Abstract
Obligate mutualistic symbioses rely on mechanisms that secure host-symbiont commitments to maximize host benefits and prevent symbiont cheating. Previous studies showed that somatic incompatibilities correlate with neutral-marker-based genetic distances between fungal symbionts of Panamanian Acromyrmex leaf-cutting ants, but the extent to which this relationship applies more generally remained unclear. Here we showed that genetic distances accurately predicted somatic incompatibility for Acromyrmex echinatior symbionts irrespective of whether neutral microsatellites or AFLP markers were used, but that such correlations were weaker or absent in sympatric Atta colombica colonies. Further analysis showed that the symbiont clades maintained by A. echinatior and A. colombica were likely to represent separate gene pools, so that neutral markers were unlikely to be similarly correlated with incompatibility loci that have experienced different selection regimes. We suggest that evolutionarily derived claustral colony founding by Atta queens may have removed selection for strong incompatibility in Atta fungi, as this condition makes the likelihood of symbiont swaps much lower than in Acromyrmex, where incipient nests stay open because queens have to forage until the first workers emerge.
Keywords: Leucoagaricus gongylophorus, Commitment, Mutualism, Fungus-growing ants, Interaction, Attini, Symbiosis, Basidiomycota
1. Introduction
Endosymbionts are normally asexual and transmitted by uniparental vertical inheritance (Sachs et al., 2011). Multicellular organisms thus have a single mitochondrial genotype and those that have photosynthesis rely on a single clone of plastids. The evolution of such obligate symbiotic mutualisms has strong elements of partner commitment driven by kin selection, because exclusive association of hosts with a single symbiont genotype ensures that its services to growth and survival of the host will benefit clone mates that are vertically transmitted when hosts reproduce (Frank, 1994, Doebeli and Knowlton, 1998, Sachs et al., 2004, Foster and Wenseleers, 2006). The same logic implies that hosts and symbionts are potentially in conflict over the mode of symbiont transmission (Frank, 1996, Douglas, 2008), as symbionts would always benefit from additional horizontal transmission. However, hosts might suffer fitness losses from this form of commitment-disloyalty and therefore suppress symbiont investments in sexual reproduction (Frank, 1996, Leigh, 2010). When, despite such host efforts, symbiont lineages manage to co-infect hosts and compete for resources, hosts will be under selection to monitor symbiont genetic diversity and eliminate additional symbiont lineages when such competition implies a net loss of cumulative symbiont service to the host (Frank, 1996).
The classical mitochondrial and plastid endosymbioses are so integrated with their host cells that their reduced genomes preclude any form of non-symbiotic life, and the same is true for many obligate and facultative endosymbionts with less reduced genomes (McCutcheon and Moran, 2012). Many of these interactions likely represent adaptive endpoints of host-symbiont coevolution, where host-symbiont conflicts were resolved in favor of the hosts (McCutcheon and Moran, 2012, Wernegreen, 2012) or symbiont (Werren et al., 2008), but their advanced stage of symbiosis normally precludes direct tests of evolutionary conflict theory over symbiont mixing because co-evolved symbionts can often not be reared in vitro. The fungus-growing ants offer a feasible model system to do such tests, because they have multiple obligate mutualisms, including fungus gardens (Schultz and Brady, 2008, Mikheyev et al., 2010) and cuticular Actinobacteria (Cafaro et al., 2011, Andersen et al., 2013) that are ectosymbionts for individual ants, but endosymbionts for the ant colonies. This implies that partners can be reared in vitro without each other's interference for sufficient periods of time to quantify antagonism between symbiont clones, monitor host reactions to alternative symbionts, and relate observed differences to the genetic characteristics of the interactants (Armitage et al., 2011, Bot et al., 2001, Seal et al., 2012).
Attine ant colonies have never been found to rear a multi-clone fungus garden (Apterostigma, Dentinger et al., 2009; Cyphomyrmex, Green et al., 2002 and Mehdiabadi et al., 2012; Atta, Mueller et al., 2010; Acromyrmex, Poulsen and Boomsma, 2005). In all studies, there was substantial genetic variation among fungus-garden clones across sympatric colonies, consistent with vertical transmission and normal patterns of variation of mitochondrial and plastid organelles across individual animals and plants (e.g. Embley and Martin, 2006). However, in contrast to these cellular endosymbionts, there may be considerable horizontal transfer of symbionts when territories of founding attine colonies overlap, consistent with species belonging to the same genus often sharing clades of symbionts (Green et al., 2002, Poulsen and Boomsma, 2005, De Fine Licht and Boomsma, 2011, De Fine Licht and Boomsma, 2014, Mehdiabadi et al., 2012), while sympatric attine ant genera normally rear distinct fungal symbiont clades (Mueller and Gerardo, 2002, Dentinger et al., 2009, Vo et al., 2009, Mehdiabadi et al., 2012, Kooij et al., 2015).
Horizontal swaps of fungus-garden symbionts between colonies of the same or closely related attine ant species may reduce the efficiency of co-evolutionary adaptation at the lowest taxonomic level, but allows ant lineages to replace an asexual crop symbiont that is compromised by genetic load or another form of maladaptation to prevailing ecological conditions (Mueller, 2002). However, as long as fungus-garden clones are thriving, they will also be under selection to actively defend their ant-care monopoly (Bot et al., 2001, Poulsen and Boomsma, 2005). Such defenses are expected to evolve when the threat to be replaced is real, i.e., any hostility of this kind should target non-self symbiont genotypes belonging to the clade of symbionts that can in fact partake in a viable symbiosis with a focal attine ant species.
The Acromyrmex echinatior and Acromyrmex octospinosus populations in Gamboa, Panama appear to co-exploit the same clade of fungus-garden symbionts in sympatry (Bot et al., 2001, Richard et al., 2007b, Poulsen et al., 2009). Resident fungus gardens of these ant species have been shown to maintain their clonal integrity by a combination of behavioral adaptations in the ants to remove and kill alternative fungus clones (Bot et al., 2001, Ivens et al., 2009) and by the expression of somatic incompatibility reactions between clones from different Acromyrmex colonies reared together on the same agar plates (Poulsen and Boomsma, 2005). These incompatibilities correlated with Amplified Fragment Length Polymorphism (AFLP) genetic distances between pairs of fungal symbionts, a pattern that also applied to the fecal fluid of Acromyrmex large workers fed with fungus from their own and other colonies (Poulsen and Boomsma, 2005). However, founding A. octospinosus queens readily accept fungal clones from other colonies, suggesting that this stage offers a special window for horizontal transfers even though signs of reduced performance with a novel symbiont taken from a mature colony were also found (Poulsen et al., 2009). This study suggested that incompatibility mechanisms might be different for sympatric Atta leaf-cutting ants, which tend to rear different fungal symbionts (Mikheyev et al., 2006, Mikheyev et al., 2007, Kooij et al., 2015) and whose queens never forage during colony founding and therefore have negligible likelihood of encountering alternative symbionts.
