Background: Twin-arginine translocases, which transport folded proteins across cellular membranes, often consist of three proteins, TatA, TatB, and TatC, but sometimes have an additional TatE available.
Results: TatE associates with active translocases and forms direct contacts with TatA, TatB, and TatC.
Conclusion: TatE is a regular constituent of twin-arginine translocases.
Significance: We elucidate a functional interplay of the paralogous proteins TatA and TatE.
Keywords: Escherichia coli (E. coli), membrane transport, protein cross-linking, protein translocation, transport, twin-arginine translocation
Abstract
Twin-arginine translocation (Tat) systems mediate the transmembrane translocation of completely folded proteins that possess a conserved twin-arginine (RR) motif in their signal sequences. Many Tat systems consist of three essential membrane components named TatA, TatB, and TatC. It is not understood why some bacteria, in addition, constitutively express a functional paralog of TatA called TatE. Here we show, in live Escherichia coli cells, that, upon expression of a Tat substrate protein, fluorescently labeled TatE-GFP relocates from a rather uniform distribution in the plasma membrane into a number of discrete clusters. Clustering strictly required an intact RR signal peptide and the presence of the TatABC subunits, suggesting that TatE-GFP associates with functional Tat translocases. In support of this notion, site-specific photo cross-linking revealed interactions of TatE with TatA, TatB, and TatC. The same approach also disclosed a pronounced tendency of TatE and TatA to hetero-oligomerize. Under in vitro conditions, we found that TatE replaces TatA inefficiently. Our collective results are consistent with TatE being a regular constituent of the Tat translocase in E. coli.
Introduction
The twin-arginine translocation (Tat)2 system is a transporter for fully folded proteins and assembled protein complexes. It has been identified in the cytoplasmic membrane of bacteria and archaea as well as in the thylakoid membrane of chloroplasts. Substrates of this translocase contain a highly conserved double arginine (RR) motif in their N-terminal signal sequence, a feature that is eponymous for the twin-arginine translocation pathway.
Enterobacteria constitutively express four components of the Tat translocase: TatA, TatB, TatC, and TatE (most recently reviewed in Refs. 1–6). Whereas TatC is a hexahelical membrane protein (7, 8), TatA, TatB, and TatE consist of only one transmembrane helix, followed by an amphipathic helix and an unstructured C terminus on the cis side of the membrane. Previous studies have revealed that TatB and TatC form a receptor complex (summarized in Ref. 9) that initially binds substrates in an energy-independent manner. TatC forms functional oligomers (10–12) and recognizes the RR-containing N terminus of the substrate precursor (13, 14). TatC also mediates the deep loop-like insertion of the signal sequence into an intramembrane binding pocket (15, 16) formed by dome-like oligomeric TatB assemblies that are surrounded by outer rings of TatC monomers (17).
How TatA assembles with the TatBC receptor complex to form a functional translocating unit is not understood. There is experimental evidence that TatA is already present during the early stages of substrate recognition (18–21). The key function of TatA seems to be to conduct substrate across the membrane. This process probably occurs rapidly and depends on the proton-motive force. One hypothesis suggests that TatA associates with the TatBC receptor complex, undergoes reorganization, and oligomerizes to form a translocation channel for the substrate (summarized in Ref. 4). In another model, it has been proposed that accumulation of TatA at the TatBC receptor might mediate the translocation of substrate proteins by destabilizing/thinning the membrane bilayer (22–24).
In addition to TatA, enterobacteria (25) and the Gram-positive corynebacteria (26) express the functional homologue TatE. In Escherichia coli, TatE is a small protein of 67 amino acids (7 kDa), 53% of which are identical to those of TatA (27). A number of in vivo studies using activity and localization assays of various Tat substrates have suggested that TatE and TatA have overlapping functions (26–31). Three-dimensional density maps of purified TatE indicated ring-shaped structures that, in contrast to what has been reported for TatA (32), would be too small to represent translocation channels, for example, for the 90-kDa E. coli Tat substrate TorA (29). Despite its striking functional and structural similarity with TatA, TatE has persisted during evolution as an individual isoform. Remarkably, tatE mRNA is equipped with a much weaker ribosomal binding site (our own observation) than tatA, which could explain why TatE is 50 times less redundant in the cell than TatA (33). To learn more about the role TatE might play in Tat-dependent protein translocation, we undertook a combined in vivo and in vitro analysis of how TatE associates with the Tat translocase of E. coli.
Experimental Procedures
Cloning of DNA Constructs
Fragments and plasmids were amplified using Pfu Ultra II Fusion HS DNA polymerase (Agilent Technologies) according to the protocol of the manufacturer. The primers are listed in supplemental Table S1. Restriction enzymes and Antarctic phosphatase were obtained from New England Biolabs, and T4 DNA ligase was from Thermo Scientific. For DNA purification, a gel extraction kit from Qiagen was used. All constructs were verified by sequencing.
Plasmids
The plasmids used for this study are listed in supplemental Table S2. For detailed cloning procedures, see supplemental data.
Microscopy
Microscopical studies were performed as detailed in Ref. 34 using BL21(DE3) (Novagen) and BL21(DE3)Δtat (ΔtatABCD) (16) cells.
Membrane Vesicles and Photo Cross-linking
Inverted inner membrane vesicles were prepared as described previously (35) from strain DADE (MC4100 ΔtatABCDE) (36) transformed with pEVol-pBpF and either pEBC_LinkRBS, pEABC_LinkRBS, or pBADxTat containing amber stop codons in tatE, tatA, or tatC. To incorporate Bpa, expression from pEVOL-pBpF was induced with 0.1% arabinose in the presence of 0.25 mm Bpa. Photo cross-linking of INV was initiated by UV irradiation as described previously (18).
Immunoblotting
To detect contacts between the Tat subunits, 5 μl of each INV (∼100 A280 units/ml) were diluted with 95 μl of INV buffer (35) and treated further as described previously (18). Proteins were separated using 9% Tricine SDS-PAGE according to Ref. 37. Blots were decorated with affinity-purified polyclonal immunoglobulins directed against peptide sequences of TatA (19), TatB (38), TatC (38), and TatE (CAMNDDDAAAKKGAD). Signals were detected by incubation in 0.1 m Tris/HCl (pH 8.6), 0.025% luminol (w/v), 54.5 μm p-coumaric acid, 0.05% (v/v) H2O2, and the Fusion-Fx7 chemiluminescence system (Peqlab Biotechnologies GmbH). Alternatively, alkaline phosphatase-linked antibodies and nitro blue tetrazolium chloride/5-bromo-4-chloro-3-indolyl phosphate (Roche) were used according to the protocol of the manufacturer.
In Vitro Reactions
Cell extracts used for in vitro synthesis of RR precursor proteins were prepared from strain SL119 (39). Coupled transcription/translation reactions were performed in 50-μl aliquots as described previously (35). INV were added 10 min after starting the synthesis reaction and incubated for 25 min at 37 °C. Protein translocation into INV was assayed as described previously (18). Quantitative analyses were obtained using the analysis toolbox of ImageQuant TL 7.0 (GE Healthcare).
