Abstract
ATP-binding cassette (ABC)-type ATPases are chemomechanical engines involved in diverse biological pathways. Recent genomic information reveals that ABC ATPase domains/subunits act not only in ABC transporters and structural maintenance of chromosome proteins, but also in iron-sulfur (Fe-S) cluster biogenesis. A novel type of ABC protein, the SufBCD complex, functions in the biosynthesis of nascent Fe-S clusters in almost all Eubacteria and Archaea, as well as eukaryotic chloroplasts. In this study, we determined the first crystal structure of the Escherichia coli SufBCD complex, which exhibits the common architecture of ABC proteins: two ABC ATPase components (SufC) with function-specific components (SufB-SufD protomers). Biochemical and physiological analyses based on this structure provided critical insights into Fe-S cluster assembly and revealed a dynamic conformational change driven by ABC ATPase activity. We propose a molecular mechanism for the biogenesis of the Fe-S cluster in the SufBCD complex.
Keywords: ABC transporter, ATPase, conformational change, iron-sulfur protein, x-ray crystallography, ABC ATPase, Suf machinery
Introduction
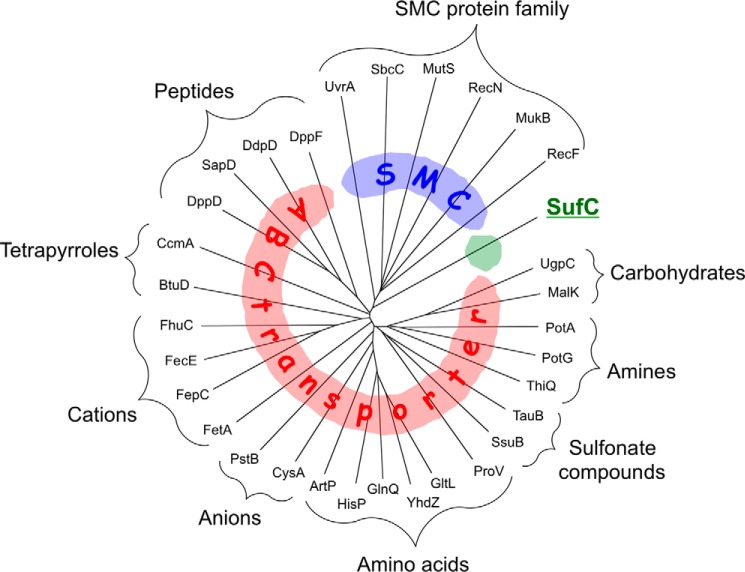
The ATP-binding cassette (ABC)3 is a ubiquitous, universally conserved ATPase domain/subunit historically defined as the nucleotide-binding domain of an ABC transporter. ABC transporters comprise a large and diverse family of membrane-spanning proteins that transport various substances, ranging from ions to proteins, across membranes (1–5). With the availability of complete genomes and the refinement of bioinformatic tools, it has become apparent that ABC type ATPase domains are present not only in ABC transporters but also in a variety of nontransporter proteins, the most well known examples of which are the structural maintenance of chromosome (SMC) proteins involved in chromosome segregation/condensation and DNA repair (6–8). Although the SMC proteins, like the ABC transporters, have attracted great interest because its members are implicated in various human diseases, there are additional types of nontransporter ABC proteins. Here, we focus on a novel type of ABC protein, the SufBCD complex, whose ABC ATPase components (SufC) segregate in a different clade from those of transporters and SMC proteins (Fig. 1).
FIGURE 1.
Dendrogram of ABC ATPases from E. coli. Among the 32 proteins, SufC belongs to clades of neither the ABC transporter nor the SMC family.
The SufBCD complex is a component in the Suf machinery that is responsible for de novo iron-sulfur (Fe-S) cluster biogenesis. This machinery is phylogenetically diverse and is present in photosynthetic organisms such as higher plants, as well as in Eubacteria and Archaea (9, 10). Fe-S clusters act as cofactors of various Fe-S proteins that are essential for maintaining fundamental biological processes such as respiratory and photosynthetic electron transfer and regulation of gene expression (11, 12). Fe-S cluster biogenesis requires a complex network of proteins that mobilize sulfur and iron, assemble nascent clusters, and transfer the assembled clusters to Fe-S proteins. The Suf machinery in Escherichia coli is composed of six proteins encoded by the sufABCDSE operon. The SufS cysteine desulfurase and SufE sulfur shuttle protein act together to provide sulfur for construction of nascent Fe-S clusters (13, 14). SufA is an Fe-S carrier protein that transfers Fe-S clusters to target apo-proteins (15). The remaining proteins, SufB, SufC, and SufD, have attracted a great deal of attention because deletion of any of them abolishes Suf function in vivo (10, 16, 17). SufB and SufD are homologues and show some sequence similarity (17% identity and 37% similarity). SufB accepts sulfur transferred from SufE (14), and SufD may play a role in iron acquisition (18). SufB, SufC, and SufD interact with each other, and three distinct states have been reported: the SufBCD ternary complex, the SufBC subcomplex (18, 19), and the SufCD subcomplex (19, 20). Recent in vitro reconstitution studies suggested that the SufBCD complex can serve as the scaffold for nascent Fe-S cluster assembly (21–24). It is clear that the SufBCD complex plays a central role in the Suf pathway, but the molecular mechanism underlying Fe-S cluster formation in the SufBCD complex remains enigmatic.
Although SufC is a member of the ABC ATPase superfamily and exhibits ATPase activity, the role of ATPase activity in Fe-S cluster biogenesis is currently unclear (21–23). In ABC transporters, energy from ATP binding/hydrolysis acts to transport specific substances across membranes (1–5, 25). In soluble SMC proteins, the ABC ATPase utilizes ATP to recognize and bind DNA (4, 26). Despite this extreme functional diversity, these proteins share a similar architecture, consisting of two ABC ATPase domains bound to substrate/function-specific partner domains; in both types of proteins, the ABC ATPase activity drives the conformational changes in partner domains required for each function (4). Therefore, it is likely that structural changes in the SufB and SufD subunits are driven by SufC ATPase activity in the SufBCD complex and that the dynamic motion of the complex should provide important clues regarding the molecular mechanism of Fe-S cluster biogenesis.