Somatic (in)compatibilities between plated fungi of Panamanian Acromyrmex species are expressed in a gradual manner that correlates with AFLP genetic distances between pairs of clones (Poulsen and Boomsma, 2005). In basidiomycetes, somatic incompatibilities are generally induced by allorecognition so that strains are increasingly likely to be incompatible when they are more genetically different (May, 1988, Worrall, 1997). These reactions tend to be stepwise (Rayner et al., 1984, Rayner, 1991, Worrall, 1997) and usually involve dark pigmentation in the interaction zone (Rayner et al., 1984), changes in septal maintenance, or blockage of septa precluding the movement of cytoplasm, and can lead to programmed cell death (Rayner, 1991). The underlying genetic mechanisms remain largely unknown, but multiple loci appear to be involved (Worrall, 1997) and their expression may be linked to sexual incompatibility genes (Van der Nest et al., 2009, Van der Nest et al., 2011). If sex occurs at all, it is extremely rare in the fungal symbionts of higher attine ants (Fisher et al., 1994a, Fisher et al., 1994b, Pagnocca et al., 2001, Mueller, 2002, Mikheyev et al., 2006). Recent work further indicated that the symbionts of Panamanian leaf-cutting ants are multi-genomic chimeras (Kooij et al., 2015), which likely explains why incompatibility patterns between Panamanian Acromyrmex symbionts appear to be gradual (Poulsen and Boomsma, 2005).
In the present study, we aimed to further our understanding of the biological factors governing somatic incompatibility among strains of attine ant fungal symbionts. We focused on comparing sympatric mature colonies of Panamanian Atta colombica and Acromyrmex echinatior leaf-cutting ants to address the following questions: (1) Do plated fungal symbionts of A. echinatior and A. colombica express similar somatic incompatibility reactions when confronted with symbionts from other colonies? (2) To what extent does the intensity of these reactions differ within and between the two genera? (3) To what extent is the intensity of these reactions correlated with genetic distance between the fungal symbionts? (4) Are sympatric colonies of A. echinatior and A. colombica rearing the same or overlapping set(s) of fungal symbionts or are they associated with distinct lineages of the Leucoagaricus gongylophorus symbiont? The same set of symbionts would be expected if horizontal symbiont transmission between the two ant genera were more frequent than natural divergence of lineages via mutation and genetic drift. In contrast, segregated lineages would be expected when horizontal transmission between genera is absent because co-adaptations in L. gongylophorus strains reared by A. echinatior and A. colombica would preclude that horizontal symbiont swaps between genera are viable. Finally, we also compared two different sets of genetic markers (AFLPs and microsatellites) and different time spans between plate inoculation and scoring of incompatibilities to evaluate the robustness of our conclusions.
2. Materials and methods
2.1. Biological material
Fungal cultivars were isolated from nine Acromyrmex echinatior colonies (Ae150A, Ae160, Ae168, Ae263, Ae266, Ae322, Ae356, Ae394, Ae488) and nine Atta colombica colonies (Ac-2006-27, Ac-2009-42, Ac-2009-46, Ac-2011-2, Ac-2011-3, Ac-2012-1, Ac-2012-2, Ac-2012-8, Ac-2012-31) living sympatrically in Gamboa, Panama, and grown on 39 g l−1 Potato Dextrose Agar (Sigma–Aldrich, St Louis, MO, USA) with the addition of 5 g l−1 yeast extract, 15 mg l−1 Tetracycline and 12 mg l−1 Streptomycin. For each of the colonies a single isolate was obtained as it has been shown before that both Acromyrmex (Poulsen and Boomsma, 2005) and Atta (Mueller et al., 2010) maintain their fungal symbiont in a monoculture. DNA of each fungal strain was extracted using the Qiagen (Venlo, The Netherlands) DNeasy Plant Tissue extraction kit and stored at −20 °C until further analysis. The Gamboa sampling site was the same as where most previous colonies of the Copenhagen fungus-growing ant research program have been collected, including the Acromyrmex colonies studied by Bot et al., 2001, Poulsen and Boomsma, 2005, Mikheyev et al., 2007, Richard et al., 2007b, Ivens et al., 2009, and Poulsen et al. (2009).
2.2. Genetic analyses
To calculate the genetic distance between each of the fungal strains we used two different methods: AFLP and ten microsatellite markers (A128, A1030, A1132, A1151, B12, B447, C101, C126, C647 and D115 developed by Scott et al. (2009)). AFLP was performed as described by Vos et al. (1995) with two selective primer combinations (Eco-ACC + Mse-CAT and Eco-ACC + Mse-CAC). Microsatellite markers were analyzed using PCR with 5 μl VWR Red Taq DNA polymerase Master Mix (VWR International, Haasrode, Belgium), 0.25 μl forward and reverse primer each, 1.5 μl ddH2O and 1 μl DNA, and a program of 5 min denaturing at 95 °C, followed by 14 cycles of 30 s denaturing at 95 °C, 30 s annealing at 68–58 °C with a touchdown of −0.5 °C per cycle, and 30 s extension at 72 °C followed by 20 cycles of 30 s denaturing at 95 °C, 30 s annealing at 58 °C and 30 s extension at 72 °C, and finally a 15 min extension at 72 °C.
Both AFLP and microsatellite amplification products were analyzed on an ABI 3130xl (Applied Biosystems, Nærum, Denmark) sequencer. Specific allele scorings (AFLP: Table S1; microsatellites: Table S2) were obtained by analyzing chromatograms in Genemapper 4.0 (Applied Biosystems, Nærum, Denmark). The program Populations 1.2.32 (Langella, 2001) was used to calculate Fst values for the microsatellite data and Nei's standard genetic distance (Ds) for the AFLP data, followed by Neighbor Joining phylogenetic analyses with 500 bootstrap replicates for each of the two types of markers. The program STRUCTURE v2.3.4 (Pritchard et al., 2000, Falush et al., 2003) was used to analyze population structure for the AFLP data with the following settings: an optimized K = 3 tested with the online program STRUCTURE HARVESTER (Earl and vonHoldt, 2011), a burn-in period of 1,000,000 iterations followed by 10,000,000 MCMC iterations, an admixture model, and independent allele frequencies among populations with λ = 0.78. The 20 runs that we obtained were merged with the Greedy analysis with 1,000,000 repeats in the CLUMPP software (Jakobsson and Rosenberg, 2007), and visualized with DISTRUCT (Rosenberg, 2004). Microsatellite data were further analyzed using the package POPPR (Kamvar et al., 2014) in R (R Core Team, 2013), to evaluate the discriminatory power of our markers and verify clonality of the symbionts.