Results
Substrate-dependent Relocation of TatE from a Uniform Distribution in the Membrane to Distinct Clusters
To investigate the localization of TatE in live E. coli cells, we linked TatE C-terminally to GFP in the same way as reported recently for TatA (34). The tatE-GFP fusion gene was constructed in plasmid pBAD33, placing it behind the ara promoter. This plasmid (pEGFP) was transformed into the E. coli tatABC deletion strain BL21(DE3)ΔTat. Two hours after inducing the expression of TatE-GFP with 0.1% arabinose in cells growing in liquid culture, the epifluorescent image depicted in Fig. 1A was obtained. Virtually all cells showed a clear rim staining, indicating that, in the absence of the other Tat components, TatABC, TatE was spread over the whole plasma membrane of E. coli. Frequently, a coarse-grained staining pattern was obtained (Fig. 1A, arrows), which might be due to the formation of TatE oligomers. The cells shown in Fig. 1A show the typical Δtat phenotype, i.e. cell elongation and a failure to separate after division (40).
FIGURE 1.
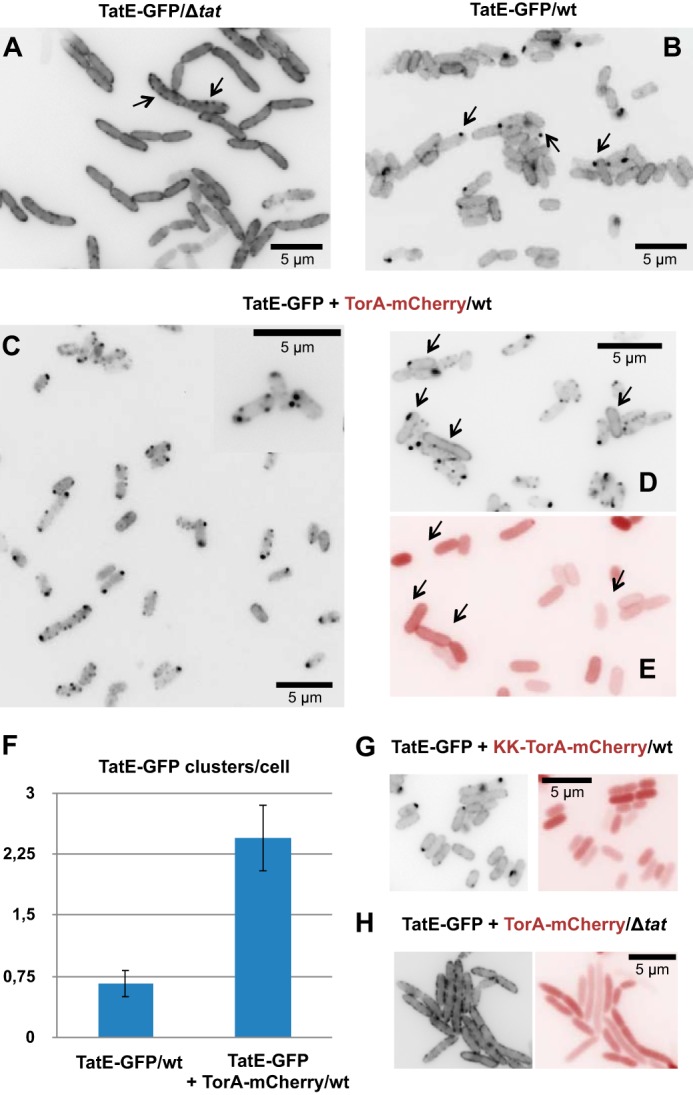
TatE-GFP relocates from a uniform membrane distribution to discrete clusters in dependence of the TatABC subunits and a functional Tat substrate. A, fluorescence micrograph of E. coli BL21(DE3)ΔtatABC cells (Δtat) expressing TatE-GFP from a derivative of pBAD33 (pEGFP) 2 h after induction with 0.1% arabinose. The cells show a clear rim staining of the GFP signal, often condensed to small grains (arrows). B, expression of TatE-GFP together with the endogenous TatABCE proteins of BL21(DE3) wild-type cells results in a smoother rim staining with few prominent clusters, often near the cell poles (arrows). C–E, intracellular distribution of TatE-GFP in BL21(DE3) wild-type E. coli cells when co-expressed with the model Tat substrate TorA-mCherry (uninduced expression from pPJ3). Export of TorA-mCherry into the periplasm can be gathered from the red rim staining (E). Expression of TorA-mCherry induces enhanced clustering of TatE-GFP (C and D). Cells that fail to express TorA-mCherry, as indicated by the loss of red stain (E, arrows), instead show the same smooth rim staining of TatE-GFP (D, arrows) as cells shown in B. The inset in C is an enlargement from an independent experiment. F, quantification of substrate-induced TatE-GFP clusters using the cell image analysis software CellProfiler. Indicated are the mean ± S.E. of the bright dots per cell obtained from counting at least 500 cells each. G, increased cluster formation of TatE-GFP is not observed for the non-functional KK variant of TorA-mCherry despite the presence of endogenous TatABC (wt). H, co-expression of TorA-mCherry and TatE-GFP in the absence of TatABC (Δtat), which does not induce cluster formation of the GFP signal. Note the cytosolic location of TorA-mCherry (no red rim staining) when its export is inhibited by the KK mutant of TorA-mCherry (G) and by a lack of TatABC (H).
When TatE-GFP was expressed in the tatABC wild-type background of E. coli strain BL21(DE3) (Fig. 1B), then it seemed to be more evenly dispersed in the plasma membrane than in the Δtat strain, with most of the wild-type cells showing a fainter rim staining. Additionally, in a number of cells, TatE-GFP was found concentrated in single clusters (Fig. 1B, arrows). A similar occasional clustering has been described previously for a GFP fusion of TatA when expressed in wild-type E. coli cells (31, 34), and it has been shown to represent Tat translocases that are engaged in the transport of the endogenous Tat substrates of the cells.
Consistent with the assumption that single TatE-GFP clusters might mark active Tat translocases, the number of clusters increased dramatically upon co-expression of the model Tat substrate TorA-mCherry (Fig. 1C). TorA-mCherry is a fusion protein consisting of the signal sequence of the natural Tat substrate TMAO (trimethylamine-N-oxide) reductase (TorA) and the fluorescent protein mCherry, which has been shown in vitro and in intact E. coli cells to be translocated across the plasma membrane in a Tat-dependent manner (16, 34). When TorA-mCherry was co-expressed with TatE-GFP together with the chromosomally encoded TatABC components, rim staining of most cells was lost, and TatE-GFP was now found concentrated in several prominent clusters scattered along the cell periphery (Fig. 1C). We noticed that increased cluster formation was not true for all cells. Some of them retained the rim staining or contained only single TatE-GFP clusters (Fig. 1D, arrows), much like cells expressing TatE-GFP without TorA-mCherry (Fig. 1B). We therefore also analyzed the cells depicted in Fig. 1D for the expression of TorA-mCherry by taking pictures through a red filter (Fig. 1E). In fact, the same cells in which TatE-GFP was found to be concentrated in a single cluster or distributed evenly along the cell periphery failed to show any red staining (Fig. 1E, arrows), proving that they did not express TorA-mCherry. Conversely, in cells that did co-express the Tat substrate, TatE had relocated from a uniform distribution into distinct membrane clusters.