In this study, we determined the crystal structure of the SufBCD (SufB1-SufC2-SufD1) complex, providing the first demonstration of the quaternary configuration of the ternary complex. Biochemical experiments based on the crystal structure demonstrated that the two SufC ABC ATPase subunits form a head to tail dimer in the complex upon ATP binding, thereby inducing a structural change in the interface between the SufB and SufD subunits. These findings, together with in vivo mutational analyses, provided insights into the mechanism of Fe-S cluster assembly in the SufBCD complex.
Experimental Procedures
Expression and Purification of the E. coli SufBCD Complex
To purify the SufBCD complex, the entire suf operon was expressed simultaneously. The plasmid pGSO164, containing the entire suf operon under the control of an arabinose-inducible promoter (22), was used to overexpress SufABCDSE in the TOP10 strain of E. coli. The cells were grown in TB medium containing ampicillin (50 μg/ml) at 37 °C. l-Arabinose was added to 0.2% (w/v) final concentration when the cultures reached an A600 of 0.4–0.6. After 3 h of SufABCDSE expression at 37 °C, the cells were harvested by centrifugation, and the cell pellets were frozen at −80 °C. Cell pellets were lysed by sonication in 50 mm Tris-HCl (pH 7.8), 100 mm NaCl, and 1 mm DTT. The soluble fraction was subjected to ammonium sulfate fractionation at 20% saturation. After centrifugation, the supernatant fraction was loaded onto a HiPrep Phenyl FF (low sub) 16/10 column (GE Healthcare), and the bound protein was eluted with a decreasing linear gradient of 20–0% ammonium sulfate. Fractions containing the SufBCD complex were pooled, dialyzed overnight in 50 mm Tris-HCl (pH 7.8) and 1 mm DTT, then loaded onto a Mono Q HR 5/50 GL column (GE Healthcare), and eluted with a linear gradient of 0–1 m NaCl. The SufBCD complex was further purified by gel filtration using a HiPrep 16/60 Sephacryl S-200 HR column (GE Healthcare) in 50 mm Tris-HCl (pH 7.8) and 150 mm NaCl. The purified SufBCD complex was concentrated and stored at −80 °C.
Site-directed mutagenesis was performed using the pGSO164 plasmid as a template and the primers listed in Table 1. Genes were expressed in mutant cells (YT2512) in which the entire sufABCDSE operon was deleted from the chromosome (10), and mutant SufBCD complexes were purified as described for the wild-type complex.
TABLE 1.
Oligonucleotides used in this study
The underlined and shaded bases comprise restriction sites and artificial promoter regions, respectively. The double underlines indicate the altered codons for site-directed mutagenesis.

Crystallization and Structure Determination
Crystallization was performed by the sitting drop vapor diffusion method. Crystals of the SufBCD complex were obtained at 4 °C using a reservoir solution containing 31% (v/v) pentaerythritol propoxylate (5/4 PO/OH), 100 mm sodium citrate (pH 5.5), and 200 mm KCl. The protein concentration was 35 mg/ml in 50 mm MES (pH 7.0). Mercury and platinum derivatives were obtained by soaking native crystals for 2 h in mother liquor containing 1 mm methylmercury(II) acetate, 5 mm methylmercury(II) chloride, or 10 mm potassium tetranitro platinate(II). All crystals were transferred to a cryo-protectant solution containing 5% (v/v) 2-methyl-2,4-pentanediol and flash cooled by immersion in liquid nitrogen. X-ray diffraction data were collected at Beamline BL44XU and BL32XU of SPring-8 and processed with the HKL2000 package (27). Experimental phases were obtained from mercury- and platinum-derivative crystals by the multiple isomorphous replacement method coupled with anomalous scattering using AutoSol in PHENIX (28). The model was built manually in COOT (29), and the structure was refined with PHENIX. Secondary structures were assigned using PROMOTIF (30), and the geometry of the final model was analyzed using PROCHECK (31). Superposition and r.m.s.d. of the structures were calculated using LSQMAN (32), and folding motifs were classified in the SCOP2 database (33). All structure figures were prepared using PyMOL (34) or UCSF Chimera (35). X-ray data and refinement statistics are given in Table 2. Coordinates of the x-ray structure of the SufBCD complex and the Hg-bound SufBCD complex have been deposited in the Protein Data Bank, under accession codes 5AWF and 5AWG.
TABLE 2.
Data collection, phasing, and refinement statistics for x-ray crystallography
The values in parentheses correspond to the highest resolution shell.
| Native | Hg derivative 1a | Hg derivative 2b | Pt derivativec | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P21 | P21 | P21 | P21 |
| Cell dimensions | ||||
| a, b, c (Å) | 119.5, 139.6, 124.7 | 119.8, 139.4, 124.4 | 120.2, 140.0, 124.6 | 119.8, 140.4, 124.5 |
| α, β, γ (°) | 90.0, 113.1, 90.0 | 90.0, 113.6, 90.0 | 90.0, 113.5, 90.0 | 90.0, 113.1, 90.0 |
| Resolution (Å) | 2.95 (3.06–2.95) | 4.30 (4.45–4.30) | 4.50 (4.66–4.50) | 3.45 (3.57–3.45) |
| Rmerge (%) | 6.0 (37.1) | 12.7 (29.5) | 14.4 (29.8) | 10.7 (34.1) |
| I/σI | 15.8 (2.8) | 6.6 (5.2) | 5.8 (5.5) | 7.6 (4.7) |
| Completeness (%) | 98.5 (98.1) | 99.3 (99.9) | 92.8 (94.5) | 99.1 (99.7) |
| Redundancy | 3.8 (3.6) | 5.6 (5.6) | 6.0 (5.9) | 5.6 (5.6) |
| Refinement | ||||
| Resolution (Å) | 41.11–2.95 | 43.94–4.30 | ||
| No. reflections | 77,254 | 50,117 | ||
| Rwork/Rfree (%) | 18.7/22.6 | 29.5/34.0 | ||
| No. atoms | ||||
| Protein | 19,914 | 18,804 | ||
| Hg2+ | 0 | 4 | ||
| B factors (Å2) | ||||
| Protein | 83.6 | 110.3 | ||
| Ion | − | 148.8 | ||
| r.m.s.d. | ||||
| Bond lengths (Å) | 0.006 | 0.007 | ||
| Bond angles (°) | 1.241 | 1.672 | ||
| Ramachandran plot | ||||
| Most favored (%) | 92.3 | 86.5 | ||
| Additionally allowed (%) | 6.9 | 12.9 | ||
| Generously allowed (%) | 0.8 | 0.6 | ||
a Hg, methylmercury(II) acetate.