2.3. Somatic incompatibility
To test whether plated fungi showed (in)compatibility, cultures of all 18 fungi were paired in all possible (171) combinations with four replicate pairings for each combination. For each pair, small tufts of mycelium (ca. 2 mm3) were placed at a distance of 1.5 cm from each other on a 5 cm Petri dish with 39 g l−1 Potato Dextrose Agar with the addition of 5 g l−1 yeast extract and 35 g l−1 Agar. The growth medium was selected in a pilot study testing somatic incompatibility reactions for control (self) encounters on this and three alternative media, which showed that the used PDYA medium most consistently avoided discolorations in controls (Fig S1).
Incompatibility reactions were assessed after 6, 8, and 10 weeks and scored using the semi-quantitative scale described by Poulsen and Boomsma (2005): 0 = demarcation zone absent, 1 = demarcation zone weak but present, 2 = demarcation zone broad and distinct, and 3 = strong demarcation zone with consistent brown or black coloration of mycelium. These four scores are consistent with the variation in somatic (in)compatibility reactions that are typically found in free-living basidiomycetes, where they usually occur in a more stepwise manner as explained above (Worrall, 1997). All scorings were done blindly by randomly assigning numbers to each plate, after which two of the authors did the initial assessment and a third author blindly checked combinations for which the first two authors did not agree on the score. Degrees of (in)compatibility were subsequently compared with genetic distances between pairs of fungal clones using Mantel and Partial Mantel Tests for Dissimilarity Matrices (“mantel”) (R Core Team, 2013) with 99,999 permutations in the Community Ecology Package: Ordination, Diversity and Dissimilarities “vegan” (Oksanen et al., 2013). Correlations were forced through the origin based on the fact that the controls were (0,0). Figures were created using Plot With Repeated Symbols by Size (“sizeplot”) in the Plotrix package (Lemon, 2006).
3. Results
For Acromyrmex symbionts, consistent scorings of somatic incompatibility were obtained 8 weeks after agar plates were inoculated. Coloration contrasts were not fully developed after 6 weeks, so Mantel correlation coefficients between incompatibility and genetic distance after 6 weeks remained low (Fig S2). Beyond 8 weeks, Mantel correlation coefficients continued to improve, but contaminations and medium desiccation problems affected scoring accuracy 10 weeks after inoculation (Fig S2) so that almost 5% of the replicates were lost. As scoring results at 8 weeks were approximately the same as the 2-months between inoculation and scoring in Poulsen and Boomsma (2005), our main results for the 8 weeks observation period are presented, to remain as comparable as possible with that previous study on somatic incompatibilities between A. echinatior and A. octospinosus fungal symbionts collected at the same sampling site more than 10 yr earlier. However, for Atta symbionts a comparable result was only obtained when using microsatellite markers, as AFLP markers produced Mantel correlation coefficients close to zero for all observation periods (Fig S2).
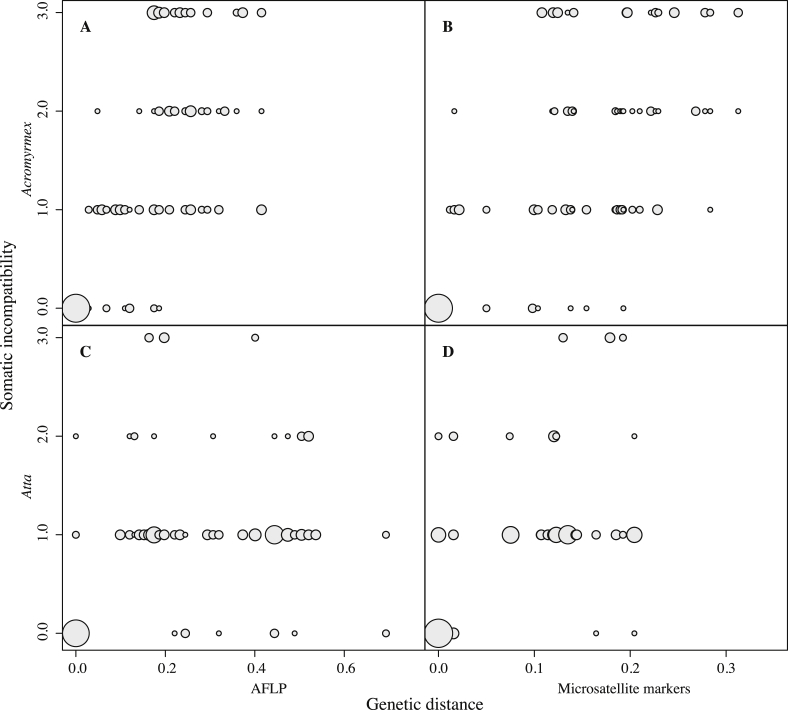
Using the 8 weeks data, somatic incompatibilities increased with increasing AFLP genetic distances between fungi (Mantel r = 0.463, p = 0.003, Fig 1A) in Acromyrmex symbionts, but not in Atta symbionts (Mantel r = −0.164, p = 0.792, Fig 1C). When using genetic distances based on microsatellite markers, both Acromyrmex (Mantel r = 0.469, p = 0.003, Fig 1B) and Atta (Mantel r = 0.312, p = 0.032, Fig 1D) symbionts had incompatibilities that increased with genetic distance, but less of the incompatibility variance was explained in Atta than in Acromyrmex. The A. echinatior results were consistent with the results obtained by Poulsen and Boomsma (2005), but in that study the variance explained by the Mantel coefficient was larger (r = 0.855; p < 0.0001). However, when the 10-weeks scorings for Acromyrmex symbiont pairings was used there was a correlation closer to the one obtained after 2 months by Poulsen and Boomsma (2005) (Microsatellites: Mantel r = 0.596, p < 0.001; AFLP: Mantel r = 0.654, p < 0.001).
Fig. 1.
Mutual somatic incompatibility reactions for Acromyrmex and Atta-associated fungal symbionts plotted against genetic distances calculated from genetic variation at AFLP's and ten microsatellite markers, with the size of each circle representing the number of times a particular combination was found. Correlations were significant for Acromyrmex with both (A) AFLP markers (Mantel r = 0.463, p = 0.003) and (B) microsatellites (Mantel r = 0.469, p = 0.003), and for Atta with (D) microsatellites (Mantel r = 0.312, p = 0.032), but not C) AFLP markers (Mantel r = −0.164, p = 0.792). All Mantel tests were performed with 99,999 permutations.
