced by subcutaneous injection of H-Ras transformed MEFs constitutively expressing S260A (non-phosphorylatable serine to alanine mutant) SIN1 in mice, have only approximately 15–20% of the volume of the corresponding cells that are expressing wild-type or S260D (phosphomimetic serine to aspartic acid mutant) SIN1, further illustrating an important role of SIN1 in cancer development.113
Concluding Remarks
mTOR is a key modulator of anabolic cellular processes and importantly, it positively controls many aspects of mRNA translation as well as other anabolic processes. mTOR exists in two major multi-protein complexes termed as mTORC1 and mTORC2. Although since the discovery of mTORC2 in 2004,14,73 a number of crosstalk mechanisms between the mTOR complexes have been described, there are still many unanswered questions within the mTORC interplay puzzle which await further investigation. First of all, it is crucial to determine whether the phosphorylation of SIN1 on Thr86 and Thr398 exerts positive or negative effects on mTORC2 in response to growth factor stimulation, and whether it plays the same role in response to other stimuli or under stress conditions. It is likely that these phosphorylation sites also affect mTORC1 activity because there is also a sustained phosphorylation of PKB at Thr308 in EGF-stimulated HeLa cells transfected with either SIN1 T86A or T398A, which should then upregulate mTORC1 activity via the phosphorylation and inhibition of TSC2. It implies that the phosphorylation of SIN1 can potentially serve as a negative feedback loop for both mTOR complexes,104 although further study is still required to determine the effect of SIN1 phosphorylation on the activity of mTORC1. Moreover, it is still unknown which of the mTOR complexes is responsible for SIN1 Ser260 phosphorylation.
Previously, insulin/IGF-mediated mTORC1 and 2 feedback mechanisms were the best characterized, whereas feedback loops in response to other stimuli were much less understood. The discovery of SIN1 phosphorylation sites (Thr86,104,105 Ser260113 and Thr398104), which can be regulated by several growth factors (Thr86 and Thr398)104,105 and energy stress (Ser260),113 opened doors in the search of mTORC feedback mechanisms in non-insulin/IGF related signaling pathways. For instance, would be an interesting field to explore mTORC crosstalk in amino acid signaling. The mechanisms by which amino acids activate mTORC1 have been extensively studied in the last decade and are now relatively well-documented (reviewed in114). One study, by Tato et al.,67 indicated that mTORC2 activity can also be acutely stimulated by amino acids in a PI3K-dependent manner, yet 1) the mechanism by which PI3K activates mTORC2, 2) also; how does mTORC1 actually sense amino acids, and 3) whether there is any similarity between the signaling pathways by which amino acids activate the two mTOR complexes, is far from understood.
Another intriguing question is how TSC1/2 and TBC1D7 positively regulate mTORC2 activity. TSC1/2 binds to mTORC2.87 Although deletion of TSC2 does not interrupt the association of RICTOR and mTOR, it severely affects mTORC2 activation in MEFs.87 There are several possible mechanisms by which the TSC complexes might contribute to mTORC2 activation, either dependent on mTORC1 (discussed in this article) or independent of it. Indeed, TSC2−/− MEFs display high levels of S6K1 stimulation and consequently it disrupts SIN1-RICTOR binding, presumably due to an increase in SIN1 phosphorylation at Thr86 and Thr398.104 Nevertheless, transient knock-down of RAPTOR, the defining mTORC1 component, in TSC2−/− MEFs, does not restore mTORC2 activity, implying that the interaction between TSC complex and mTORC2 can also be mTORC1-independent.87 A recent computational model based on experimental data obtained by Dalle Pezze et al. suggests that TSC1/2 has no input into mTORC2, and a PI3K variant insensitive to negative feedback loops involving mTORC1 is responsible for mTORC2 activation.115 It is important to note that the model proposed by these authors is mainly based on research data using mTOR Ser2481 phosphorylation as a specific readout of mTORC2 activity,115 yet mTOR Ser2481 phosphorylation also occurs in mTORC1 and can therefore be used as a marker of intrinsic activity for either of the mTOR complexes.116 Consequently, the conclusions of this study must be treated with caution,117 and questions remain regarding the existence of such a PI3K variant.
Deregulation of the translational control resulting from disruptions in the interplay between the mTORC1 and mTORC2 circuitry may have a profound impact on disease pathogenesis. For instance, chronic excess nutrients, which gives rise to obesity, causes chronic activation of mTORC1 and subsequently S6K1, which results in the downregulation of IRS and therefore the inhibition of insulin signaling, leading to the development of insulin resistance.93 On the other hand, mTORC2 activation is cytoprotective to cardiomyocytes.118,119 mTORC1 stimulation which causes cardiac hypertrophy as well as downregulation of mTORC2 signaling, leads to increased cardiomyocyte apoptosis and tissue damage after myocardial infarction.118,119 Conversely, blocking mTORC1 activation by overexpressing PRAS40 can promote mTORC2 signaling and hence myocardial protection.118,119 In addition, as mentioned before, many components within the complex cross-talk network between mTORC1 and mTORC2, such as SIN1104,113 and TSC2 (reviewed in120), play important roles in the development of cancer.
In summary, here we have described the crosstalk mechanisms between the two mTOR complexes. Each mTOR complex regulates a unique subset of downstream targets involving in many essential cellular functions, especially cell growth, proliferation and survival. Abnormal regulation of the mTOR complexes is implicated in the development of a variety of human illnesses such as diabetes, cardiovascular diseases (cardiac hypertrophy) and cancer. Interestingly, the regulation of mTOR complexes is tightly linked, and therefore it is of great interest and importance to understand the implications of mTORC crosstalk mechanisms in the pathogenesis and treatment of these diseases. We expect that future studies will shed more light on the yet-to-be completed picture of mTORC cross-regulation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Work on mTOR signaling in the author’s laboratory is supported by funding (to CGP) from the Biotechnology and Biological Sciences Research Council, the British Heart Foundation and the Wellcome Trust.