We quantified the TatE-GFP dots in cells that either expressed or lacked TorA-mCherry (cf. Fig. 1, B and C). In each case, more than 500 individual cells derived from at least three parallel experiments were used. Thereby, the Tat substrate TorA-mCherry could be shown to increase the clustering of TatE-GFP by a factor of 5 (Fig. 1F). This compares well with the substrate-induced increase of TatA-GFP clusters determined previously (31, 34).
To further demonstrate that these clusters reflect a functionally relevant reorganization of TatE-GFP during Tat transport, we also tested a non-functional variant of TorA-mCherry in which the characteristic double arginine motif (RR) in the TorA signal sequence had been replaced by a double lysine (KK). E. coli cells that expressed this transport-defective KK variant of TorA-mCherry diffusely stained red and were separated by non-stained gaps because of the retention of KK-TorA-mCherry in the cytosol (Fig. 1G, right panel). In contrast, active TorA-mCherry accumulated in the periplasm of cells, as manifested by the appearance of red rims (Fig. 1E). When inspecting the GFP signal in cells expressing KK-TorA-mCherry, TatE-GFP was not found to be concentrated in multiple clusters but, rather, showed the same staining of the cell periphery and the occurrence of occasional single clusters (Fig. 1G, left panel) as in cells that did not express any TorA-mCherry (Fig. 1B).
TorA-mCherry-dependent clustering of TatE-GFP also strictly required the presence of the TatABC subunits. This is shown in Fig. 1H (left panel), in which ΔTat cells expressing TatE-GFP together with TorA-mCherry showed the same coarse-grained TatE-GFP-specific rim staining and lack of prominent clusters as cells that did not contain any TorA-mCherry (Fig. 1A). Therefore, TorA-mCherry was unable to induce clustering of TatE-GFP when the TatABC subunits were missing, lending strong support to the idea that the prominent TatE-GFP clusters represent Tat translocases.
To further rule out that the varying membrane distribution of TatE-GFP was caused by different expression levels, we determined, by Western blotting, the TatE content of the cells used above for fluorescent microscopy. Overexpression of TatE went along with the emergence of an approximately 9-kDa band recognized by anti-TatE antibodies (Fig. 2A, lane 1). In cells overexpressing TatE-GFP, the major TatE-immunoreactive material had the expected molecular mass of the TatE-GFP fusion (approximately 37 kDa; Fig. 2A, lanes 3–6). Expression of TatE-GFP did not noticeably influence the amount of the 9-kDa TatE band, demonstrating stability of the TatE-GFP construct. Importantly, the expression level of TatE-GFP did not change upon co-expression of TorA-mCherry (Fig. 2A, compare lane 3 with lane 4 and lane 5 with lane 6), indicating that the different locations of TatE-GFP observed in Fig. 1 were, in fact, only related to the presence or absence of the Tat substrate TorA-mCherry and the other Tat components.
FIGURE 2.
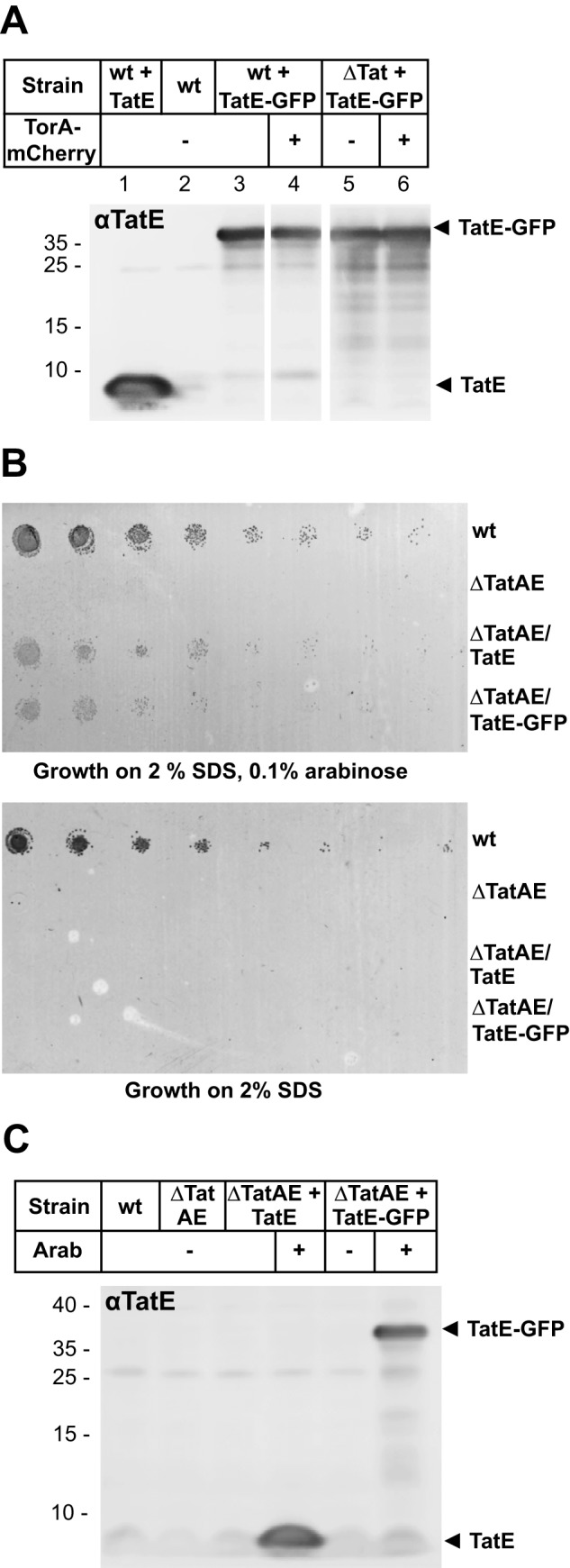
Expression levels of TatE-GFP in different cells and functionality of TatE-GFP. A, E. coli strains used for fluorescence imaging in Fig. 1 were analyzed via Western blotting. TatE (pEKI+) and TatE-GFP (pEGFP) expression was induced by 0.1% arabinose. Two hours after induction, whole cells derived from 1 ml of culture and adjusted to an A600 of 0.2 (per lane) were precipitated with 5% trichloroacetic acid, separated by 9% Tricine SDS-PAGE, and analyzed on immunoblots decorated with anti-TatE antibodies. Control (Top10) E. coli cells overexpressing TatE (wt + TatE) show a TatE signal at ∼9 kDa (lane 1). BL21(DE3) wild-type cells express very little endogenous TatE. TatE-GFP overexpression in BL21(DE3) (wt + TatE-GFP, lanes 3 and 4) and in BL21(DE3)Δtat (ΔTat + TatE-GFP, lanes 5 and 6) is comparable and not influenced by the simultaneous co-expression of TorA-mCherry (lanes 4 and 6). B, cell growth on agar plates containing 2% SDS and 0.1% arabinose where indicated. E. coli MC4100 cells (wt), the ΔtatAE mutant strain JARV16 (ΔTatAE), JARV16 expressing TatE from plasmid pEKI+ (ΔTatAE/TatE), and JARV16 expressing TatE-GFP from plasmid pEGFP (ΔTatAE/TatE-GFP) were grown in liquid culture with or without 0.1% arabinose to an A600 of 2.0. Prior to plating, cells were diluted to an A600 of 0.1 and serially diluted 5-fold. Cells lacking TatE or TatE-GFP did not grow on SDS. C, cells used for the complementation assay shown in B were analyzed via Western blotting. Arab, arabinose.