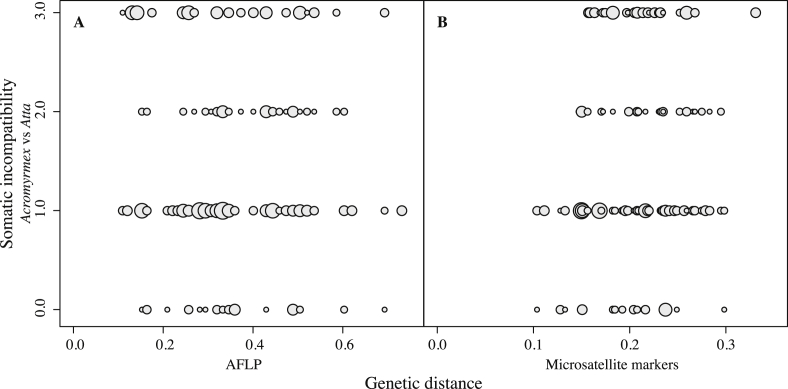
Overall, the Atta fungi had smaller genetic distances when calculated from the microsatellite marker data (0.09 ± 0.01 SE), but larger genetic distances when using AFLP markers (0.26 ± 0.03 SE) compared to Acromyrmex (0.13 ± 0.01 SE and 0.17 ± 0.02 SE, respectively), which is reflected in the average number of AFLP bands observed (Atta: 37.3 ± 1.8 SE; Acromyrmex 30.9 ± 1.5 SE; t15.49 = −2.724, p = 0.015). This implied that AFLP and microsatellite genetic distances (Fst) were only comparable for Acromyrmex symbionts (r = 0.932; Fig S3), whereas correlations decreased in comparisons between Acromyrmex and Atta symbionts and became very low in comparisons involving only colonies of Atta colombica (Fig S3). Independent of the type of genetic marker used, mean genetic distances were higher in comparisons between Acromyrmex and Atta (Microsatellites: 0.20 ± 0.01 SE; AFLP: 0.36 ± 0.02 SE) than in comparisons within the ant genera (means ± SEs given above; microsatellite markers: F2,168 = 45.127, p < 0.0001; AFLP: F2,168 = 22, p < 0.0001). Also the average incompatibility scores were different for comparisons within and across ant species (genera) (χ2 = 12.236; df = 2; p < 0.01). This difference was mostly due to less strongly expressed somatic incompatibilities between Atta symbionts, because the stronger mean reactions among Acromyrmex symbionts alone and between Acromyrmex and Atta symbionts were not significantly different from each other (W = 1635, p = 0.338). After pooling data across the entire range of genetic distances, the increase in somatic incompatibility with genetic distance was completely absent (AFLP: Mantel r = −0.045, p = 0.595, Fig 2A) or no longer significant (Microsatellites: Mantel r = 0.153, p = 0.213, Fig 2B).
Fig. 2.
Somatic incompatibilities in confrontations between fungal symbionts from sympatric Acromyrmex and Atta colonies plotted against genetic distances calculated using AFLP (A) and microsatellite (B) markers. The size of circles is proportional to the number of times that particular combinations were found. Correlations were not significant (microsatellites: Mantel r = 0.1518, p = 0.2064; AFLP: Mantel r = −0.1034, p = 0.7063). All tests were performed with 99,999 permutations.
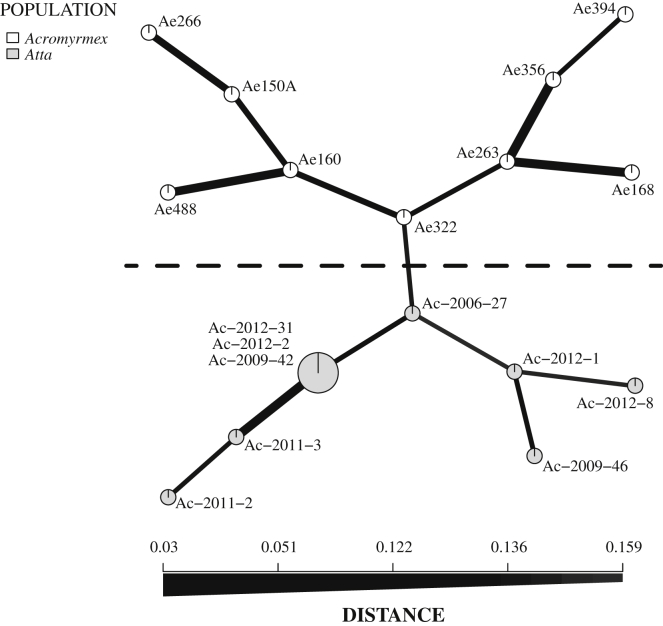
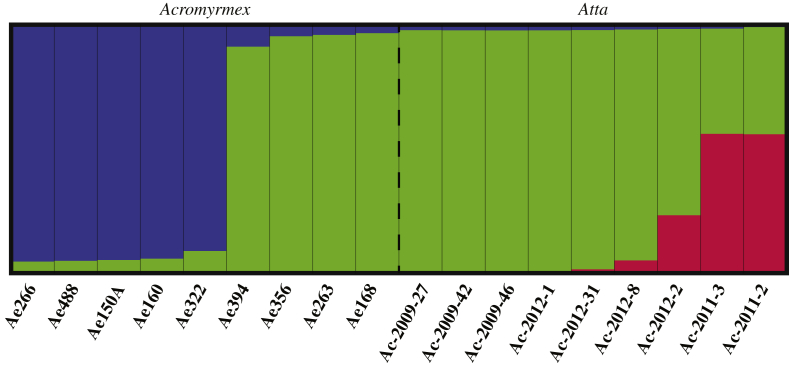
Specific evaluation of the microsatellite genetic differences between the fungal symbionts of Atta and Acromyrmex colonies showed that they were completely separated (Fig 3, Fig S4 and Supplementary Information), consistent with earlier findings by Mikheyev et al. (2007) for the same sampling site. Calculations for the Standardized Indexes of Association, a linkage test (, with 999 permutations), accounting for the number of loci sampled, were significant for the Acromyrmex symbionts (IA = 1.451, p = 0.001; = 0.184, p = 0.001) and for the Atta symbionts (IA = 1.923, p = 0.001; = 0.319, p = 0.001). However, because only seven out of nine Atta symbionts had independent multi-locus genotypes, with three being identical (see Supplementary Information for details), a clone-correction was applied. This showed that the Standardized Index of Association for the Atta symbionts remained highly significant (IA = 0.744, p = 0.037; = 0.119, p = 0.009), consistent with all multi-locus genotypes being clonal. Rooting the symbiont tree with a sympatric fungal symbiont of Trachymyrmex zeteki (Fig S5) and by constructing a Minimum Spanning Network (Fig 3) and a UPGMA tree (Supplementary Information), both based on Bruvo genetic distances, confirmed that the symbionts belonged to separate clades, although bootstrap values for the rooted tree were low. Mirror imaging of trees obtained by microsatellite and AFLP markers showed almost complete congruence for the Acromyrmex fungi, but more noisy correspondence for the Atta fungal symbionts (Fig S6), confirming that these markers were less reliable predictors of somatic incompatibilities for Atta symbionts. Furthermore, STRUCTURE analyses of the AFLP data showed that four of the nine Acromyrmex symbionts were similar to the Atta symbionts (Fig 4), and these were the same four symbionts that were most closely related to Atta symbionts in the rooted tree based on microsatellites (Fig S5).