Glossary
Abbreviations:
- 4E-BP
eIF4E-binding protein
- 5′-TOP
5′-terminal oligopyrimidines
- AL
activation loop
- AMPK
5′-AMP-activated protein kinase
- aPKC
atypical PKC
- CK1α
casein kinase 1α
- cPKC
conventional PKC
- DEPTOR
dishevelled, eEF, eukaryotic elongation factors
- eEF2K
eEF2 kinase
- EGF
epidermal growth factor
- EGFR
EGF receptor
- egl-10
pleckstrin domain-containing mTOR interacting protein
- eIF
eukaryotic initiation factor
- ELT3
Eker rat uterine leiomyoma cell line 3
- ERK
extracellular signal-regulated kinase
- Fbw8
F-box 8, also termed as Fbxw8 or Fbx29
- FKBP12
FK506-binding protein, 12 kDa
- FoxO
forkhead box protein
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchange factor
- Grb10
growth factor bound-receptor protein 10
- GRp58
58 kDa glucose-regulated protein
- GSK3
glycogen synthase kinase 3
- HM
hydrophobic motif
- IGF
insulin-like growth factor
- IGF1R
IGF1 receptor
- IKK
inhibitor of NF-κB (nuclear factor κ-light-chain-enhancer of activated B cells) kinase
- IMP
IGF2 mRNA-binding protein
- InsR
insulin receptor
- IRS
insulin receptor substrate
- Maf1
matrix attachment region binding filament–like protein 1 associated factor 1
- mNIP7
mammalian nuclear import protein 7
- mLST8
mammalian ortholog of lethal with sec thirteen, also known as Gβ-like protein or GβL
- mTOR
mammalian target of rapamycin
- mTORC1
mTOR complex 1
- mTORC2
mTOR complex 2
- NBS1
Nijmegen breakage syndrome protein 1
- nPKC
novel PKC
- PDCD4
programmed cell death protein 4
- PDGF
platelet-derived growth factor
- PDK1
phosphoinositide dependent protein kinase 1
- PI3K
phosphatidylinositol 3-phosphate kinase
- PIP2
phosphatidylinositol (4,5)-bisphosphate
- PIP3
phosphatidylinositol (3,4,5)-trisphosphate
- PKB
protein kinase B
- PKC
protein kinase C
- PRAS40
Pro-rich Akt substrate of 40 kDa
- PROTOR
protein observed with RICTOR
- Rac1
Ras-related C3 botulinum toxin substrate 1
- RAPTOR
regulatory-associated protein of mTOR
- Rbx1
RING-box protein 1
- rDNA
ribosomal DNA
- Rheb
Ras homolog enriched in brain
- Rhes
Ras homolog enriched in striatum
- RhoA
Ras homolog gene family, member A
- RICTOR
rapamycin-insensitive companion of mTOR
- RP
ribosomal protein
- rRNA
ribosomal RNA
- RSK
p90 RP S6 kinase
- RTK
receptor tyrosine kinase
- S6K
p70 RP S6 kinase
- SCFβTrCP
Skp1/Cul1/F box protein adaptor β-transducin repeat-containing protein
- SGK
serum- and glucocorticoid-induced protein kinase
- SIN1
stress activated protein kinase interacting protein 1
- siRNA
small interfering RNA
- TBC1D7
Tre2-Bub2-Cdc16 1 domain family, member 7
- Tel2
telomere maintenance 2
- TFIIIC
transcription factor 3C
- TM
turn motif
- 5′-TOP
5′-terminal oligopyrimidine tract
- Tti1
Tel2 interacting protein 1
- VEGFR
vascular epithelial growth factor receptor
- XPLN
exchange factor found in platelets, leukemic, and neuronal tissues
References
- 1.Kim DH, Sarbassov DD, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, Tempst P, Sabatini DM.. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell 2003; 11:895 - 904; http://dx.doi.org/ 10.1016/S1097-2765(03)00114-X; PMID: 12718876 [DOI] [PubMed] [Google Scholar]
- 2.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM.. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 2009; 137:873 - 86; http://dx.doi.org/ 10.1016/j.cell.2009.03.046; PMID: 19446321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takai H, Wang RC, Takai KK, Yang H, de Lange T.. Tel2 regulates the stability of PI3K-related protein kinases. Cell 2007; 131:1248 - 59; http://dx.doi.org/ 10.1016/j.cell.2007.10.052; PMID: 18160036 [DOI] [PubMed] [Google Scholar]
- 4.Kaizuka T, Hara T, Oshiro N, Kikkawa U, Yonezawa K, Takehana K, Iemura S, Natsume T, Mizushima N.. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J Biol Chem 2010; 285:20109 - 16; http://dx.doi.org/ 10.1074/jbc.M110.121699; PMID: 20427287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramírez-Rangel I, Bracho-Valdés I, Vázquez-Macías A, Carretero-Ortega J, Reyes-Cruz G, Vázquez-Prado J.. Regulation of mTORC1 complex assembly and signaling by GRp58/ERp57. Mol Cell Biol 2011; 31:1657 - 71; http://dx.doi.org/ 10.1128/MCB.00824-10; PMID: 21321085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saci A, Cantley LC, Carpenter CL.. Rac1 regulates the activity of mTORC1 and mTORC2 and controls cellular size. Mol Cell 2011; 42:50 - 61; http://dx.doi.org/ 10.1016/j.molcel.2011.03.017; PMID: 21474067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM.. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002; 110:163 - 75; http://dx.doi.org/ 10.1016/S0092-8674(02)00808-5; PMID: 12150925 [DOI] [PubMed] [Google Scholar]
- 8.Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K.. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 2002; 110:177 - 89; http://dx.doi.org/ 10.1016/S0092-8674(02)00833-4; PMID: 12150926 [DOI] [PubMed] [Google Scholar]
- 9.Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM.. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell 2007; 25:903 - 15; http://dx.doi.org/ 10.1016/j.molcel.2007.03.003; PMID: 17386266 [DOI] [PubMed] [Google Scholar]
