Abstract
To cause disease, bacterial pathogens need to be able to adapt to the physiological conditions found within the host, including an osmolality of approximately 290 mosmol kg−1. While investigating Streptococcus pneumoniae genes contained within pneumococcal pathogenicity island 1, we identified a three-gene operon of unknown function termed phgABC. PhgC has a domain with similarity to diacylglycerol kinases of eukaryotes and is the first described member of a family of related proteins found in many gram-positive bacteria. phgA and phgC mutant strains were constructed by insertional duplication mutagenesis and found to have impaired growth under conditions of high osmotic and oxidative stress. The compatible solutes proline and glycine betaine improved growth of the wild-type and the phgA mutant strains in hyperosmolar medium, and when analyzed by electron microscopy, the cellular morphology of the phgA mutant strain was unaffected by osmotic stress. The phgA and phgC mutant strains were reduced in virulence in models of both systemic and pulmonary infection. As the virulence of the phgA mutant strain was not restored in gp91phox−/− mice and the phgA and phgC mutant strains had reduced growth in both blood and serum, the reduced virulence of these strains is unlikely to be due to increased sensitivity to the respiratory burst of phagocytes but is, instead, due to impaired growth at physiological osmolality.
A fundamental requirement for invasive bacterial pathogens is an ability to adapt to the physiological conditions found within the host, including a temperature of 37°C, pH 7.4, a restricted availability of certain essential nutrients such as iron, and an osmolality of around 290 mosmol kg−1. In addition, growth in certain organs involves exposure to particular environmental stresses, such as the high oxygen tension found within the lungs. Bacteria unable to grow efficiently in these conditions will be compromised in virulence, a point demonstrated by several signature-tagged mutagenesis screens which have identified many bacterial genes involved in basic physiological processes, including the stress response (8, 12, 14). An understanding of the mechanisms by which pathogens adjust to physiological conditions in vivo may identify new therapeutic possibilities to either prevent or treat disease due to microbial pathogens (5).
To maintain turgor and to prevent plasmolysis (separation of the outer membrane from the inner aspect of the bacterial cell wall), bacteria have an intracellular osmotic potential which is greater than the extracellular osmotic potential (9). The mechanisms by which Escherichia coli and Bacillus subtilis adjust to an increase in the osmotic potential of their environments have been defined and are broadly similar (9, 10, 13). Initially there is a rapid influx of K+ ions and then a slower influx of compatible solutes (compounds which can accumulate in high concentrations intracellularly without affecting cellular function), such as proline and glycine betaine (9, 10, 25, 26). In addition, E. coli probably counters the increase in the intracellular positive charge associated with the influx of K+ by an efflux of polyamines (9). The similarities in the responses of E. coli and B. subtilis to changes in the environmental osmotic potential suggest that many bacterial pathogens probably use the same mechanisms. This has been confirmed for the food-borne pathogens Staphylococcus aureus and Listeria monocytogenes, both of which have a high tolerance of high-salt environments (1, 6, 11, 23). However, in contrast to other physiological requirements for growth in the host such as efficient iron uptake mechanisms (3, 4, 18), there is little information on how many common bacterial pathogens respond to changes in the osmolality of their environment and the importance of these responses for virulence.
The general features of a 27-kb S. pneumoniae pathogenicity island called pneumococcal pathogenicity island 1 (PPI1) have previously been described (3). PPI1 contains 28 genes, including the piaABCD operon, which encodes an iron uptake ABC transporter required for in vivo growth (3). During further investigation of PPI1, we identified a three-gene operon termed phgABC which is required for the full virulence of S. pneumoniae in animal models of systemic and pneumonic infection. phgA and phgC mutant strains have impaired growth in conditions of high osmotic and oxidative stress, and PhgC is the first described member of a group of related proteins found in gram-positive bacteria. Further evaluation of phg mutant strains in animal models of infection and during growth in blood or heat-treated serum suggests that these mutant strains are reduced in virulence due to their poor growth at physiological osmolality.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
S. pneumoniae strains used for this work are listed in Table 1. All mutant strains are derived from a capsular serotype 3 S. pneumoniae strain, 0100993, isolated from a patient with pneumonia and obtained from GlaxoSmithKline (12). S. pneumoniae strains were cultured at 37°C and 5% CO2 on Columbia agar supplemented with 5% horse blood or in Todd-Hewitt broth supplemented with 0.5% yeast extract (THY). The following stress media were used for the experiments described in this paper: modified RPMI (RPMIm), a medium with an osmolality similar to that of blood (3); Chelex-THY, a cation-depleted medium (3); high pH, achieved by adjusting the pH of THY with NaOH to pH 8.0; low pH, achieved by adjusting the pH of THY to pH 5.5 with HCl; and oxidative stress, achieved by the addition of up to 5 mM paraquat (Sigma-Aldrich) to THY. When necessary, medium was supplemented with the following: chloramphenicol at a concentration of 4 μg ml−1; 50, 100, or 200 mM NaCl; 100 or 400 mM sucrose; 0.6 M glycine betaine (Sigma-Aldrich); or 0.6 M proline (Sigma-Aldrich). Data for growth curves were collected either by measuring at 1-h intervals the optical density at 580 nm (OD580) of 1-ml cultures grown in sterilized 1.5-ml cuvettes or, for growth under oxidative stress conditions, by using 96-well microtiter dishes (200 μl of culture per well) incubated at 37°C with no added CO2 in a Multiskan Ascent (Labsystems) which had been programmed to measure the OD540 at 1-h intervals. Heparinized blood was obtained from a human volunteer and used for growth experiments immediately. For growth experiments in serum, fresh serum was obtained by centrifugation of an unheparinized human blood sample for 5 min at 10,000 × g. After the addition of heparin, the serum was heat treated for 20 min at 65°C to denature complement components and used immediately. For growth in blood and serum, 106 CFU of each strain was added to 1 ml of blood or serum and incubated with agitation at 37°C, and aliquots were taken for the calculation of CFU per milliliter by plating serial dilutions. Strains were stored at −70°C as aliquots of THY culture (OD580 of 0.3 to 0.4) containing 10% glycerol. Plasmids were amplified in the E. coli strain DH5α, grown at 37°C on Luria-Bertani medium with appropriate selection (17).