Fig. 3.
Minimum Spanning Network based on Bruvo genetic distances. Fungal symbionts from Acromyrmex colonies (white circles) are separated distinctly from those of Atta colonies (gray circles). The size of the nodes is proportional to the number of represented clones (normally one but in one case three), and the thickness of the lines represents the Bruvo genetic distance between two nodes (thicker lines mean larger genetic distance).
Fig. 4.
DISTRUCT plot for the nine Acromyrmex and nine Atta symbionts separated with a dashed line. Each bar represents an individual fungus clone, and the three colors represents different clusters, determined by STRUCTURE analysis. Four Acromyrmex symbionts cluster with the dominant in Atta symbionts.
4. Discussion
The results of our study show that sympatric Panamanian colonies of A. echinatior and A. colombica rear genetically different lineages of the leaf-cutting ant garden symbiont L. gongylophorus and that microsatellite markers appear to predict genetic (in)compatibility better than AFLP markers. However, even when using microsatellite markers, the correlation between somatic incompatibility and neutral-marker-based genetic distance in Atta is noisier than in Acromyrmex. This shows that incompatibility reactions correlate only with genetic distances among fungal strains that have a realistic probability of being horizontally transferred, and not between more distant clades that are apparently unsuitable as symbionts for the sister genus of leaf-cutting ants. We discuss these findings in more detail below.
4.1. Is somatic incompatibility only between fungal symbionts that may be exchanged?
The results of our study confirm the earlier findings by Poulsen and Boomsma (2005) showing that somatic (in)compatibility of Panamanian Acromyrmex symbionts is predictable from pairwise genetic distances for AFLP markers (Figs 1A, C) and that the same result can be obtained with more specific microsatellite markers (Figs 1B, D). They also indicate that incompatibility reactions between separate clades of fungal symbionts, maintained by the two different genera of leaf-cutting ants, can no longer be predicted by neutral genetic markers. This lack of genetic signal may be due to incompatibility being ultimately caused by allelic variation at unknown loci (Worrall, 1997) that only correlate with neutral markers when there is recent common ancestry. This is only likely for local fungal symbiont lineages that are exploited by a single metapopulation of attine ants that share a joint pool of symbionts because each ant colony can in principle establish a viable symbiosis with each of these fungal genotypes.
We expect somatic incompatibility to be actively maintained by selection only in populations where the ants have the possibility to acquire multiple genetically different symbionts that are viable alternatives. Such selection would then be driven by resident fungus-garden symbionts being under selection to defend their monopoly against alternative strains that might be secondarily introduced, and with the active support of the farming ants that would lose fitness when maintaining multiple lineages of the same symbiont that compete for their attention rather than serving their hosts unconditionally (Frank, 1996, Bot et al., 2001). When symbionts belong to different clades that no longer mix or exchange genes, each symbiont lineage is only a viable symbiont for one lineage of ant farmers or the other, but not for both. Our finding that A. colombica and A. echinatior maintain separated clades of fungal symbionts (Figs 3 and S5) suggests that L. gongylophorus has, in fact, been split into an Atta and Acromyrmex clade after its monophyletic origin 2–3 MYA (Mikheyev et al., 2010). The two species of leaf-cutting ants that we investigated rear representatives of these symbiont lineages that appear adapted to being, respectively, an Atta and Acromyrmex symbiont.
The fact that the fungal symbionts of Panamanian leaf-cutting ants appear to have split in two monophyletic clades, is consistent with the results of a recent study that experimentally swapped fungus-garden symbionts between sympatric Trachymyrmex septentrionalis and Atta texana from Texas, USA (Seal et al., 2012). Although these ants are at the northern edge of the attine ant distribution (Mueller et al., 2011a), and likely to have lower genetic variation among their symbionts, they share even less common ancestry than the Panamanian fungal symbionts of our present study (Mueller et al., 2011b). The swapped fungal symbionts thus could only serve as viable mutualists for either Trachymyrmex or Atta, but not both, which was confirmed in the published experiments showing that: (1) Alternative fungus gardens were not always rejected by the ants but were never adopted as a viable alternative symbiont, because the ants were able to grow their original symbiont back from minuscule remnants that the authors had been unable to remove. (2) None of the T. septentrionalis colonies ever produced virgin queens when they maintained an A. texana fungal symbiont, consistent with the new combination being non-viable for transmitting ant or fungal genes to future generations. A follow-up study transplanting A. texana fungus to colonies of both T. septentrionalis and Trachymyrmex turrifex confirmed that these Trachymyrmex species cannot enter into viable symbiosis with L. gongylophorus symbionts and that the virgin queens produced on swapped gardens had poor fat reserves making it unlikely that they could successfully found colonies (Seal and Mueller, 2013).
To make further progress, it would be desirable to identify the genes that are directly responsible for somatic incompatibility, as this would allow direct studies on the signatures of selection and specialization across the clades of higher attine ant symbionts. In another, non-eusocial model system of fungus-growing insects, the Sirex wood wasp, a range of genes are involved in somatic incompatibility reactions among lineages of the associated fungus, including fusion and recognition genes and genes that mediate cellular damage, stress response, and programmed cell death (Van der Nest et al., 2011). Whether these genes have homologs or analogs in attine ant fungal symbionts remains to be explored, as the Sirex symbiont Amylostereum areolatum belongs to a distantly related clade of basidiomycetes (Binder and Hibbett, 2002), and their respective domestication histories may have implied that recognition systems were lost and gained over evolutionary time.