To finally demonstrate the functionality of our TatE-GFP fusion, we assessed its ability to restore growth of a ΔtatAE strain in the presence of 2% SDS (Fig. 2B). Compared with wild-type tat+ cells, the TatAE-lacking mutant did not grow on agar plates containing 2% SDS unless expression of TatE or TatE-GFP was induced by the addition of arabinose (Fig. 2B, compare the top and bottom panel, ± arabinose). TatE-GFP was almost as active as wild-type TatE, and this effect was not due to cleavage of TatE-GFP into TatE (Fig. 2C).
Collectively, these data indicate that the location of TatE-GFP is reorganized from a rather uniform distribution in the plasma membrane to a number of clusters, provided that cells co-express an active Tat substrate. A virtually undistinguishable staining pattern has been reported recently for fluorescently labeled TatA variants when challenged with active Tat substrates (31, 34). We therefore propose that the substrate-dependent clustering of TatE-GFP reflects its population and concomitant labeling of active Tat translocases.
Marked Propensity of TatE to Hetero-oligomerize with TatA
To further analyze the interaction of TatE with the other Tat components, the photo-activatable cross-linker p-benzoyl-l-phenylalanine (Bpa) was introduced by an amber stop codon approach into TatE at several positions. For this purpose, we constructed derivatives of the vector pBAD33 in which either the tatE gene alone (TatE) or tatE in front of tatBC (TatEBC) or in front of tatABC (TatEABC) was cloned behind the ara promoter. The corresponding plasmid carrying tatABC in the absence of tatE was also constructed (TatABC). Classical Shine-Dalgarno sequences were engineered in front of tatE and the downstream tat operons. Using those vectors, amber stop codon mutants of tatE were generated by mutagenizing PCR and transformed into ΔtatABCDE cells of E. coli, which, upon growth in the presence of Bpa, yielded the corresponding Bpa variants of TatE. For comparison, variants of TatA that carried Bpa at the corresponding positions as in TatE were constructed in plasmids expressing tatABC and tatEABC.
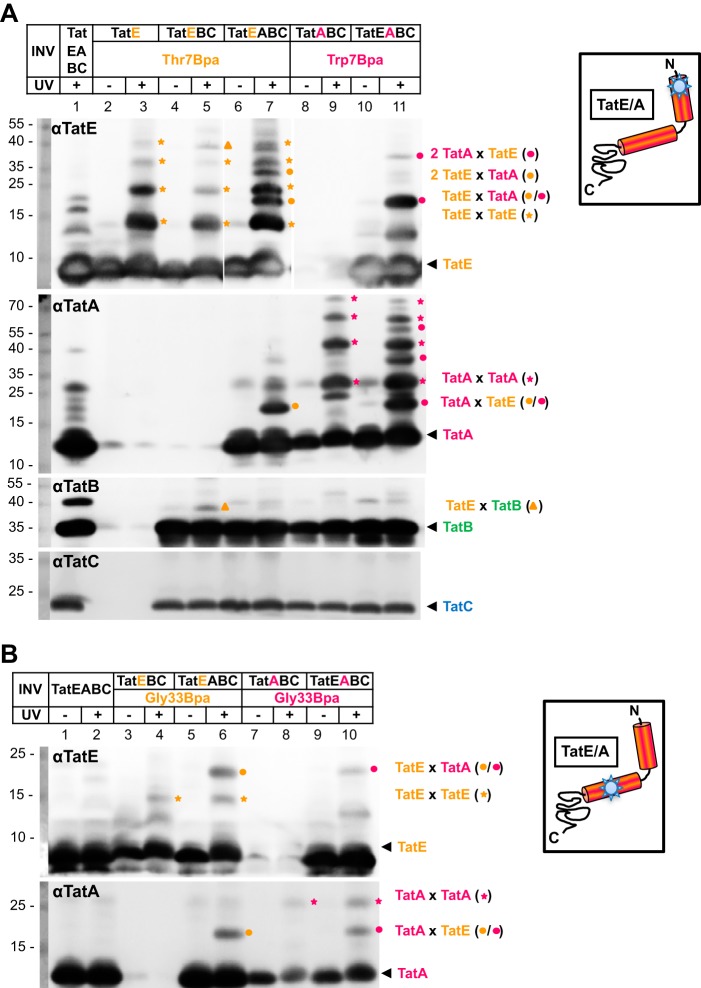
Inside-out inner membrane vesicles (INV) were then prepared from those strains and subjected to irradiation with UV light to initiate photo cross-linking. Adducts to TatE were analyzed via Tricine SDS-PAGE and anti-TatE Western blotting. Fig. 3A, top panel, αTatE, shows the results obtained when the cross-linker was incorporated at position Thr-7 in the transmembrane helix of TatE. TatE-cross-reactive bands appearing only after UV exposure of INV that contained the Thr-7Bpa variant of TatE should represent adducts to TatE (Fig. 3A, compare lane 3 with lanes 1 and 2). In fact, in vesicles containing TatEThr-7Bpa alone (Fig. 3A, lane 3) or together with TatB and TatC (Fig. 3A, lane 5), homo-oligomers of TatE up to presumed pentamers could be detected in isolated vesicles (Fig. 3A, orange stars). However, when vesicles contained TatE, TatB, TatC, and TatA (Fig. 3A, lane 7), additional adducts were recognized by anti-TatE antibodies (Fig. 3A, orange dots). The most prominent of those had a molecular mass of about 19 kDa. It was indistinguishable in size from a UV-dependent adduct obtained from INV that contained the cross-linker in TatA at the equivalent position (Trp-7) rather than in TatE (Fig. 3A, lane 11, magenta dot). Co-migration of both adducts strongly suggests that they represent a TatA-TatE heterodimer. In full agreement, a 19-kDa adduct was also recognized by anti-TatA antibodies irrespective of whether Bpa was incorporated into TatE or TatA (Fig. 3A, αTatA, lanes 7 and 11).
FIGURE 3.
Hetero-oligomerization of TatE and TatA. A, INV containing the indicated Tat components were UV-irradiated (+) to initiate cross-linking of the Thr-7Bpa variant of TatE and the Trp-7Bpa variant of TatA. Homo- and hetero-oligomeric TatE adducts were resolved by Tricine SDS-PAGE and detected by immunoblots using anti-TatE (αTatE), anti-TatA (αTatA), anti-TatB (αTatB), and anti-TatC (αTatC) antibodies. Orange stars mark TatE oligomers. The approximately 19-kDa adducts to TatEThr-7Bpa (orange dots) and TatATrp-7Bpa (magenta dots) are recognized by both antibodies and therefore indicate 1:1 complexes between TatE and TatA. TatE-TatB cross-links are labeled with orange triangles. The αTatC immunoblot confirms equal loads of all vesicles analyzed. Numbers to the left indicate the molecular masses of protein markers in kilodalton. B, as in A, except that INV contained the Bpa variants of TatE and TatA at position Gly-33. TatEGly-33Bpa yielded more prominent heterodimers with TatA than homodimers with TatE.