b Hg, methylmercury(II) chloride.
c Pt, potassium tetranitro platinate(II).
The crystallographic asymmetric unit contained two SufBCD complexes, termed Complex1 and Complex2. Although the electron density for the SufBCD complex was mostly continuous, the densities for some regions were poorly defined: in Complex1, SufB residues 1–33 and 80–156, SufD residues 1–7 and 422–423, SufCSufB residues 244–248, and SufCSufD residues 236–248; and in Complex2, SufB residues 1–35 and 79–157, SufD residues 1–7 and 422–423, SufCSufB residues 244–248, and SufCSufD residues 237–248. Accordingly, these residues were not included in the model.
Single-particle Electron Microscopy Reconstruction
The SufBCD complex was prepared from a peak fraction of gel filtration in 50 mm MES (pH 6.5), 150 mm NaCl, and 5 mm MgCl2. The negatively stained SufBCD complex was examined using an H9500SD transmission electron microscope (Hitachi High-Tech) operated at 200 kV at room temperature. The images were acquired on a 2k × 2k charge-coupled device camera (TVIPS) with a physical pixel size of 0.24 nm. Random conical tilt reconstruction was performed using the software package SPIDER (36). The obtained three-dimensional structure from random conical tilt was refined by using the EMAN1 software suite (37). The final reconstruction of the SufBCD complex was computed from ∼7,146 particles. The particle images were low pass filtered at 30 Å before refinement, and therefore the Fourier shell correlation that was calculated using eotest of EMAN1 shows higher values than 0.5 in every frequency ranges. The EM structure of the SufBCD complex has been deposited in the Electron Microscopy Data Bank under accession number EMD-3163.
Solution Scattering Data Collection and Analysis
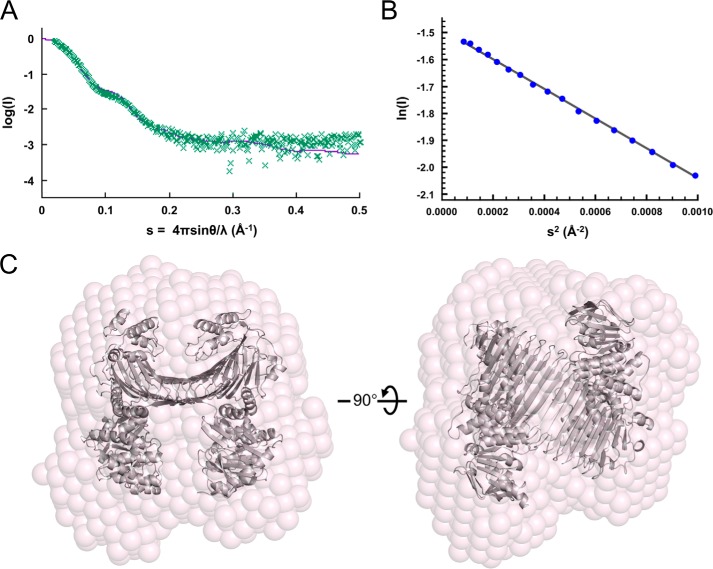
The SufBCD complex (2–18 mg/ml) for small angle x-ray scattering (SAXS) experiments was prepared in 50 mm Tris-HCl (pH 7.8) and 150 mm NaCl. SAXS experiments were performed at room temperature on a Rigaku BioSAXS-1000, using CuKα radiation from the Rigaku FR-X rotating anode x-ray generator. The scattering vector range was set from qmin = 0.009 Å−1 to qmax = 0.69 Å−1 (q = 4πsinθ/λ). Protein samples were placed in a quartz capillary with a diameter of 1.0 mm using an exposure time of 15 min/frame. The final scattering curve was radially averaged from eight frames with the program SAXSLab (Rigaku). Subsequent data were analyzed by the ATSAS program package (38). Data quality was assessed on the basis of the linearity of Guinier plots. Scattering profile simulations from the crystal structure were carried out using CRYSOL (39). Ab initio models were generated using DAMMIF (40). 10 individual reconstructions were aligned and averaged, and the most typical model was generated using DAMAVER (41). The crystal structure was fitted to the dummy model by manually. The SAXS data at 8 mg/ml measurement were used for Fig. 4. Data collection and structural parameters are summarized in Table 3.
FIGURE 4.
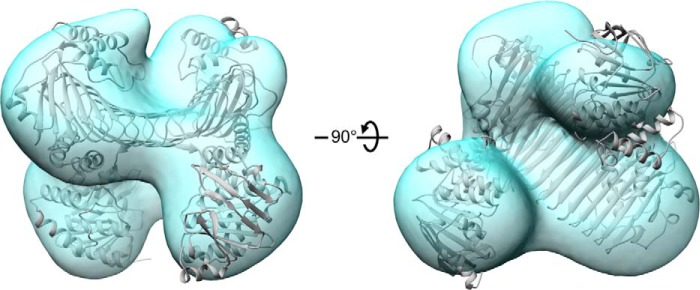
SAXS analyses of the SufBCD complex. A, comparison of the crystal structure and SAXS result. Experimental x-ray scattering curve from the SufBCD complex (green dotted line) and the theoretical curve estimated from the crystal structure (purple solid line) are shown. B, the Guinier plots for the low angle region of the experimental scattering curve at (A). Its linearity indicates the absence of protein aggregation. C, molecular modeling of the SufBCD complex in solution. Left panel, ribbon representation of the crystal structure of the SufBCD complex (gray) is superimposed on the transparent ab initio dummy atom model (pink). Right panel, view rotated by 90° about the horizontal axis relative to the left panel.