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description or sequence | Reference or source |
|---|---|---|
| Strains | ||
| 0100993 | Capsular serotype 3 clinical isolate | 12 |
| phgA mutant | 0100993 containing an insertion in phgA (Sp1043); Cmr | This study |
| phgC mutant | 0100993 containing an insertion in phgC (Sp1045); Cmr | This study |
| PPC50 | 0100993 containing an insertion downstream of phgC; Cmr | This study |
| Plasmids | ||
| pID701 | Disruption vector for S. pneumoniae derived from pEVP3; Cmr | 12 |
| pPC33 | pID701 with ORF3.1/ORF3.2 PCR product ligated into the XbaI site; Cmr | This study |
| pPC49 | pID701 with ORF5.6/ORF5.7 PCR product ligated into the XbaI site; Cmr | This study |
| pPC50 | pID701 with ORF5.8/ORF5.9 PCR product ligated into the XbaI site; Cmr | This study |
| Primers (5′ to 3′) | ||
| ORF3.1 | GCT CTA GAG TAA ATT ACC AAG TGA GG | |
| ORF3.2 | CGC TCT AGA TCA CCT GTA TAG GGT CG | |
| ORF5.6 | GCT CTA GAA CCC TAC TTC TGG TGG C | |
| ORF5.7 | CGC TCT AGA TTG TGA ATC GCC TCA GGC | |
| ORF5.8 | GCT CTA GAT CGG TTC TCT GCC TGA GGC | |
| ORF5.9 | CGC TCT AGA TTT GCA AGG AAT GTC CGT TG | |
| O2.1 | GAA GTG AAG ACG GTG AAG CG | |
| O3.3 | CCT TGT AAG TAG GCA CCT AC | |
| O3.4 | CTC AAG TAG GTG CCT ACT TAC | |
| O4.1 | CTC CAA TAA TTG CTC CAA GC | |
| O4.2 | GCT TGG AGC AAT TAT TGG AG | |
| O5.1 | GCC ACC AGA AGT AGG GTT G | |
| O5.3 | GGG GAT GTT GTC GCA AAT G | |
| O5.4 | CAT ACT CCG ACT CAG GCC | |
| PsaA.1 | CGT TCC GAT TGG GCA AGA C | |
| PsaA.2 | GCA CTT GGA ACA CCA TAG |
Nucleic acid isolation manipulations and analysis.
The following kits were used for the isolation of nucleic acids: for S. pneumoniae chromosomal DNA, Wizard genomic DNA isolation kits (Promega); for plasmid DNA from E. coli, QIAGEN plasmid kits; and for S. pneumoniae RNA, the SV Total RNA Isolation System (Promega). Prepared RNA samples were stored as single-use aliquots at −70°C and protected from degradation by the addition of 0.5% RNasin (Promega). The Access reverse transcription (RT)-PCR system (Promega) was used to amplify cDNA from RNA by using 400 pmol of target-specific primers. Prior to use for RNA transcript analysis, each RNA preparation was tested for DNA contamination by RT-PCR by using heat-treated reverse transcriptase. Semiquantitative RT-PCRs for psaA expression were performed as previously described by using primers PsaA.1 and PsaA.2 (Table 1) (4). RT-PCR with primers designed to amplify 16S RNA was used to correct for differences in the quality of the RNA template obtained from the wild-type and phgA mutant strains (15). Cloning, transformation, restriction digests, and ligations of plasmid DNA were performed according to standard protocols (17). S. pneumoniae sequence data were obtained from The Institute for Genomic Research (TIGR) website (http://www.tigr.org) and analyzed and manipulated by using the programs MacVector (International Biotechnologies, Inc.) and Artemis3 (Genome Research Ltd.). BLAST searches of the available nucleotide and protein databases (including unfinished microbial genomes) for sequence similarities and for conserved protein domains were performed through the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST/), and ClustalW and SeqVu were used to align sequences. Dye terminator chemistry (Applied Biosystems) and cycle sequencing, performed by the Medical Research Council DNA Sequencing Service, Hammersmith Hospital, London, United Kingdom, were used to obtain plasmid nucleotide sequences.
Construction of mutant strains.
Plasmids, primers, and S. pneumoniae strains constructed and used for this work are described in Table 1. To construct Sp1043/phgA and Sp1045/phgC disruption vectors, internal portions of the genes (bp 22 to 495 for Sp1043 and bp 12 to 428 for Sp1045) were amplified by PCR (by using the primers open reading frame 3.1 [ORF3.1]/ORF3.2 and ORF5.6/ORF5.7, respectively) and ligated into the suicide vector pID701 (12) to make pPC33 and pPC49, respectively (both Cmr). Plasmid insert identities were confirmed by DNA sequencing, and S. pneumoniae mutant strains containing disrupted copies of Sp1043 and Sp1045 were constructed by insertion-duplication mutagenesis with pPC33 and pPC49 according to the previously described transformation protocol utilizing competence-stimulating peptide 1 (3, 12). A strain containing an insertion of 83 bp 3′ to Sp1045, termed PPC50, was made by the transformation of S. pneumoniae with pPC50 (constructed by amplification of DNA homologous to the terminal portion of Sp1045 and 83 bp 3′ to the stop codon by PCR using primers ORF5.8/ORF5.9 and ligating the product into pID701). Mutant constructs were confirmed by PCR. All mutations were stable after two 8-h growth cycles (each representing approximately 12 rounds of cell division) in THY without antibiotic selection, with 100% of the 100 colonies tested retaining Cmr.