Adducts larger than 19 kDa were also obtained for the TatEThr-7Bpa and TatATrp-7Bpa variants (Fig. 3A, lanes 7 and 11, orange and magenta dots). Because they were each missing in vesicles that contained only TatE or TatA (Fig. 3A, compare lanes 5 and 7 and lanes 9 and 11), they likely represent higher-order TatE-TatA oligomers. Because those TatAE oligomers can form only from homomers of the Bpa-containing Tat subunit with maximally one Bpa-less heteromer cross-linked to it, they differ in size depending on whether they contain more TatA or TatE. The adduct labeled with an orange triangle (Fig. 3A) will be discussed below.
TatE did not only form cross-links with TatA when Bpa was present in the N-terminal position Thr-7 of TatE. Cross-linking to TatA also occurred through position Gly-33 located in the amphipathic helix of TatE (Fig. 3B). Compared with Thr-7Bpa, the Gly-33Bpa variant of TatE did not give rise to extensive oligomerization but yielded only one UV-dependent adduct of about 15 kDa corresponding to the size of a TatE dimer (Fig. 3B, orange stars). However, a more prominent 19-kDa adduct appeared on TatE and TatA blots, provided that INV contained both TatE and TatA and carried Bpa at the conserved position Gly-33 of either Tat subunit (Fig. 3B, orange and magenta dots). Collectively, the results depicted in Fig. 3 imply that, under our experimental conditions, TatE and TatA both exhibit a strong propensity to assemble into hetero-oligomeric complexes.
TatE Also Cross-links to TatB and TatC
When UV-irradiated vesicles carrying the TatEThr-7Bpa variant were also probed for TatB-cross-reactive species, a 38-kDa adduct became visible (Fig. 3A, αTatB, orange triangle). This adduct was also recognized by the TatE antibodies but was absent from INV lacking TatB (Fig. 3A, compare lanes 3 and 5), suggesting that it is a TatE-TatB dimer.
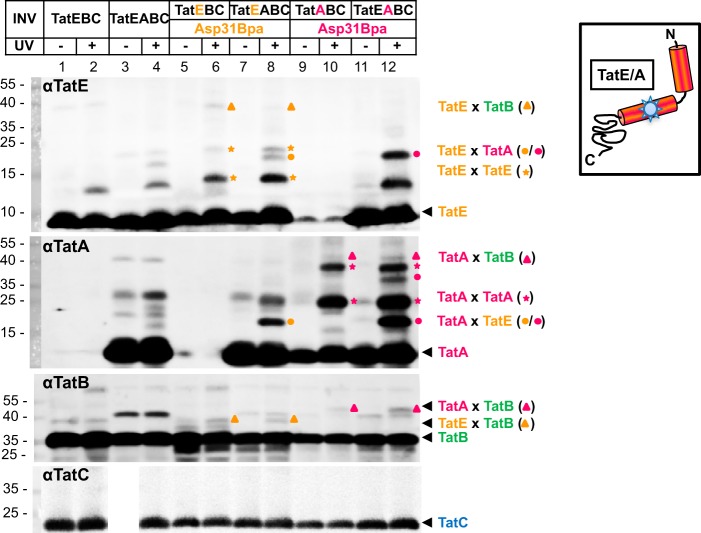
We have reported previously that position Asp-31 of TatA, which is conserved in TatE, is a rather specific contact site for TatB (18). We therefore generated the Asp-31Bpa mutant of TatE and analyzed its cross-linking behavior in the presence and absence of TatA and also directly compared it with the Asp-31Bpa variant of TatA (Fig. 4). As described above for the TatE variants carrying Bpa at positions Thr-7 and Gly-33, UV irradiation of INV harboring TatEAsp-31Bpa gave rise to the formation of TatE homodimers (Fig. 4, lanes 6 and 8, orange stars). In the presence of TatA, the 19-kDa TatE-TatA heterodimer that was recognized by antibodies against TatE (αTatE) and TatA (αTatA) was additionally obtained (Fig. 4, lane 8, orange dots). An equally sized adduct resulted from the UV irradiation of INV that had Bpa incorporated at position Asp-31 of TatA rather than TatE (Fig. 4, magenta dots, compare lanes 8 and 12). When these vesicles were also probed with anti-TatB antibodies (αTatB), UV-dependent adducts of TatEAsp-31Bpa (Fig. 4, orange triangles) and TatAAsp-31Bpa (Fig. 4, magenta triangles) became visible, the sizes of which correspond to those expected for TatE-TatB and TatA-TatB heterodimers, respectively. Therefore, TatE and TatA both contact TatB at a conserved residue in their amphipathic helices.
FIGURE 4.
TatE also interacts with TatB. INV were prepared from E. coli strains expressing the Asp-31Bpa variants of TatA and TatE in combination with the indicated Tat components. After UV irradiation, INV were probed for TatA- and TatE-cross-reactive material by immunoblotting using anti-TatE (αTatE), anti-TatA (αTatA), anti-TatB (αTatB), and anti-TatC (αTatC) antibodies. The 38-kDa TatE-TatB complexes are labeled with orange triangles and those between TatA and TatB with magenta triangles. Marked are homo-oligomers of TatA (magenta stars) and TatE (orange stars) and heterodimers formed between TatE and TatA and between TatA and TatE (orange and magenta dots, respectively). The TatA-TatE oligomer of ∼38 kDa (lane 12, αTatA, top magenta dot) is presumably a TatA dimer cross-linked to one TatE molecule and, as such, is not visible in the absence of TatE (lane 10).
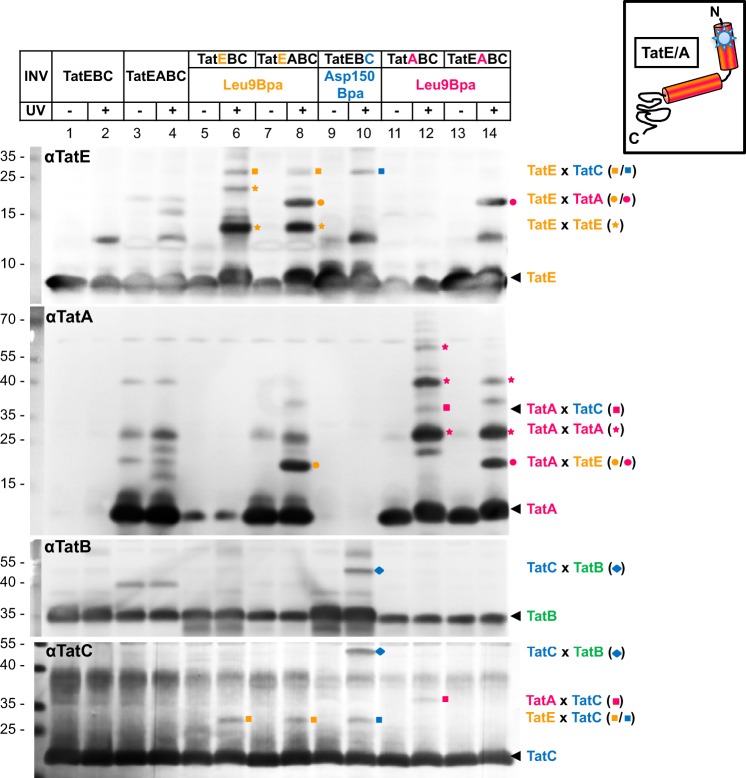
Interactions between TatE and TatC became evident using the Leu-9Bpa mutant of TatE. Upon UV irradiation, membrane vesicles containing this TatE variant again yielded TatE oligomers (Fig. 5, lanes 6 and 8, orange stars) as well as the 19-kDa TatE-TatA heterodimer when INV also contained TatA (Fig. 5, lane 8, orange dots). The latter was again verified by its co-migration with an UV-dependent cross-link of INV containing the Leu-9Bpa mutation in TatA instead of TatE (Fig. 5, compare lanes 8 and 14, orange and magenta dots).
