Background: FYCO1 binds to LC3 and is involved in transport of autophagosomes.
Results: FYCO1 uses a C-terminally extended LIR motif for specific interaction with LC3A/B stimulating autophagosome maturation during basal autophagy.
Conclusion: The transport adaptor FYCO1 engages in a specific interaction with LC3A/B to stimulate autophagosome maturation.
Significance: This work will lead to an increased understanding of LIR interactions and the role of FYCO1 in autophagy.
Keywords: autophagy, crystal structure, protein degradation, protein-protein interaction, vesicles, FYCO1, LC3, LIR
Abstract
FYCO1 (FYVE and coiled-coil protein 1) is a transport adaptor that binds to phosphatidylinositol 3-phosphate, to Rab7, and to LC3 (microtubule-associated protein 1 light chain 3) to mediate transport of late endosomes and autophagosomes along microtubules in the plus end direction. We have previously shown that FYCO1 binds to LC3B via a 19-amino acid sequence containing a putative core LC3-interacting region (LIR) motif. Here, we show that FYCO1 preferentially binds to LC3A and -B. By peptide array-based two-dimensional mutational scans of the binding to LC3B, we found FYCO1 to contain a C-terminally extended LIR domain. We determined the crystal structure of a complex between a 13-amino acid LIR peptide from FYCO1 and LC3B at 1.53 Å resolution. By combining the structural information with mutational analyses, both the basis for the C-terminally extended LIR and the specificity for LC3A/B binding were revealed. FYCO1 contains a 9-amino acid-long F-type LIR motif. In addition to the canonical aromatic residue at position 1 and the hydrophobic residue at position 3, an acidic residue and a hydrophobic residue at positions 8 and 9, respectively, are important for efficient binding to LC3B explaining the C-terminal extension. The specificity for binding to LC3A/B is due to the interaction between Asp1285 in FYCO1 and His57 in LC3B. To address the functional significance of the LIR motif of FYCO1, we generated FYCO1 knock-out cells that subsequently were reconstituted with GFP-FYCO1 WT and LIR mutant constructs. Our data show that FYCO1 requires a functional LIR motif to facilitate efficient maturation of autophagosomes under basal conditions, whereas starvation-induced autophagy was unaffected.
Introduction
Macroautophagy (hereafter autophagy) is an evolutionary conserved degradation pathway that directs surplus or damaged cytosolic components to lysosomal degradation to maintain cellular homeostasis (1). A double membrane structure, the phagophore, grows around part of the cytoplasm, or targeted components, and closes upon itself to form the autophagosome (1). The autophagosome matures by fusion, either with a late endosome, or directly with a lysosome.
The ATG8/LC3/GABARAP2 proteins are globular proteins with a C-terminal ubiquitin-like core containing a five-stranded β-sheet wrapped around a central α-helix. Distinct from ubiquitin, they harbor an N-terminal arm with two α-helices. Six human ATG8 proteins form three phylogenetic groups: one containing LC3A, -B, and -C, a second harboring γ-aminobutyric acid receptor-associated protein (GABARAP) and GABARAP-like 1 (GABARAPL1), with GABARAPL2/GATE16 alone in the third group (2). The Atg8 proteins are synthesized as precursor proteins that are processed by the cysteine protease ATG4. LC3s are processed to LC3-I, which is lipidated to give LC3-II anchored on both sides of the phagophore membrane. The lipidated Atg8 molecules act in recruitment of cargo (via selective autophagy receptors) (3, 4), autophagy components, and regulatory proteins (5), as well as facilitating phagophore expansion (6). Atg8 proteins are also required in the closure of the phagophore to form the autophagosome (7, 8). Their retention inside autophagosomes is widely used as a autophagosome marker.
The interaction between autophagy receptors and ATG8 ubiquitin-like proteins, first described for p62/SQSTM1, involves a short sequence motif named the LC3-interacting region (LIR) (9). The core LIR motif, (W/F/Y)XX(L/I/V), contains two absolutely conserved positions occupied by an aromatic residue and a hydrophobic residue separated by two variable positions. The aromatic residue is often flanked N-terminally by one or more acidic residues or phosphorylatable Ser or Thr residues. There is also a strong tendency for either acidic or hydrophobic residues to occupy the position immediately C-terminal to the aromatic residue (5, 10). Structural studies revealed the details of the LIR-Atg8 interaction. The aromatic residue occupies one hydrophobic pocket (HP1), and the hydrophobic residue occupies another (HP2) (11, 12). The N-terminal arms of the Atg8 molecules often engage in electrostatic interactions with acidic or phosphorylated residues preceding the aromatic residue of the LIR motif (5). The following structures of LIR peptides in complex with Atg8s have been reported: p62/SQSTM1-LC3B (11, 12); yeast Atg19-Atg8 (12); ATG4B-LC3B (13); NBR1-GABARAP (14); NDP52-LC3C (15); Optineurin-LC3B (4); Bcl-2-GABARAP (16); ATG13-LC3A and ATG13-LC3C (17); ALFY-GABARAP (18); PLEKHM1-LC3B (19); and KBTBD6-GABARAP (20).
Proteins interacting with Atg8 family proteins via a LIR motif are either selective autophagy receptors or proteins interacting with Atg8 proteins on the outside of autophagic structures and other vesicles. Only two LIR-containing proteins, MAPK8IP1/JIP1 and FYCO1 (FYVE and coiled-coil protein 1), are reported to be involved in transport of autophagosomes (21, 22). FYCO1 is a 1478-amino acid-long phosphatidylinositol 3-phosphate-binding protein and Rab7 effector that interacts with LC3, and is involved in the transport of autophagosomes along microtubules in the plus end direction (21). Mutations in FYCO1 cause autosomal-recessive congenital cataracts, suggesting that FYCO1 is required for human lens development, transparency, or both (23). FYCO1 is recruited by LC3 to Dectin-1 phagosomes during LC3-associated phagocytosis to facilitate maturation of early p40phox-containing phagosomes into late LAMP1-positive phagosomes (24). In macrophage cell lines exposed to LPS, tubular lysosomes form. Rab7 and its effectors RILP (Rab7-interacting lysosomal protein) and FYCO1 are required for this formation through modulation of dynein- and kinesin-driven transport along microtubules, respectively (25). Intriguingly, FYCO1 is involved, together with the endoplasmic reticulum protein protrudin, in mediating microtubule-dependent transport of late endosomes via endoplasmic reticulum-endosome contact sites to produce cell protrusions and neurite outgrowth (26).
Here, we describe the details of the interaction between FYCO1 and LC3B based on x-ray crystallography and mutational analyses. FYCO1 binds LC3 via a canonical F-type core LIR motif. However, a C-terminal extension of the LIR is required because a vital interaction for the binding is between Glu1287 (position 8) and Arg70 of LC3. FYCO1 preferably interacts with LC3A and -B. This is, at least in part, due to a specific interaction between His57 in LC3B and Asp1285 in the FYCO1 LIR. His57 is found in LC3A and -B, but not in other ATG8 family members. By reconstituting FYCO1 KO cells with FYCO1 WT and FYCO1 with the core LIR mutation F1280A/I1283A, we found that a functional LIR is required for efficient maturation of autophagosomes under basal conditions.
Experimental Procedures
Antibodies and Reagents
Primary antibodies used were mouse anti-FYCO1 (H00079443-B01P, Abnova), rabbit anti-FYCO1 (HPA035526, Sigma-Aldrich), rabbit anti LC3B (L7543, Sigma-Aldrich), mouse monoclonal anti-Myc (9B11, Cell Signaling), rabbit anti-GFP (ab290, Abcam), and mouse monoclonal anti-LAMP1 (G1/139/5, Developmental Studies Hybridoma Bank (DSHB)). HRP-conjugated anti-GST antibody (clone RPN1236) was purchased from GE Healthcare. Secondary antibodies used were HRP-conjugated goat anti-rabbit IgG (554021, BD PharmingenTM), Alexa Fluor® 555-conjugated goat anti-rabbit IgG (A-21428, Life Technologies), Alexa Fluor® 488-conjugated goat anti-mouse IgG (A-11029, Life Technologies), Alexa Fluor® 488-conjugated goat anti-rabbit IgG (A-11008, Life Technologies), Alexa Fluor® 647-conjugated goat anti-rabbit IgG (A-21245, Life Technologies), IRDye® 680LT goat anti-mouse IgG (926-68020, LI-COR), and IRDye®800CW goat anti-rabbit IgG (926-32211, LI-COR).
Plasmids
The Gateway entry clones pENTR-FYCO1 (21), encoding human FYCO1, and pENTR-LC3B (9), encoding human LC3B, have been described previously. Point mutants of pENTR-FYCO1 (D1276A, D1277A, D1276A/D1277A, F1280A/I1283A, D1281A, D1285A, E1287A, L1288A) and pENTR-LC3B (H57D, R70A, R10A) were done using the QuikChange site-directed mutagenesis kit (Stratagene). Gateway destination vectors used were pDestEGFP-C1 (mammalian expression of EGFP fusions), pDest15 (Invitrogen) (bacterial expression of GST fusions), pDestMyc (mammalian expression and/or in vitro translation of Myc-tagged fusions) (27), and pDest-Flp-In-EGFP-C1 (10)(mammalian Flp-In vector for stable and inducible expression of EGFP fusions). Transfer from entry clones of LC3B point mutants into pDest15, FYCO1 point mutants into pDestMyc, wild type FYCO1 into pDest-Flp-In-EGFP-C1, and FYCO1 Phe1280/Ile1283 into pDest-Flp-In-EGFP-C1 was done by Gateway LR reactions using the Gateway recombination system (Invitrogen). Other cDNA expression constructs used in this study (pDestEGFP-FYCO1, pDestMyc-FYCO1, pDest15-LC3A, pDest15-LC3B, pDest15-LC3C, pDest15-GABARAP, pDest15-GABARAP-L1,pDest15-GABARAP-L2, pDestEGFP-LC3A, pDestEGFP-LC3B,pDestEGFP-LC3C, pDestEGFP-GABARAP, pDestEGFP-GABARAP-L1, pDestEGFP-GABARAP-L2) have been described previously (9, 10, 21, 28). Oligonucleotides for mutagenesis, PCR, and DNA sequencing reactions were obtained from Invitrogen and Sigma-Aldrich. Plasmid constructs were verified by DNA sequencing (BigDye, Applied Biosystems).