Electron microscopy.
Bacteria were fixed in 4% paraformaldehyde and 2.5% glutaraldehyde in phosphate-buffered saline on ice for 10 min, pelleted and fixed again for 20 min, gently rinsed in phosphate-buffered saline followed by sodium cacodylate buffer, and then placed in 1% osmium tetroxide in sodium cacodylate buffer for 1 h at room temperature. The samples were dehydrated through an increasing ethanol series (staining en bloc in 2% uranyl acetate at the 30% ethanol stage) and embedded in TAAB 812 resin (TAAB Laboratories Equipment Ltd). Ultrathin sections (60-nm), cut on a Leica UCT ultramicrotome onto Formvar-supported grids, were contrasted with uranyl acetate and lead citrate and examined on a Philips CM100 transmission electron microscope.
In vivo studies of mouse models of S. pneumoniae infection.
Experiments were performed according to the institutional guidelines for animal use and care by using outbred male white mice (strain CD1), wild-type C57B/6 mice obtained from Charles Rivers Breeders, or gp91phox−/− mice bred within Imperial College (gift from G. Dougan) (16), all weighing from 20 to 25 g. For mixed infections, mice were inoculated with approximately equivalent numbers of cells from defrosted stocks of the wild-type and mutant bacterial strains being investigated after appropriate dilution in 0.9% saline. Mice were inoculated either by intraperitoneal (i.p.) injection (100-μl inoculum containing 5 × 103 CFU; systemic infection model) or by intranasal (i.n.) inhalation under halothane (Zeneca) general anesthesia (40-μl inoculum containing 1 ×106 CFU; pneumonia model) (3). Mice were sacrificed at appropriate time points, and the target organs were recovered and homogenized in 0.5 ml of 0.9% saline. Dilutions of the homogenized organs were plated onto nonselective medium, and at least 100 colonies were transferred to selective medium after an overnight incubation to allow calculation of the competitive index (CI), defined as the ratio of the number of CFU recovered from mice infected with the mutant strain to that from mice infected with the wild-type strain divided by the ratio of the number of mutant CFU to the number of wild-type strain CFU in the inoculum (2, 3). For survival curves mice were inoculated i.n. with a pure inoculum of 106 CFU of the wild-type or mutant strains and observed for the development of clinical infection. Mice were sacrificed when they exhibited the following signs of disease: hunched posture, poor mobility, weight loss, and (for i.n. inoculation only) coughing and tachypnea (3).
Statistical analysis.
Growth data presented are representative results for experiments performed two or three times, and all data points are the mean values ± standard deviations (SD) of three samples. Results for ODs, CFU in blood and serum, and CIs were compared by using two-tailed Student's t tests. Survival curve data were compared by using the log rank method.
RESULTS
Identification of an operon contained within PPI1 required for virulence.
As pathogenicity islands often contain several operons whose products are required for virulence (7), we investigated if genes other than piaABCD (3) within PPI1 might be required for growth in vivo. Isogenic mutant strains containing disruptions of genes within PPI1 were constructed and investigated for loss of virulence by using mixed infections with the wild-type strain. One such mutant strain, containing a disruption in the gene Sp1043 (TIGR genome designation) (20), previously identified as ORF3 (3), had a CI of 0.007 ± 0.004 (number of mice, 4) compared to the wild-type strain in a mouse model of pneumonia, indicating that this strain is severely defective for growth in vivo. Sp1043 was investigated further to identify the basis for this attenuation in virulence.
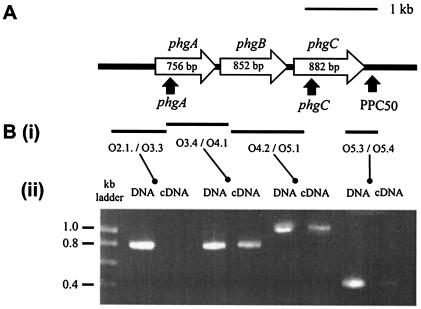
Sp1043 encodes a protein with a predicted length of 252 amino acid residues and is likely to be transcribed as an operon with two adjacent ORFs, Sp1044 (predicted length of 284 amino acid residues) and Sp1045 (predicted length of 294 amino acid residues) (Fig. 1A) (20). The genes (phg for pneumococcal hyperosmotic growth) were phgA (Sp1043), phgB (Sp1044), and phgC (Sp1045). To confirm that these three genes are cotranscribed, the transcript structure of this region was analyzed by using RT-PCR. PCR with primers which span the junctions of phgA/phgB and phgB/phgC amplified identical products when either DNA or cDNA made from total RNA was used as the target (Fig. 1B). However, PCR using primers designed to amplify the region from upstream of the phgA start codon to within phgA or from an internal portion of phgC to 209 bp 3′ to its stop codon failed to amplify products from cDNA, although products were amplified from DNA. Hence, the phgABC genes are cotranscribed beginning with phgA and terminating after phgC. Therefore, an insertion of heterologous DNA containing a stop codon (such as the plasmid pID701) within phgA would result in disruption of the expression of all three genes within the operon.
FIG. 1.
(A) Genetic organization of the phg locus. Thick black line, chromosomal DNA; open arrows, phg ORFs (phgA is Sp1043, phgB is Sp1044, and phgC is Sp1045) with corresponding gene sizes; filled arrows, sites of insertions in mutant strains. (B) Transcriptional analysis of the phg locus. Ethidium bromide-stained agarose gels containing products with the same primer pairs for PCR using S. pneumoniae chromosomal DNA as a template (on the left) and RT-PCR using S. pneumoniae RNA as the template (on the right) are shown in panel ii. RT-PCRs containing no reverse transcriptase generated no products. Bars marked in panel i represent the corresponding target products for each pair of primers used.