4.2. Why do Atta fungal symbionts express weaker incompatibility reactions?
Incompatibility reactions among Atta symbionts were significantly weaker and less predictable from neutral Fst marker values than similar reactions between Acromyrmex-associated fungi. One possible explanation for this difference could be that there is a fundamental difference in colony founding in the sense that Acromyrmex queens forage during colony founding similar to all more basal attine ants, whereas Atta queens have secondarily evolved claustral colony founding. This implies that newly-mated Atta queens close off their nest cavity to raise the first worker cohort purely on their body reserves, so that new colonies will only be opened by these workers 80–100 d after they were founded (Weber, 1969, Fernandez-Marin and Wcislo, 2005). However, Acromyrmex queens not only forage for leaf fragments to manure their incipient fungus garden but, particularly when they have lost their garden, also for a replacement garden of another incipient colony whose queen is out foraging (Poulsen et al., 2009), an option that is unavailable for founding Atta queens. Although swapping of incipient fungus gardens with a fungus garden fragment from a mature Acromyrmex colony was relatively unconstrained during early colony founding, it is likely that stronger mutual commitment between a founding queen and her resident fungus garden builds up in a matter of weeks, including stronger incompatibility reactions in case one of the first workers brings in an unrelated fungus garden fragment from a neighboring nest (Poulsen et al., 2009). This can never happen in the 80–100 d during which Atta colonies remain closed, removing selection for expressing incompatibility mechanisms during colony founding.
Why neutral markers should be weaker predictors of somatic incompatibility in Atta colonies after workers start foraging remains unclear. Imprinting of workers on the odor of a resident fungus garden is a possibility (Seal et al., 2012), but it seems unclear why such mechanisms should differ between Atta and Acromyrmex symbionts and why that should reduce selection for more direct defenses by fungal symbionts against being replaced. Fungus gardens of Panamanian Acromyrmex colonies differ in chemical profiles (Richard et al., 2007a, Richard et al., 2007b), but these differences do not correlate with genetic distances and comparable data for sympatric Atta colonies are lacking. Another hypothesis may be that mature Atta colonies have hundreds of fungus gardens, whereas sympatric mature Acromyrmex colonies have one or a few at best. This may make a difference in the likelihood of a resident fungus garden symbiont being replaced by an accidentally imported small fragment of fungus garden from a neighboring colony, so that less accurate recognition systems suffice in mature colonies of Atta but not in Acromyrmex.
Finally, there could also be a technical explanation for the incompatibility differences between the fungal symbionts of Atta and Acromyrmex. We found similar variation for the symbiont-specific microsatellite markers obtained from Panamanian symbiont samples (Scott et al., 2009) and the general AFLP markers for Acromyrmex symbionts, but enhanced variation in AFLP peaks for Atta symbionts, relative to Acromyrmex symbionts (Fig S3). This suggests that DNA from other organisms may have been amplified with the AFLPs and that such other organisms were only present in Atta symbiont cultures. In principle, this could be viral (Pearson et al., 2009), bacterial (Suen et al., 2010) or prion (Wickner et al., 2007) DNA. However, universal bacterial 16S primers did not amplify the DNA samples (P.W. Kooij, unpublished data), making bacterial contaminations unlikely.
Acknowledgments
We thank the Smithsonian Tropical Research Institute (STRI), Panama, for providing logistic help and facilities to work in Gamboa, the Autoridad Nacional del Ambiente y el Mar (ANAM) for permission to sample ant colonies in Panama and export them to Denmark, and three anonymous reviewers for constructive comments on the manuscript. The work was supported by a grant from the Danish National Research Foundation (DNRF57) and an Advanced ERC grant (323085) to JJB.
Footnotes
Supplementary information related to this article can be found at http://dx.doi.org/10.1016/j.funeco.2015.08.003.
Supplementary Information
The following are the supplementary information related to this article:
Binary allele score (A) and allele fragment sizes (B) for 106 identified alleles from AFLP analyses on nine Acromyrmex and nine Atta fungal symbionts. Cutoff for allele identification was set at minimum 50 bp and maximum 500 bp. Columns show colony ID, host ant species, and allele number.
Microsatellite allele scores for nine Acromyrmex and nine Atta fungal symbionts for each of the ten different loci. Columns show colony ID, host ant species, and locus name with respective allele sizes.
References
- Andersen S.B., Hansen L.H., Sapountzis P., Sørensen S.J., Boomsma J.J. Specificity and stability of the Acromyrmex-Pseudonocardia symbiosis. Mol. Ecol. 2013;22:4307–4321. doi: 10.1111/mec.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage S.A.O., Broch J.F., Fernández-Marín H., Nash D.R., Boomsma J.J. Immune defense in leaf-cutting ants: a cross-fostering approach. Evolution. 2011;65:1791–1799. doi: 10.1111/j.1558-5646.2011.01241.x. [DOI] [PubMed] [Google Scholar]
- Binder M., Hibbett D.S. Higher-level phylogenetic relationships of homobasidiomycetes (mushroom-forming fungi) inferred from four rDNA regions. Mol. Phylogenet Evol. 2002;22:76–90. doi: 10.1006/mpev.2001.1043. [DOI] [PubMed] [Google Scholar]
- Bot A.N.M., Rehner S.A., Boomsma J.J. Partial incompatibility between ants and symbiotic fungi in two sympatric species of Acromyrmex leaf-cutting ants. Evolution. 2001;55:1980–1991. doi: 10.1111/j.0014-3820.2001.tb01315.x. [DOI] [PubMed] [Google Scholar]
- Cafaro M.J., Poulsen M., Little A.E.F., Price S.L., Gerardo N.M., Wong B., Stuart A.E., Larget B., Abbot P., Currie C.R. Specificity in the symbiotic association between fungus-growing ants and protective Pseudonocardia bacteria. Proc. Biol. Sci. 2011;278:1814–1822. doi: 10.1098/rspb.2010.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fine Licht H.H., Boomsma J.J. 13th Congress of the European Society of Evolutionary Biology. 2011. Variation in fungal enzyme spectra may affect mutualistic division of labour between ants and fungus gardens. [Google Scholar]