Cell Culture and Transfections
HeLa cells were grown in Eagle's minimum essential medium supplemented with 10% fetal bovine serum (Biochrom AG, S0615), non-essential amino acids, 2 mm l-glutamine, and 1% streptomycin-penicillin (Sigma, P4333). HEK293 cells were maintained in Dulbecco's modified Eagle's medium with the same supplements as described above. Subconfluent cells were transfected with plasmids using TransIT-LT1 (Mirus, MIR2300) (for microscopy analysis) or Metafectene PRO (Biontex) (for immunoprecipitation and zinc finger nucleases) following the supplier's instructions. Twenty-four hours after transfection, cells were fixed and permeabilized in pre-chilled (−20 °C) methanol for 10 min and washed two times in PBS. Fixed cells were blocked with 3% pre-immune goat serum in PBS for 30 min at room temperature before incubation for 1 h at room temperature with primary antibodies diluted in PBS with 1% goat serum. Cells were washed five times in PBS before incubation with Alexa Fluor® secondary antibodies (Life Technologies) diluted 1:500 supplemented with 1% goat serum. Before imaging, cells were washed five times in PBS. Stable Flp-In GFP-FYCO1 and GFP-FYCO1 LIRmut cell lines were generated using the Flp-In recombination system following the manufacturer's protocol (Invitrogen). Cells were treated as indicated with 0.2 μm bafilomycin A1 (Sigma, B1793).
Immunoprecipitation
Transfected cells were rinsed twice with PBS prior to lysis in a modified radioimmunoprecipitation assay buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 2 mm EDTA, 0.25% sodium deoxycholate, 1% Nonidet P-40) supplemented with cOmplete Mini EDTA-free protease inhibitor mixture tablets (1 tablet/10 ml) (11836170001, Roche Applied Science). Lysates were centrifuged for 5 min at 13,000 rpm 4 °C followed by incubation with anti-Myc affinity gel (B23401 (Biotool)) for 18 h at 4 °C. Beads were washed five times with lysis buffer and eluted with Myc peptide (B23411 (Biotool)) while shaking at 4 °C for 30 min. Supernatant was added to SDS-PAGE loading buffer with 1 mm DTT and boiled for 5 min. Samples were resolved by SDS-PAGE and transferred to Nitrocellulose membrane (LI-COR). Membranes were blocked with Odyssey chemical blocking buffer (LI-COR) (diluted 50% in PBS) for 30 min.
Generation of FYCO1 Knock-out Cell Lines
Flp-In T-REx HEK293 cells were seeded in 60-mm plates. Subconfluent cells were transfected with plasmids encoding zinc finger nucleases targeting FYCO1 (Sigma-Aldrich). After 5 days, cells were sorted singularly in 96-well plates using a FACSAria cell sorter (BD Biosciences). Genomic DNA was isolated from each subsequent cell line and probed for nuclease cleavage with the Surveyor® mutation detection kit (Transgenomic). Cleavage was visualized by polyacrylamide-Tris borate-EDTA gel electrophoresis. Cell lines positive for nuclease cleavage were subjected to Western blotting and probed with antibodies against FYCO1.
Fluorescence Confocal Microscopy Analyses
Cells were examined using a Zeiss Axio Observer.Z1 LSM780 CLSM system (Carl Zeiss Microscopy GmbH, Jena, Germany) with a plan-Apochromat 63× NA1.4 objective or a C-Apochromat 40× NA1.2W objective, running the ZEN 2012 (black edition) software. Quantifications were performed using the Volocity software (PerkinElmer).
GST Pulldown Experiments
All GST-tagged proteins were expressed in Escherichia coli SoluBL21 (Genlantis). GST fusion proteins were purified on glutathione-Sepharose 4 Fast Flow beads (GE Healthcare 17-5132-01). 35S-labeled Myc-tagged proteins were synthesized in vitro using the TnT T7-coupled reticulocyte lysate system (Promega). Translation reaction products from 0.25 μg of plasmid DNA were incubated with GST-labeled proteins on glutathione-Sepharose beads in NETN-E buffer (50 mm Tris, pH 8.0, 100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40) supplemented with cOmplete Mini EDTA-free protease inhibitor mixture tablets (1 tablet/10 ml) (11836170001, Roche Applied Science) for 1 h at 4 °C. The beads were washed five times with 400 μl of NETN-E buffer, boiled with 2× SDS-PAGE gel loading buffer with 1 mm DTT, and subjected to SDS-PAGE. Gels were stained with Coomassie Brilliant Blue and vacuum-dried. 35S-labeled proteins were detected using a Fujifilm bioimaging analyzer BAS-5000 (Fuji), and quantifications were performed using the Image Gauge software (Fuji).
Protein Expression and Purification for X-ray Crystallography
LC3B was expressed from pGEX4T plasmid in BL21 (DE3). LC3 was expressed as a GST-tagged protein. The protein was purified using glutathione-Sepharose 4B, cation exchange, and gel filtration chromatography. The GST moiety was proteolytically removed by thrombin protease. The protein solution was concentrated to 21.2 mg/ml by ultrafiltration in 25 mm Tris-HCl (pH 7.5) and 1 mm dithiothreitol. The concentrated LC3 was mixed with a peptide representing residues 1276–1288 from FYCO1 with 1:2 molar ratio and incubated for 24 h at 4 °C.
Crystallization and Data Collection, Structure Determination, and Refinement
Crystals of LC3-FYCO1 peptide complex were obtained by the sitting-drop vapor diffusion method at 293 K in drops containing a mixture of 1 μl of protein and the 1 μl of reservoir solution, which consisted of 0.1 m potassium thiocyanate and 30% w/v polyethylene glycol monomethyl ether 2000. Crystals were flash-cooled in a nitrogen gas stream. X-ray diffraction data sets for LC3-FYCO1 peptide complex were collected at 100 K on beamline BL44XU (SPring-8, Hyogo, Japan). Data processing and reduction were carried out with HKL2000 (29). The crystals belonged to space group P21 with one molecule in the asymmetric unit. Data collection, phasing, and refinement statistics are summarized in Table 1. The structure of the LC3-FYCO1 peptide complex was determined by molecular replacement using MOLREP (30) with LC3 (Protein Data Bank (PDB) ID code 1UGM) (31) as a search model. Models were subsequently improved through an alternate cycle of manual rebuilding using COOT (32), and refinement with the program REFMAC5 (33). The final refined model consists of residues 5–123 of LC3 in the asymmetric unit. For the FYCO1 peptide, the density allowed building on residues 1277–1288 complexed to the molecule. Refinement statistics are summarized in Table 1. There are no residues in disallowed regions of the Ramachandran plot. Structure figures were generated using PyMOL.
TABLE 1.
Data collection, phasing, and refinement statistics for x-ray crystallography of the FYCO1 LIR peptide-LC3B complex
| Data collection | |
| Wave length (Å) | 0.9 |
| Unit cell a, b, c (Å) | 40.7, 39.1, 42.8 |
| Unit cell α, β, γ (°) | 90.00, 115.1, 90.00 |
| Space group | P21 |
| Resolution range (Å) | 50.0–1.53 (1.56–1.53) |
| No. of unique reflections | 17,994 |
| Completeness (%) | 96.4 (97.9) |
| Rmerge (%) | 5.2 (9.8) |
| 〈I/σ(I)〉 | 49.7 (27.5) |
| Redundancy | 3.6 (3.7) |
| Wilson B-factor (Å2) | 12.0 |
| Refinement statistics | |
| Resolution range (Å) | 19.58–1.53 |
| No. of reflections used | 17,088 |
| Free R reflections (%) | 5 |
| R/Rfree | 0.175/0.217 |
| RMSDa bond length (Å) | 0.023 |
| RMSDa bond angle (°) | 2.010 |
| Average atomic B-factor of protein (Å2) | 11.7 |
| Average atomic B-factor of ligand (Å2) | 20.4 |
| Average atomic B-factor of solvent atoms (Å2) | 20.6 |
| Ramachandran analysis (%) | |
| Residues in most favored regions | 94.1 |
| Residues in additional allowed regions | 5.9 |
| Residues in generously allowed regions | 0.0 |
| Residues in disallowed regions | 0.0 |
a RMSD, root mean square deviation.
SPOT Synthesis and GST Overlay Assays
Peptides were synthesized on cellulose membranes using a MultiPrep automated peptide synthesizer (INTAVIS Bioanalytical Instruments AG, Cologne, Germany), as described previously (34). Membranes were blocked in Tris-buffered saline-Tween 20 with 5% nonfat dry milk and probed by overlaying with GST fusion of LC3B at 1 μg/ml for 2 h at room temperature. Membranes were washed in Tris-buffered saline-Tween 20, and bound proteins were detected with HRP-conjugated anti-GST antibody (1:5000; clone RPN1236; GE Healthcare).