- 10.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH.. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol 2007; 9:316 - 23; http://dx.doi.org/ 10.1038/ncb1547; PMID: 17277771 [DOI] [PubMed] [Google Scholar]
- 11.Wang L, Harris TE, Roth RA, Lawrence JC Jr... PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J Biol Chem 2007; 282:20036 - 44; http://dx.doi.org/ 10.1074/jbc.M702376200; PMID: 17510057 [DOI] [PubMed] [Google Scholar]
- 12.Thedieck K, Polak P, Kim ML, Molle KD, Cohen A, Jenö P, Arrieumerlou C, Hall MN.. PRAS40 and PRR5-like protein are new mTOR interactors that regulate apoptosis. PLoS One 2007; 2:e1217; http://dx.doi.org/ 10.1371/journal.pone.0001217; PMID: 18030348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fonseca BD, Smith EM, Lee VH, MacKintosh C, Proud CG.. PRAS40 is a target for mammalian target of rapamycin complex 1 and is required for signaling downstream of this complex. J Biol Chem 2007; 282:24514 - 24; http://dx.doi.org/ 10.1074/jbc.M704406200; PMID: 17604271 [DOI] [PubMed] [Google Scholar]
- 14.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM.. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol 2004; 14:1296 - 302; http://dx.doi.org/ 10.1016/j.cub.2004.06.054; PMID: 15268862 [DOI] [PubMed] [Google Scholar]
- 15.Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B.. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 2006; 127:125 - 37; http://dx.doi.org/ 10.1016/j.cell.2006.08.033; PMID: 16962653 [DOI] [PubMed] [Google Scholar]
- 16.Yang Q, Inoki K, Ikenoue T, Guan KL.. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev 2006; 20:2820 - 32; http://dx.doi.org/ 10.1101/gad.1461206; PMID: 17043309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frias MA, Thoreen CC, Jaffe JD, Schroder W, Sculley T, Carr SA, Sabatini DM.. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr Biol 2006; 16:1865 - 70; http://dx.doi.org/ 10.1016/j.cub.2006.08.001; PMID: 16919458 [DOI] [PubMed] [Google Scholar]
- 18.Pearce LR, Huang X, Boudeau J, Pawłowski R, Wullschleger S, Deak M, Ibrahim AF, Gourlay R, Magnuson MA, Alessi DR.. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem J 2007; 405:513 - 22; http://dx.doi.org/ 10.1042/BJ20070540; PMID: 17461779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khanna N, Fang Y, Yoon MS, Chen J.. XPLN is an endogenous inhibitor of mTORC2. Proc Natl Acad Sci U S A 2013; 110:15979 - 84; http://dx.doi.org/ 10.1073/pnas.1310434110; PMID: 24043828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang JQ, Chen JH, Chen YC, Chen MY, Hsieh CY, Teng SC, Wu KJ.. Interaction between NBS1 and the mTOR/Rictor/SIN1 complex through specific domains. PLoS One 2013; 8:e65586; http://dx.doi.org/ 10.1371/journal.pone.0065586; PMID: 23762398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y, Lai E, Liu J, Lin J, Yang C, Jia C, Li Y, Bai X, Li M.. IKK interacts with rictor and regulates mTORC2. Cell Signal 2013; 25:2239 - 45; http://dx.doi.org/ 10.1016/j.cellsig.2013.07.008; PMID: 23872070 [DOI] [PubMed] [Google Scholar]
- 22.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM.. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell 2006; 22:159 - 68; http://dx.doi.org/ 10.1016/j.molcel.2006.03.029; PMID: 16603397 [DOI] [PubMed] [Google Scholar]
- 23.Zeng Z, Sarbassov D, Samudio IJ, Yee KW, Munsell MF, Ellen Jackson C, Giles FJ, Sabatini DM, Andreeff M, Konopleva M.. Rapamycin derivatives reduce mTORC2 signaling and inhibit AKT activation in AML. Blood 2007; 109:3509 - 12; http://dx.doi.org/ 10.1182/blood-2006-06-030833; PMID: 17179228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barlow AD, Xie J, Moore CE, Campbell SC, Shaw JA, Nicholson ML, Herbert TP.. Rapamycin toxicity in MIN6 cells and rat and human islets is mediated by the inhibition of mTOR complex 2 (mTORC2). Diabetologia 2012; 55:1355 - 65; http://dx.doi.org/ 10.1007/s00125-012-2475-7; PMID: 22314813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG.. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem 2005; 280:18717 - 27; http://dx.doi.org/ 10.1074/jbc.M414499200; PMID: 15772076 [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, Cicchetti G, Onda H, Koon HB, Asrican K, Bajraszewski N, Vazquez F, Carpenter CL, Kwiatkowski DJ.. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J Clin Invest 2003; 112:1223 - 33; http://dx.doi.org/ 10.1172/JCI200317222; PMID: 14561707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA.. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol 2003; 5:566 - 71; http://dx.doi.org/ 10.1038/ncb996; PMID: 12766776 [DOI] [PubMed] [Google Scholar]
- 28.Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G.. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell 2003; 11:1457 - 66; http://dx.doi.org/ 10.1016/S1097-2765(03)00220-X; PMID: 12820960 [DOI] [PubMed] [Google Scholar]