- De Fine Licht H.H., Boomsma J.J. Variable interaction specificity and symbiont performance in Panamanian Trachymyrmex and Sericomyrmex fungus-growing ants. BMC Evol. Biol. 2014;14:244. doi: 10.1186/s12862-014-0244-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentinger B.T.M., Lodge D.J., Munkacsi A.B., Desjardin D.E., McLaughlin D.J. Phylogenetic placement of an unusual coral mushroom challenges the classic hypothesis of strict coevolution in the Apterostigma pilosum group ant-fungus mutualism. Evolution. 2009;63:2172–2178. doi: 10.1111/j.1558-5646.2009.00697.x. [DOI] [PubMed] [Google Scholar]
- Doebeli M., Knowlton N. The evolution of interspecific mutualisms. P. Natl. Acad. Sci. U.S.A. 1998;95:8676–8680. doi: 10.1073/pnas.95.15.8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas A.E. Conflict, cheats and the persistence of symbioses. New Phytol. 2008;177:849–858. doi: 10.1111/j.1469-8137.2007.02326.x. [DOI] [PubMed] [Google Scholar]
- Earl D.A., vonHoldt B.M. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2011;4:359–361. [Google Scholar]
- Embley T.M., Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- Falush D., Stephens M., Pritchard J.K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Marín H., Wcislo W.T. Production of minima workers by gynes of Atta colombica Guérin-Ménéville (Hymenoptera: Formicidae: Attini) that lack a fungal pellet. J. Kans. Entomol. Soc. 2005;78:290–292. [Google Scholar]
- Fisher P.J., Stradling D.J., Pegler D.N. Leaf cutting ants, their fungus gardens and the formation of basidiomata of Leucoagaricus gongylophorus. Mycologist. 1994;8:128–131. [Google Scholar]
- Fisher P.J., Stradling D.J., Pegler D.N. Leucoagaricus basidiomata from a live nest of the leaf-cutting ant Atta cephalotes. Mycol. Res. 1994;98:884–888. [Google Scholar]
- Foster K.R., Wenseleers T. A general model for the evolution of mutualisms. J. Evol. Biol. 2006;19:1283–1293. doi: 10.1111/j.1420-9101.2005.01073.x. [DOI] [PubMed] [Google Scholar]
- Frank S.A. Genetics of mutualism: the evolution of altruism between species. J. Theor. Biol. 1994;170:393–400. doi: 10.1006/jtbi.1994.1200. [DOI] [PubMed] [Google Scholar]
- Frank S.A. Host-symbiont conflict over the mixing of symbiotic lineages. Proc. Biol. Sci. 1996;263:339–344. doi: 10.1098/rspb.1996.0052. [DOI] [PubMed] [Google Scholar]
- Green A.M., Mueller U.G., Adams R.M.M. Extensive exchange of fungal cultivars between sympatric species of fungus-growing ants. Mol. Ecol. 2002;11:191–195. doi: 10.1046/j.1365-294x.2002.01433.x. [DOI] [PubMed] [Google Scholar]
- Ivens A.B., Nash D.R., Poulsen M., Boomsma J.J. Caste-specific symbiont policing by workers of Acromyrmex fungus-growing ants. Behav. Ecol. 2009;20:378–384. [Google Scholar]
- Jakobsson M., Rosenberg N.A. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Kamvar Z.N., Tabima J.F., Grunwald N.J. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ. 2014;2:e281. doi: 10.7717/peerj.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooij P.W., Aanen D.K., Schiøtt M., Boomsma J.J. Evolutionarily advanced ant farmers rear polyploid fungal crops. J. Evol. Biol. 2015 doi: 10.1111/jeb.12718. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langella O. 2001. Populations.http://bioinformatics.org/populations/ Available at: [Google Scholar]
- Leigh E.G. The evolution of mutualism. J. Evol. Biol. 2010;23:2507–2528. doi: 10.1111/j.1420-9101.2010.02114.x. [DOI] [PubMed] [Google Scholar]
- Lemon J. 2006. Plotrix: a Package in the Red Light District of R. R-news; pp. 8–12. [Google Scholar]
- May G. Somatic incompatibility and individualism in the coprophilous Basidiomycete, Coprinus cinereus. T. Brit Mycol. Soc. 1988;91:443–451. [Google Scholar]
- McCutcheon J.P., Moran N.A. Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 2012;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- Mehdiabadi N.J., Mueller U.G., Brady S.G., Himler A.G., Schultz T.R. Symbiont fidelity and the origin of species in fungus-growing ants. Nat. Commun. 2012;3:840. doi: 10.1038/ncomms1844. [DOI] [PubMed] [Google Scholar]
- Mikheyev A.S., Mueller U.G., Abbot P. Cryptic sex and many-to-one coevolution in the fungus-growing ant symbiosis. P. Natl. Acad. Sci. U.S.A. 2006;103:10702–10706. doi: 10.1073/pnas.0601441103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikheyev A.S., Mueller U.G., Abbot P. Comparative dating of attine ant and lepiotaceous cultivar phylogenies reveals coevolutionary synchrony and discord. Am. Nat. 2010;175:E126–E133. doi: 10.1086/652472. [DOI] [PubMed] [Google Scholar]
- Mikheyev A.S., Mueller U.G., Boomsma J.J. Population genetic signatures of diffuse co-evolution between leaf-cutting ants and their cultivar fungi. Mol. Ecol. 2007;16:209–216. doi: 10.1111/j.1365-294X.2006.03134.x. [DOI] [PubMed] [Google Scholar]
- Mueller U.G. Ant versus fungus versus mutualism: ant-cultivar conflict and the deconstruction of the attine ant-fungus symbiosis. Am. Nat. 2002;160:S67–S98. doi: 10.1086/342084. [DOI] [PubMed] [Google Scholar]
- Mueller U.G., Gerardo N.M. Fungus-farming insects: multiple origins and diverse evolutionary histories. P. Natl. Acad. Sci. U.S.A. 2002;99:15247–15249. doi: 10.1073/pnas.242594799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller U.G., Mikheyev A.S., Hong E., Sen R., Warren D.L., Solomon S.E., Ishak H.D., Cooper M., Miller J.L., Shaffer K.A., Juenger T.E. Evolution of cold-tolerant fungal symbionts permits winter fungiculture by leafcutter ants at the northern frontier of a tropical ant-fungus symbiosis. P. Natl. Acad. Sci. U.S.A. 2011;108:4053–4056. doi: 10.1073/pnas.1015806108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller U.G., Mikheyev A.S., Solomon S.E., Cooper M. Frontier mutualism: coevolutionary patterns at the northern range limit of the leaf-cutter ant-fungus symbiosis. Proc. Biol. Sci. 2011;278:3050–3059. doi: 10.1098/rspb.2011.0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller U.G., Scott J.J., Ishak H.D., Cooper M., Rodrigues A. Monoculture of leafcutter ant gardens. PLoS ONE. 2010;5:e12668. doi: 10.1371/journal.pone.0012668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F.G., Kindt R., Legendre P., Minchin P.R., O'Hara R.B., Simpson G.L., Solymos P., Stevens M.H.H., Wagner H. 2013. Vegan: Community Ecology Package.http://CRAN.R-project.org/package=vegan Available at: [Google Scholar]
