Background: Sterile α-motif/histidine-aspartate domain-containing protein (SAMHD1), a GTP/dGTP-activated dNTPase, plays an important role in human innate immunity, autoimmunity, and cell cycle control.
Results: We have determined a novel tetrameric SAMHD1 structure bound to GTP alone.
Conclusion: GTP-bound SAMHD1 exists as a tetramer complex that is immediately activated upon the addition of a dNTP substrate.
Significance: The present study elucidates the efficient mechanism of dNTP self-regulation of SAMHD1.
Keywords: enzyme catalysis, human immunodeficiency virus (HIV), innate immunity, lentivirus, SAM domain- and HD domain-containing protein 1 (SAMHD1), dNTPase, restriction factor
Abstract
Sterile α-motif/histidine-aspartate domain-containing protein (SAMHD1), a homo-tetrameric GTP/dGTP-dependent dNTP triphosphohydrolase, catalyzes the conversion of dNTP into deoxynucleoside and triphosphate. As the only characterized dNTP triphosphohydrolase in human cells, SAMHD1 plays an important role in human innate immunity, autoimmunity, and cell cycle control. Previous biochemical studies and crystal structures have revealed that SAMHD1 interconverts between an inactive monomeric or dimeric form and a dGTP/GTP-induced active tetrameric form. Here, we describe a novel state of SAMHD1 (109–626 amino acids, SAMHD1C) that is characterized by a rapid initial hydrolysis rate. Interestingly, the crystal structure showed that this novel SAMHD1 tetramer contains only GTP and has structural features distinct from the GTP/dNTP-bound SAMHD1 tetramer. Our work thus reveals structural features of SAMHD1 that may represent one of its biological assembly states in cells. The biochemical and structural information generated by the present study not only provides an ordered pathway for the assembly and activation of SAMHD1 but also provides insights into the potential mechanisms of the high-efficiency catalytic activity of this enzyme family in vivo.
Introduction
In dividing cells, the appropriate balance of dNTP pools is critical in minimizing mutations during DNA replication to maintain genome stability (1). However, in quiescent immune cells, the concentration of dNTP pools tends to be about 10-fold lower than that of dividing cells (2). This severe depression in the concentration of dNTP pools efficiently restricts retroviral infections such as HIV (3–6). The expression or activation of enzymes responsible for the synthesis and degradation of dNTPs influences the regulation of canonical dNTP levels. In humans, ribonucleotide triphosphate reductases catalyze the formation of deoxyribonucleotides from ribonucleotides (7, 8). Conversely, sterile α-motif/histidine-aspartate domain-containing protein (SAMHD1)4 hydrolyzes the dNTP into a deoxynucleoside and a triphosphate molecule (9–11); this protein is ubiquitously expressed in various human organs. The regulation of these enzymes is likely to contribute to the maintenance of dNTPs at different stages of the cell cycle or in specific cell types (12–14).
SAMHD1 blocks infection by retroviruses, including HIV (15, 16), and transposition of endogenous retroelements (17); it also prevents infection with certain DNA viruses such as herpes simplex virus type 1 (HSV-1) (18) and vaccinia virus (19). SAMHD1-like dNTPases are conserved from bacteria to humans and maintain dNTP pools at low levels to prevent microbial infections (9). Interestingly, the dNTPase antagonists in pathogens also exist in both prokaryotes and eukaryotes. The HIV-1 and simian immunodeficiency virus accessory protein Vpx targets SAMHD1 for ubiquitination and subsequent degradation (15, 16, 19–24). A similar antagonistic mechanism has been observed in bacteriophage T7 against Escherichia coli dGTPase (25). Furthermore, mutations in the SAMHD1 gene are associated with chronic lymphocytic leukemia (26) and autoimmunity disorders, including Aicardi-Goutières syndrome (27, 28) and systemic lupus erythematosus (29). The mutations apparently disrupt the dNTPase activity of SAMHD1, thereby causing the accumulation of nucleic acids, which in turn triggers an autoimmune response (27).
The antiviral activity of SAMHD1, particularly against HIV-1, has sparked interest because of its assembly and related catalytic mechanism (30, 31). Previous structural studies have shown that SAMHD1 is regulated by two allosteric sites, namely, a guanine-specific activator site (A1) and a dNTP-binding activator site (A2), which require GTP/dGTP as an activator to induce the formation of a tetrameric quaternary structure that hydrolyzes the substrates dATP, dTTP, dCTP, dGTP, and even dUTP (32–35). The crystal structure of the SAMHD1 catalytic core (HD domain) is in complex with mixtures of nucleotides, providing a compelling mechanism for the regulation of SAMHD1 enzymatic activity (36). It has recently been proposed that SAMHD1 can also be activated by GTP, resulting in the generation of a long-lived activated state that plays a key role in efficient dNTP depletion (37). In the present study, we discovered the loose steady state of the SAMHD1C tetramer, which has a rapid initial rate of dNTPase activity and does not require additional free GTP/dGTP to maintain its conformation in vitro. Here we present the crystal structure of SAMHD1C in complex with GTP at a resolution of 2.6 Å. The structure is the first demonstration of the existence of a relatively stable tetrameric form as reported previously (37). The present study has shown that SAMHD1C undergoes similar tetramerization and activation to the full-length wild-type protein, thereby suggesting that this tetrameric form may represent one of the main biological assembly states of SAMHD1 in cells and may be responsible for efficient dNTP depletion.
Experimental Procedures
Plasmid Construction of SAMHD1C
The DNA sequence encoding human SAMHD1 residues 109–626 (SAMHD1C) was amplified by PCR and inserted into the pET28a-Plus vector with an N-terminal His6-tagged fusion. SAMHD1C mutations were generated from the wild-type SAMHD1C plasmid using a DpnI-mediated site-directed mutagenesis kit, and mutations were confirmed by sequence analysis.