- 29.Potter CJ, Pedraza LG, Xu T.. Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol 2002; 4:658 - 65; http://dx.doi.org/ 10.1038/ncb840; PMID: 12172554 [DOI] [PubMed] [Google Scholar]
- 30.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC.. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell 2002; 10:151 - 62; http://dx.doi.org/ 10.1016/S1097-2765(02)00568-3; PMID: 12150915 [DOI] [PubMed] [Google Scholar]
- 31.Dan HC, Sun M, Yang L, Feldman RI, Sui XM, Ou CC, Nellist M, Yeung RS, Halley DJ, Nicosia SV, et al.. Phosphatidylinositol 3-kinase/Akt pathway regulates tuberous sclerosis tumor suppressor complex by phosphorylation of tuberin. J Biol Chem 2002; 277:35364 - 70; http://dx.doi.org/ 10.1074/jbc.M205838200; PMID: 12167664 [DOI] [PubMed] [Google Scholar]
- 32.Inoki K, Li Y, Zhu T, Wu J, Guan KL.. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 2002; 4:648 - 57; http://dx.doi.org/ 10.1038/ncb839; PMID: 12172553 [DOI] [PubMed] [Google Scholar]
- 33.Irvine RF, Letcher AJ, Heslop JP, Berridge MJ.. The inositol tris/tetrakisphosphate pathway--demonstration of Ins(1,4,5)P3 3-kinase activity in animal tissues. Nature 1986; 320:631 - 4; http://dx.doi.org/ 10.1038/320631a0; PMID: 3010126 [DOI] [PubMed] [Google Scholar]
- 34.Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PR, Reese CB, Cohen P.. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol 1997; 7:261 - 9; http://dx.doi.org/ 10.1016/S0960-9822(06)00122-9; PMID: 9094314 [DOI] [PubMed] [Google Scholar]
- 35.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM.. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005; 307:1098 - 101; http://dx.doi.org/ 10.1126/science.1106148; PMID: 15718470 [DOI] [PubMed] [Google Scholar]
- 36.Subramaniam S, Napolitano F, Mealer RG, Kim S, Errico F, Barrow R, Shahani N, Tyagi R, Snyder SH, Usiello A.. Rhes, a striatal-enriched small G protein, mediates mTOR signaling and L-DOPA-induced dyskinesia. Nat Neurosci 2012; 15:191 - 3; http://dx.doi.org/ 10.1038/nn.2994; PMID: 22179112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Tsang CK, Watkins M, Bertram PG, Zheng XF.. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature 2006; 442:1058 - 61; http://dx.doi.org/ 10.1038/nature05020; PMID: 16900101 [DOI] [PubMed] [Google Scholar]
- 38.Mayer C, Grummt I.. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 2006; 25:6384 - 91; http://dx.doi.org/ 10.1038/sj.onc.1209883; PMID: 17041624 [DOI] [PubMed] [Google Scholar]
- 39.Kantidakis T, Ramsbottom BA, Birch JL, Dowding SN, White RJ.. mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc Natl Acad Sci U S A 2010; 107:11823 - 8; http://dx.doi.org/ 10.1073/pnas.1005188107; PMID: 20543138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shor B, Wu J, Shakey Q, Toral-Barza L, Shi C, Follettie M, Yu K.. Requirement of the mTOR kinase for the regulation of Maf1 phosphorylation and control of RNA polymerase III-dependent transcription in cancer cells. J Biol Chem 2010; 285:15380 - 92; http://dx.doi.org/ 10.1074/jbc.M109.071639; PMID: 20233713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsang CK, Liu H, Zheng XF.. mTOR binds to the promoters of RNA polymerase I- and III-transcribed genes. Cell Cycle 2010; 9:953 - 7; http://dx.doi.org/ 10.4161/cc.9.5.10876; PMID: 20038818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iadevaia V, Zhang Z, Jan E, Proud CG.. mTOR signaling regulates the processing of pre-rRNA in human cells. Nucleic Acids Res 2012; 40:2527 - 39; http://dx.doi.org/ 10.1093/nar/gkr1040; PMID: 22121221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM.. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 2012; 485:109 - 13; http://dx.doi.org/ 10.1038/nature11083; PMID: 22552098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al.. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 2012; 485:55 - 61; http://dx.doi.org/ 10.1038/nature10912; PMID: 22367541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huo Y, Iadevaia V, Yao Z, Kelly I, Cosulich S, Guichard S, Foster LJ, Proud CG.. Stable isotope-labelling analysis of the impact of inhibition of the mammalian target of rapamycin on protein synthesis. Biochem J 2012; 444:141 - 51; http://dx.doi.org/ 10.1042/BJ20112107; PMID: 22428559 [DOI] [PubMed] [Google Scholar]
- 46.Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, et al.. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 2010; 328:1172 - 6; http://dx.doi.org/ 10.1126/science.1187532; PMID: 20508131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larsson O, Morita M, Topisirovic I, Alain T, Blouin MJ, Pollak M, Sonenberg N.. Distinct perturbation of the translatome by the antidiabetic drug metformin. Proc Natl Acad Sci U S A 2012; 109:8977 - 82; http://dx.doi.org/ 10.1073/pnas.1201689109; PMID: 22611195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stumpf CR, Moreno MV, Olshen AB, Taylor BS, Ruggero D.. The translational landscape of the mammalian cell cycle. Mol Cell 2013; 52:574 - 82; http://dx.doi.org/ 10.1016/j.molcel.2013.09.018; PMID: 24120665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sonenberg N, Hinnebusch AG.. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 2009; 136:731 - 45; http://dx.doi.org/ 10.1016/j.cell.2009.01.042; PMID: 19239892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jackson RJ, Hellen CU, Pestova TV.. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 2010; 11:113 - 27; http://dx.doi.org/ 10.1038/nrm2838; PMID: 20094052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brunn GJ, Hudson CC, Sekulić A, Williams JM, Hosoi H, Houghton PJ, Lawrence JC Jr., Abraham RT.. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 1997; 277:99 - 101; http://dx.doi.org/ 10.1126/science.277.5322.99; PMID: 9204908 [DOI] [PubMed] [Google Scholar]