Sequence similarity searches with BLAST were performed for each of the derived amino acid sequences of phgA, phgB, and phgC. The closest homologue to PhgA is an open reading frame within the incomplete genome sequence of Acidithiobacillus ferrooxidans, to which PhgA has 36% identity over 112 amino acids. In contrast, the predicted protein product of phgB has almost identical homologues in Streptococcus mitis (91% identity over 244 amino acids) and Streptococcus gordonii (85% identity over 254 amino acids), species which are closely related to S. pneumoniae, as well as significant similarity to several proteins from unrelated bacterial species. An open reading frame in the S. mitis genome also has very high levels of identity at the amino acid level to PhgC (98% identity over 293 amino acids) and forms a probable operon with the S. mitis homologue of phgB. In addition PhgC has 50% or greater similarity over 293 amino acids to proteins predicted from the genome sequences of numerous gram-positive bacteria, forming a previously undescribed protein family. phgC is predicted to encode a cytoplasmic protein and was previously identified by a signature-tagged mutagenesis screen for virulence genes (8), but no other function(s) has been assigned to the homologues of phgABC. However, the derived amino acid sequences of PhgC and related proteins are similar to part of the consensus diacylglycerol kinase (DGK) catalytic domain for eukaryotic organisms (48% over 123 amino acids for Sp1045) (Fig. 2A), including the proposed ATP binding site (Fig. 2B) (22), suggesting that PhgC might function as a kinase.
FIG. 2.
(A) Alignment of the DGK domain consensus sequence with the N terminus of PhgC and related proteins from other gram-positive bacteria. Residues highlighted in gray are identical between proteins. (B) ATP binding motifs from the consensus DGK sequence and PhgC.
phgA and phgC mutant strains are more susceptible to osmotic and oxidative stress.
Growth of the phgA mutant strain was identical to that of the wild-type strain in THY when measured by OD (Fig. 3A), demonstrating that the virulence defect of this strain is not due to a growth defect which is detectable during culture in complete medium. To further characterize the basis of the attenuated virulence of the phgA mutant strain, this strain was screened for growth defects in medium designed to partially replicate some of the conditions found during infection (osmololality around 290 mosmol kg−1, restricted availability of certain cations, and high oxidative stress) and under other high-stress conditions. Growth of the phgA mutant strain in cation-depleted medium (Chelex-THY), under high- or low-pH conditions, or when cultured at 40°C was identical to the wild-type strain (data not shown). However, growth of the phgA mutant was delayed in conditions of hyperosmotic stress. The phgA mutant strain had no discernible growth defect in THY (calculated osmolality, 133 mosmol kg−1) and THY containing 50 mM NaCl (calculated osmolality, 233 mosmol kg−1), a moderate growth defect in THY containing 100 mM NaCl (calculated osmolality, 333 mosmol kg−1), and a severe growth defect in THY containing 200 mM NaCl (calculated osmolality, 533 mosmol kg−1) (Fig. 3B and C). The phgA mutant strain also had delayed growth in THY supplemented with 100 or 400 mM sucrose (Fig. 3D), indicating that the growth defect in medium supplemented with NaCl was due to the increased osmolality rather than a specific effect of NaCl. In RPMIm, a medium with the same osmolality as blood and extracellular fluid, the phgA mutant strain showed a consistent growth defect during late-log-phase growth, with ODs at 7 and 9 h of 0.271 (SD, 0.022) and 0.500 (SD, 0.034), respectively, compared to 0.343 (SD, 0.005) and 0.607 (SD, 0.004) for the wild-type strain, respectively (P = 0.041 and P = 0.025, respectively). A strain containing a mutation in phgC had a growth defect identical to that of the phgA mutant strain in conditions of high osmotic stress (Fig. 3C and data not shown). Therefore, the phenotype associated with phgA may have been due to a polar effect on transcription of phgC. Strain PPC50, containing a mutation 83 bp 3′ to phgC, had no growth defect in hyperosmotic medium, confirming that the phenotype was linked specifically to the phgABC operon (Fig. 3C and data not shown). In further experiments the phg mutant strains were also found to have impaired growth compared to the wild-type and PPC50 strains when cultured in THY supplemented with paraquat, indicating a role for these genes in S. pneumoniae adaptation to oxidative stress (Fig. 4). Hence, the phgABC operon is required for the full resistance of S. pneumoniae to both osmotic and oxidative stress.
FIG. 3.
Growth curves as measured by the ODs of the phg mutant and wild-type strains in THY (A), THY plus 50 (open symbols) or 100 (filled symbols) mM NaCl (B), THY plus 200 mM NaCl (C), and THY plus 100 mM (open symbols) or 400 mM (filled symbols) sucrose (D). Diamond, wild-type strain; square, phgA mutant (Sp1043); triangle, phgC mutant (Sp1045); circle, PPC50. Data points marked with asterisks have a P value of <0.05 compared to the results for the wild-type strain at the same time point and under the same growth conditions (comparisons are shown only for later time points).
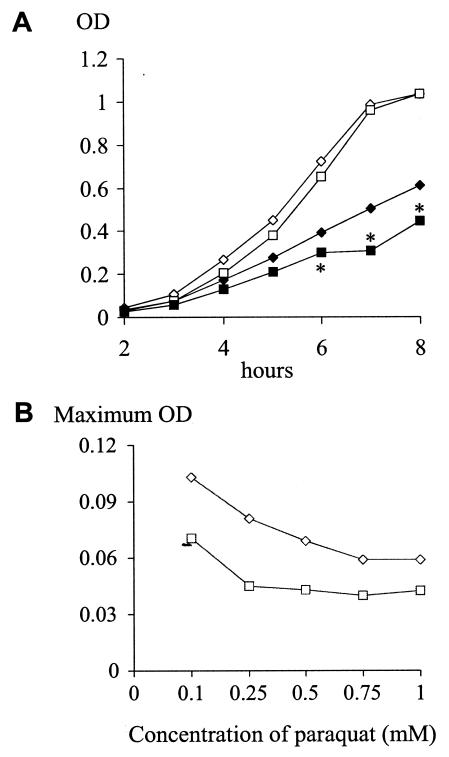
FIG. 4.