- Pagnocca F.C., Bacci M., Jr., Fungaro M.H., Bueno O.C., Hebling M.J.A., Sant'Anna A., Capelari M. RAPD analysis of the sexual state and sterile mycelium of the fungus cultivated by the leaf-cutting ant Acromyrmex hispidus fallax. Mycol. Res. 2001;105:173–176. [Google Scholar]
- Pearson M.N., Beever R.E., Boine B., Arthur K. Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol. Plant Pathol. 2009;10:115–128. doi: 10.1111/j.1364-3703.2008.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen M., Boomsma J.J. Mutualistic fungi control crop diversity in fungus-growing ants. Science. 2005;307:741–744. doi: 10.1126/science.1106688. [DOI] [PubMed] [Google Scholar]
- Poulsen M., Fernandez-Marin H., Currie C.R., Boomsma J.J. Ephemeral windows of opportunity for horizontal transmission of fungal symbionts in leaf-cutting ants. Evolution. 2009;63:2235–2247. doi: 10.1111/j.1558-5646.2009.00704.x. [DOI] [PubMed] [Google Scholar]
- Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2013. R: a Language and Environment for Statistical Computing.http://www.R-project.org/ Available at: [Google Scholar]
- Rayner A.D.M. The challenge of the individualistic mycelium. Mycologia. 1991;83:48–71. [Google Scholar]
- Rayner A.D.M., Coates D., Ainsworth A.M., Adams T.J.H., Williams E.N.D., Todd N.K. The biological consequences of the individualistic mycelium. In: Jennings D.H., Rayner A.D.M., editors. The Ecology and Physiology of the Fungal Mycelium. Cambridge University Press; 1984. pp. 509–540. [Google Scholar]
- Richard F.-J., Poulsen M., Drijfhout F., Jones G., Boomsma J.J. Specificity in chemical profiles of workers, brood and mutualistic fungi in Atta, Acromyrmex, and Sericomyrmex fungus-growing ants. J. Chem. Ecol. 2007;33:2281–2292. doi: 10.1007/s10886-007-9385-z. [DOI] [PubMed] [Google Scholar]
- Richard F.-J., Poulsen M., Hefetz A., Errard C., Nash D.R., Boomsma J.J. The origin of the chemical profiles of fungal symbionts and their significance for nestmate recognition in Acromyrmex leaf-cutting ants. Behav. Ecol. Sociobiol. 2007;61:1637–1649. [Google Scholar]
- Rosenberg N.A. DISTRUCT: a program for the graphical display of population structure. Mol. Ecol. Notes. 2004 [Google Scholar]
- Sachs J.L., Essenberg C.J., Turcotte M.M. New paradigms for the evolution of beneficial infections. Trends Ecol. Evol. 2011;26:202–209. doi: 10.1016/j.tree.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Sachs J.L., Mueller U.G., Wilcox T.P., Bull J.J. The evolution of cooperation. Q. Rev. Biol. 2004;79:135–160. doi: 10.1086/383541. [DOI] [PubMed] [Google Scholar]
- Schultz T.R., Brady S.G. Major evolutionary transitions in ant agriculture. P. Natl. Acad. Sci. U.S.A. 2008;105:5435–5440. doi: 10.1073/pnas.0711024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J.J., Weskin M.K., Cooper M., Mueller U.G. Polymorphic microsatellite markers for the symbiotic fungi cultivated by leaf cutter ants (Attini, Formicidae) Mol. Ecol. Resour. 2009;9:1391–1394. doi: 10.1111/j.1755-0998.2009.02684.x. [DOI] [PubMed] [Google Scholar]
- Seal J.N., Gus J., Mueller U.G. Fungus-gardening ants prefer native fungal species: do ants control their crops? Behav. Ecol. 2012;23:1250–1256. [Google Scholar]
- Seal J.N., Mueller U.G. Instability of novel ant-fungal associations constrains horizontal exchange of fungal symbionts. Evol. Ecol. 2013 [Google Scholar]
- Suen G., Scott J.J., Aylward F.O., Adams S.M., Tringe S.G., Pinto-Tomás A.A., Foster C.E., Pauly M., Weimer P.J., Barry K.W., Goodwin L.A., Bouffard P., Li L., Osterberger J., Harkins T.T., Slater S.C., Donohue T.J., Currie C.R. An insect herbivore microbiome with high plant biomass-degrading capacity. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Nest M.A., Slippers B., Steenkamp E.T., de Vos L., van Zyl K., Stenlid J., Wingfield M.J., Wingfield B.D. Genetic linkage map for Amylostereum areolatum reveals an association between vegetative growth and sexual and self-recognition. Fungal Genet. Biol. 2009;46:632–641. doi: 10.1016/j.fgb.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Van der Nest M.A., Steenkamp E.T., Slippers B., Mongae A., van Zyl K., Stenlid J., Wingfield M.J., Wingfield B.D. Gene expression associated with vegetative incompatibility in Amylostereum areolatum. Fungal Genet. Biol. 2011;48:1034–1043. doi: 10.1016/j.fgb.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Vo T.L., Mueller U.G., Mikheyev A.S. Free-living fungal symbionts (Lepiotaceae) of fungus-growing ants (Attini: Formicidae) Mycologia. 2009;101:206–210. doi: 10.3852/07-055. [DOI] [PubMed] [Google Scholar]
- Vos P., Hogers R., Bleeker M., Reijans M., Van de Lee T., Hornes M., Frijters A., Pot J., Peleman J., Kuiper M., Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber N.A. Ecological relations of three Atta species in Panama. Ecology. 1969;50:141–147. [Google Scholar]
- Wernegreen J.J. Strategies of genomic integration within insect-bacterial mutualisms. Biol. Bull. 2012;223:112–122. doi: 10.1086/BBLv223n1p112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J.H., Baldo L., Clark M.E. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- Wickner R.B., Edskes H.K., Shewmaker F., Nakayashiki T. Prions of fungi: inherited structures and biological roles. Nat. Rev. Microbiol. 2007;5:611–618. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall J.J. Somatic incompatibility in basidiomycetes. Mycologia. 1997;89:24–36. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Binary allele score (A) and allele fragment sizes (B) for 106 identified alleles from AFLP analyses on nine Acromyrmex and nine Atta fungal symbionts. Cutoff for allele identification was set at minimum 50 bp and maximum 500 bp. Columns show colony ID, host ant species, and allele number.
Microsatellite allele scores for nine Acromyrmex and nine Atta fungal symbionts for each of the ten different loci. Columns show colony ID, host ant species, and locus name with respective allele sizes.