Protein Expression and Purification
BL21(DE3)Transetta E. coli cells were transformed with the SAMHD1C plasmids. A single colony of resulting cells was grown in 5 ml of LB medium containing 50 μg/ml kanamycin at 37 °C overnight. The culture was then inoculated into 1 liter of LB medium containing 50 μg/ml kanamycin and grown with shaking at 37 °C to A600 = ∼1.0. Protein expression was induced by adding isopropyl-β-d-thiogalactoside to a final concentration of 0.1 mm. Cells were grown for an additional 16 h at 20 °C. The recombinant proteins were purified by two-step liquid column chromatography, using a nickel-nitrilotriacetic acid column and a Superdex200 column (GE Healthcare). Purified proteins were stored in a gel filtration column chromatography buffer containing 20 mm Tris-HCl, pH 8.0, with 200 mm NaCl, 5 mm MgCl2, and 0.5 mm TCEP.
Crystallization and Structure Determination
SAMHD1C protein (15 mg/ml, in 20 mm Tris-HCl, pH 8.0, 200 mm NaCl, 5 mm MgCl2, and 0.5 mm TCEP) was crystallized by the hanging-drop vapor diffusion method, equilibrated against a reservoir solution of 0.2 m lithium citrate tribasic tetrahydrate, 24% PEG3350. Crystals appeared after 4 days of incubation at 20 °C. The crystals were dehydrated and frozen in a cryoprotectant solution, which consisted of Paratone-N mixed with paraffin oil at a 3:7 volume ratio. Diffraction data were collected on beam station 3W1A of the Beijing Synchrotron Radiation Facility (BSRF). The data were processed and scaled to 2.6 Å using the HKL2000 software package. The crystals of SAMHD1C were found to belong to the space group P212121, with cell dimensions of a = 80.943 Å, b = 142.765 Å, c = 199.501 Å, with four copies of SAMHD1C in the unit cell.
Structure Determination and Refinement
The initial phase was obtained by molecular replacement using one monomer of 3U1N as a template. The Phaser program (38) was able to locate four molecules in the asymmetric unit. The model was built manually using the Coot program (39) and refined using the Phenix program (40). The final structure had an Rcryst value of 18.9% and an Rfree value of 24.4%. The qualities of the final structures were evaluated using PROCHECK (41). The Ramachandran plot showed that >95% of the residues were in the most favorable region. Detailed data collection and statistics are summarized in Table 1. All figures were prepared using PyMOL. Sequence homology analysis was performed using ClustalW (42).
TABLE 1.
Data collection and refinement statistics
| SAMHD1C(109–626) | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Unit cell (Å) | a = 80.9, b = 142.8, c = 199.5 |
| Wavelength (Å) | 0.9793 |
| Resolution range (Å) | 50–2.6 (2.69–2.60) |
| No. of unique reflections | 68,937 |
| Redundancy | 6.2 (5.3)a |
| Rsym (%)b | 8.1 (49.8)a |
| I/σ | 20.0 (2.4)a |
| Completeness (%) | 95.1 (92.9)a |
| Refinement | |
| Resolution range (Å) | 30–2.6 |
| Rcrystal (%)c | 18.9 |
| Rfree (%)d | 24.4 |
| RMSDbond (Å) | 0.009 |
| RMSDangle(°) | 1.159 |
| No. of atoms | |
| Protein atoms | 14,281 |
| Ligand atoms | 100 |
| Solvent atoms | 56 |
| Ramachandran plot (%) | |
| Favored | 96.4 |
| Allowed | 3.6 |
| Disallowed | 0 |
| Average B factor (Å2) | |
| Protein | 51.7 |
| Ligand/ion | 43.9 |
| Solvent atoms | 35.6 |
a The highest resolution shell is in parentheses.
b Σj|〈I〉 − Ij/Σ〈I〉.
c Rcrystal = Σhkl|Fobs − Fcalc|/ΣhklFobs.
d Rfree, calculated the same as Rcrystal, but from a test set containing 5% of the data excluded from the refinement calculation.
dNTPase Activity Assays
dNTPase assays were carried out in a reaction buffer containing 20 mm Tris-HCl, pH 7.8, 50 mm NaCl, 5 mm MgCl2, and 500 μm dNTPs, each at 0.8 μm protein. Reactions were incubated for the indicated times at 25 °C and terminated by the addition of 0.5 m EDTA to a final concentration of 10 mm. The volume of the entire reaction system was 250 μl. Protein was separated by using an Amicon Ultra 0.5-ml 10-kDa filter (Millipore) at 12,000 × g for 20 min. The nucleotide hydrolysis reactions were analyzed by using a Venusil MP-C18 (150 × 4.6 mm) column (Agela Technologies) on a Waters HPLC system. The column was equilibrated at 40 °C in a 10 mm solution of triethylamine in water, pH 5.0 (buffer A). Injected samples were eluted with an 8-min linear gradient of methanol (buffer B) from 0 to 35%, followed by an isocratic phase of 35% buffer B over 10 min at a flow rate of 1 ml min−1. The absorbance data recorded at 260 nm were employed to calculate the amount of product in all cases.