Results
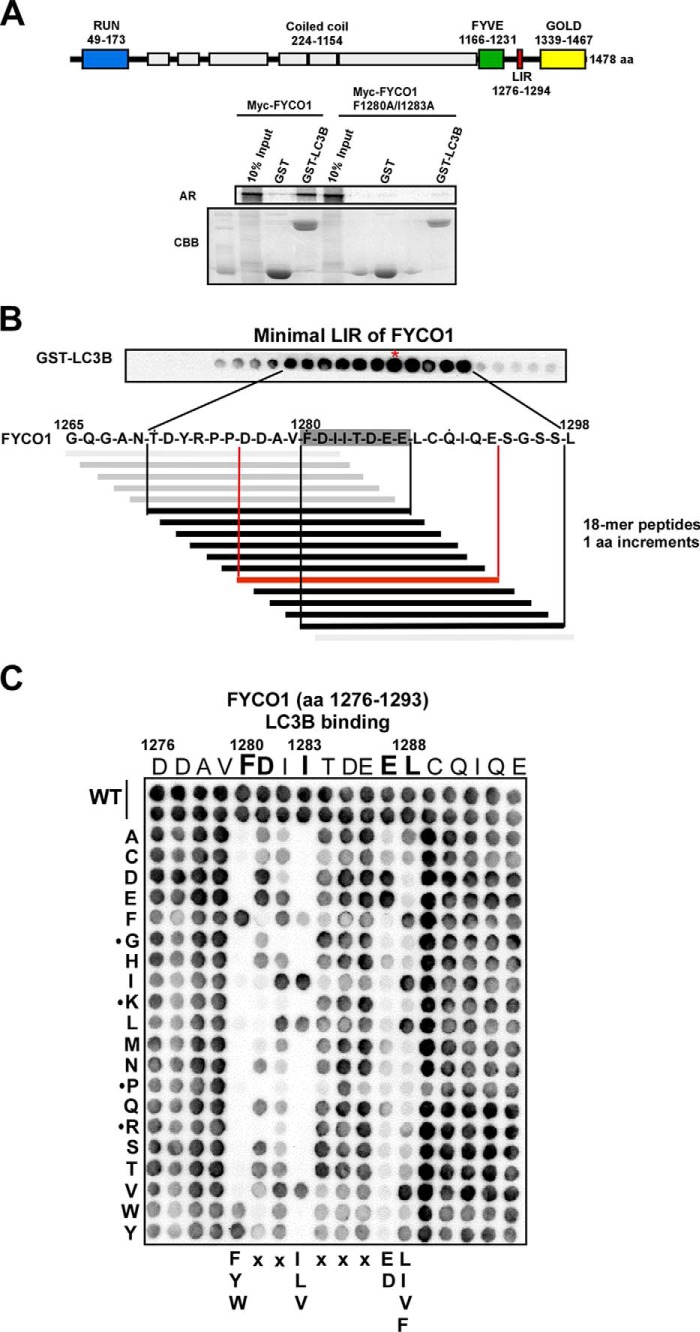
FYCO1 Contains a C-terminally Extended LIR Motif
We have previously reported that FYCO1 contains a LIR motif that we functionally mapped to the 19-amino acid-long region 1276–1294, located between the FYVE and GOLD domains (21). This region contains the sequence FDII confined to the core LIR motif consensus sequence (W/F/Y)XX(L/I/V) (5). To test whether this is the core LIR motif of an F-type LIR, we mutated the Phe1280 and Ile1283 residues to alanine and performed pulldown assays with GST-LC3B bound to beads and in vitro translated Myc-tagged, full-length WT and the F1280A/I1283A double mutant. As seen in Fig. 1A, the LIR mutant completely lost binding to GST-LC3B. To identify the minimal peptide sequence required for efficient binding to LC3B, we employed a peptide array strategy with 18-mer peptides spanning amino acids 1265–1298 of FYCO1. The peptide walk was done with steps of one amino acid to allow high resolution mapping (10). Interestingly, the minimal binding motif was determined to encompass the octapeptide FDIITDEE starting with the aromatic residue of the core motif, but extending 4 amino acids C-terminal to the core (Fig. 1B). Weak binding was also seen for peptides shortened from the C-terminal end down to the conserved hydrophobic position of the core motif (here represented by Ile1283). However, strong, binding required the four-amino acid C-terminal extension. In comparison, for ULK1 and ATG13, we have previously defined the minimal motifs to the pentameric sequences DFVMV and DFVMI, respectively (10). To study the extent of the functional LIR motif, and to assess the relative importance of specific residues involved in the interaction, we performed a peptide array with two-dimensional amino acid substitutions of peptides covering the region between 1276 and 1293 (Fig. 1C). This approach confirmed the C-terminally extended LIR motif and even suggested that Leu1288 is part of a nonameric LIR sequence. Strikingly, Glu1287 can only be substituted with another acidic residue, Asp, to restore binding. The most C-terminal Leu1288 is only successfully replaced by the hydrophobic, Ile, Val, or Phe residues. As expected, the conserved aromatic Phe1280 in the core LIR can only be substituted with the two other aromatic residues, Trp and Tyr. Similarly, in the hydrophobic position of the core LIR only Leu or Val, and to some extent Phe, can replace Ile1283. There is also a tendency for preference of acidic charge in the residue immediately C-terminal to Phe1280. As noted previously for the ATG13 and ULK1 LIRs and apparent from alignments of known LIR motifs (10), Gly or Pro residues affecting secondary structure were not tolerated at the core positions. Basic Lys or Arg residues were not tolerated, either (Fig. 1C). In conclusion, whereas the previously identified canonical LIR motifs are 4 amino acids long, FYCO1 contains a C-terminally extended 9-amino acid-long F-type LIR motif with an acidic residue and a hydrophobic residue at positions 8 and 9 important for efficient binding to LC3B.
FIGURE 1.
FYCO1 contains a C-terminally extended LIR motif required for the interaction with LC3B. A, schematic representation of the domain architecture of human FYCO1 with RUN (RPIP8 (RaP2 interacting protein 8), UNC-14, and NESCA (new molecule containing SH3 at the carboxyl terminus), FYVE, LIR, and GOLD (Golgi dynamics) domains indicated. The predicted coiled-coil regions are shown as light gray boxes. Myc-tagged FYCO1 WT and F1280A/I1283A constructs in vitro translated in the presence of [35S]methionine were analyzed for binding in GST pulldown assays. Bound proteins were detected by autoradiography (AR). The integrity and relative amounts of immobilized GST and GST-LC3B used in the binding assays are shown in the Coomassie Brilliant Blue-stained gel (CBB). aa, amino acids. B, peptide array with 18-mers of FYCO1 spanning amino acids 1265–1298 to define the minimal peptide of FYCO1 able to interact with recombinant GST-LC3B. The peptide walk was done with steps of one amino acid from one spot to the next. C, a two-dimensional peptide array scan analyzing the effects of single amino acid substitutions at all positions of the indicated 18-mer peptides from FYCO1 (amino acids 1276–1293). Each position of the 18-mer peptides was replaced with all 20 amino acids. The peptide arrays were probed with 1 μg/ml GST-LC3B for 2 h, and binding to GST-LC3B was detected with anti-GST antibodies.
FYCO1 Interacts Preferentially with LC3A and -B
When a peptide spanning FYCO1 amino acids 1276–1294 was fused to the C terminus of GFP, in vitro translated and assayed against all human ATG8 homologues in a GST pulldown assay, only LC3A and -B bound strongly to the FYCO1 LIR (Fig. 2A, upper panel) (10). In vitro translated full-length FYCO1 also shows preference for LC3A and -B in GST pulldown assays (Fig. 2A, lower panel), although we see some binding to LC3C and GABARAP in vitro. However, we have earlier observed a stronger tendency for unspecific binding by GABARAP in such assays (10). The preferential binding to LC3A and -B was confirmed in vivo by co-immunoprecipitation experiments of Myc-tagged FYCO1 and GFP fusions of human ATG8 family proteins expressed in HEK293 cells (Fig. 2B). GFP-LC3A and GFP-LC3B were efficiently co-precipitated with Myc-FYCO1, whereas GFP-LC3C, GFP-GABARAP, GFP-GABARAPL1 and -L2 were not. We conclude that FYCO1 preferentially binds to LC3A and -B via its C-terminally extended F-type LIR motif. Consistently, we could also show that endogenous FYCO1 co-localized with endogenous LC3A/B in cells (Fig. 2C).
FIGURE 2.
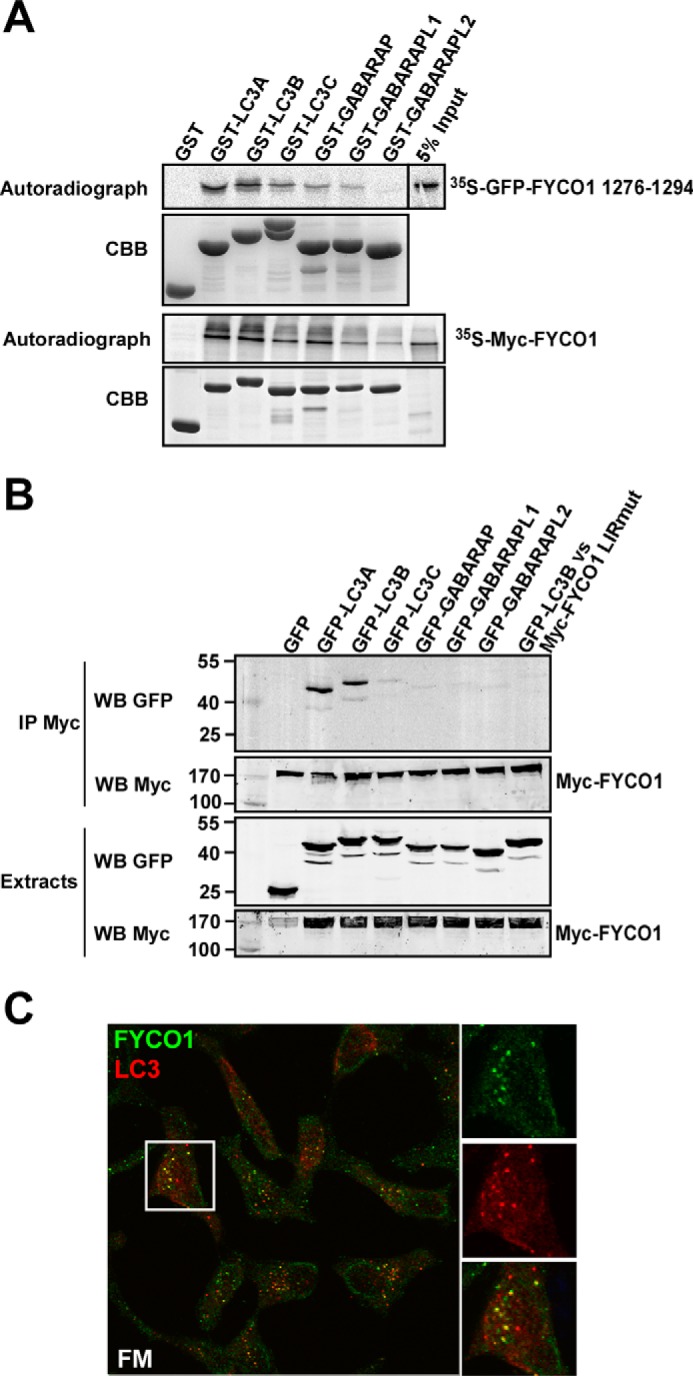
FYCO1 preferably interacts with the ATG8 homologues LC3A and -B. A, GST pulldown assays to analyze binding of an in vitro translated GFP-tagged peptide spanning FYCO1 residues 1276–1294 (upper panel) and full-length Myc-tagged FYCO1 (lower panel) against recombinant human GST-ATG8 family members. The immobilized GST fusion proteins used are displayed on the Coomassie Brilliant Blue-stained gels (CBB) below the autoradiographs. B, immunoprecipitation of GFP-tagged human ATG8 proteins co-expressed with Myc-FYCO1 or Myc-FYCO1 LIR point mutant (LIRmut) in human HEK293 cells. Myc-tagged full-length FYCO1 was immunoprecipitated (IP) with Myc antibodies. The inputs of the various GFP-tagged ATG8 proteins (lower panel), the expression level and precipitated Myc-FYCO1 (middle panels), and the co-precipitated GFP-ATG8 proteins (upper panel) were analyzed by Western blotting (WB) using the indicated anti-GFP and anti-Myc antibodies. C, endogenous FYCO1 and LC3 co-localize in puncta in HeLa cells (FM; full medium).