(A) Growth curves as measured by the ODs of the phgA mutant (square) and wild-type (diamond) strains in THY (open symbols) and THY plus 0.5 mM paraquat (filled symbols). Asterisks mark the time points when P is ≤0.05 for the differences between the wild-type and phgA mutant strains. (B) Maximum OD in THY plus increasing concentrations of paraquat of the phgA mutant (□) and wild-type (⋄) strains when measured under aerobic conditions in a microtiter plate.
phg mutant strains respond normally to the osmotic protectants glycine betaine and proline.
For B. subtilis and E. coli a normal response to hyperosmotic stress is the active uptake of the compatible solutes glycine betaine and proline (9, 10), and addition of these reagents to the growth medium partially protects these bacteria from hyperosmotic stress (9, 10, 24). To determine whether the growth defect of phg mutant strains under hyperosmotic conditions could be due to an inability to utilize glycine betaine or proline as compatible solutes, we investigated whether the addition of these reagents to the growth medium improves S. pneumoniae growth in hyperosmotic medium and whether this is affected by disruption of phgA (Fig. 5). The addition of either 0.6 M glycine betaine or 0.6 M proline improved the growth of both the wild-type and the phgA mutant strain in THY supplemented with 100 mM NaCl. Under these conditions the OD of the S. pneumoniae wild-type strain was increased by 139% if glycine betaine or proline was added to the medium, while growth of the phgA mutant strain was increased by 168 and 155%, respectively (at the 8-h time point) (Fig. 5). Hence, as for other bacteria, glycine betaine and proline assist S. pneumoniae growth in hyperosmotic medium. However, this osmoprotective effect was not reduced in the phgA mutant strain, and the reduced growth of the phg mutant strains under hyperosmotic conditions is unlikely to be due to an inability to utilize glycine betaine and proline as compatible solutes.
FIG. 5.
Growth curves as measured by the ODs of the phgA mutant (square) and wild-type (diamond) strains in THY plus 100 mM NaCl with (filled symbols) or without (open symbols) supplementation with 0.6 M glycine betaine (A) and THY plus 100 mM NaCl with (filled symbols) or without (open symbols) supplementation with 0.6 M proline (B).
Cellular morphology is unaffected in a phgA mutant strain.
To assess whether the growth delay of the phg mutant strains in hyperosmotic medium is associated with visible changes in the morphology of S. pneumoniae cells, these strains were examined by electron microscopy after exposure to hyperosmotic medium. The wild-type and phgA mutant strains were cultured in THY to mid-log phase and then subjected to various degrees of osmotic shock by transfer to THY containing 0, 100, or 400 mM or 2 M NaCl for 30 min. The bacteria were then washed and fixed immediately for analysis. No visible differences were identified in the overall cellular morphology, thickness and regularity of the cell wall, and size of the phgA mutant strain compared to the wild-type strain under these conditions. These results indicate that mutation of phgA does not cause gross morphological alterations to bacterial cells after exposure to hyperosmolar conditions.
Expression of psaA is not affected in a phgA mutant strain.
The ABC transporter encoded by psaBCA is known to be important for protection of S. pneumoniae against oxidative stress (21). As the phgA mutant strain was more sensitive to oxidative stress, we investigated whether the expression of psaA was affected in this strain by using semiquantitative RT-PCR. After 20, 24, 28, and 32 RT-PCR cycles, the quantities of amplified psaA product as measured by densitometry were similar when RNA extracted from either the phgA mutant or the wild-type strain was used as the template. This demonstrated that there were no major differences in the expression of psaA in the phgA mutant strain compared to the wild-type strain and that this is probably not the explanation for this strain's impaired growth in conditions of high oxidative stress (data not shown).
phg mutant strains are attenuated in models of both systemic and pulmonary infection and in gp91phox−/− mice.
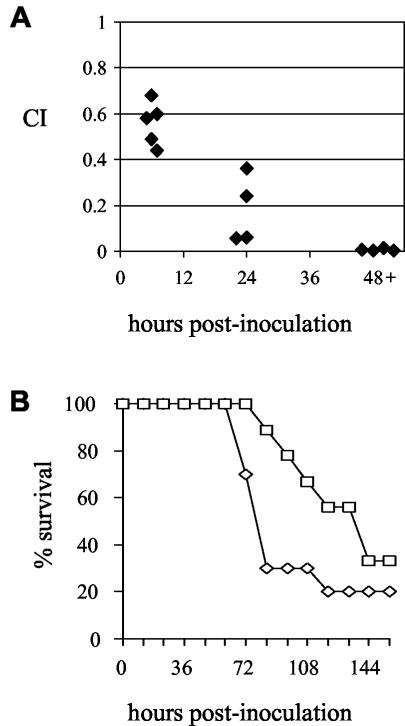
The phgA mutant strain was further investigated to determine the cause of its decreased virulence. Mixed infections with the wild-type strain in mice inoculated i.p. demonstrated that the phgA mutant strain was also attenuated in models of systemic infection, although to a lesser degree than in the pneumonia model (Table 2), showing that the loss of virulence of this strain is not specific to infection of the lung. When measured at different time points after i.n. inoculation, the CI of the phgA mutant strain compared to the wild-type strain decreased as the infection progressed, resulting in a very low CI after 48 h (Fig. 6A and Table 2) and thus indicating that this strain has a severe competitive in vivo growth defect compared to the wild-type strain. In keeping with the growth defects in hyperosmotic medium, the effect on virulence of the disruption of phgC was similar to that of disruption of phgA, and the PPC50 strain was not reduced in virulence. These results confirm that the virulence defect was due to mutations affecting the phg operon and not to polar effects of the disruptions on neighboring genes. Mixed infections are a sensitive method of identifying virulence defects, but whether mutant strains are capable of causing progressive infection is better assessed by assessing the development of infection in groups of animals infected with a pure inoculum of either the wild-type or a mutant strain. We therefore inoculated groups of mice i.n. with either the wild-type or phgA mutant strain. There was a delay in the time taken for symptoms of terminal infection to develop for mice given the phgA mutant strain (median time to terminal infection, 160 h) compared to the wild-type strain (90 h) (Fig. 6B), although this did not reach statistical significance (P = 0.14 by a log rank test). These results show that despite the low CI, the phgA mutant strain is still capable of causing infection, albeit probably a more slowly progressive infection than that seen in mice infected with the wild-type strain.