Results
A Novel High-efficiency State of SAMHD1C
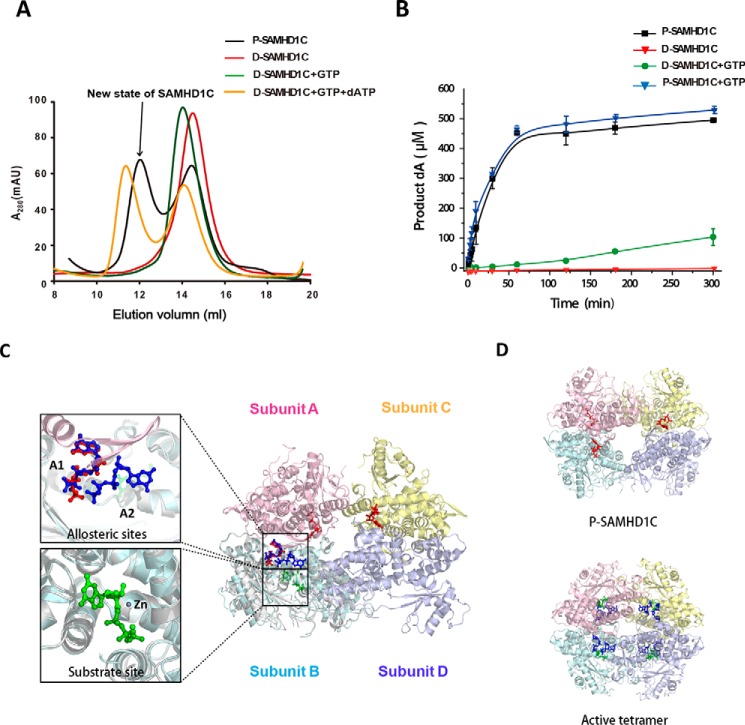
Based on the complexity of the mechanism of allosteric regulation of SAMHD1, it is possible that multiple active oligomeric states exist under physiological conditions. We observed that the purified SAMHD1C (referred to P-SAMHD1C) from E. coli was in equilibrium with monomers, dimers, and novel tetrameric forms, even without the addition of dNTP/NTP. After dialysis into the buffer (20 mm Tris-HCl, pH 8.0, 200 mm NaCl, 5 mm MgCl2, 0.5 mm TCEP) overnight, the protein (referred to D-SAMHD1C) was mainly in its monomeric form, mixed with a few dimers, depending on the concentration (Fig. 1A). The addition of GTP strongly shifted the equilibrium of D-SAMHD1C to its dimeric form. Each D-SAMHD1C tetramer bound to GTP through a dATP substrate, and the hydrolysis of dATP followed, whereas the control experiment demonstrated D-SAMHD1C in a monomeric/dimeric form that was incapable of enzymatically acting without the addition of GTP. We also observed that the P-SAMHD1C underwent a rapid initial burst of dATP hydrolysis, which resulted in an almost-complete hydrolysis of dATP after 1 h, whereas ∼80% of the substrate still remained after 5 h of incubation in the presence of the D-SAMHD1C, even after the addition of the activator of GTP (Fig. 1B). Here, to P-SAMHD1C, the enzyme hydrolyzed dATP at a similar rate of ∼9 μm/min whether GTP was present or not. However, the D-SAMHD1C activity was about 90-fold lower, ∼0.1 μm/min, in the presence of GTP. Therefore, the results show that P-SAMHD1C from E. coli has a much higher catalytic activity than does a standard reaction of the dimeric form of SAMHD1C induced by GTP under similar conditions.
FIGURE 1.
The new state of SAMHD1C possesses a more efficient dNTPase activity than the dimer induced by GTP. A, size-exclusion column profiles of different states of SAMHD1C. P-SAMDH1C (purified SAMDH1 109–626) was purified from E. coli without nucleotides in the solution. D-SAMDH1C was obtained by dialysis of P-SAMDH1C into buffer (20 mm Tris-HCl, pH 8.0, with 200 mm NaCl, 5 mm MgCl2, and 0.5 mm TCEP) overnight to remove the nucleotides bound to the protein in vivo. mAU, milliabsorbance units. B, P-SAMDH1C has a more efficient dNTPase activity than the induced dimer of D-SAMDH1C+GTP. The amount of dA generated at each indicated time point, as described under “Experimental Procedures,” is shown. The experiment was repeated three times (n = 3). The error bars indicate the S.D. of three replicates. C, graphic representation of the new form of SAMHD1C. One subunit from 4QFX overlapping the subunit B is shown to indicate the positions of the two allosteric sites (A1, A2) and a substrate site. Subunits A, B, C, and D are colored pink, light blue, light yellow, and light purple, respectively. Three GTP activators occupying the A1 sites are shown as red sticks. In 4QFX, two dGTP molecules occupying the allosteric sites (A1, A2) are colored deep blue, and a dGTP molecule occupying the substrate site is in green. D, overall structure comparison of the active tetramer 4QFX and our structure.
The present study also demonstrated that GTP was unable to stimulate the hydrolysis of dATP beyond the activation provided by P-SAMHD1C (0.8 μm) alone. The addition of a saturating 0.1 mm of GTP resulted in an enzyme kinetic progress curve similar to that of P-SAMHD1C (Fig. 1B). This finding suggests that P-SAMHD1C was already tightly bound to GTP during the heterologous expression in E. coli.
The Overall Structure of the High-efficiency Catalytic Form of P-SAMHD1C
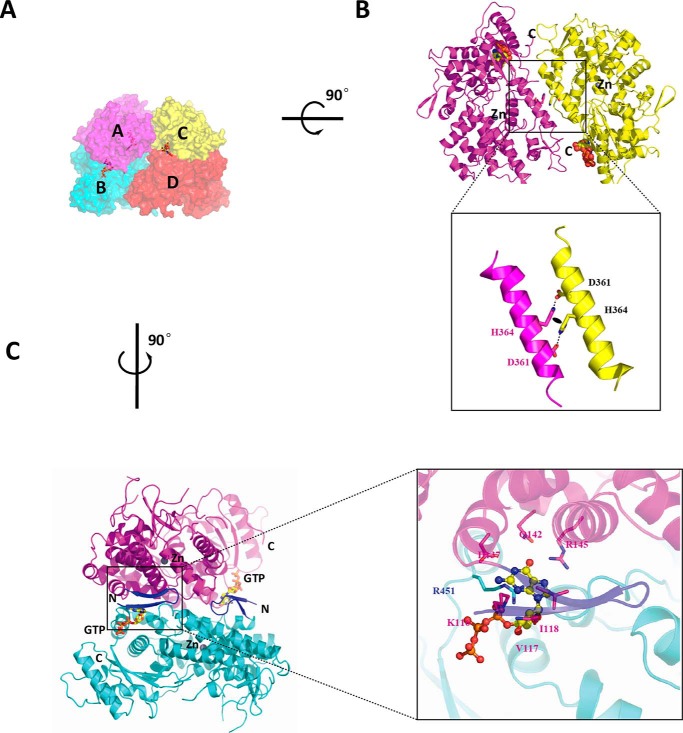
To illustrate the mechanism of the instantaneous reaction caused by P-SAMHD1C, we determined the crystal structure of P-SAMHD1C. Interestingly, P-SAMHD1C was bound to GTP (Protein Data Bank (PDB) ID: 4Q7H) in a novel loosely associated tetrameric form (Figs. 1C and 2A). Only three GTP molecules were clearly identified as binding to the A1 sites, whereas the electron density of GTP in subunit D was invisible. This loose tetrameric structure of SAMHD1C thus revealed relatively loose protein-protein interfaces that have not been previously observed. The buried interface between subunits A and B (∼1,800 Å2), as well as the interface between subunits B and D (464 Å2), was highly similar to that of the structure of SAMHD1c (120–626) (PDB ID: 3U1N). The interface between subunits A and C (540 Å2) was also a novel feature (Fig. 2B). The interactions between the two dimers might play an important role in the stabilization of the tetramer, because the mutations on the interface impaired the tetramerization of SAMHD1C (Fig. 3).