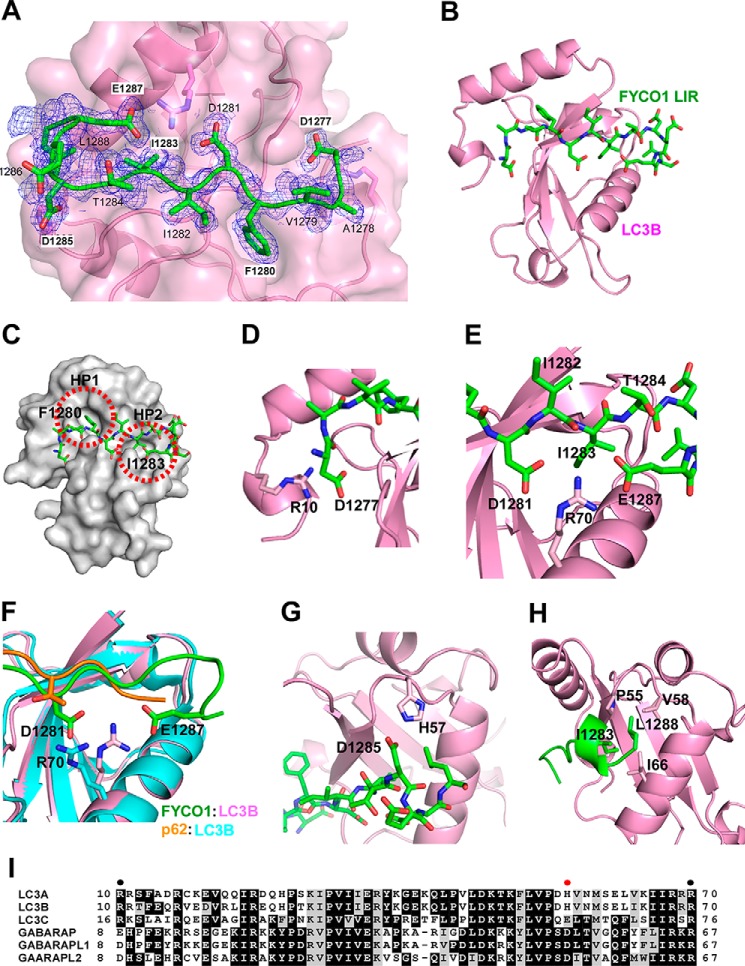
Structure of the LC3B-FYCO1 LIR Complex
To further explore the molecular mechanism responsible for the preferred binding of FYCO1 to LC3A and -B, we determined the structure of the LC3B-FYCO1 LIR complex by x-ray crystallography. The complex consists of full-length LC3B-(1–125) bound to a 13-amino acid LIR peptide of FYCO1 (residues 1276–1288). The crystal structure of the LC3B-FYCO1 LIR complex was determined by molecular replacement using the crystal structure of the LC3B monomer (residues 5–120, PDB code 1UGM), and refined to 1.53 Å resolution. The structure of LIR-bound LC3B (Fig. 3, A and B), which consists of a five-stranded β-sheet and five α-helices, is essentially identical to the previously reported structures of peptide-free LC3B and p62 LIR-bound LC3B (11, 31, 35); these structures have an average 0.7 and 1.0 Å root mean square deviation for the C-α positions, respectively. FYCO1 LIR binds within the LC3 groove in an extended conformation and forms a short 310 helix at the C-terminal containing residues Asp1285 to Glu1287 (Fig. 3, A and B). The FYCO1-LIR-binding surface of LC3B consists of two loops (β1-β2 and β2-α3), two α-helices (α2 and α3), and two β-strands (β1 and β2). The side chains of the core FYCO1-LIR residues (Phe1280 and Ile1283) are bound deeply into the two hydrophobic pockets (HP1 and HP2) of LC3B (Fig. 3C), similar to that observed previously for other canonical LIR interactions (5, 36). Moreover, the LC3B-FYCO1 LIR complex structure displays three remarkable electrostatic interactions: 1) Arg10 of LC3B with Asp1277 of FYCO1 (Fig. 3D); 2) Arg70 of LC3B with Asp1281 and Glu1287 of FYCO1 (Fig. 3, E and F); and 3) His57 of LC3B with Asp1285 of FYCO1 (Fig. 3G). These would potentially form a series of salt bridges between the two molecules. In particular, the structure suggests that Arg70 plays an important role for the interaction between LC3B and FYCO1. However, Arg70 is conserved in GABARAP, -L1, -L2, LC3A, -B, and -C and cannot therefore explain the preferential binding of FYCO1 to LC3A and -B. It should be noted that the conformation of Arg70 in the LC3B-FYCO1 LIR complex is different from that of the LC3B-p62 LIR complex (Fig. 3F). Arg70 does not form strong interactions with the acidic residues of the p62 LIR. Contrary to Arg70, His57 is only conserved in LC3A and -B. Hence, the His57 residue is likely also contributing to the specific interaction of FYCO1 with LC3A and -B. Finally, the most C-terminally located important residue of the FYCO1 LIR determined from the two-dimensional peptide array mutational analysis, Leu1288, makes hydrophobic interactions with Pro55, Val58, and Ile66 of LC3B (Fig. 3H). A sequence alignment of human ATG8 proteins shows that His57 is found only in LC3A and -B and that Arg10 is not present in GABARAP proteins (Fig. 3I).
FIGURE 3.
Structure of FYCO1 LIR peptide bound to LC3B. A, electron density map of FYCO1 peptide on LC3. The Fo − Fc omit map of the FYCO1 peptide is contoured at 0.6 σ. B, overview of the LIR peptide (residues 1276–1288)(green) bound to LC3B (1–125)(magenta). Note that the structure in B is rotated 180 degrees relative to A. C, the side chains of the aromatic Phe1280 residue and the hydrophobic Ile1283 residues dock into hydrophobic pockets HP1 and HP2 of the LIR docking site of LC3B. D, detail of the electrostatic interaction between Arg10 in the N-terminal arm of LC3B and Asp1277 located N-terminal to the core LIR. E, both Glu1287 and Asp1281 of the LIR engage in electrostatic interactions with Arg70 of LC3B. F, this interaction involving Arg70 is not seen in the p62 LIR-LC3B complex as visualized in the structural comparison. G, the electrostatic interaction between FYCO1 Asp1285 and His57 in LC3B is likely a specificity determinant important for the preferential binding of FYCO1 to LC3A and -B. H, the most C-terminal Leu1288 residue in the crystalized LIR peptide engages in hydrophobic interactions with Pro55, Val58, and Ile66 of LC3B. I, sequence alignment of human ATG8 proteins generated with Clustal Omega (46). EPS image was generated with BOXSHADE. Shading indicates similarity. Solid circles indicate residues in LC3B required for interaction with FYCO1 LIR. Red solid circle indicates residue in LC3A/B conferring preference toward FYCO1 interaction.
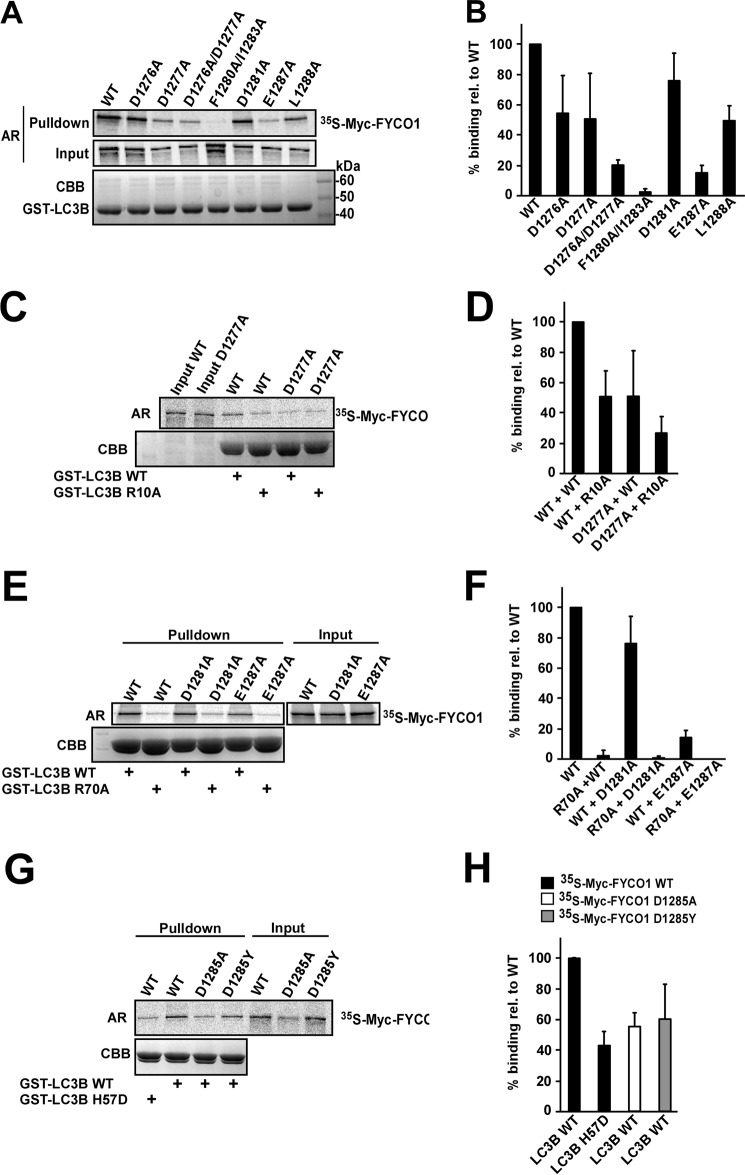
The C-terminally Extended FYCO1 LIR Is Required for Both Binding Strength and Selective Binding to LC3A/B
Analyses of more than 40 verified LIR motifs show that the LIR motifs (W/F/Y)XX(L/I/V) frequently contain acidic residues (Glu or Asp) or phosphorylatable residues (Ser or Thr) N- or C-terminally (or both) to the core aromatic residue (5). For the FYCO1 LIR, there is no acidic residue directly N-terminal to the core aromatic Phe1280 residue. However, mutation of Asp1277 engaging in an electrostatic interaction with Arg10 in the N-terminal arm of LC3B (Fig. 4C), and also its neighbor Asp1276, had a negative effect on binding. This was particularly evident for the D1276A/D1277A double mutant, as assayed by GST pulldown assays (Fig. 4, A and B). The importance of the electrostatic interaction between LC3B Arg10 and Asp1277 revealed from the structure was confirmed by GST pulldown analyses showing that mutation of either Arg10 or Asp1277 to Ala had a similar negative effect (50% of WT), whereas mutation of both residues to Ala reduced the binding proportionally to about 25% of WT (Fig. 4, C and D). The binding of D1276A and D1277A mutants varied in several experiments. A possible explanation may be that they compensate for each other. This may explain why the double mutant had a more dramatic effect than the single mutants.