- 52.Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N.. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev 1999; 13:1422 - 37; http://dx.doi.org/ 10.1101/gad.13.11.1422; PMID: 10364159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang D, Brunn GJ, Lawrence JC Jr... Mutational analysis of sites in the translational regulator, PHAS-I, that are selectively phosphorylated by mTOR. FEBS Lett 1999; 453:387 - 90; http://dx.doi.org/ 10.1016/S0014-5793(99)00762-0; PMID: 10405182 [DOI] [PubMed] [Google Scholar]
- 54.Mothe-Satney I, Brunn GJ, McMahon LP, Capaldo CT, Abraham RT, Lawrence JC Jr... Mammalian target of rapamycin-dependent phosphorylation of PHAS-I in four (S/T)P sites detected by phospho-specific antibodies. J Biol Chem 2000; 275:33836 - 43; http://dx.doi.org/ 10.1074/jbc.M006005200; PMID: 10942774 [DOI] [PubMed] [Google Scholar]
- 55.Mothe-Satney I, Yang D, Fadden P, Haystead TA, Lawrence JC Jr... Multiple mechanisms control phosphorylation of PHAS-I in five (S/T)P sites that govern translational repression. Mol Cell Biol 2000; 20:3558 - 67; http://dx.doi.org/ 10.1128/MCB.20.10.3558-3567.2000; PMID: 10779345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gingras AC, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, Polakiewicz RD, Wyslouch-Cieszynska A, Aebersold R, Sonenberg N.. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev 2001; 15:2852 - 64; PMID: 11691836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chung J, Kuo CJ, Crabtree GR, Blenis J.. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell 1992; 69:1227 - 36; http://dx.doi.org/ 10.1016/0092-8674(92)90643-Q; PMID: 1377606 [DOI] [PubMed] [Google Scholar]
- 58.Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL, Polakiewicz RD, Sonenberg N, Hershey JW.. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J 2004; 23:1761 - 9; http://dx.doi.org/ 10.1038/sj.emboj.7600193; PMID: 15071500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M.. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 2006; 314:467 - 71; http://dx.doi.org/ 10.1126/science.1130276; PMID: 17053147 [DOI] [PubMed] [Google Scholar]
- 60.Holz MK, Ballif BA, Gygi SP, Blenis J.. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 2005; 123:569 - 80; http://dx.doi.org/ 10.1016/j.cell.2005.10.024; PMID: 16286006 [DOI] [PubMed] [Google Scholar]
- 61.Harris TE, Chi A, Shabanowitz J, Hunt DF, Rhoads RE, Lawrence JC Jr... mTOR-dependent stimulation of the association of eIF4G and eIF3 by insulin. EMBO J 2006; 25:1659 - 68; http://dx.doi.org/ 10.1038/sj.emboj.7601047; PMID: 16541103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nairn AC, Bhagat B, Palfrey HC.. Identification of calmodulin-dependent protein kinase III and its major Mr 100,000 substrate in mammalian tissues. Proc Natl Acad Sci U S A 1985; 82:7939 - 43; http://dx.doi.org/ 10.1073/pnas.82.23.7939; PMID: 3906654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Knebel A, Morrice N, Cohen P.. A novel method to identify protein kinase substrates: eEF2 kinase is phosphorylated and inhibited by SAPK4/p38delta. EMBO J 2001; 20:4360 - 9; http://dx.doi.org/ 10.1093/emboj/20.16.4360; PMID: 11500363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG.. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J 2001; 20:4370 - 9; http://dx.doi.org/ 10.1093/emboj/20.16.4370; PMID: 11500364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Browne GJ, Proud CG.. A novel mTOR-regulated phosphorylation site in elongation factor 2 kinase modulates the activity of the kinase and its binding to calmodulin. Mol Cell Biol 2004; 24:2986 - 97; http://dx.doi.org/ 10.1128/MCB.24.7.2986-2997.2004; PMID: 15024086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gan X, Wang J, Su B, Wu D.. Evidence for direct activation of mTORC2 kinase activity by phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem 2011; 286:10998 - 1002; http://dx.doi.org/ 10.1074/jbc.M110.195016; PMID: 21310961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tato I, Bartrons R, Ventura F, Rosa JL.. Amino acids activate mammalian target of rapamycin complex 2 (mTORC2) via PI3K/Akt signaling. J Biol Chem 2011; 286:6128 - 42; http://dx.doi.org/ 10.1074/jbc.M110.166991; PMID: 21131356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zinzalla V, Stracka D, Oppliger W, Hall MN.. Activation of mTORC2 by association with the ribosome. Cell 2011; 144:757 - 68; http://dx.doi.org/ 10.1016/j.cell.2011.02.014; PMID: 21376236 [DOI] [PubMed] [Google Scholar]