TABLE 3.
Data collection and structural parameters for SAXS analysis
| Data collection parameters | |
| Instrument | Rigaku BioSAXS-1000 |
| X-ray source | Rigaku FR-X |
| Wavelength (Å) | 1.54 |
| q range (Å−1) | 0.009–0.69 |
| Exposure time (min) | 15 |
| Concentration range (mg/ml) | 2–18 |
| Temperature | Room temperature |
| Structural parametersa | |
| I(0) from P(r) | 0.225 ± 0.001 |
| Rg from P(r) (Å) | 40. 8 ± 0.6 |
| I(0) from Guinier | 0.222 ± 0.001 |
| Rg from Guinier (Å) | 39.9 ± 0.1 |
| Dmax (Å) | 138.5 |
| x2 of DAMMIF models | 1.22 |
| Molecular mass determination | |
| Mr from I(0) (kDa) | 150.2 |
| Mr from sequence (kDa) | 156.7 |
a Reported for 8 mg/ml measurement.
In Vivo Complementation Assay with Mutated SufC
Site-directed mutations were generated in plasmid pBBR-sufC and introduced into E. coli mutant strain UT109 (16) harboring two plasmids, pUMV22 and pRK-sufAB-DSE (ΔsufCp). UT109 contains deletions of the chromosomal suf (ΔsufABCDSE) and isc (ΔiscUA-hscBA) operons. Normally, deletion of both pathways is lethal because of the lack of the biosynthetic apparatus for Fe-S clusters (10). However, plasmid pUMV22, which harbors genes for mevalonate kinase, phosphomevalonate kinase, and diphosphomevalonate decarboxylase cloned from Streptomyces sp., allows UT109 to grow in the presence of d-mevalonate because the essential Fe-S enzymes IspG and IspH involved in the 2-C-methyl-d-erythritol 4-phosphate pathway for isoprenoid biosynthesis can be bypassed by the foreign mevalonate pathway.4 Upon shift to the absence of mevalonate, the cells are unable to grow without introduction of a functional sufC gene (in this case from pBBR-sufC) to complete the partial Suf system provided by pRK-sufAB-DSE (ΔsufCp).
For the construction of plasmid pRK-sufAB-DSE (ΔsufCp), the sufAB fragment was amplified using primers SufAF and SufB-RSc3, and sufDSE was amplified using primers SufD-FSc5 and SufER (Table 1). Because the coding region of sufC contains the promoter elements for sufDSE, an artificial promoter sequence was added to the upstream region of sufD in the SufD-FSc5 primer. After digestion with restriction enzymes, the two PCR fragments were cloned simultaneously into the XhoI/NheI sites of pRKNSE (42). The expression plasmid pBBR-sufC was constructed by transferring the XbaI-SacI fragment carrying the ribosome-binding sequence and the SufC coding region from the pET-21a (+) derivative (20) to the pBBR1MCS-4 plasmid (43), in which expression was driven by the lac promoter. Mutagenesis of SufC was performed using the pBBR-sufC plasmid as a template, and the primers are listed in Table 1.
ATP Hydrolysis Measurement
ATP hydrolysis rates were determined by a linked enzyme assay that coupled the formation of ADP to the oxidation of NADH, as described previously (44).
Disulfide Cross-linking Experiment
The purified mutant complexes (1 mg/ml) were incubated at room temperature for 30 min in the presence of 5 mm ATP, 5 mm MgCl2, and 0.05 mm CuSO4, and the resultant products were analyzed by Western blot of native PAGE (7.5% gel) and nonreducing SDS-PAGE (12.5% gel) using antibodies against SufB, SufC, and SufD.
Fluorescence Labeling Experiment
For assays using 1-anilinonaphthalene-8-sulfonate (ANS), the purified mutant complexes (1 mg/ml) were mixed with 50 μm ATP, 50 μm MgCl2, 5 μm CuSO4, and 30 μm ANS, and time-dependent changes in fluorescence at room temperature was measured for 30 min. For the N-(7-dimethylamino-4-methylcoumarinyl)-maleimide (DACM) assays, the purified mutant complexes (1 mg/ml) were incubated at room temperature for 30 min in the presence of 50 μm ATP, 50 μm MgCl2, 5 μm CuSO4, and 10 μm DACM, and then their fluorescence was measured. All fluorescence spectra were recorded on a FP-8200 fluorescence spectrometer (JASCO).
In Vivo Iron-Sulfur Cluster Formation Analysis
Site-directed mutagenesis was performed using the pGSO164 plasmid as a template and the primers listed in Table 1, and the genes were expressed in YT2512 (10). The cells were grown in LB medium containing ampicillin (50 μg/ml) and ferric ammonium citrate (0.1 mg/ml) at 37 °C. l-Arabinose was added to 0.2% (w/v) final concentration when the cultures obtained an A600 of 0.4–0.6. After 3 h of expression of SufABCDSE at 37 °C, the cells were harvested by centrifugation. UV-visible absorption spectra were recorded at room temperature on a V-630 spectrophotometer (JASCO).
Protein Sequences
The multiple sequence alignments in Figs. 1 and 6 were performed using Clustal Omega (45), and the figures were prepared with NJplot (46) and ESPript (47). EcoGene (48) accession numbers for the proteins from E. coli K-12 aligned in Figs. 1 and 6 are as follows: SufC, EG13964; UgpC, EG11048; MalK, EG10558; PotA, EG10749; PotG, EG11630; ThiQ, EG11572; TauB, EG13299; SsuB, EG12358; ProV, EG10771; GltL, EG12663; YhdZ, EG12837; GlnQ, EG10389; HisP, EG10452; ArtP, EG11624; CysA, EG10183; PstB, EG10783; FetA, EG13259; FepC, EG10295; FecE, EG10290; FhuC, EG10304; BtuD, EG10128; CcmA, EG12059; DppD, EG12627; SapD, EG12304; DdpD, EG13787; DppF, EG12628; UvrA, EG11061; SbcC, EG10927; MutS, EG10625; RecN, EG10831; MukB, EG10618; and RecF, EG10828.