FIGURE 4.
Mutational analyses reveal the Glu1287--Arg70 interaction to be crucial for binding of the FYCO1 LIR to LC3B. A, GST pulldown assays of in vitro translated, [35S]methionine-labeled, full-length, Myc-tagged WT FYCO1 and FYCO1 LIR mutants against recombinant WT GST-LC3B. The autoradiograph (AR) of the gel with 10% of the inputs (Input) to the binding reactions is shown below the autoradiograph of the gel displaying the result of the pulldowns (Pulldown). The Coomassie Brilliant Blue-stained gel of the immobilized GST-LC3B is shown in the lower panel (CBB). B, quantifications of the experiments shown in A. C, GST pulldown assays of in vitro translated, [35S]methionine-labeled, full-length, Myc-tagged FYCO1 WT or D1277A mutant against recombinant GST-LC3B WT or the R10A mutant. D, quantifications of the interactions of the mutants in C relative to WT. E, GST pulldown assays of in vitro translated FYCO1 WT or D1281A or E1287A mutants against recombinant GST-LC3B WT or the R70A mutant. F, quantifications of the interactions of the mutants in E relative to WT. G, GST pulldown assays of in vitro translated FYCO1 WT or D1285A mutant against recombinant GST-LC3B WT or the H57D mutant. H, quantifications of the interactions of the mutants in G relative to WT. For all quantifications shown in B, D, F, and H, mean percentage of binding relative to WT with S.D. from at least three independent experiments is shown for each mutant analyzed. Input in A, C, E, and G, always refers to 10% of the total input to the binding reactions.
Directly C-terminal to Phe1280 lies also the acidic Asp1281 residue. The charge of this residue is not absolutely crucial for the LC3B binding because the D1281A mutant still retained binding in the pulldown assay (Fig. 4, E and F). However, the two-dimensional peptide array clearly suggested that Asp or Glu is preferred at this position (Fig. 1C). The requirement for the C-terminally extended LIR and crucial role of Glu1287 is clearly seen because the E1287A mutant was almost as detrimental as the double mutant of the core LIR F1280A/I1283A (Fig. 4, A and B). The L1288A mutant also had a clear negative effect on the binding, confirming its importance as the most C-terminal residue in the 9-amino acid-long FYCO1 LIR (Fig. 4, A and B). It is also likely that the Ala mutant is the least severe mutation in this position as judged from the peptide array and structure of the LC3B-FYCO1 LIR complex (Figs. 1C and 3H). The crystal structure shows that both Asp1281 and Glu1287 engage in electrostatic interactions and form hydrogen bonds with Arg70 in the LIR docking site of LC3B (Fig. 3, E and F). The R70A mutant of LC3B completely abolished binding to FYCO1 in the GST pulldown assays. The D1281A mutant was less affected, whereas E1287A only retained minimal binding and no binding at all was measured with LC3B R70A versus FYCO1 E1287A (Fig. 4, E and F). Because FYCO1 Asp1285 and LC3B His57 engage in an electrostatic interaction, we tested the importance of this interaction by pulldown assays. His57 was mutated to Asp, which is present in the corresponding position of GABARAP family proteins. An ∼50% reduction in binding was observed for both FYCO1 D1285A versus WT LC3B and -B H57D versus FYCO1 WT (Fig. 4, G and H). Taken together, the mutational analyses show the Glu1287 residue of the extended LIR motif to be essential for binding to LC3B via its interaction with Arg70, and His57 is essential for the preferred binding to LC3A/B.
Generation of Flp-In T-REx HEK293 Cells Knocked Out for FYCO1
We employed the zinc finger nuclease strategy targeting the large exon 8 of the human FYCO1 gene on chromosome 3p21.3 to generate a HEK293 Flp-In T-REx cell line lacking expression of FYCO1 (Fig. 5A). The nuclease cut is at amino acid position 295 of the coding sequence. The presence of a deletion in the FYCO1 gene was verified using the surveyor kit from Transgenomic, Inc. The expected bands with sizes of 310, 182, and 128 bp were present, confirming cleavage by the CEL-1 enzyme (Fig. 5B). Loss of FYCO1 protein expression was confirmed by Western blotting with an antibody against FYCO1 (Fig. 5C). Three cell lines lacking FYCO1 expression were generated. We chose clone 12 for our further studies and denoted it FYCO1 KO.
FIGURE 5.
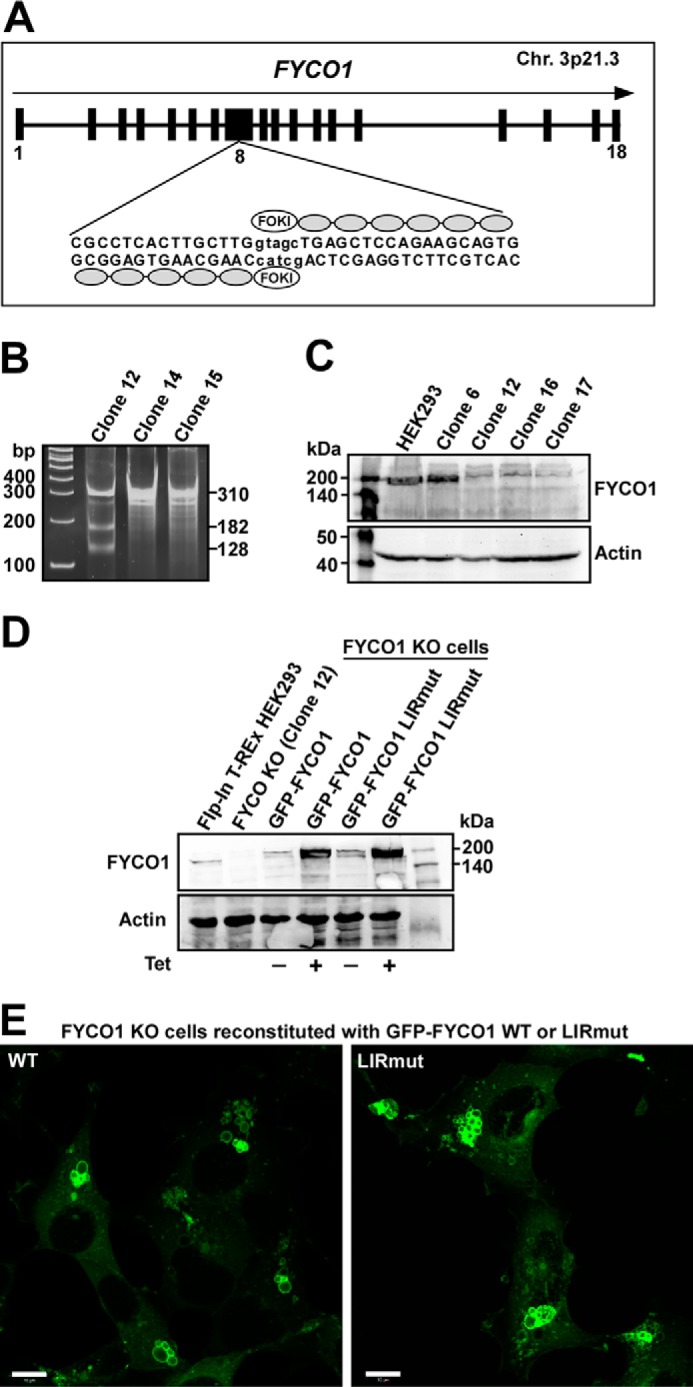
Generation of FYCO1 knock-out HEK293 Flp-In T-Rex cell lines using zinc finger nuclease technology. A, exon-intron structure of the human FYCO1 gene, showing the zinc finger nuclease recognition sequence and FokI cutting site in exon number 8. Chr. 3p21.3, chromosome 3p21.3. B, Surveyor assay (CelI cleavage) indicating deletion in the FYCO1 gene in different cell clones. C, Western blot of control and FYCO1 knock-out cell lines probed with antibodies against FYCO1. D, Western blot of control and FYCO1 knock-out (KO) cell lines reconstituted with tetracycline-inducible GFP-FYCO1 WT and GFP-FYCO1 F1280A/1283A (GFP-FYCO1 LIRmut). Levels of FYCO1 both with and without tetracycline (1 μg/ml) added were measured. E, confocal fluorescence microscopy images of FYCO1 KO cells reconstituted with GFP-FYCO1 WT or GFP-FYCO1 LIRmut. Scale bars, 10 μm.