FIGURE 2.
Detailed view of the interactions between the subunits of SAMHD1C. A, surface representation. Subunits A, B, C, and D are colored magenta, cyan, yellow, and red, respectively. Three GTP activators occupying the A1 sites are shown as red sticks. B, rotated around the horizontal axis by 90°. The interface between subunits A and C can now be observed. Details of the interactions between subunits A and C are shown below. The black oval represents a hydrophobic interaction between two His-364 amino acids. C, rotated around the vertical axis by 90°. The interface between chains A and B is similar to that existing in the 3U1N dimer structure. The additional two-stranded anti-parallel β sheet in the N-terminal of our structure, which interacts with the GTP activator (shown as yellow sticks), is colored blue.
FIGURE 3.
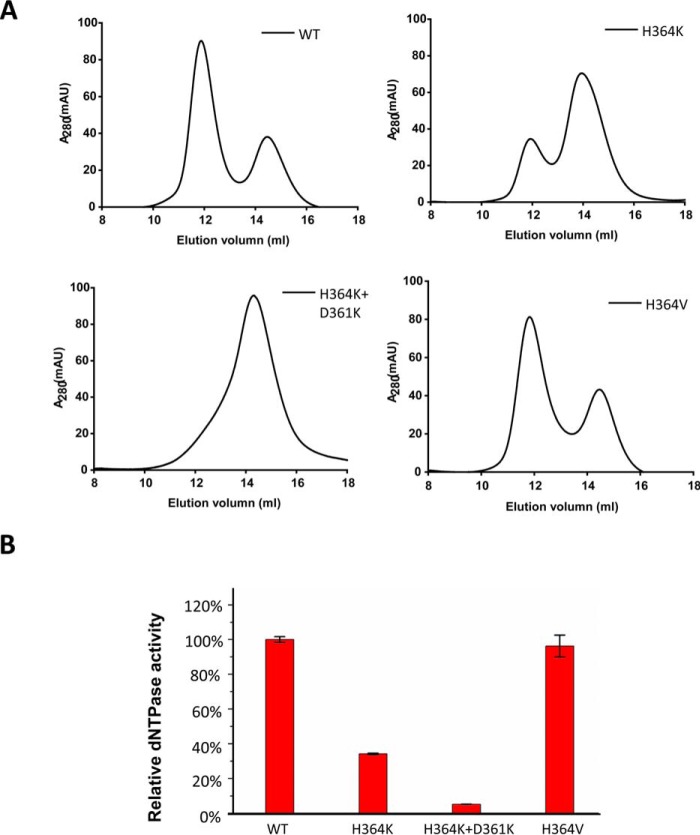
The interface between the A and C subunits is essential for the tetramer formation of SAMHD1C. A, mutations of residues involved in the interactions between subunits A and C of SAMHD1 affect tetramer formation. Purified recombinant SAMHD1C and SAMHD1C mutant proteins were incubated with 50 mm dGTP and separated by size-exclusion chromatography. The elution profiles were monitored by ultraviolet absorbance at A280 and compared with known molecular size markers. mAU, milliabsorbance units. B, mutations of residues involved the interactions between subunits A and C of SAMHD1C affect dNTPase activity. Purified recombinant SAMHD1C and SAMHD1C mutant proteins were analyzed for dNTPase activity as described under “Experimental Procedures,” and the activity of SAMHD1C was set to 100%. The S.D. from triplicate samples (n = 3) is shown.
Occupancy of the A1 Site by GTP Stabilizes the Dimeric Form
In our structure, two GTP molecules were observed at the interface between subunits A and B, and another GTP molecule was found between subunits C and D (Fig. 2C). A detailed view of the GTP-binding site provides an explanation for the stabilization of the SAMHD1C dimer. Considering that residues Lys-116, Val-117, Asp-137, Gln-142, and Arg-145 from subunit A formed extensive hydrogen bonds with GTP, it is possible that GTP could also bind to monomer SAMHD1C, which would be in agreement with the results of kinetic analysis. Residues Arg-451 and Lys-455 of subunit B also formed hydrogen bonds with GTP. Reciprocal interactions exist on the other side of the dimeric form. Hence, GTP molecules bound to the A1 site of each monomer served as clamps that affix these monomers together.
Cooperation of the dNTPs at the A2 Sites and the First GTP at the A1 Sites Induces the Activation of SAMHD1C
Comparison of the new interface formed between subunits A and C and that of the dGTP-bound tetramer complex (4QFX) revealed interesting structural differences. Only GTP molecules occupied the A1 sites, helix α13 from neighboring subunits was loosely bound through hydrophobic interactions that were formed by two His-364 residues. One side of the helix (Asp-361) from subunit A forms hydrogen bond interactions with the helix (His-364) from subunit C. Reciprocal interactions also exist between these two subunits. Mutation of H364K resulted in significant defective tetramer formation and reduced dNTPase activity (Fig. 3), which may be due to the destruction of the hydrophobic interaction. In contrast, valine, which is present in the EF1143, more easily adapted to the interaction site, resulting in the maintenance of SAMHD1 activity. Mutation of D361K,H364K abolished tetramerization and impaired the hydrolysis of dNTP in vitro. When occupying the A2 allosteric site in the 4QFX structure, dGTP/dATP interacted with the first allosteric GTP/dGTP molecule through Mg2+. Two pairs of allosteric molecules at both ends of helix α13 from subunits A and C served as a hinge that allowed subunit C to move toward the core of the tetramer (∼20°) when subunit A was affixed. The flat surface formed by the two subunits thereby adopted a bowl-shaped conformation in 4QFX, which enabled closer interactions and facilitated the tight folding of the tetramer (Fig. 4).
FIGURE 4.