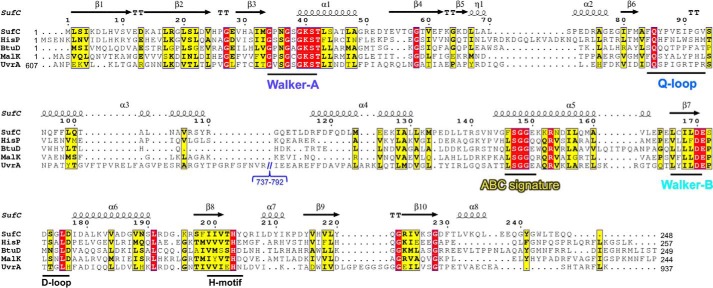
FIGURE 6.
Sequence alignment of SufC with various ABC ATPases from E. coli: HisP, BtuD, and MalK as ABC transporters and UvrA as a SMC protein. Red and yellow indicate identical and similar residues, respectively. Secondary structures of SufC are shown above the alignment with spirals (α-helices) and arrows (β-strands). Motifs conserved in ABC ATPases are shown below the alignment. Residues 737–792 in UvrA, an unrelated region, are omitted from the sequence.
Results
Structure of the SufBCD Complex
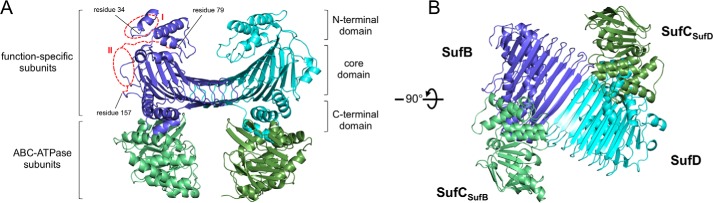
We determined the first crystal structure of the SufBCD complex from E. coli at 2.95 Å resolution (Fig. 2, A and B) by the multiple isomorphous replacement method coupled with anomalous scattering phasing from Hg/Pt derivatives. The SufBCD complex consists of one SufB subunit, two SufC subunits, and one SufD subunit with a stoichiometry of 1:2:1, consistent with previous biochemical experiments (14, 20). Each of the SufC subunits is bound to a subunit of the SufB-SufD protomers and is accordingly termed SufCSufB and SufCSufD. This overall configuration is common among ABC proteins, in which two ABC ATPase subunits bind to function-specific subunits with their ATP-binding motifs facing each other. The two bound SufC subunits, however, are spatially separated, in contrast to the analogous domains/subunits of canonical ABC transporters; the distance between the SufC subunits within the SufBCD complex is more than 40 Å (Fig. 2B). The asymmetric unit contains two complexes with almost identical structures; superposition of them showed some mobility of two SufC subunits that shifted <1 Å toward each other. The r.m.s.d. is less than 0.71 Å for the main chain Cα atoms.
FIGURE 2.
Overall structure of the SufBCD complex from E. coli. A, ribbon representation of the crystal structure of the SufBCD complex. Individual subunits are shown in purple (SufB), cyan (SufD), and green (SufC). The two disordered regions of SufB: residues 1–33 (region I) and 80–156 (region II), are depicted by the red dotted lines and ovals. B, view rotated by 90° about the horizontal axis relative to A.
The structure of the SufBCD complex was further examined by three-dimensional reconstruction imaging based on negative stain electron microscopy (Fig. 3). The structures obtained by both methods agreed closely, confirming the quaternary structure of the SufBCD complex. In addition, the crystal structure was consistent with SAXS data from the as-isolated SufBCD complex in solution (Fig. 4), indicating that the configuration of the SufBCD complex in the crystalline state was not affected by crystal packing.
FIGURE 3.
Three-dimensional reconstruction image of the SufBCD complex obtained by electron microscopy. Left panel, ribbon representation of the crystal structure of the SufBCD complex (gray) is superimposed on the transparent EM structure (light blue). Right panel, view rotated by 90° about the horizontal axis relative to the left panel.
The structures of SufB and SufD are similar and share a common domain organization: an N-terminal helical domain, a core domain consisting of a right-handed parallel β-helix, and a C-terminal helical domain that contains the SufC binding site (Fig. 2A). The β-helix in the core domain of SufB is partly composed of shorter strands than the corresponding domain of SufD, whereas the C-terminal helical domain and the mode of SufC binding are strikingly similar between SufB and SufD. Intriguingly, the mode of binding between the SufC and SufB/SufD subunits is conserved in ABC transporters, an interaction termed the “transmission interface” (49) (discussed below). The heterodimer interface of SufB-SufD protomers consists primarily of 25 hydrogen bonds that form two anti-parallel β-sheets. Although the structure of the SufD subunit in the SufBCD complex was almost identical to that of the previously reported SufD homodimer crystallized alone (50), some structural difference was observed around the interaction site with SufC. The SufD monomers are superimposable, with an r.m.s.d. of 0.59 Å between Cα atoms. Previous biochemical study has suggested that the SufBCD complex binds a flavin as redox cofactor and proposed four motifs involved in FAD binding (24). One proposed motif is located in the disordered region and could not be observed. The remaining motifs were spatially separated in the SufBCD structure, and no conclusions could be drawn about their putative role in flavin binding.
The SufC subunit has two domains, as observed in the members of the ABC ATPase family: a catalytic α/β domain that contains the nucleotide-binding Walker A and Walker B motifs, and a helical domain specific to ABC ATPases containing an ABC signature motif (Fig. 5). The two domains are connected by a Q-loop that contains a strictly conserved glutamine residue (Fig. 6). SufCSufB and SufCSufD have almost identical structures, but some minor structural differences occurred in one loop region that were likely due to crystal packing effects (r.m.s.d. < 0.63 Å).