A Functional LIR Motif of FYCO1 Is Required for Efficient Maturation of Autophagosomes
The generation of the FYCO1 KO cells allowed us to stably express inducible mutants of FYCO1 in a FYCO1 null background. FYCO1 KO cells were reconstituted with tetracycline-inducible Flp-In GFP-FYCO1 WT and the F1280A/I1283A LIR mutant (LIRmut) constructs (Fig. 5D). Without tetracycline induction, the expression levels of GFP-FYCO1 WT and GFP-FYCO1 LIRmut constructs were comparable with that of endogenous FYCO1 in the parental Flp-In T-REx HEK293 cells (Fig. 5D).
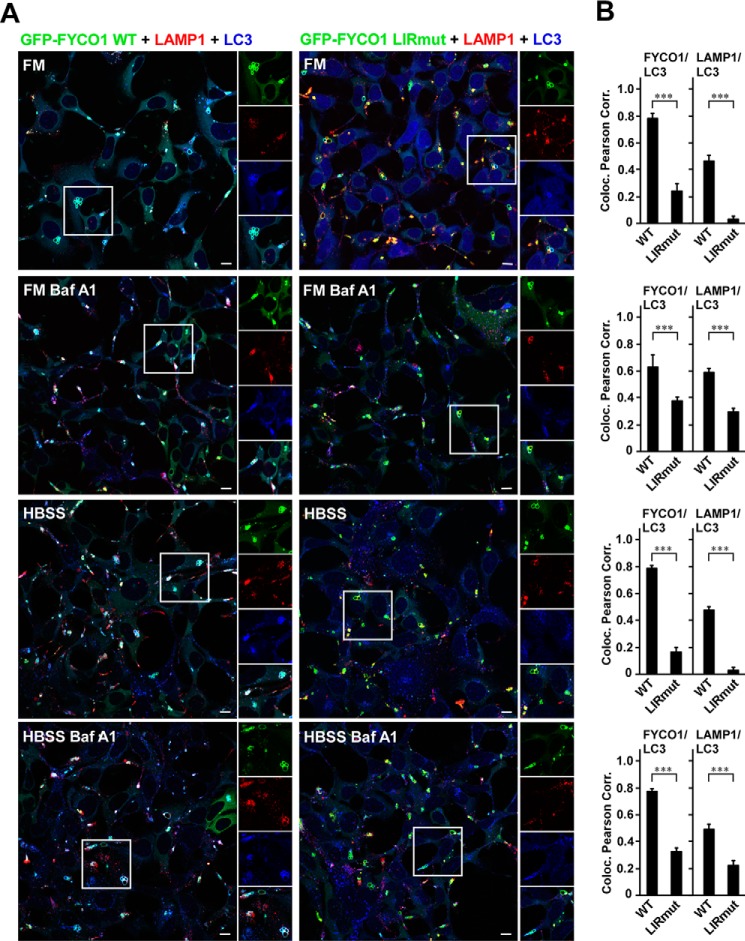
The interaction of FYCO1 with LC3 is essential for its role in the maturation of LC3-positive phagosomes into LAMP1-positive structures (24). Because the LIR motif is also used for binding of FYCO1 to autophagosomes (21), we next examined the subcellular localization pattern of GFP-FYCO1 WT and GFP-FYCO1 LIRmut in the reconstituted KO cell lines without the interference of endogenous FYCO1. To be able to clearly see the GFP signal in these cells, we induced the expression with tetracycline. WT and LIRmut GFP-FYCO1 both localized to clusters of relatively large vesicular structures (Fig. 5E), likely resulting from homotypic fusion events. FYCO1 is localized on the ring perimeters, and the vesicles vary in diameter from 2 μm in HeLa cells to 15 μm in HEK293 cells. Also, the co-localization with LAMP1 was unchanged for WT and LIR mutant GFP-FYCO1 (data not shown). However, both the co-localization of FYCO1 with LC3 and that of LC3 with LAMP1 were dramatically reduced in the LIRmut reconstituted cells relative to the WT reconstituted cells (Fig. 6, A and B). This was the case both in full medium and upon serum and amino acid starvation for 2 h. When the cells were treated with bafilomycin A1, to inhibit fusions between autophagosomes and lysosomes and the lysosomal acidification, the differences between FYCO1 WT and LIRmut were less prominent. This is likely due to accumulation of LC3-positive autophagosomes that co-cluster with FYCO1-positive vesicles in a LIR-independent manner.
FIGURE 6.
LC3 co-localization with FYCO1 and with LAMP1 is dependent on a functional LIR motif in FYCO1. A, FYCO1 KO HEK293 cells reconstituted with tetracycline-inducible GFP-FYCO1 WT or GFP-FYCO1 LIRmut were stained with antibodies against LC3 and LAMP1. B, co-localization between FYCO1 versus LC3, and LAMP1 versus LC3 shown as Pearson correlation (Coloc. Pearson Corr.). Quantifications of co-localization were based on >100 cells and performed using the Volocity (PerkinElmer) software. HBSS, Hank's balanced salt solution. Error bars denote S.D. ***, p < 0.001 (unpaired t test). Scale bars, 10 μm.
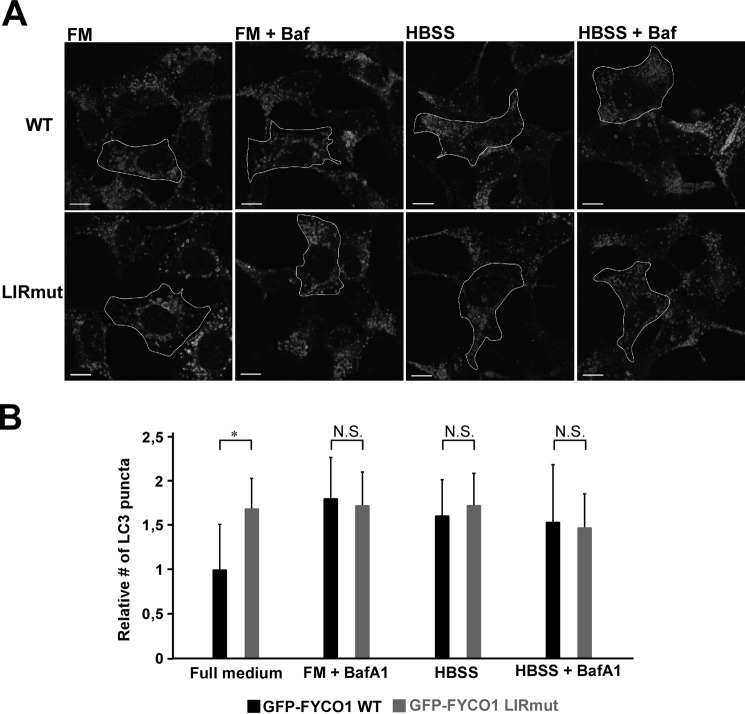
We then compared the levels of the lipidated LC3-II form in the four different cell lines by Western blotting. For these experiments, the reconstituted cell lines were not induced with tetracycline to keep the expression levels as close to endogenous levels as possible. Both in the FYCO1 KO cell line and in the LIRmut cell line, LC3-II levels were increased in the full medium situation (Fig. 7, A and B). This may suggest either an increase in formation of lipidated LC3 or an inhibition of clearance. To distinguish between these possibilities, we treated the cells with bafilomycin A1 for 6 h to block lysosomal acidification and halt degradation. This resulted in a lower increase in LC3-II levels in the KO and LIRmut cell lines as compared with parent HEK293 and KO cells reconstituted with GFP-FYCO1 WT (Fig. 7, A and B). Therefore, the increased LC3-II level in full medium was not caused by increased formation of LC3-II, but rather inhibition of clearance. To visualize this effect on autophagic flux more clearly, we calculated the amount of LC3-II accumulated upon bafilomycin A1 treatment by subtracting the LC3B-II level without bafilomycin A1 from the LC3-II level in the bafilomycin A1-treated sample. Strikingly, in cells lacking FYCO1, the flux was greatly inhibited, seen as strongly reduced accumulation of LC3-II. The same effect was also seen in FYCO1 KO cells reconstituted with GFP-FYCO1, either WT or LIRmut (Fig. 7C). Additionally, we quantified the amount of endogenous LC3 puncta in the uninduced cells. Here, we found that for the LIRmut cells, the number of puncta in the full medium situation was higher than for the WT cells. Because the number of LC3 puncta of FYCO1 WT and LIRmut cells was equal upon bafilomycin A1 treatment, we conclude that there is a maturation defect in the LIRmut cells (Fig. 8). Taken together, these data indicate that, in the full medium situation, the LIRmut cells have a reduced ability to clear autophagosomes.
FIGURE 7.
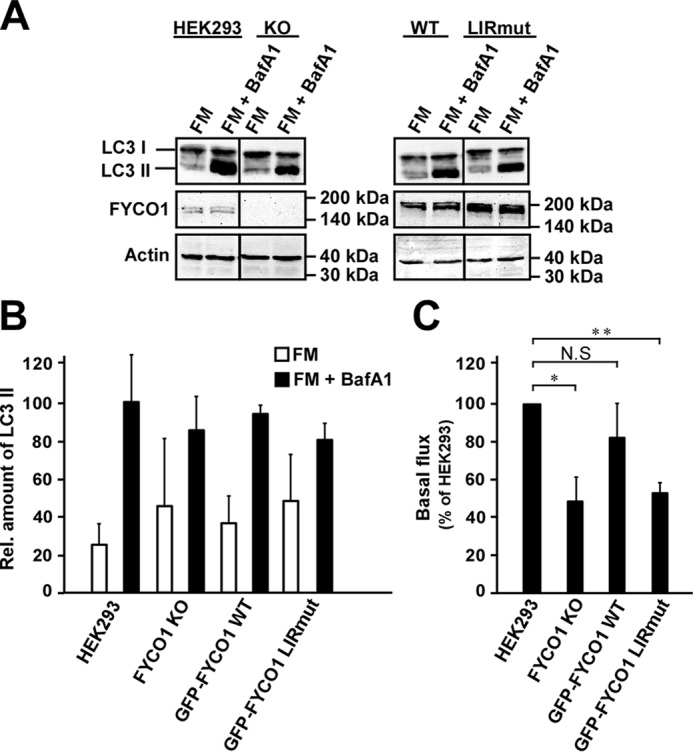
FYCO1 is required for efficient maturation of autophagosomes in a LIR-dependent manner during basal autophagy. A, Western blots of LC3A/B in total extracts from HEK293 WT, FYCO1 KO, and reconstituted KO cells with GFP-FYCO1 WT and LIRmut, respectively. FM, full medium; BafA1, bafilomycin A1. B, relative levels of LC3 II normalized to the actin loading control. The blots were quantified using ImageJ. Error bars denote S.D. from three independent experiments. C, autophagy flux in full medium calculated by subtracting the LC3 II level without bafilomycin A1 from the LC3 II level in the bafilomycin A1-treated samples. Flux for each experiment was displayed as the percentage of the flux in the control HEK293 cell line. *, p < 0.05, **p < 0.01 (one-sample t test). NS, not significant.