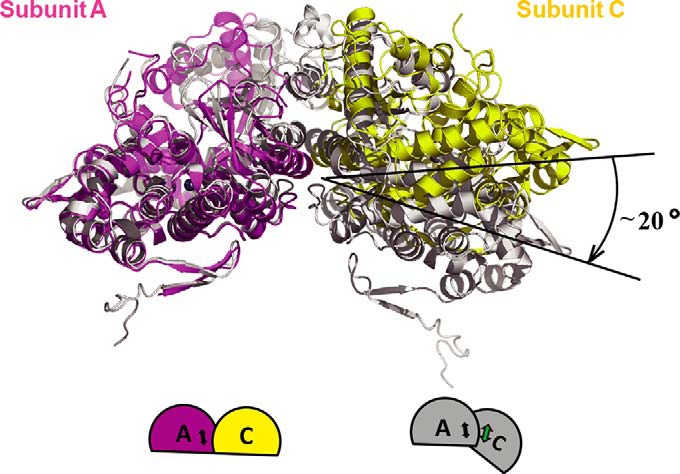
Occupancy of the secondary allosteric site by dNTP induces the formation of a tight SAMHD1 tetramerization. Shown is the superposition of subunit A (magenta) and subunit C (yellow) from our structure on the subunits from the tetramer structure 4QFX (gray). Subunit C moves toward the core of the tetramer (∼20°) in 4QFX when subunit A is fixed, enabling closer tetramer formation. Bottom, schematic graphic of the conformational change, in the same color scheme.
The Loose Tetrameric Form of SAMHD1-C Is Closed
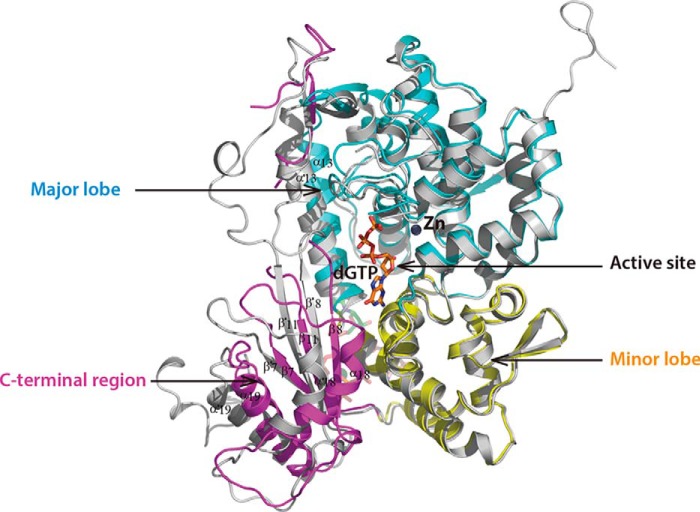
Detailed comparison of one subunit from our structure and that from the inactive dimer structure 3U1N showed that the overall average main chain conformation was virtually identical (RMSD 0.52 Å for 435 Cα atoms). However, significant differences were observed in the catalytic site when we compared the subunit of our structure with that of the active structure 4QFX (Figs. 1C and 5). In addition, α18, β8, β11, β7, and α19 formed a fan-shaped structure and entirely moved away from the active center of 4QFX. However, this C-terminal region in both the dimer structure 3U1N and our structure was much closer to the minor lobe. The observed rate of inactive/active state turnover suggests that the C-terminal region might be responsible for blocking the channel for substrate access: it would rotate to “open” in the active tetramer state, or “closed” in the inactive state. Taken together, these observations suggest that this new tetrameric structure in complex with GTP remains in an inactive state.
FIGURE 5.
Superposition of one chain of our structure with that of tetramer structure 4QFX. The major lobe, minor lobe, and C-terminal region of our structure are colored cyan, yellow, and magenta, respectively. The monomer of 4QFX is gray, and the substrate in the active site of 4QFX is shown as orange sticks.
Discussion
In this study, we have determined a novel tetramer structure of SAMHD1 containing only GTP. The corresponding SAMHD1 complex has a more rapid initial hydrolysis rate for dNTP than either monomer or dimer SAMHD1. Furthermore, the crystal structure showed that this novel SAMHD1 tetramer has structural features distinct from the previously reported (36) GTP/dNTP-bound SAMHD1 tetramer.
Given that the concentration of intracellular GTP in E. coli (43) is similar to that in human macrophages (2), we infer that the recombinant SAMHD1 expressed in E. coli harbors GTP molecules as it does in vivo. The loose inactive tetramer form of SAMHD1C could therefore be a natural state. GTP (not dGTP) apparently occupied the A1 sites because the GTP could not be hydrolyzed by the active site, and the concentration of GTP is much higher than that of dGTP in organisms (20). We effectively isolated a high-efficiency tetramer form of SAMHD1C that remained stable for a relatively long time. In addition, the process of crystallization would also homogenize and stabilize this particular state. We detected both GTP and dGTP in the solution of crystallized protein by MALDI-TOF mass spectrometry (data not shown). Because GTP alone could not efficiently induce SAMHD1 tetramerization, we therefore propose that low dNTP levels facilitate the maintenance of the inactive state of SAMHD1 instead of depleting it completely.
The oligomerization of SAMHD1 is concentration-dependent. Therefore, even SAMHD1c (120–626) was incapable of binding GTP, and the dimer still formed during the crystallization of 3U1N. The interface in the dimer was exactly similar to that in the active tetramer structure and in our inactive tetramer structure, indicating that the crystal structure represents the optimal mechanism of interaction for the protein in solution because it tends to pack in the most comfortable manner. Limited interactions were observed in the relatively remote, solvent-exposed interface between subunits B and D, and these interactions were also detected in 3U1N. This finding suggests that the interactions between subunits B and D might also provide an optimal mechanism for dimer association.