- 69.García-Martínez JM, Alessi DR.. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1). Biochem J 2008; 416:375 - 85; http://dx.doi.org/ 10.1042/BJ20081668; PMID: 18925875 [DOI] [PubMed] [Google Scholar]
- 70.Ikenoue T, Inoki K, Yang Q, Zhou X, Guan KL.. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J 2008; 27:1919 - 31; http://dx.doi.org/ 10.1038/emboj.2008.119; PMID: 18566587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Facchinetti V, Ouyang W, Wei H, Soto N, Lazorchak A, Gould C, Lowry C, Newton AC, Mao Y, Miao RQ, et al.. The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J 2008; 27:1932 - 43; http://dx.doi.org/ 10.1038/emboj.2008.120; PMID: 18566586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oh WJ, Wu CC, Kim SJ, Facchinetti V, Julien LA, Finlan M, Roux PP, Su B, Jacinto E.. mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. EMBO J 2010; 29:3939 - 51; http://dx.doi.org/ 10.1038/emboj.2010.271; PMID: 21045808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jacinto E, Loewith R, Schmidt A, Lin S, Rüegg MA, Hall A, Hall MN.. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol 2004; 6:1122 - 8; http://dx.doi.org/ 10.1038/ncb1183; PMID: 15467718 [DOI] [PubMed] [Google Scholar]
- 74.Gulati N, Karsy M, Albert L, Murali R, Jhanwar-Uniyal M.. Involvement of mTORC1 and mTORC2 in regulation of glioblastoma multiforme growth and motility. Int J Oncol 2009; 35:731 - 40; PMID: 19724909 [DOI] [PubMed] [Google Scholar]
- 75.Liu L, Das S, Losert W, Parent CA.. mTORC2 regulates neutrophil chemotaxis in a cAMP- and RhoA-dependent fashion. Dev Cell 2010; 19:845 - 57; http://dx.doi.org/ 10.1016/j.devcel.2010.11.004; PMID: 21145500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rosner M, Fuchs C, Siegel N, Valli A, Hengstschläger M.. Functional interaction of mammalian target of rapamycin complexes in regulating mammalian cell size and cell cycle. Hum Mol Genet 2009; 18:3298 - 310; http://dx.doi.org/ 10.1093/hmg/ddp271; PMID: 19505958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang T, Blumhagen R, Lao U, Kuo Y, Edgar BA.. LST8 regulates cell growth via target-of-rapamycin complex 2 (TORC2). Mol Cell Biol 2012; 32:2203 - 13; http://dx.doi.org/ 10.1128/MCB.06474-11; PMID: 22493059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stumpf CR, Moreno MV, Olshen AB, Taylor BS, Ruggero D.. The translational landscape of the mammalian cell cycle. Mol Cell 2013; 52:574 - 82; http://dx.doi.org/ 10.1016/j.molcel.2013.09.018; PMID: 24120665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shiota C, Woo JT, Lindner J, Shelton KD, Magnuson MA.. Multiallelic disruption of the rictor gene in mice reveals that mTOR complex 2 is essential for fetal growth and viability. Dev Cell 2006; 11:583 - 9; http://dx.doi.org/ 10.1016/j.devcel.2006.08.013; PMID: 16962829 [DOI] [PubMed] [Google Scholar]
- 80.Matheny RW Jr., Lynch CM, Leandry LA.. Enhanced Akt phosphorylation and myogenic differentiation in PI3K p110β-deficient myoblasts is mediated by PI3K p110α and mTORC2. Growth Factors 2012; 30:367 - 84; http://dx.doi.org/ 10.3109/08977194.2012.734507; PMID: 23137199 [DOI] [PubMed] [Google Scholar]
- 81.Esen E, Chen J, Karner CM, Okunade AL, Patterson BW, Long F.. WNT-LRP5 signaling induces Warburg effect through mTORC2 activation during osteoblast differentiation. Cell Metab 2013; 17:745 - 55; http://dx.doi.org/ 10.1016/j.cmet.2013.03.017; PMID: 23623748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yao Y, Suraokar M, Darnay BG, Hollier BG, Shaiken TE, Asano T, Chen CH, Chang BH, Lu Y, Mills GB, et al.. BSTA promotes mTORC2-mediated phosphorylation of Akt1 to suppress expression of FoxC2 and stimulate adipocyte differentiation. Sci Signal 2013; 6:ra2; http://dx.doi.org/ 10.1126/scisignal.2003295; PMID: 23300339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Case N, Thomas J, Sen B, Styner M, Xie Z, Galior K, Rubin J.. Mechanical regulation of glycogen synthase kinase 3β (GSK3β) in mesenchymal stem cells is dependent on Akt protein serine 473 phosphorylation via mTORC2 protein. J Biol Chem 2011; 286:39450 - 6; http://dx.doi.org/ 10.1074/jbc.M111.265330; PMID: 21956113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM.. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell 2006; 11:859 - 71; http://dx.doi.org/ 10.1016/j.devcel.2006.10.007; PMID: 17141160 [DOI] [PubMed] [Google Scholar]