TABLE 2.
Comparison of the virulence of phg mutant strains to the wild-type strain in mouse models of pneumonia or systemic infection in wild-type and gp91phox−/− mice
| Strains compared | Mouse strain | Time point (h) | Routea | CI (SD) | n |
|---|---|---|---|---|---|
| Wild type vs phgA mutant | Wild-type CD1 | 24 | i.p. | 0.10 (0.03)b | 5 |
| Wild type vs phgC mutant | Wild-type CD1 | 24 | i.p. | 0.16 (0.11)b | 5 |
| Wild type vs PPC50 | Wild-type CD1 | 24 | i.p. | 1.24 (0.36)b | 5 |
| Wild type vs phgA mutant | Wild-type CD1 | 6 | i.p. | 0.20 (0.10) | 4 |
| Wild type vs phgA mutant | Wild-type C57B/6 | 24 | i.p. | 0.30 (0.10)c | 4 |
| Wild type vs phgA mutant | gp91phox−/− C57B/6 | 24 | i.p. | 0.35 (0.16)c | 5 |
| Wild type vs phgA mutant | Wild-type CD1 | 48+ | i.n. | 0.007 (0.004)b | 4 |
| Wild type vs phgC mutant | Wild-type CD1 | 48+ | i.n. | 0.03 (0.014)b | 5 |
| Wild type vs PPC50 mutant | Wild-type CD1 | 48+ | i.n. | 0.73 (0.22)b | 4 |
| Wild type vs phgA mutant | gp91phox−/− C57B/6 | 48+ | i.n. | 0.011 (0)d | 3 |
i.n., pneumonia model of infection; i.p., systemic model of infection.
For the differences between the CIs for the phgA mutant or phgC mutant strain versus the wild-type strain compared to the CIs for the PPC50 strain versus the wild-type strain, P is ≤0.05 for both routes of inoculation.
For the difference in CIs from the wild-type and gp91phox−/− C57B/6 mice inoculated i.p., P = 0.53. No results are available for wild-type C57B/6 mice inoculated i.n. due to the relative resistance of this strain to i.n. S. pneumoniae.
No Cmr colonies recovered (CI calculated as one Cmr colony in 100).
FIG. 6.
(A) CIs for mixed infections comparing the wild-type and phgA mutant strains in a mouse model of S. pneumoniae pneumonia. Each diamond represents results for one mouse at each time point (6, 24, and 48 to 72 h). For the differences between 24 and 48 to 72 h versus 6 h, P = 0.005 and 0.002, respectively (two-tailed Student's t test). (B) Survival of groups of 9 or 10 mice inoculated i.n. with 3 × 106 CFU of the phgA mutant (□) or wild-type strains (⋄) (P = 0.14, log rank test).
During infection S. pneumoniae is exposed to oxidative stress mediated by the respiratory burst of professional phagocytes and the high oxygen tension in the respiratory tract, and the organism also has to be able to grow in extracellular fluid and blood with an osmolality of approximately 290 mosmol kg−1. Therefore, the growth defect of the phg mutant strains under conditions of either oxidative stress or high osmolarity could result in decreased virulence. To help distinguish between these two possibilities, mixed infections of the phgA mutant and wild-type strains were repeated in gp91phox−/− mice. Granulocytes from gp91phox−/− mice cannot generate the respiratory burst due to a disruption in the gene encoding the cytochrome oxidase of the phagosome NADPH oxidase complex (16). Hence, if the reduced resistance of the phgA mutant strain to oxidative stress results in a greater susceptibility to the phagocyte respiratory burst, the competitive advantage of the wild-type bacterial strain over the phg mutants might be reduced in gp91phox−/− mice, and the CI would be closer to 1.0. However, the CIs for infection with the phgA mutant versus the wild-type S. pneumoniae strain were similar in gp91phox−/− mice to those for infection in wild-tye mice in both systemic and pneumonia models of infection (Table 2). These results indicate that phg mutants are not attenuated in virulence because of increased sensitivity to the oxidative stress created by the phagocyte respiratory burst, although they could still be attenuated due to impaired growth in the respiratory tract because of exposure to the relatively high levels of environmental oxidative stress.
phg mutants have reduced growth in blood and serum.