Structural Comparison of Hs-SAMHD1 and EF1143
SAMHD1 inhibits the retroviral infection of quiescent cells in the immune system by maintaining low levels of dNTP pools. Interestingly, a similar strategy for inhibiting bacteriophage infection is employed by prokaryotes. SAMHD1 from humans has a high sequence identity with that of Mus musculus (75.2%), Xenopus laevis (68.2%), and Danio rerio (67.5%) (Fig. 6). Although Hs-SAMHD1 and EF1143 from Enterococcus faecalis have low sequence similarity (21.5%), their overall structures are nevertheless similar. Superposition of Hs-SAMHD1C onto EF1143 gives an RMSD value of 2.28 Å for 321 Cα atoms. Further analysis has shown that the conformation is highly conserved in the major lobe, but less so in the minor lobe and the C-terminal region. The residues responsible for binding and the hydrolytic substrates are highly conserved in the active site, indicating that these enzymes could identify and hydrolyze substrates in a similar manner. The position of GTP in our structure was exactly overlaid by the dGTP at the A1 site of EF1143 (PDB ID: 3IRH). As expected, all the critical residues of the A1 sites for pre-activation were 100% conserved throughout the enzyme family, in agreement with the high guanine specificity of the A1 sites. The residues involved in the binding to A2 sites were less conserved. Although the residues Phe-157, Arg-333, Arg-336, and His-376 of SAMHD1 were conserved (corresponding to Phe-56, Arg-206, Arg-209, and His-245 in EF1143), Met-115, Asn-119, and Val-156 in SAMHD1 were not, as their counterparts were Glu-13, Arg-17, and Thr-55 in EF1143. Variations in the residues between the A2 sites could result in differences in the regulation of the two enzymes. When the A2 sites were occupied by dATP/dCTP, EF1143 could be catalytically induced to assume its active state, whereas binding to dGTP/dTTP resulted in negative regulation of EF1143. No analogous regulatory mechanism was observed in SAMHD1. However, given the global similarities of the enzymes in terms of structure and activity, the protein family shares a general allosteric catalytic mechanism.
FIGURE 6.
Comparison of the enzymes from various species. Shown is sequence alignment of SAMHD1s from various species. Secondary structure and residue numbers are based on the structure of human SAMHD1. The major lobe, minor lobe, and C-terminal region of human SAMHD1 are shown in cyan, yellow, and magenta, respectively. Hs, Homo sapiens; Mum, M. musculus; Xl, X. laevis; Dr, D. rerio; Ef, E. faecalis. Red stars indicate residues involved in the primary allosteric site. Red dots indicate residues involved in the secondary allosteric site. Black dots indicate residues involved in the active site, and black stars indicate the key residues of the H … HD … D motif in the active center. Dotted lines indicate that amino acids are not included in our current structure because of conformational flexibility. Conserved residues in the four species are highlighted in red.
Ordered Assembly and Activation Pathway of SAMHD1 in Vivo
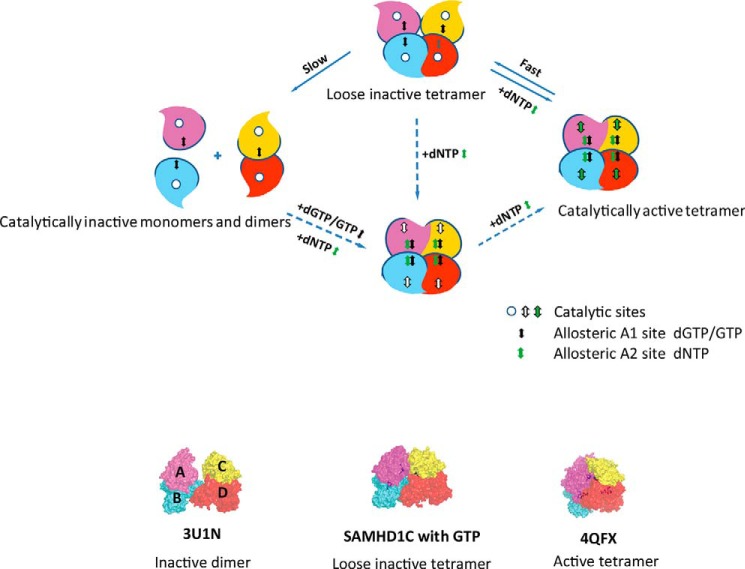
The combined structures and oligomerization data facilitated the construction of a model for an ordered activation pathway of SAMHD1 in vivo (Fig. 7). Under physiological conditions, the occupation of the A1 site by GTP maintained the dimerization of SAMHD1. Occupation of the A2 site by the coactivator dNTP induced a drastic relative movement of the dimers, which in turn promoted the formation of the tightly folded tetrameric form of SAMHD1, as well as an efficient active pocket. Most importantly, after activation, the tetrameric form of SAMHD1 did not immediately dissociate into inactive dimer or monomer states under low dNTP conditions and instead remained in a loosely associated tetramer form, as shown in our structure. Once the concentration of dNTP increased to a sufficient level, this form of SAMHD1 was immediately activated, thereby causing an instantaneous reaction and the restoration of the dNTP pool to a low level. We propose that the level of the dNTP pools is elegantly regulated by the self-sensing regulation of SAMHD1. The series of states of SAMHD1 crystal structures provides information on the ordered assembly and activation pathway of SAMHD1. Fig. 7 outlines the qualitative model for SAMHD1 allosteric regulation.
FIGURE 7.
Ordered pathway for activation of SAMHD1. Shown is a model of the dGTP/GTP-dependent allosteric regulation of SAMHD1 catalysis. Subunits of SAMHD1 are labeled A, B, C, and D, and the catalytically inactive monomers and dimers are shown with empty substrate-binding pockets. dGTP/GTP+dNTP could induce an active tetrameric formation of SAMHD1 to hydrolyze the substrates. After the reaction, the tetramer form of SAMHD1 would not immediately dissociate to inactive dimer or monomer states, but instead would prefer to stay in a loose inactive tetramer form that would immediately turn into an active tetramer state in response to the addition of dNTP. The dashed arrows indicate that a form might exist in the pathway. The loose inactive tetramer form containing four GTPs may exist under physiological conditions, and the GTP that might exist in subunit D was shown in gray. Bottom, surface representation of the 3U1N inactive dimer structure, our new loose tetrameric structure, and the 4QFX active tetramer structure. Subunits are in the same color scheme as in Fig. 2.
In summary, activation by GTP and dNTP results in a loosely associated tetrameric SAMHD1 that could exist stably even when activating nucleotides fall below a certain threshold. However, this form of SAMHD1 complex can be quickly activated when dNTP levels are elevated. Our structural and functional studies shed light on the efficient dNTP self-regulation mechanism in organisms. We also provide new insights into the assembly mechanism and activation pathway of the SAMHD1 protein family. The long-lived loose tetrameric form of SAMHD1 may play an important role in the maintenance of nanomolar range of dNTPs that is commonly observed in macrophages and resting CD4+ T cells.