FIGURE 5.
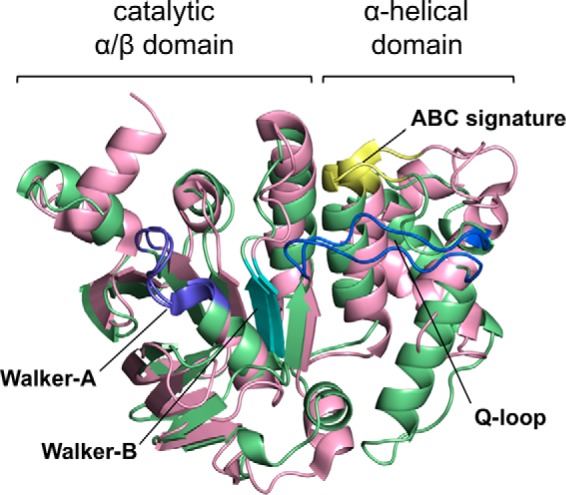
Superposition of the overall structures of E. coli SufC and HlyB (Protein Data Bank code 1MT0). Green and light pink denote the SufC and HlyB structures, respectively. Motifs conserved in ABC ATPases are depicted by different colors: purple (Walker A motif), cyan (Walker B motif), yellow (ABC signature motif), and blue (Q-loop).
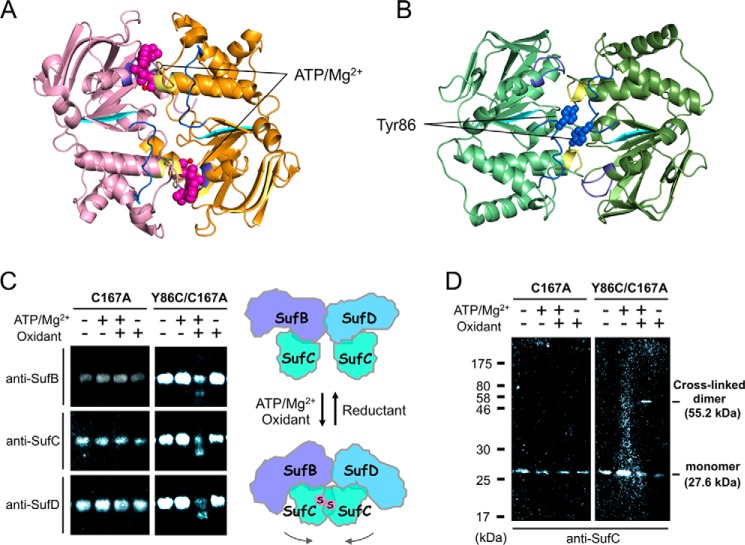
Although the overall structure of SufC subunits in the SufBCD complex is similar to that of monomeric SufC (51), significant structural changes occur around the ATP-binding pocket upon complex formation (Fig. 7). The unique salt bridge observed in the monomeric SufC between Glu-171 (an invariant catalytic residue among typical ABC ATPases (52)) and Lys-152 (Fig. 7A) is cleaved in the complex, allowing the rotation of the Glu-171 side chain toward the ATP-binding pocket (Fig. 7, B and C). Furthermore, His-203, another key residue of ABC ATPase activity (53), is shifted about 4 Å toward Glu-171 in the complex (Fig. 7, A–C). These structural changes rearrange the catalytic pocket of SufC to be suitable for ATP binding and hydrolysis; consequently, the local structure of SufC more closely resembles that of active ABC ATPases. These findings are consistent with recent kinetic experiments showing that the ATPase activity of SufC is enhanced by complex formation with SufB/SufD (19).
FIGURE 7.
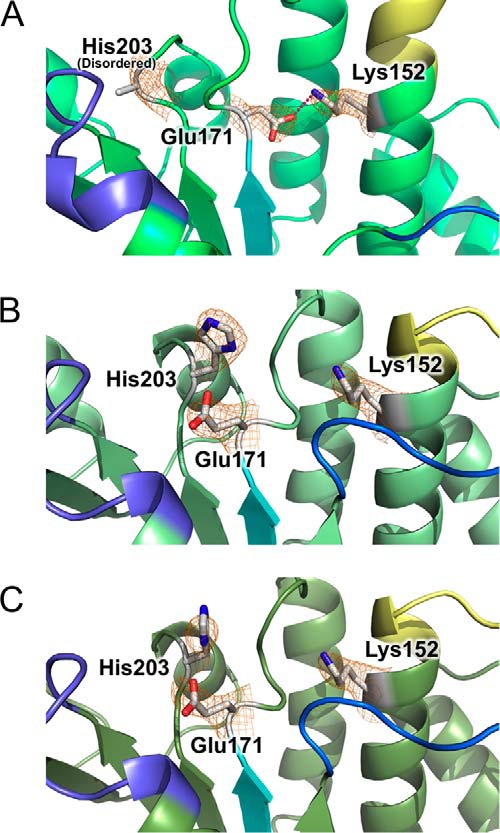
Structural changes and rearrangements of the ATP-binding pocket in SufC. Comparison of the active site structures of SufC among the SufC monomer (Protein Data Bank code 2D3W) (A), the SufCSufB subunit (B), and the SufCSufD subunit (C). The orientation and color coding for the conserved motifs in ABC ATPases are the same as in Fig. 5. Lys-152, Glu-171, and His-203 residues are shown in the stick models, and Fo − Fc maps omitting the side chains of these residues, contoured at 2.0 σ (orange), are overlaid on the stick models. The red broken line in A indicates a salt bridge.