FIGURE 8.
Cells expressing FYCO1 with mutated LIR show reduced autophagosome maturation. A, immunofluorescence images of FYCO1 KO HEK293 Flp-In T-Rex cells reconstituted with tetracycline-inducible GFP-FYCO1 WT or GFP-FYCO1 LIRmut stained with antibodies against LC3. To keep the expression levels close to the endogenous level, the cells were not induced by tetracycline. Cells were left in full medium (FM) with and without bafilomycin A1 (Baf) for 6 h, or Hanks' balanced salt solution (HBSS) with and without bafilomycin A1 for 2 h. B, quantification of relative number of LC3 puncta per cell area based on scoring >70 cells using the Volocity (PerkinElmer) software. BafA1, bafilomycin A1. Error bars denote S.D. *, p < 0.05 (unpaired t test). Scale bars, 10 μm. N.S., not significant.
Discussion
We showed previously that FYCO1 induces microtubule plus end transport of LC3-positive autophagic vesicles (21). In that study, we also noted that FYCO1 has a canonical F-type core LIR motif with a phenylalanine at the aromatic position and an isoleucine at the hydrophobic position. The LIR motif is generally defined as having the core consensus sequence (W/F/Y)XX(L/I/V) (5). Here, we show that FYCO1 preferentially interacts with LC3A and -B, and that the functional LIR extends with five residues C-terminal to the hydrophobic position of the core LIR. Detailed analyses, including data obtained from two-dimensional peptide array-based mutation scanning, x-ray crystallography, and mutation analyses by GST pulldown assays, allow us to firmly conclude that the C-terminal extension is involved in determining both binding strength and specificity toward LC3A and -B. In particular, Asp1285 of FYCO1 provide specificity toward binding to LC3A and -B by interacting with His57 in LC3B. The corresponding residue Asp54 of GABARAP, GABARAPL1 and -L2, and Glu63 of LC3C would lead to charge repulsion and act unfavorably for interaction with Asp1285 of FYCO1. ALFY (autophagy-linked FYVE protein, also called WDFY3) is a large phosphatidyl-inositol 3-phosphate-binding protein interacting with the autophagy receptors p62/SQSTM1 and NBR1 and helps to clear ubiquitinated protein aggregates by autophagy (37, 38). ALFY binds preferentially to the GABARAP subfamily of ATG8 proteins (18). This specificity is in part mediated by a tyrosine residue (Tyr3351) C-terminal to the core LIR at the corresponding positon of Asp1285 in FYCO1. Tyr3351 interacts with Asp54 in GABARAPs, but is sterically hindered by His57 of LC3B (18). Hence, in both these LIRs, a residue located two residues C-terminal to the conserved hydrophobic position of the core LIR is involved in distinguishing between the LC3A and -B and GABARAP subfamilies.
Many LIR motifs contain acidic residues N-terminally to the core aromatic residue that engage in electrostatic interactions with basic residues in the N-terminal arm of ATG8s (5). Asp3344 of the ALFY-LIR is able to form ionic interactions with Lys24 and Tyr25 of GABARAP, but not with the corresponding Gln26 and His27 of LC3B. This way these residues also contribute to the preferential binding to GABARAPs (18). For FYCO1, Asp1277 binds to Arg10 in the N-terminal arm of LC3B, and a double mutation of both Asp1277 and Asp1276 reduced binding to LC3B by 80%. Clearly, these two N-terminal residues are important for the binding affinity and may also impact on the specificity as seen for the ALFY-LIR.
The most dramatic mutation affecting the binding of FYCO1 to LC3B, apart from the core LIR F1280A/I1283A double mutation, was the E1287A mutant. This residue is located four residues C-terminal to the conserved core hydrophobic residue and interacts electrostatically with Arg70 in LC3B. The E1287A mutation resulted in an 85% reduction in binding. The crucial importance of this ionic interaction is underscored by the peptide array scan showing that the only allowed substitution was to the other acidic residue, i.e. E1287D. Because the Arg70 residue is conserved in GABARAPs too, this is not an interaction contributing to the specificity, but it is crucial for binding strength for the FYCO1-LC3B interaction. Our two-dimensional mutation analysis by peptide array scan also showed that Leu1288, the most C-terminal residue interacting with LC3B, can only be substituted with the other hydrophobic residues, Ile, Val, and Phe, and this residue interacts with other hydrophobic residues in LC3B. However, based on sequence conservation between ATG8 family members at these positions, it is not likely that these interactions contribute to the binding preference.
The presence of at least six different human ATG8 homologues is intriguing, as yeast has only one Atg8 and Caenorhabditis elegans and Drosophila have two. The degree of redundancy relative to specific individual roles for the mammalian ATG8 family members is only beginning to be studied. Based on knockdown and overexpression studies in HeLa cells, a division of labor between LC3B for initiation and GABARAPL2 for completion of autophagosomes has been proposed (8). On the other hand, studies in C. elegans indicate that the GABARAP-like ATG8 homologue LGG-1 is required for autophagosome formation and the LC3 homologue LGG-2 is required for fusion with the lysosome (39). In knockdown studies, the LC3 subfamily has been found to be dispensable for starvation-induced autophagy in hepatocytes and prostate cancer cells, whereas the GABARAP subfamily is required (40). This, as well as the fact that most autophagy studies in mammals have had a focus on LC3B, has motivated us to ask whether LIR-containing proteins have any specificity toward any of the six human ATG8 homologues. A handful of studies have identified LIR-containing proteins with a preference for the GABARAP subfamily, such as the scaffold protein ALFY involved in selective autophagy (18), and proteins of the Unc-51 like kinase (ULK) complex (10). These proteins interact with ATG8s on the outer surface of the phagophore/autophagosome. Also, Rab effectors such as FYCO1 and Rab GTPase-activating proteins interact with LC3/GABARAP on the outer surface (21, 22, 41, 42). The preference of FYCO1 for the LC3 subfamily is therefore interesting, and FYCO1 is as far as we know the only LIR-containing protein with a preference for LC3A and -B. Another group of LIR-containing proteins are the autophagy receptors that interact with LC3/GABARAP on the inside of the forming phagophore, and end up being degraded by autophagy (reviewed in Refs. 4 and 5). Among the autophagy receptors, there are different binding preferences, arguing against the idea that a specific subset of ATG8 homologues is responsible for the attachment of cargos to the inner membrane. The autophagy receptor p62/SQSTM1 binds very well to all ATG8 family proteins in in vitro binding studies. However, for selective autophagy of p62/SQSTM1, it has been found that LC3B is required and not the GABARAP subfamily (43, 44).
We previously showed that FYCO1 uses its LIR motif to bind to autophagosomes (21). Knockdown of FYCO1 in HeLa cells stably expressing GFP-LC3B leads to a perinuclear clustering of GFP-LC3B under basal conditions (21). This indicated a role for FYCO1 in plus end-directed transport of phagophores and/or autophagosomes. To look more specifically at the importance of the LIR motif, in this study, we made FYCO1 knock-out cell lines stably expressing wild type or LIR mutated FYCO1. Our data confirmed that the LIR motif is required for co-localization of FYCO1 with autophagosomes, and expression of a LIR mutant construct negatively affected maturation of autophagosomes under basal conditions. The effects we see on late steps of basal autophagy on mutation of the FYCO1 LIR motif correlate with a role of FYCO1 in regulating kinesin-mediated transport of LC3-positive autophagic structures. The effects we observed by mutating the FYCO1 LIR motif were seen only for cells grown in full medium. We have recently shown that FYCO1 interacts with kinesin 1 (26), and it has been demonstrated that anterograde movement of autophagosomes is dependent on kinesin 1 only when cells are grown in full medium (45). This supports our finding that we see the effect on maturation in basal autophagy when cells are fed, and not during starvation.
Author Contributions
H. L. O. and T. J. designed and analyzed experiments, conceived and coordinated the study, and wrote the paper. H. L. O. also performed experiments. T. L. designed and analyzed experiments and contributed to writing and editing of the paper. K. B. L., G. E., and A. Ø performed and analyzed experiments. K. T. and T. M. designed and performed experiments, analyzed data, and contributed to the writing and editing of the paper.
Acknowledgments
We thank Masaaki Komatsu (Niigata University) for helpful discussions and the Bioimaging core facility at the Institute of Medical Biology (UiT – The Arctic University of Norway) for the use of instrumentation and expert assistance. X-ray data collection was performed on synchrotron beamline BL44XU at SPring-8 under the Cooperative Research Program of the Institute for Protein Research, Osaka University (proposals 2014A6952 and 2014B6952).
This work was supported by grants from the FRIBIO and FRIBIOMED programs of the Norwegian Research Council (Grants 196898 and 214448) and by the Norwegian Cancer Society (Grant 71043-PR-2006-0320) (to T. J.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 5D94) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- ATG
- autophagy-related
- FYCO1
- FYVE and coiled-coil protein 1
- GABARAP
- γ-aminobutyric acid receptor-associated protein
- GABARAPL1–2
- γ-aminobutyric acid receptor-associated protein-like 1–2
- LAMP1
- lysosome-associated membrane protein 1
- LC3A–C
- microtubule-associated protein 1 light chain 3A–C
- LIR
- LC3-interacting region
- ALFY
- autophagy-linked FYVE protein
- HP
- hydrophobic pocket.