Author Contributions
X. Q. and X. Y. designed the study and wrote the paper. Y. L. purified and crystallized the protein and determined its X-ray structure. X. P. characterized the enzyme activity in vitro. W. H. provided helpful comments on the text. J. K. designed and constructed vectors for expression of mutant proteins. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank Yong Gong for providing technical assistance. We also thank the staff of Beamline 3W1A of the Beijing Synchrotron Radiation Facility for excellent technical assistance during data collection and Dr. Deborah McClellan for editorial assistance.
This work was supported in part by funding from the Chinese Ministry of Science and Technology (Grants 2012CB911100 and 2013ZX0001-005), the National Natural Science Foundation of China (Grant 31400645), and the Natural Science Foundation of Tianjin (Grants 15JCQNJC09800 and 15JCQNJC06200). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 4Q7H) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- SAMHD1
- sterile α-motif/histidine-aspartate domain-containing protein
- TCEP
- tris(2-carboxyethyl)phosphine
- RMSD
- root mean square deviation
- P-SAMHD1C
- purified SAMHD1C
- D-SAMHD1C
- dialyzed SAMHD1C
- Hs-SAMHD1
- Homo sapiens SAMHD1.
References
- 1.Bebenek K., Roberts J. D., and Kunkel T. A. (1992) The effects of dNTP pool imbalances on frameshift fidelity during DNA replication. J. Biol. Chem. 267, 3589–3596 [PubMed] [Google Scholar]
- 2.Kennedy E. M., Gavegnano C., Nguyen L., Slater R., Lucas A., Fromentin E., Schinazi R. F., and Kim B. (2010) Ribonucleoside Triphosphates as substrate of human immunodeficiency virus type 1 reverse transcriptase in human macrophages. J. Biol. Chem. 285, 39380–39391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger A., Sommer A. F. R., Zwarg J., Hamdorf M., Welzel K., Esly N., Panitz S., Reuter A., Ramos I., Jatiani A., Mulder L. C. F., Fernandez-Sesma A., Rutsch F., Simon V., König R., and Flory E. (2011) SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 7, e1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldauf H. M., Pan X., Erikson E., Schmidt S., Daddacha W., Burggraf M., Schenkova K., Ambiel I., Wabnitz G., Gramberg T., Panitz S., Flory E., Landau N. R., Sertel S., Rutsch F., Lasitschka F., Kim B., König R., Fackler O. T., and Keppler O. T. (2012) SAMHD1 restricts HIV-1 infection in resting CD4+ T cells. Nat. Med. 18, 1682–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Descours B., Cribier A., Chable-Bessia C., Ayinde D., Rice G., Crow Y., Yatim A., Schwartz O., Laguette N., and Benkirane M. (2012) SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4+ T-cells. Retrovirology 9, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lahouassa H., Daddacha W., Hofmann H., Ayinde D., Logue E. C., Dragin L., Bloch N., Maudet C., Bertrand M., Gramberg T., Pancino G., Priet S., Canard B., Laguette N., Benkirane M., Transy C., Landau N. R., Kim B., and Margottin-Goguet F. (2012) SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13, 223–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engström Y., Eriksson S., Jildevik I., Skog S., Thelander L., and Tribukait B. (1985) Cell cycle-dependent expression of mammalian ribonucleotide reductase. Differential regulation of the two subunits. J. Biol. Chem. 260, 9114–9116 [PubMed] [Google Scholar]
- 8.Nordlund P., and Reichard P. (2006) Ribonucleotide reductases. Annu. Rev. Biochem. 75, 681–706 [DOI] [PubMed] [Google Scholar]
- 9.Goldstone D. C., Ennis-Adeniran V., Hedden J. J., Groom H. C. T., Rice G. I., Christodoulou E., Walker P. A., Kelly G., Haire L. F., Yap M. W., de Carvalho L. P. S., Stoye J. P., Crow Y. J., Taylor I. A., and Webb M. (2011) HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 [DOI] [PubMed] [Google Scholar]
- 10.Powell R. D., Holland P. J., Hollis T., and Perrino F. W. (2011) Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 286, 43596–43600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franzolin E., Pontarin G., Rampazzo C., Miazzi C., Ferraro P., Palumbo E., Reichard P., and Bianchi V. (2013) The deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of DNA precursor pools in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 110, 14272–14277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cribier A., Descours B., Valadão A. L. C., Laguette N., and Benkirane M. (2013) Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 3, 1036–1043 [DOI] [PubMed] [Google Scholar]
- 13.Li N., Zhang W., and Cao X. (2000) Identification of human homologue of mouse IFN-γ induced protein from human dendritic cells. Immunol. Lett. 74, 221–224 [DOI] [PubMed] [Google Scholar]
- 14.White T. E., Brandariz-Nuñez A., Valle-Casuso J. C., Amie S., Nguyen L. A., Kim B., Tuzova M., and Diaz-Griffero F. (2013) The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13, 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hrecka K., Hao C., Gierszewska M., Swanson S. K., Kesik-Brodacka M., Srivastava S., Florens L., Washburn M. P., and Skowronski J. (2011) Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Ségéral E., Yatim A., Emiliani S., Schwartz O., and Benkirane M. (2011) SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao K., Du J., Han X., Goodier J. L., Li P., Zhou X., Wei W., Evans S. L., Li L., Zhang W., Cheung L. E., Wang G., Kazazian H. H. Jr., and Yu X. F. (2013) Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutieres syndrome-related SAMHD1. Cell Rep. 4, 1108–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim E. T., White T. E., Brandariz-Núñez A., Diaz-Griffero F., and Weitzman M. D. (2013) SAMHD1 restricts herpes simplex virus 1 in macrophages by limiting DNA replication. J. Virol. 87, 12949–12956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollenbaugh J. A., Gee P., Baker J., Daly M. B., Amie S. M., Tate J., Kasai N., Kanemura Y., Kim D. H., Ward B. M., Koyanagi Y., and Kim B. (2013) Host factor SAMHD1 restricts DNA viruses in non-dividing myeloid cells. PLoS Pathog. 