Key ABC ATPase Motifs of SufC Are Critical for Suf Function
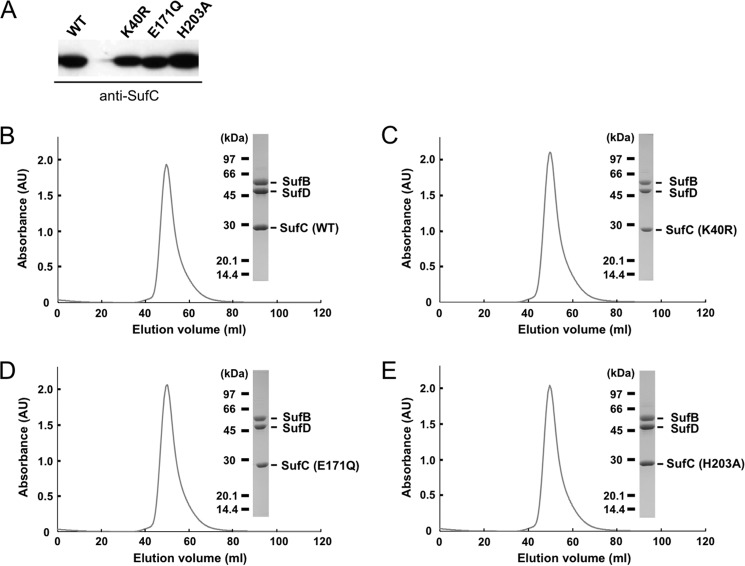
Superposing the structure of E. coli SufC on the typical ABC ATPase, E. coli HlyB of the α-hemolysin export protein (53), reveals very similar overall topologies (Fig. 5). SufC contains highly conserved sets of amino acid residues including an ABC signature motif, Q-loop, D-loop, and H-motif in addition to the Walker A and Walker B motifs, all of which are characteristic of ABC ATPases (Fig. 6). We focused on three strictly conserved amino acid residues considered to be essential for ATP hydrolysis in the ABC ATPases (52–54): the Lys residue in the Walker A motif (corresponding to the Lys-40 in SufC), the Glu residue immediately following the Walker B motif (the Glu-171 in SufC), and the His residue in the H-motif (the His-203 in SufC). In vitro measurements of ATPase activity clearly demonstrated that SufBCD complexes containing mutated SufC proteins (K40R, E171Q, and H203A) almost completely lacked activity (Fig. 8). These mutations did not impair the structural stability of SufC or its interaction with partner proteins (Fig. 9). These results prove that as in the canonical ABC ATPase, the residues of the ABC sequence motifs are responsible for ATPase activity in SufC.
FIGURE 8.
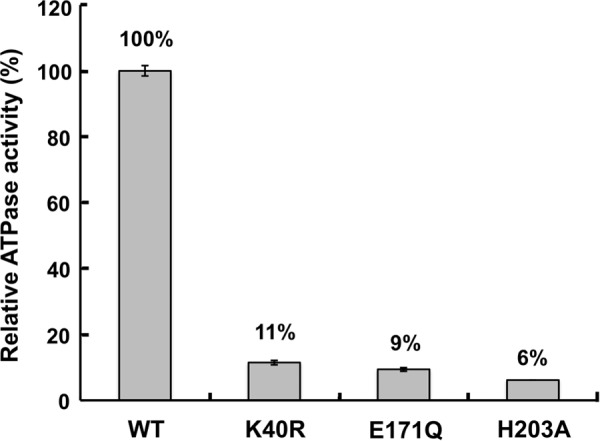
In vitro ATPase activity measurements of the SufBCD complex. Percentages indicate the ratios relative to wild-type complex (∼0.07 μmol of ATP hydrolyzed min−1 mg−1). Error bars, S.D. (n = 3).
FIGURE 9.
Mutational analyses of the SufC protein. A, comparison of the expression in wild-type and mutant SufC proteins by immunoblot analysis using an antibody against SufC. In the cell, the wild-type and SufC variants are expressed equally. B–E, comparison of the size exclusion chromatograms of the native SufBCD complex (B) and the SufC mutant complex of K40R (C), E171Q (D), and H203A (E). These SufC mutants form a stable SufB1-SufC2-SufD1 complex similar to the wild-type complex. Elution curves from the gel filtration column (Sephacryl S-200) are monitored by the absorbance at 280 nm. The inset shows SDS-PAGE analysis of each peak fraction.
To determine whether the ABC ATPase activity of SufC is necessary for the Fe-S cluster biogenesis, we assessed its in vivo function using a recently established method4 in the E. coli Δisc Δsuf mutant strain UT109 (16). The site-directed mutants of SufC, K40R, E171Q, and H203A were not able to complement mutant cells, indicating that these residues are indispensable for in vivo Fe-S cluster biosynthesis (Fig. 10A). Our results are in good agreement with previous experiments regarding the SufC K40R mutant (18). Thus, SufC can behave as an ABC-type ATPase, and the activity is indispensable for in vivo Fe-S cluster assembly.
FIGURE 10.
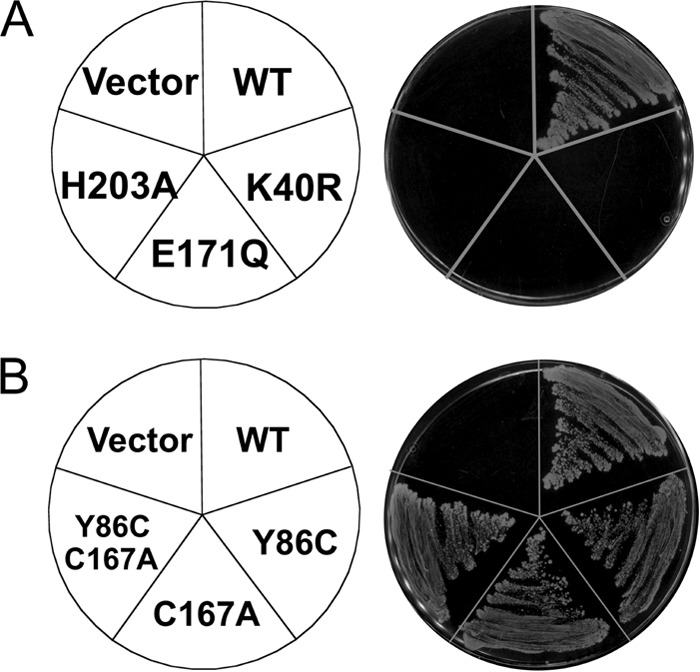
Phenotypic characterization of the SufC mutations. Growth of the mutant cells (Δisc Δsuf) indicates complementation for the loss of sufC. A, site-directed mutants K40R, E171Q, and H203A of SufC cannot complement the E. coli UT109 mutant strain, indicating these residues are indispensable for in vivo Fe-S cluster biosynthesis. B, Site-directed mutants Y86C, C167A, and Y86C/C167A of SufC can complement mutant cells, indicating that these mutations do not affect the in vivo Fe-S cluster biosynthesis.