FIGURE 5.
The transmembrane part of TatE is in close proximity to TatC. INV containing, in addition to the indicated Tat components, the Leu-9Bpa variants of TatE and TatA as well as the Asp-150Bpa variant of TatC were UV irradiated and analyzed by immunoblotting using antibodies against all Tat components (αTatE, αTatA, αTatB, and αTatC). The ∼30 kDa adduct of TatELeu-9Bpa to TatC is marked with an orange square and that of TatCAsp150Bpa to TatE with a blue square. Cross-links between TatALeu-9Bpa and TatC are indicated by magenta squares. They disappear in the presence of TatE (lanes 12 and 14). The TatE-TatA dimer is marked with an orange dot and the TatA-TatE dimer with a magenta dot. Homo-oligomers of TatE and TatA are labeled with orange and magenta stars, respectively. Note complexes of the size of a TatA dimer appearing even when TatA was Bpa-free (e.g. lanes 4 and 8) because of SDS-resistant oligomerization of TatA. The blue diamond indicates the interaction of TatB with position Asp-150 in TatC.
In addition, TatELeu-9Bpa gave rise to a cross-link of about 30 kDa (Fig. 5, lanes 6 and 8, orange squares), which was also recognized by anti-TatC antibodies (Fig. 5, αTatC, lanes 6, 8, and 10, orange and blue squares). Furthermore, it co-migrated with a UV-dependent adduct of the Asp-150Bpa variant of TatC (Fig. 5, lane 10, blue square). These results do not only demonstrate the interaction of TatE with TatC but also identify Asp-150 of TatC, which has been shown previously to interact with TatA (13, 17), as a binding site for TatE. As reported previously (13, 17), Asp-150 of TatC also cross-links to TatB. This is again illustrated on the TatC and TatB blots in Fig. 5 (αTatB, αTatC), revealing a prominent, ∼50 kDa adduct that was recognized simultaneously by both antibodies (blue diamond, lane 10).
Finally, a cross-link of ∼37 kDa was detected by antibodies directed against both TatC and TatA when vesicles contained the Leu-9Bpa variant of TatA (Fig. 5, αTatA, αTatC, lane 12, magenta square), reconfirming contacts reported previously between TatC and the transmembrane helix of TatA (15, 18). Therefore, TatA and TatE both cross-link to TatC via the same conserved residue (Leu-9) and interact with TatC around its residue Asp-150. Consistent with a common binding site of TatA and TatE on TatC, the TatA-TatC adduct was largely impaired in the presence of TatE (Fig. 5, magenta squares, lanes 12 and 14).
In Vitro Translocation Activity of TatE
Finally, we also wanted to study the involvement of TatE in TatABC-based translocation events on a functional level. To this end, we in vitro-synthesized and radioactively labeled the two natural Tat substrates SufI and AmiC and the artificial TorA-MalE and incubated them with INV to allow transport into the lumen of the vesicles. Afterward, samples were treated with proteinase K to digest all non-translocated proteins. Fig. 6A shows the results obtained with vesicles whose Tat translocases were composed either of TatABC, TatEBC, or TatEABC. The maximal transport of all three substrates, as judged by the amounts of proteinase K-resistant precursor (p) and signal sequence-less, mature forms (m), was achieved when using vesicles that contained TatA, TatB, and TatC (Fig. 6, lane 4). This is also illustrated in Fig. 6B, which depicts quantified data obtained from several independent experiments. As expected from previous in vivo studies (26, 27, 29–31), TatE was also able to compensate for a lack of TatA in our in vitro assay (TatEBC) but with much reduced efficiency (Fig. 6, A, compare lane 6 with lanes 2 and 4, and B). When present simultaneously in the vesicles with TatA at grossly similar levels (Fig. 6C), TatE did not significantly alter the TatA-mediated translocation efficiency (Fig. 6B). The in vitro translocation assays, therefore, seem to support the idea that TatE cooperates with TatA in the regular translocation process rather than being a fully functional surrogate of it.
FIGURE 6.
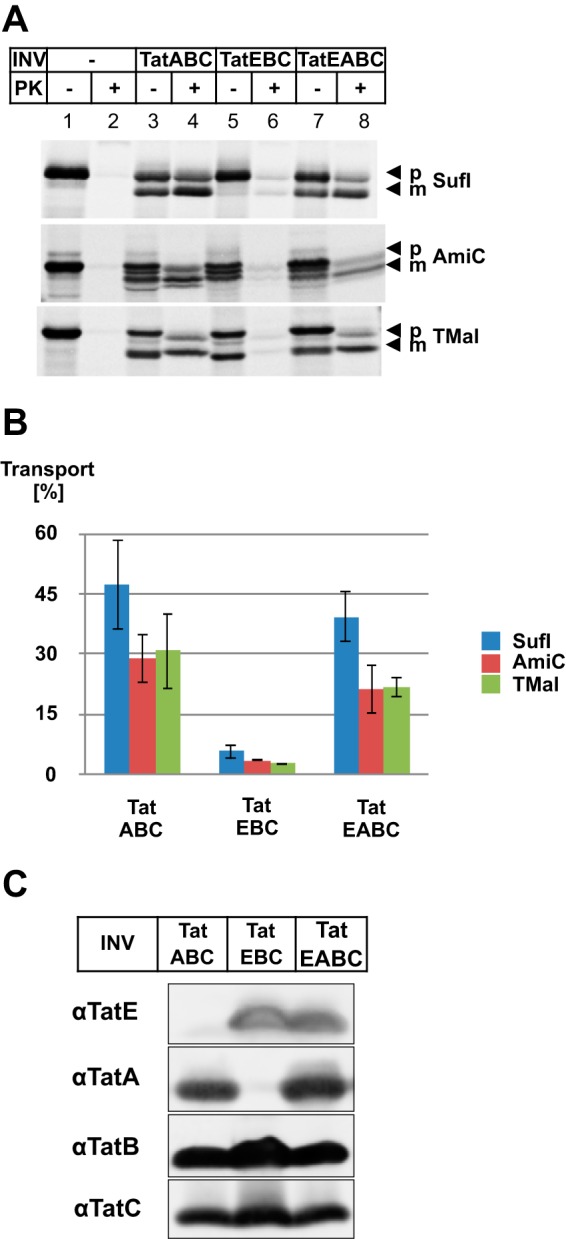
Influence of TatE on the translocation of in vitro-synthesized Tat-dependent substrates. A, the RR precursor proteins pSufI, TorA-MalE (TMal), and AmiC were in vitro-synthesized and radiolabeled by cell-free transcription/translation in the absence or presence of INV containing, in addition to TatB and TatC, either TatA (TatABC), TatE (TatEBC), or both (TatEABC). Where indicated, samples were digested with proteinase K (PK) to visualize protease-resistant, i.e. translocated, precursor (p) and mature (m) forms of the indicated proteins. Radiolabeled proteins were separated by SDS-PAGE and visualized by phosphorimaging. B, quantitative data (mean ± S.E.) obtained from seven parallel experiments for the precursor protein pSufI (blue columns) and three parallel experiments for the precursors AmiC (red columns) and TorA-MalE (green columns) shown in A. Transport efficiency of the indicated INV was calculated using analyzing tools of ImageQuantTL. INV containing only TatEBC showed significantly reduced translocation activity compared with INV containing TatABC and TatEABC. C, the Tat components of the indicated vesicles were separated by Tricine SDS-PAGE and visualized by immunoblotting using polyclonal antibodies against TatE (αTatE), TatA (αTatA), TatB (αTatB), and TatC (αTatC).