References
- 1.Mizushima N., and Komatsu M. (2011) Autophagy: renovation of cells and tissues. Cell 147, 728–741 [DOI] [PubMed] [Google Scholar]
- 2.Shpilka T., Weidberg H., Pietrokovski S., and Elazar Z. (2011) Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol. 12, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johansen T., and Lamark T. (2011) Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogov V., Dötsch V., Johansen T., and Kirkin V. (2014) Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell 53, 167–178 [DOI] [PubMed] [Google Scholar]
- 5.Birgisdottir Å. B., Lamark T., and Johansen T. (2013) The LIR motif: crucial for selective autophagy. J. Cell Sci. 126, 3237–3247 [DOI] [PubMed] [Google Scholar]
- 6.Xie Z., Nair U., and Klionsky D. J. (2008) Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 19, 3290–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujita N., Hayashi-Nishino M., Fukumoto H., Omori H., Yamamoto A., Noda T., and Yoshimori T. (2008) An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol. Biol. Cell 19, 4651–4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weidberg H., Shvets E., Shpilka T., Shimron F., Shinder V., and Elazar Z. (2010) LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 29, 1792–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., and Johansen T. (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 10.Alemu E. A., Lamark T., Torgersen K. M., Birgisdottir A. B., Larsen K. B., Jain A., Olsvik H., Øvervatn A., Kirkin V., and Johansen T. (2012) ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs. J. Biol. Chem. 287, 39275–39290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ichimura Y., Kumanomidou T., Sou Y. S., Mizushima T., Ezaki J., Ueno T., Kominami E., Yamane T., Tanaka K., and Komatsu M. (2008) Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 283, 22847–22857 [DOI] [PubMed] [Google Scholar]
- 12.Noda N. N., Kumeta H., Nakatogawa H., Satoo K., Adachi W., Ishii J., Fujioka Y., Ohsumi Y., and Inagaki F. (2008) Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 13, 1211–1218 [DOI] [PubMed] [Google Scholar]
- 13.Satoo K., Noda N. N., Kumeta H., Fujioka Y., Mizushima N., Ohsumi Y., and Inagaki F. (2009) The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 28, 1341–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rozenknop A., Rogov V. V., Rogova N. Y., Löhr F., Güntert P., Dikic I., and Dötsch V. (2011) Characterization of the interaction of GABARAPL-1 with the LIR motif of NBR1. J. Mol. Biol. 410, 477–487 [DOI] [PubMed] [Google Scholar]
- 15.von Muhlinen N., Akutsu M., Ravenhill B. J., Foeglein Á., Bloor S., Rutherford T. J., Freund S. M., Komander D., and Randow F. (2012) LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol. Cell 48, 329–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma P., Schwarten M., Schneider L., Boeske A., Henke N., Lisak D., Weber S., Mohrlüder J., Stoldt M., Strodel B., Methner A., Hoffmann S., Weiergräber O. H., and Willbold D. (2013) Interaction of Bcl-2 with the autophagy-related GABAA receptor-associated protein (GABARAP): biophysical characterization and functional implications. J. Biol. Chem. 288, 37204–37215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki H., Tabata K., Morita E., Kawasaki M., Kato R., Dobson R. C., Yoshimori T., and Wakatsuki S. (2014) Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism. Structure 22, 47–58 [DOI] [PubMed] [Google Scholar]
- 18.Lystad A. H., Ichimura Y., Takagi K., Yang Y., Pankiv S., Kanegae Y., Kageyama S., Suzuki M., Saito I., Mizushima T., Komatsu M., and Simonsen A. (2014) Structural determinants in GABARAP required for the selective binding and recruitment of ALFY to LC3B-positive structures. EMBO Rep. 15, 557–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McEwan D. G., Popovic D., Gubas A., Terawaki S., Suzuki H., Stadel D., Coxon F. P., Miranda de Stegmann D., Bhogaraju S., Maddi K., Kirchof A., Gatti E., Helfrich M. H., Wakatsuki S., Behrends C., Pierre P., and Dikic I. (2015) PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell 57, 39–54 [DOI] [PubMed] [Google Scholar]
- 20.Genau H. M., Huber J., Baschieri F., Akutsu M., Dötsch V., Farhan H., Rogov V., and Behrends C. (2015) CUL3-KBTBD6/KBTBD7 ubiquitin ligase cooperates with GABARAP proteins to spatially restrict TIAM1-RAC1 signaling. Mol. Cell 57, 995–1010 [DOI] [PubMed] [Google Scholar]
- 21.Pankiv S., Alemu E. A., Brech A., Bruun J. A., Lamark T., Øvervatn A., Bjørkøy G., and Johansen T. (2010) FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 188, 253–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu M. M., Nirschl J. J., and Holzbaur E. L. (2014) LC3 binding to the scaffolding protein JIP1 regulates processive dynein-driven transport of autophagosomes. Dev. Cell 29, 577–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J., Ma Z., Jiao X., Fariss R., Kantorow W. L., Kantorow M., Pras E., Frydman M., Pras E., Riazuddin S., Riazuddin S. A., and Hejtmancik J. F. (2011) Mutations in FYCO1 cause autosomal-recessive congenital cataracts. Am. J. Hum. Genet. 88, 827–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma J., Becker C., Reyes C., and Underhill D. M. (2014) Cutting edge: FYCO1 recruitment to dectin-1 phagosomes is accelerated by light chain 3 protein and regulates phagosome maturation and reactive oxygen production. J. Immunol. 192, 1356–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mrakovic A., Kay J. G., Furuya W., Brumell J. H., and Botelho R. J. (2012) Rab7 and Arl8 GTPases are necessary for lysosome tubulation in macrophages. Traffic 13, 1667–1679 [DOI] [PubMed] [Google Scholar]
- 26.Raiborg C., Wenzel E. M., Pedersen N. M., Olsvik H., Schink K. O., Schultz S. W., Vietri M., Nisi V., Bucci C., Brech A., Johansen T., and Stenmark H. (2015) Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature 520, 234–238 [DOI] [PubMed] [Google Scholar]
- 27.Lamark T., Perander M., Outzen H., Kristiansen K., Øvervatn A., Michaelsen E., Bjørkøy G., and Johansen T. (2003) Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J. Biol. Chem. 278, 34568–34581 [DOI] [PubMed] [Google Scholar]
- 28.Kirkin V., Lamark T., Sou Y. S., Bjørkøy G., Nunn J. L., Bruun J. A., Shvets E., McEwan D. G., Clausen T. H., Wild P., Bilusic I., Theurillat J. P., Øvervatn A., Ishii T., Elazar Z., Komatsu M., Dikic I., and Johansen T. (2009) A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516 [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 30.Vagin A., and Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 31.Sugawara K., Suzuki N. N., Fujioka Y., Mizushima N., Ohsumi Y., and Inagaki F. (2004) The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells 9, 611–618 [DOI] [PubMed] [Google Scholar]
- 32.Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 33.Murshudov G. N., Vagin A. A., and Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 34.Kramer R. M., Roberts E. F., Um S. L., Börsch-Haubold A. G., Watson S. P., Fisher M. J., and Jakubowski J. A. (1996) p38 mitogen-activated protein kinase phosphorylates cytosolic phospholipase A2 (cPLA2) in thrombin-stimulated platelets: evidence that proline-directed phosphorylation is not required for mobilization of arachidonic acid by cPLA2. J. Biol. Chem. 271, 27723–27729 [DOI] [PubMed] [Google Scholar]
- 35.Rogov V. V., Suzuki H., Fiskin E., Wild P., Kniss A., Rozenknop A., Kato R., Kawasaki M., McEwan D. G., Löhr F., Güntert P., Dikic I., Wakatsuki S., and Dötsch V. (2013) Structural basis for phosphorylation-triggered autophagic clearance of Salmonella. Biochem. J. 454, 459–466 [DOI] [PubMed] [Google Scholar]
- 36.Noda N. N., Ohsumi Y., and Inagaki F. (2010) Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 584, 1379–1385 [DOI] [PubMed] [Google Scholar]
- 37.Clausen T. H., Lamark T., Isakson P., Finley K., Larsen K. B., Brech A., Øvervatn A., Stenmark H., Bjørkøy G., Simonsen A., and Johansen T. (2010) p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy 6, 330–344 [DOI] [PubMed] [Google Scholar]
- 38.Filimonenko M., Isakson P., Finley K. D., Anderson M., Jeong H., Melia T. J., Bartlett B. J., Myers K. M., Birkeland H. C., Lamark T., Krainc D., Brech A., Stenmark H., Simonsen A., and Yamamoto A. (2010) The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol. Cell 38, 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manil-Ségalen M., Lefebvre C., Jenzer C., Trichet M., Boulogne C., Satiat-Jeunemaitre B., and Legouis R. (2014) The C. elegans LC3 acts downstream of GABARAP to degrade autophagosomes by interacting with the HOPS subunit VPS39. Dev. Cell 28, 43–55 [DOI] [PubMed] [Google Scholar]
- 40.Szalai P., Hagen L. K., Sætre F., Luhr M., Sponheim M., Øverbye A., Mills I. G., Seglen P. O., and Engedal N. (2015) Autophagic bulk sequestration of cytosolic cargo is independent of LC3, but requires GABARAPs. Exp. Cell Res. 333, 21–38 [DOI] [PubMed] [Google Scholar]
- 41.Itoh T., Kanno E., Uemura T., Waguri S., and Fukuda M. (2011) OATL1, a novel autophagosome-resident Rab33B-GAP, regulates autophagosomal maturation. J. Cell Biol. 192, 839–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Popovic D., Akutsu M., Novak I., Harper J. W., Behrends C., and Dikic I. (2012) Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol. Cell. Biol. 32, 1733–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maruyama Y., Sou Y. S., Kageyama S., Takahashi T., Ueno T., Tanaka K., Komatsu M., and Ichimura Y. (2014) LC3B is indispensable for selective autophagy of p62 but not basal autophagy. Biochem. Biophys. Res. Commun. 446, 309–315 [DOI] [PubMed] [Google Scholar]
- 44.Shvets E., Abada A., Weidberg H., and Elazar Z. (2011) Dissecting the involvement of LC3B and GATE-16 in p62 recruitment into autophagosomes. Autophagy 7, 683–688 [DOI] [PubMed] [Google Scholar]
- 45.Geeraert C., Ratier A., Pfisterer S. G., Perdiz D., Cantaloube I., Rouault A., Pattingre S., Proikas-Cezanne T., Codogno P., and Poüs C. (2010) Starvation-induced hyperacetylation of tubulin is required for the stimulation of autophagy by nutrient deprivation. J. Biol. Chem. 285, 24184–24194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]