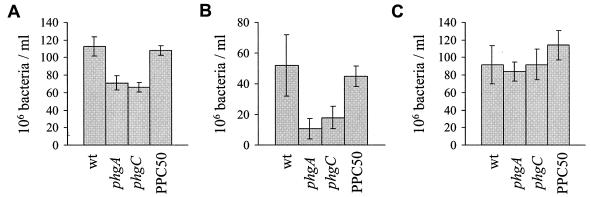
To test if the virulence defect of phg mutants is due to a growth defect at physiological osmolality, the growth of the phg mutants in blood and serum (which have an osmolality similar to that of extracellular fluid) was investigated. After a 4-h incubation at 37°C in blood, the numbers of CFU of phg mutants were consistently around 60% of the number of CFU of the wild-type strain (Fig. 7A). To establish that this growth defect was not dependent on the cellular component of blood or complement proteins, the growth of phg mutants was also compared to that of the wild-type strain in heat-treated serum. After 4 h of incubation in heat-treated serum, the numbers of CFU of the phg mutants were consistently less than 33% of the number of the wild-type strain (Fig. 7B). When serum was diluted by the addition of distilled water to an osmolality of 200 mosmol kg−1, the difference in the numbers of CFU between the phg mutant and wild-type strains was abolished (Fig. 7C). These results demonstrate that the phg mutants have a growth defect in physiological fluids due to their impaired growth in hyperosmolar medium, and this probably explains their reduced virulence.
FIG. 7.
Comparison of the growth of wild-type and phg mutant bacteria (106 CFU/ml) after being cultured for 4 h in heparinized human blood (A), human heat-treated serum (B), and human heat-treated serum diluted with distilled water to an osmolality of 200 mosmol kg−1(C). Error bars represent the standard deviation (for three samples per data point), and data are presented from one representative experiment of two (graphs B and C) or five (graph A) performed. For the differences between the phgA mutant and the wild-type strains, P has a value of 0.02 in undiluted serum and 0.001 in blood, and for the differences between the phgC mutant and the wild-type strain, P is 0.036 in undiluted serum and 0.001 in blood.
DISCUSSION
While investigating genes carried within S. pneumoniae PPI1, we have identified a three-gene operon, phgABC, whose products are required for normal growth under conditions of osmotic and oxidative stress. Disruption of the terminal gene phgC resulted in a phenotype similar to that caused by loss of the whole operon, indicating that PhgC is a key component for the function of the phgABC operon. Interestingly, PhgC belongs to a group of related proteins found in gram-positive bacteria which have not previously been investigated. These proteins all contain a 5′ region with similarity to part of the consensus DGK catalytic domain, including a glycine-rich motif thought to be part of the ATP binding site (22). Eukaryotic DGKs modulate the level of the second messenger diacylglycerol and participate in the control of many cellular and physiological processes but are not known to affect cell responses to osmotic stress (22). However, diacylglycerol is unlikely to be the substrate for PhgC as PhgC has partial similarity to only part of the catalytic domain of eukaryotic DGKs and is otherwise unrelated to DGKs. Furthermore, in prokaryotes phosphorylation of diacylglycerol is catalyzed by an enzyme unrelated to eukaryotic DGKs, and a homolog of this enzyme exists within the S. pneumoniae genome (Sp0968) (22). The relationship between the similarity of the 5′ region of PhgC to eukaryotic DGK and the function of this protein in protecting S. pneumoniae from osmotic and oxidative stress is at present unclear.
How S. pneumoniae adapts to hyperosmolar conditions has not previously been investigated, but it likely utilizes similar mechanisms to those used by other bacteria such as the rapid accumulation of K+ and the uptake or synthesis of compatible solutes (9, 10, 25, 26). Indeed, we have demonstrated that proline and glycine betaine protect S. pneumoniae from hyperosmotic shock, suggesting that these compounds can act as compatible solutes for S. pneumoniae as well as B. subtilis, L. monocytogenes, S. aureus, and E. coli (6, 10, 11, 24, 25). However, the phgA mutant has no defect in proline- or glycine betaine-mediated osmoprotection and has no detectable defect in gross cellular morphology in high osmotic medium compared to the wild-type strain. In addition to their growth defect in hyperosmolar conditions, phg mutants are also more susceptible to oxidative stress, and the function of PhgABC might not be specific to osmotic stress but could regulate S. pneumoniae responses to stress in general. Such a role would be compatible with the predicted cytoplasmic localization of PhgC and with its possible kinase domain. The osmolality of physiological fluids is between that of THY supplemented with 50 or 100 mM NaCl. As growth of the phg mutants is impaired in RPMIm and physiological fluid and the virulence of the phgA mutant is not restored in gp91phox−/− mice, the reduced virulence of these strains is likely due to their impaired growth at physiological osmolality rather than increased sensitivity to oxidative burst-dependent killing by host phagocytes. In addition, the high oxygen tension in the respiratory tract may also affect the virulence of the phg mutants and could partially explain why they have lower CI values in the pneumonia model than in the septicemia model. The ability of bacterial pathogens to adjust to physiological osmolality has rarely been investigated, but the identification of potassium, glycine betaine, and proline transporters by signature-tagged mutagenesis screens suggests that bacterial osmotic responses are often important for bacterial growth in the host and, hence, for virulence (8, 12, 19).
PPI1 was initially identified due to the low G+C content of this region (32.6%) compared to the mean for the S. pneumoniae genome (3, 7). However, the G+C content of the region of PPI1 containing phgABC is 37.9%, which is closer to the normal level for the S. pneumoniae genome (39.7%) (20), and the description of the TIGR S. pneumoniae genome suggests that PPI1 consists of two regions of atypical nucleotide composition at the 5′ and 3′ ends of PPI1 but not including the phg locus (20). In addition, with the availability of sequence data for many more bacteria since our initial description of PPI1, we have been able to identify genes encoding predicted proteins which are almost identical to PhgB and PhgC in the genome of S. mitis, a close relative of S. pneumoniae. Therefore, it is likely that at least phgB and phgC were not acquired horizontally, and PPI1 may actually represent two regions of horizontally acquired DNA which integrated into the S. pneumoniae genome on separate occasions and which flank phgBC.