- 85.Wang X, Paulin FE, Campbell LE, Gomez E, O’Brien K, Morrice N, Proud CG.. Eukaryotic initiation factor 2B: identification of multiple phosphorylation sites in the epsilon-subunit and their functions in vivo. EMBO J 2001; 20:4349 - 59; http://dx.doi.org/ 10.1093/emboj/20.16.4349; PMID: 11500362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gao D, Inuzuka H, Tan MK, Fukushima H, Locasale JW, Liu P, Wan L, Zhai B, Chin YR, Shaik S, et al.. mTOR drives its own activation via SCF(βTrCP)-dependent degradation of the mTOR inhibitor DEPTOR. Mol Cell 2011; 44:290 - 303; http://dx.doi.org/ 10.1016/j.molcel.2011.08.030; PMID: 22017875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang J, Dibble CC, Matsuzaki M, Manning BD.. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol 2008; 28:4104 - 15; http://dx.doi.org/ 10.1128/MCB.00289-08; PMID: 18411301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, Finan PM, Kwiatkowski DJ, Murphy LO, Manning BD.. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell 2012; 47:535 - 46; http://dx.doi.org/ 10.1016/j.molcel.2012.06.009; PMID: 22795129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goncharova EA, Goncharov DA, Li H, Pimtong W, Lu S, Khavin I, Krymskaya VP.. mTORC2 is required for proliferation and survival of TSC2-null cells. Mol Cell Biol 2011; 31:2484 - 98; http://dx.doi.org/ 10.1128/MCB.01061-10; PMID: 21482669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen CC, Jeon SM, Bhaskar PT, Nogueira V, Sundararajan D, Tonic I, Park Y, Hay N.. FoxOs inhibit mTORC1 and activate Akt by inducing the expression of Sestrin3 and Rictor. Dev Cell 2010; 18:592 - 604; http://dx.doi.org/ 10.1016/j.devcel.2010.03.008; PMID: 20412774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peeters H, Debeer P, Bairoch A, Wilquet V, Huysmans C, Parthoens E, Fryns JP, Gewillig M, Nakamura Y, Niikawa N, et al.. PA26 is a candidate gene for heterotaxia in humans: identification of a novel PA26-related gene family in human and mouse. Hum Genet 2003; 112:573 - 80; PMID: 12607115 [DOI] [PubMed] [Google Scholar]
- 92.Tanti JF, Jager J.. Cellular mechanisms of insulin resistance: role of stress-regulated serine kinases and insulin receptor substrates (IRS) serine phosphorylation. Curr Opin Pharmacol 2009; 9:753 - 62; http://dx.doi.org/ 10.1016/j.coph.2009.07.004; PMID: 19683471 [DOI] [PubMed] [Google Scholar]
- 93.Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, et al.. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 2004; 431:200 - 5; http://dx.doi.org/ 10.1038/nature02866; PMID: 15306821 [DOI] [PubMed] [Google Scholar]
- 94.Kim SJ, DeStefano MA, Oh WJ, Wu CC, Vega-Cotto NM, Finlan M, Liu D, Su B, Jacinto E.. mTOR complex 2 regulates proper turnover of insulin receptor substrate-1 via the ubiquitin ligase subunit Fbw8. Mol Cell 2012; 48:875 - 87; http://dx.doi.org/ 10.1016/j.molcel.2012.09.029; PMID: 23142081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sarikas A, Xu X, Field LJ, Pan ZQ.. The cullin7 E3 ubiquitin ligase: a novel player in growth control. Cell Cycle 2008; 7:3154 - 61; http://dx.doi.org/ 10.4161/cc.7.20.6922; PMID: 18927510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al.. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 2011; 332:1317 - 22; http://dx.doi.org/ 10.1126/science.1199498; PMID: 21659604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villén J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, et al.. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 2011; 332:1322 - 6; http://dx.doi.org/ 10.1126/science.1199484; PMID: 21659605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, Davis JG, Salmon AB, Richardson A, Ahima RS, et al.. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 2012; 335:1638 - 43; http://dx.doi.org/ 10.1126/science.1215135; PMID: 22461615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dibble CC, Asara JM, Manning BD.. Characterization of Rictor phosphorylation sites reveals direct regulation of mTOR complex 2 by S6K1. Mol Cell Biol 2009; 29:5657 - 70; http://dx.doi.org/ 10.1128/MCB.00735-09; PMID: 19720745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Boulbes D, Chen CH, Shaikenov T, Agarwal NK, Peterson TR, Addona TA, Keshishian H, Carr SA, Magnuson MA, Sabatini DM, et al.. Rictor phosphorylation on the Thr-1135 site does not require mammalian target of rapamycin complex 2. Mol Cancer Res 2010; 8:896 - 906; http://dx.doi.org/ 10.1158/1541-7786.MCR-09-0409; PMID: 20501647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Julien LA, Carriere A, Moreau J, Roux PP.. mTORC1-activated S6K1 phosphorylates Rictor on threonine 1135 and regulates mTORC2 signaling. Mol Cell Biol 2010; 30:908 - 21; http://dx.doi.org/ 10.1128/MCB.00601-09; PMID: 19995915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Treins C, Warne PH, Magnuson MA, Pende M, Downward J.. Rictor is a novel target of p70 S6 kinase-1. Oncogene 2010; 29:1003 - 16; http://dx.doi.org/ 10.1038/onc.2009.401; PMID: 19935711 [DOI] [PubMed] [Google Scholar]