Discussion
Our in vivo studies show that the induced expression exclusively of an intact Tat substrate causes TatE to form multiple clusters in the membrane as long as the other Tat components are present, suggesting that those clusters represent active Tat translocases. Consistent with this assumption, cells that did not overexpress a Tat substrate frequently showed TatE-GFP accumulated in only one single cluster, often near one of the cell poles. These single clusters most likely mark individual Tat translocases involved in the housekeeping export of endogenous Tat substrates because they were not seen when TatE-GFP was expressed in the absence of TatABC. The intracellular TatE-GFP levels were unchanged whether or not the TatABC subunits and a recombinant Tat substrate were co-expressed. Therefore, clustering of TatE-GFP obtained only in the presence of the TatABC subunits and an intact Tat substrate cannot be due to the formation of inclusion bodies from overexpressed TatE-GFP. Rather, the results suggest that, in live E. coli cells, TatE is recruited to sites where Tat substrates are exported across the plasma membrane. They do, however, not allow assessment of whether TatE joins TatABC translocases and, therefore, whether it is involved regularly in all Tat-specific translocation events of E. coli or whether TatA-less translocases composed of TatEBC occur.
Association of TatE with TatABC translocases, however, is strongly suggested by our cross-linking studies. They revealed interactions of TatE with all other Tat subunits, TatA, TatB, and TatC, of which the most prominent ones were those with TatA. The formation of composite TatABCE translocases is actually supported by several past findings. Thus, the TorA activity was found to be mislocalized drastically to the cytoplasm both in the ΔtatA and ΔtatE mutant strains (27), from which the authors inferred that the Tat translocase might contain copies of both proteins. In another study, a TatA-YFP construct was found to be distributed over the whole plasma membrane of E. coli cells, provided that the cells also contained TatE, whereas it accumulated in a few spots in the absence of TatE (31). The authors concluded that the largely inactive TatA-YFP caused a transport arrest of the Tat translocases from which it could only be released after the concomitant binding of active TatE molecules. In addition, the inactive TatA-YFP fusion markedly enhanced TatE-mediated transport, also supporting an interaction of TatA and TatE at the active Tat translocases (31).
Because the molecular mechanism by which TatA mediates the translocation step is not known, one can only speculate as to the possible roles that TatE might fulfil. In vivo, TatE seems to be expressed at an approximately 50 times lower level than TatA (33). A few TatE molecules could therefore interdigitate between TatA protomers and, thereby, enhance the translocation-promoting function of TatA, be it the formation of translocation pores or the destabilization of the lipid bilayer. In general, TatE could influence the extent to which TatA oligomerizes. Because we found TatE cross-linked to TatC, it might also affect the way in which TatA and/or TatB associate with TatC. Common binding sites of TatA and TatB on TatC have recently been demonstrated, and it has been suggested that TatB might control the access and oligomerization of TatA on the inside of concentric TatBC assemblies (6, 15, 17). As we have shown here, TatE, at high concentrations, seems to be able to displace TatA from TatC. TatE, therefore, seems to be another player in influencing the association and, possibly, assembly of TatA and TatB on TatC. Our collective data suggest that TatE, rather than totally replacing TatA, is a regular constituent of TatABC-based translocases.
Author Contributions
E. E. performed the experiments and helped with the writing of the manuscript. J. F. supervised the project. A. S. B. helped with the transport assays. M. M. supervised the project and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Tracy Palmer for strain BL21(DE3)ΔTat.
This study was supported by SFB746 Grant P13 and the Excellence Initiative (Grant GSC-4, Spemann Graduate School) of the German Research Foundation. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Tables S1 and S2, additional data, and references.
- Tat
- twin-arginine translocation
- INV
- inverted inner membrane vesicle(s)
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- Bpa
- p-benzoyl-l-phenylalanine.
References
- 1.Celedon J. M., and Cline K. (2013) Intra-plastid protein trafficking: how plant cells adapted prokaryotic mechanisms to the eukaryotic condition. Biochim. Biophys. Acta 1833, 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berks B. C., Lea S. M., and Stansfeld P. J. (2014) Structural biology of Tat protein transport. Curr. Opin. Struct. Biol. 27C, 32–37 [DOI] [PubMed] [Google Scholar]
- 3.Goosens V. J., Monteferrante C. G., and van Dijl J. M. (2014) The Tat system of Gram-positive bacteria. Biochim. Biophys. Acta 1843, 1698–1706 [DOI] [PubMed] [Google Scholar]
- 4.Kudva R., Denks K., Kuhn P., Vogt A., Müller M., and Koch H. G. (2013) Protein translocation across the inner membrane of Gram-negative bacteria: the Sec and Tat dependent protein transport pathways. Res. Microbiol. 164, 505–534 [DOI] [PubMed] [Google Scholar]
- 5.Patel R., Smith S. M., and Robinson C. (2014) Protein transport by the bacterial Tat pathway. Biochim. Biophys. Acta 1843, 1620–1628 [DOI] [PubMed] [Google Scholar]
- 6.Cline K. (2015) Mechanistic Aspects of folded protein transport by the twin arginine translocase (Tat). J. Biol. Chem. 290, 16530–16538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramasamy S., Abrol R., Suloway C. J., and Clemons W. M. Jr. (2013) The glove-like structure of the conserved membrane protein TatC provides insight into signal sequence recognition in twin-arginine translocation. Structure 21, 777–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rollauer S. E., Tarry M. J., Graham J. E., Jääskeläinen M., Jäger F., Johnson S., Krehenbrink M., Liu S. M., Lukey M. J., Marcoux J., McDowell M. A., Rodriguez F., Roversi P., Stansfeld P. J., Robinson C. V., Sansom M. S., Palmer T., Högbom M., Berks B. C., and Lea S. M. (2012) Structure of the TatC core of the twin-arginine protein transport system. Nature 492, 210–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fröbel J., Rose P., and Müller M. (2012) Twin-arginine-dependent translocation of folded proteins. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 367, 1029–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma X., and Cline K. (2010) Multiple precursor proteins bind individual Tat receptor complexes and are collectively transported. EMBO J. 29, 1477–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Celedon J. M., and Cline K. (2012) Stoichiometry for binding and transport by the twin arginine translocation system. J. Cell Biol. 197, 523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cléon F., Habersetzer J., Alcock F., Kneuper H., Stansfeld P. J., Basit H., Wallace M. I., Berks B. C., and Palmer T. (2015) The TatC component of the twin-arginine protein translocase functions as an obligate oligomer. Mol. Microbiol. 