In conclusion, we have identified a three-gene operon, phgABC, whose genes encode proteins required for normal S. pneumoniae growth in hyperosmotic medium and under conditions of oxidative stress and, therefore, for virulence in mice models of pneumonia and systemic infection. The last gene of this operon, phgC, encodes a protein which is the first described member of a new family of proteins in gram-positive bacteria. Further investigation is required to ascertain how the phg operon influences growth in hyperosmotic medium.
Acknowledgments
This work was supported by a Wellcome Trust Advanced Fellowship for Medical and Dental Graduates to J. S. Brown (056586) and by grants from the Wellcome Trust and the Medical Research Council to D. W. Holden.
Editor: J. N. Weiser
REFERENCES
- 1.Bayles, D. O., and B. J. Wilkinson. 2000. Osmoprotectants and cryoprotectants for Listeria monocytogenes. Lett. Appl. Microbiol. 30:23-27. [DOI] [PubMed] [Google Scholar]
- 2.Beuzon, C. R., S. Meresse, K. E. Unsworth, J. Ruiz-Albert, S. Garvis, S. R. Waterman, T. A. Ryder, E. Boucrot, and D. W. Holden. 2000. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 19:3235-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, J. S., S. M. Gilliland, and D. W. Holden. 2001. A Streptococcus pneumoniae pathogenicity island encoding an ABC transporter involved in iron uptake and virulence. Mol. Microbiol. 40:572-585. [DOI] [PubMed] [Google Scholar]
- 4.Brown, J. S., S. M. Gilliland, and D. W. Holden. 2002. Characterization of Pit, a Streptococcus pneumoniae iron uptake transporter. Infect. Immun. 70:4389-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, J. S., A. D. Ogunniyi, M. C. Woodrow, D. W. Holden, J. C. Paton. 2001. Immunization with components of two iron uptake ABC transporters protects mice against systemic infection with Streptococcus pneumoniae. Infect. Immun. 69:6702-6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham, J. E., and B. J. Wilkinson. 1992. Staphylococcus aureus osmoregulation: roles for choline, glycine betaine, proline, and taurine. J. Bacteriol. 174:2711-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 8.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389-1406. [PMC free article] [PubMed] [Google Scholar]
- 9.Ingraham, J. L. 1987. Effect of temperature, pH, water activity, and pressure on growth, p. 1548-1554. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella typhimurium cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 10.Kempf, B., and E. Bremer. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 170:319-330. [DOI] [PubMed] [Google Scholar]
- 11.Ko, R., L. T. Smith, and G. M. Smith. 1994. Glycine betaine confers enhanced osmotolerance and cryotolerance on Listeria monocytogenes. J. Bacteriol. 176:426-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau, G. W., S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison, and D. W. Holden. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40:555-571. [DOI] [PubMed] [Google Scholar]
- 13.Measures, J. C. 1975. Role of amino acids in osmoregulation of non-halophilic bacteria. Nature 257:398-400. [DOI] [PubMed] [Google Scholar]
- 14.Mei, J. M., F. Nourbakhsh, C. W. Ford, and D. W. Holden. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26:399-407. [DOI] [PubMed] [Google Scholar]
- 15.Ogunniyi, A. D., P. Giammarinaro, and J. C. Paton. 2002. The genes encoding virulence-associated proteins and the capsule of Streptococcus pneumoniae are upregulated and differentially expressed in vivo. Microbiology 148:2045-2053. [DOI] [PubMed] [Google Scholar]
- 16.Pollock, J. D., D. A. Williams, M. A. Gifford, L. L. Li, X. Du, J. Fisherman, S. H. Orkin, C. M. Doerschuk, and M. C. Dinauer. 1995. Mouse model of X-linked chronic granulomatous disease, an inherited defect in phagocyte superoxide production. Nat. Genet. 9:202-209. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Schryvers, A. B., and I. Stojiljkovic. 1999. Iron acquisition systems in the pathogenic Neisseria. Mol. Microbiol. 32:1117-1123. [DOI] [PubMed] [Google Scholar]
- 19.Schwan, W. R., S. N. Coulter, E. Y. Ng, M. H. Langhorne, H. D. Ritchie, L. L. Brody, S. Westbrock-Wadman, A. S. Bayer, K. R. Folger, and C. K. Stover. 1998. Identification and characterization of the PutP proline permease that contributes to in vivo survival of Staphylococcus aureus in animal models. Infect. Immun. 66:567-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 21.Tseng, H. J., A. G. McEwan, J. C. Paton, and M. P. Jennings. 2002. Virulence of Streptococcus pneumoniae: PsaA mutants are hypersensitive to oxidative stress. Infect. Immun. 70:1635-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Blitterswijk, W. J., and B. Houssa. 2000. Properties and functions of diacylglycerol kinases. Cell. Signal. 12:595-605. [DOI] [PubMed] [Google Scholar]
- 23.Vijaranakul, U., M. J. Nadakavukaren, D. O. Bayles, B. J. Wilkinson, and R. K. Jayaswal. 1997. Characterization of an NaCl-sensitive Staphylococcus aureus mutant and rescue of the NaCl-sensitive phenotype by glycine betaine but not by other compatible solutes. Appl. Environ. Microbiol. 63:1889-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Blohn, C., B. Kempf, R. M. Kappes, and E. Bremer. 1997. Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B. Mol. Microbiol. 25:175-187. [DOI] [PubMed] [Google Scholar]
- 25.Whatmore, A. M., J. A. Chudek, and R. H. Reed. 1990. The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J. Gen. Microbiol. 136:2527-2535. [DOI] [PubMed] [Google Scholar]
- 26.Whatmore, A. M., and R. H. Reed. 1990. Determination of turgor pressure in Bacillus subtilis: a possible role for K+ in turgor regulation. J. Gen. Microbiol. 136:2521-2526. [DOI] [PubMed] [Google Scholar]