9, e1003481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amie S. M., Bambara R. A., and Kim B. (2013) GTP is the primary activator of the anti-HIV restriction factor SAMHD1. J. Biol. Chem. 288, 25001–25006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahn J., Hao C., Yan J., DeLucia M., Mehrens J., Wang C., Gronenborn A. M., and Skowronski J. (2012) HIV/simian immunodeficiency virus (SIV) accessory virulence factor Vpx loads the host cell restriction factor SAMHD1 onto the E3 ubiquitin ligase complex CRL4DCAF1. J. Biol. Chem. 287, 12550–12558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goujon C., Rivière L., Jarrosson-Wuilleme L., Bernaud J., Rigal D., Darlix J. L., and Cimarelli A. (2007) SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology 4, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laguette N., Rahm N., Sobhian B., Chable-Bessia C., Münch J., Snoeck J., Sauter D., Switzer W. M., Heneine W., Kirchhoff F., Delsuc F., Telenti A., and Benkirane M. (2012) Evolutionary and functional analyses of the interaction between the myeloid restriction factor SAMHD1 and the lentiviral Vpx protein. Cell Host Microbe 11, 205–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welbourn S., Dutta S. M., Semmes O. J., and Strebel K. (2013) Restriction of virus infection but not catalytic dNTPase activity is regulated by phosphorylation of SAMHD1. J. Virol. 87, 11516–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakai H., and Richardson C. C. (1990) The gene 1.2 protein of bacteriophage T7 interacts with the Escherichia coli dGTP triphosphohydrolase to form a GTP-binding protein. J. Biol. Chem. 265, 4411–4419 [PubMed] [Google Scholar]
- 26.Clifford R., Louis T., Robbe P., Ackroyd S., Burns A., Timbs A. T., Wright Colopy G., Dreau H., Sigaux F., Judde J. G., Rotger M., Telenti A., Lin Y. L., Pasero P., Maelfait J., Titsias M., Cohen D. R., Henderson S. J., Ross M. T., Bentley D., Hillmen P., Pettitt A., Rehwinkel J., Knight S. J. L., Taylor J. C., Crow Y. J., Benkirane M., and Schuh A. (2014) SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage. Blood 123, 1021–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice G. I., Bond J., Asipu A., Brunette R. L., Manfield I. W., Carr I. M., Fuller J. C., Jackson R. M., Lamb T., Briggs T. A., Ali M., Gornall H., Couthard L. R., Aeby A., Attard-Montalto S. P., Bertini E., Bodemer C., Brockmann K., Brueton L. A., Corry P. C., Desguerre I., Fazzi E., Cazorla A. G., Gener B., Hamel B. C. J., Heiberg A., Hunter M., van der Knaap M. S., Kumar R., Lagae L., Landrieu P. G., Lourenco C. M., Marom D., McDermott M. F., van der Merwe W., Orcesi S., Prendiville J. S., Rasmussen M., Shalev S. A., Soler D. M., Shinawi M., Spiegel R., Tan T. Y., Vanderver A., Wakeling E. L., Wassmer E., Whittaker E., Lebon P., Stetson D. B., Bonthron D. T., and Crow Y. J. (2009) Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat. Genet. 41, 829–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crow Y. J. (2013) Aicardi-Goutieres syndrome. Handb. Clin. Neurol. 113, 1629–1635 [DOI] [PubMed] [Google Scholar]
- 29.Ramantani G., Häusler M., Niggemann P., Wessling B., Guttmann H., Mull M., Tenbrock K., and Lee-Kirsch M. A. (2011) Aicardi-Goutieres syndrome and systemic lupus erythematosus (SLE) in a 12-year-old boy with SAMHD1 mutations. J. Child Neurol. 26, 1425–1428 [DOI] [PubMed] [Google Scholar]
- 30.Gramberg T., Kahle T., Bloch N., Wittmann S., Müllers E., Daddacha W., Hofmann H., Kim B., Lindemann D., and Landau N. R. (2013) Restriction of diverse retroviruses by SAMHD1. Retrovirology 10, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim B., Nguyen L. A., Daddacha W., and Hollenbaugh J. A. (2012) Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J. Biol. Chem. 287, 21570–21574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu C., Gao W., Zhao K., Qin X., Zhang Y., Peng X., Zhang L., Dong Y., Zhang W., Li P., Wei W., Gong Y., and Yu X. F. (2013) Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase. Nat. Commun. 4, 2722. [DOI] [PubMed] [Google Scholar]
- 33.Ji X., Wu Y., Yan J., Mehrens J., Yang H., DeLucia M., Hao C., Gronenborn A. M., Skowronski J., Ahn J., and Xiong Y. (2013) Mechanism of allosteric activation of SAMHD1 by dGTP. Nat. Struct. Mol. Biol. 20, 1304–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan J., Kaur S., DeLucia M., Hao C., Mehrens J., Wang C., Golczak M., Palczewski K., Gronenborn A. M., Ahn J., and Skowronski J. (2013) Tetramerization of SAMHD1 is required for biological activity and inhibition of HIV infection. J. Biol. Chem. 288, 10406–10417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koharudin L. M., Wu Y., DeLucia M., Mehrens J., Gronenborn A. M., and Ahn J. (2014) Structural basis of allosteric activation of sterile α motif and histidine-aspartate domain-containing protein 1 (SAMHD1) by nucleoside triphosphates. J. Biol. Chem. 289, 32617–32627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji X., Tang C., Zhao Q., Wang W., and Xiong Y. (2014) Structural basis of cellular dNTP regulation by SAMHD1. Proc. Natl. Acad. Sci. U.S.A. 111, E4305–E4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansen E. C., Seamon K. J., Cravens S. L., and Stivers J. T. (2014) GTP activator and dNTP substrates of HIV-1 restriction factor SAMHD1 generate a long-lived activated state. Proc. Natl. Acad. Sci. U.S.A. 111, E1843–E1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 40.Zwart P. H., Afonine P. V., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., McKee E., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Storoni L. C., Terwilliger T. C., and Adams P. D. (2008) Automated structure solution with the PHENIX suite. Methods Mol. Biol. 426, 419–435 [DOI] [PubMed] [Google Scholar]
- 41.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., and Thornton J. M. (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 42.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., and Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 43.Buckstein M. H., He J., and Rubin H. (2008) Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J. Bacteriol. 190, 718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]