98, 111–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zoufaly S., Fröbel J., Rose P., Flecken T., Maurer C., Moser M., and Müller M. (2012) Mapping precursor-binding site on TatC subunit of twin arginine-specific protein translocase by site-specific photo cross-linking. J. Biol. Chem. 287, 13430–13441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma X., and Cline K. (2013) Mapping the signal peptide binding and oligomer contact sites of the core subunit of the pea twin arginine protein translocase. Plant Cell 25, 999–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aldridge C., Ma X., Gerard F., and Cline K. (2014) Substrate-gated docking of pore subunit Tha4 in the TatC cavity initiates Tat translocase assembly. J. Cell Biol. 205, 51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fröbel J., Rose P., Lausberg F., Blümmel A. S., Freudl R., and Müller M. (2012) Transmembrane insertion of twin-arginine signal peptides is driven by TatC and regulated by TatB. Nat. Commun. 3, 1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blümmel A. S., Haag L. A., Eimer E., Müller M., and Fröbel J. (2015) Initial assembly steps of a translocase for folded proteins. Nat. Commun. 6, 7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fröbel J., Rose P., and Müller M. (2011) Early contacts between substrate proteins and TatA translocase component in twin-arginine translocation. J. Biol. Chem. 286, 43679–43689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alami M., Lüke I., Deitermann S., Eisner G., Koch H. G., Brunner J., and Müller M. (2003) Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol. Cell 12, 937–946 [DOI] [PubMed] [Google Scholar]
- 20.Taubert J., Hou B., Risselada H. J., Mehner D., Lünsdorf H., Grubmüller H., and Brüser T. (2015) TatBC-independent TatA/Tat substrate interactions contribute to transport efficiency. PLoS ONE 10, e0119761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Behrendt J., and Brüser T. (2014) The TatBC complex of the Tat protein translocase in Escherichia coli and its transition to the substrate-bound TatABC complex. Biochemistry 53, 2344–2354 [DOI] [PubMed] [Google Scholar]
- 22.Brüser T., and Sanders C. (2003) An alternative model of the twin arginine translocation system. Microbiol. Res. 158, 7–17 [DOI] [PubMed] [Google Scholar]
- 23.Aldridge C., Storm A., Cline K., and Dabney-Smith C. (2012) The chloroplast twin arginine transport (tat) component, tha4, undergoes conformational changes leading to tat protein transport. J. Biol. Chem. 287, 34752–34763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez F., Rouse S. L., Tait C. E., Harmer J., De Riso A., Timmel C. R., Sansom M. S., Berks B. C., and Schnell J. R. (2013) Structural model for the protein-translocating element of the twin-arginine transport system. Proc. Natl. Acad. Sci. U.S.A. 110, E1092–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yen M. R., Tseng Y. H., Nguyen E. H., Wu L. F., and Saier M. H. Jr. (2002) Sequence and phylogenetic analyses of the twin-arginine targeting (Tat) protein export system. Arch. Microbiol. 177, 441–450 [DOI] [PubMed] [Google Scholar]
- 26.Kikuchi Y., Date M., Itaya H., Matsui K., and Wu L. F. (2006) Functional analysis of the twin-arginine translocation pathway in Corynebacterium glutamicum ATCC 13869. Appl. Environ. Microbiol. 72, 7183–7192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sargent F., Bogsch E. G., Stanley N. R., Wexler M., Robinson C., Berks B. C., and Palmer T. (1998) Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 17, 3640–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heikkilä M. P., Honisch U., Wunsch P., and Zumft W. G. (2001) Role of the Tat transport system in nitrous oxide reductase translocation and cytochrome cd1 biosynthesis in Pseudomonas stutzeri. J. Bacteriol. 183, 1663–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baglieri J., Beck D., Vasisht N., Smith C. J., and Robinson C. (2012) Structure of TatA paralog, TatE, suggests a structurally homogeneous form of Tat protein translocase that transports folded proteins of differing diameter. J. Biol. Chem. 287, 7335–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sargent F., Stanley N. R., Berks B. C., and Palmer T. (1999) Sec-independent protein translocation in Escherichia coli: a distinct and pivotal role for the TatB protein. J. Biol. Chem. 274, 36073–36082 [DOI] [PubMed] [Google Scholar]
- 31.Alcock F., Baker M. A., Greene N. P., Palmer T., Wallace M. I., and Berks B. C. (2013) Live cell imaging shows reversible assembly of the TatA component of the twin-arginine protein transport system. Proc. Natl. Acad. Sci. U.S.A. 110, E3650–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gohlke U., Pullan L., McDevitt C. A., Porcelli I., de Leeuw E., Palmer T., Saibil H. R., and Berks B. C. (2005) The TatA component of the twin-arginine protein transport system forms channel complexes of variable diameter. Proc. Natl. Acad. Sci. U.S.A. 102, 10482–10486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jack R. L., Sargent F., Berks B. C., Sawers G., and Palmer T. (2001) Constitutive expression of Escherichia coli tat genes indicates an important role for the twin-arginine translocase during aerobic and anaerobic growth. J. Bacteriol. 183, 1801–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rose P., Fröbel J., Graumann P. L., and Müller M. (2013) Substrate-dependent assembly of the Tat translocase as observed in live Escherichia coli cells. PLoS ONE 8, e69488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moser M., Panahandeh S., Holzapfel E., and Müller M. (2007) In vitro analysis of the bacterial twin-arginine-dependent protein export. Methods Mol. Biol. 390, 63–79 [DOI] [PubMed] [Google Scholar]
- 36.Wexler M., Sargent F., Jack R. L., Stanley N. R., Bogsch E. G., Robinson C., Berks B. C., and Palmer T. (2000) TatD is a cytoplasmic protein with DNase activity: no requirement for TatD family proteins in Sec-independent protein export. J. Biol. Chem. 275, 16717–16722 [DOI] [PubMed] [Google Scholar]
- 37.Schägger H. (2006) Tricine-SDS-PAGE. Nat. Protoc. 1, 16–22 [DOI] [PubMed] [Google Scholar]
- 38.Alami M., Trescher D., Wu L. F., and Müller M. (2002) Separate analysis of twin-arginine translocation (Tat)-specific membrane binding and translocation in Escherichia coli. J. Biol. Chem. 277, 20499–20503 [DOI] [PubMed] [Google Scholar]
- 39.Lesley S. A., Brow M. A., and Burgess R. R. (1991) Use of in vitro protein synthesis from polymerase chain reaction-generated templates to study interaction of Escherichia coli transcription factors with core RNA polymerase and for epitope mapping of monoclonal antibodies. J. Biol. Chem. 266, 2632–2638 [PubMed] [Google Scholar]
- 40.Stanley N. R., Findlay K., Berks B. C., and Palmer T. (2001) Escherichia coli strains blocked in Tat-dependent protein export exhibit pleiotropic defects in the cell envelope. J. Bacteriol. 183, 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.