- 103.Gao D, Wan L, Inuzuka H, Berg AH, Tseng A, Zhai B, Shaik S, Bennett E, Tron AE, Gasser JA, et al.. Rictor forms a complex with Cullin-1 to promote SGK1 ubiquitination and destruction. Mol Cell 2010; 39:797 - 808; http://dx.doi.org/ 10.1016/j.molcel.2010.08.016; PMID: 20832730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu P, Gan W, Inuzuka H, Lazorchak AS, Gao D, Arojo O, Liu D, Wan L, Zhai B, Yu Y, et al.. Sin1 phosphorylation impairs mTORC2 complex integrity and inhibits downstream Akt signalling to suppress tumorigenesis. Nat Cell Biol 2013; 15:1340 - 50; http://dx.doi.org/ 10.1038/ncb2860; PMID: 24161930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Humphrey SJ, Yang G, Yang P, Fazakerley DJ, Stöckli J, Yang JY, James DE.. Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab 2013; 17:1009 - 20; http://dx.doi.org/ 10.1016/j.cmet.2013.04.010; PMID: 23684622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cancer Genome Atlas Research Network.. Integrated genomic analyses of ovarian carcinoma. Nature 2011; 474:609 - 15; http://dx.doi.org/ 10.1038/nature10166; PMID: 21720365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rodrik-Outmezguine VS, Chandarlapaty S, Pagano NC, Poulikakos PI, Scaltriti M, Moskatel E, Baselga J, Guichard S, Rosen N.. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer Discov 2011; 1:248 - 59; http://dx.doi.org/ 10.1158/2159-8290.CD-11-0085; PMID: 22140653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chaturvedi D, Gao X, Cohen MS, Taunton J, Patel TB.. Rapamycin induces transactivation of the EGFR and increases cell survival. Oncogene 2009; 28:1187 - 96; http://dx.doi.org/ 10.1038/onc.2008.490; PMID: 19151764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhuang G, Yu K, Jiang Z, Chung A, Yao J, Ha C, Toy K, Soriano R, Haley B, Blackwood E, et al.. Phosphoproteomic analysis implicates the mTORC2-FoxO1 axis in VEGF signaling and feedback activation of receptor tyrosine kinases. Sci Signal 2013; 6:ra25; http://dx.doi.org/ 10.1126/scisignal.2003572; PMID: 23592840 [DOI] [PubMed] [Google Scholar]
- 110.Dai N, Rapley J, Angel M, Yanik MF, Blower MD, Avruch J.. mTOR phosphorylates IMP2 to promote IGF2 mRNA translation by internal ribosomal entry. Genes Dev 2011; 25:1159 - 72; http://dx.doi.org/ 10.1101/gad.2042311; PMID: 21576258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dai N, Christiansen J, Nielsen FC, Avruch J.. mTOR complex 2 phosphorylates IMP1 cotranslationally to promote IGF2 production and the proliferation of mouse embryonic fibroblasts. Genes Dev 2013; 27:301 - 12; http://dx.doi.org/ 10.1101/gad.209130.112; PMID: 23388827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zanchin NI, Roberts P, DeSilva A, Sherman F, Goldfarb DS.. Saccharomyces cerevisiae Nip7p is required for efficient 60S ribosome subunit biogenesis. Mol Cell Biol 1997; 17:5001 - 15; PMID: 9271378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen CH, Kiyan V, Zhylkibayev AA, Kazyken D, Bulgakova O, Page KE, Bersimbaev RI, Spooner E, Sarbassov D.. Autoregulation of the mechanistic target of rapamycin (mTOR) complex 2 integrity is controlled by an ATP-dependent mechanism. J Biol Chem 2013; 288:27019 - 30; http://dx.doi.org/ 10.1074/jbc.M113.498055; PMID: 23928304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jewell JL, Guan KL.. Nutrient signaling to mTOR and cell growth. Trends Biochem Sci 2013; 38:233 - 42; http://dx.doi.org/ 10.1016/j.tibs.2013.01.004; PMID: 23465396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dalle Pezze P, Sonntag AG, Thien A, Prentzell MT, Gödel M, Fischer S, Neumann-Haefelin E, Huber TB, Baumeister R, Shanley DP, et al.. A dynamic network model of mTOR signaling reveals TSC-independent mTORC2 regulation. Sci Signal 2012; 5:ra25; PMID: 22457331 [DOI] [PubMed] [Google Scholar]
- 116.Soliman GA, Acosta-Jaquez HA, Dunlop EA, Ekim B, Maj NE, Tee AR, Fingar DC.. mTOR Ser-2481 autophosphorylation monitors mTORC-specific catalytic activity and clarifies rapamycin mechanism of action. J Biol Chem 2010; 285:7866 - 79; http://dx.doi.org/ 10.1074/jbc.M109.096222; PMID: 20022946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Manning BD. Comment on “A dynamic network model of mTOR signaling reveals TSC-independent mTORC2 regulation”: building a model of the mTOR signaling network with a potentially faulty tool. Sci Signal 2012; 5:lc3; author reply lc4. [DOI] [PubMed] [Google Scholar]
- 118.Völkers M, Konstandin MH, Doroudgar S, Toko H, Quijada P, Din S, Joyo A, Ornelas L, Samse K, Thuerauf DJ, et al.. Mechanistic target of rapamycin complex 2 protects the heart from ischemic damage. Circulation 2013; 128:2132 - 44; http://dx.doi.org/ 10.1161/CIRCULATIONAHA.113.003638; PMID: 24008870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Völkers M, Toko H, Doroudgar S, Din S, Quijada P, Joyo AY, Ornelas L, Joyo E, Thuerauf DJ, Konstandin MH, et al.. Pathological hypertrophy amelioration by PRAS40-mediated inhibition of mTORC1. Proc Natl Acad Sci U S A 2013; 110:12661 - 6; http://dx.doi.org/ 10.1073/pnas.1301455110; PMID: 23842089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Huang J, Manning BD.. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem Soc Trans 2009; 37:217 - 22; http://dx.doi.org/ 10.1042/BST0370217; PMID: 19143635 [DOI] [PMC free article] [PubMed] [Google Scholar]