SufC Forms the Transient Head to Tail Dimer
The structure of the SufB1-SufC2-SufD1 complex revealed the configuration of each subunit: the SufC subunit of the ABC ATPase binds to the C-terminal helical domains of the SufB/SufD subunits, and the two SufCs are oriented face to face. According to the current consensus model, ABC ATPases form a transient head to tail dimer in which two nucleotides are sandwiched at the dimer interface between the Walker motifs of one subunit and the ABC signature motif of the other subunit (Fig. 11A). Based on this concept, we generated a putative dimer model of SufC by superimposing the structure of SufCSufB and SufCSufD onto the dimeric form of the ATP-bound HlyB (H662A) ABC ATPase (53). The resulting model showed that the local structural changes in SufC (mentioned above) enable an ideal association for the head to tail dimer without steric hindrances (Fig. 11B). Despite the favorable modeling results, SufCSufB and SufCSufD subunits are spatially separated in the SufBCD complex, with their ATP-binding motifs facing one another (Fig. 2B); they would have to move ∼20 Å closer to each other to form the head to tail dimer, a distance that is unusually long compared with other structurally characterized ABC proteins.
FIGURE 11.
Disulfide cross-linking analyses of the mutated SufBCD complex. A, dimeric structure of ABC ATPase HlyB (H662A) (Protein Data Bank code 1XEF). Light pink and orange indicate individual subunits. Pink and red denote bound ATPs with van der Waals surfaces and Mg2+ ions, respectively. B, putative dimer model of SufC. Tyr-86 residues are depicted with their van der Waals surfaces. C, disulfide bond formation between two mutated SufC subunits in the SufBCD complex detected by native PAGE/Western blot analyses using antibodies against SufB, SufC, and SufD. D, nonreducing SDS-PAGE/Western blot analyses using an antibody against SufC.
We conducted disulfide cross-linking experiments to determine whether the separated SufC subunits could transiently associate with each other in the SufBCD complex. In the putative dimer model, the Cβ atoms of Tyr-86 in each SufC subunit are in close proximity (< 5.8 Å) (Fig. 11B). Hence, we replaced Tyr-86 with a cysteine to allow for covalent trapping of the transient SufC dimer via disulfide bond formation between the subunits. To simplify analysis, we also replaced the sole native cysteine residue on SufC, Cys-167, with an alanine. These mutations did not affect the function of the SufBCD complex (Fig. 10B). After the mutated complex (SufC-Y86C/C167A) was incubated in the presence of ATP/Mg2+ and an oxidant (CuSO4) to enhance disulfide-bond formation, disulfide-bond formation was assessed by native PAGE analysis. The results revealed an additional band on the gel that migrated more quickly than the as-isolated SufBCD complex (Fig. 11C). No such band was observed when a reducing agent (DTT) was incubated with the sample (data not shown), indicating that disulfide-bond gives the new band. Because Western blot analyses using antibodies against SufB, SufC, and SufD revealed all of the corresponding signals (Fig. 11C), we conclude that the novel band represents a conformationally distinct form of the SufBCD complex. In addition, nonreducing SDS-PAGE/Western blot analyses using an antibody against SufC also revealed an additional band whose molecular size was consistent with a molecule 2-fold larger than SufC (Fig. 11D). These findings strongly support the idea that SufC can form a transient dimer, even within the SufBCD complex, in the presence of ATP/Mg2+. The mobility shift on native PAGE demonstrates that a structural change occurs in the SufBCD complex upon SufC dimerization.
Gross Structural Change of SufB-SufD Protomers upon SufC Dimerization
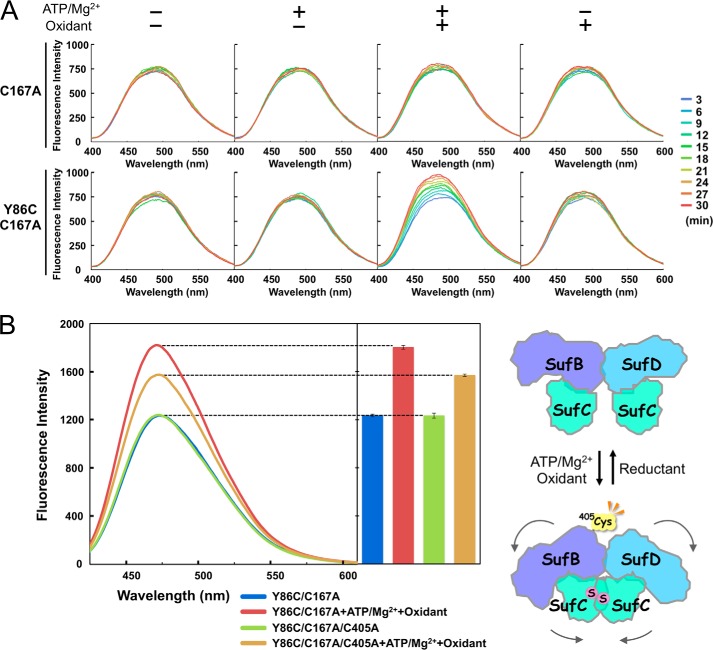
We detected the conformational change of the SufBCD complex, initiated by SufC dimerization, in fluorescent labeling experiments using ANS. ANS, which is poorly fluorescent in an aqueous environment, is highly fluorescent upon binding to hydrophobic regions on protein surfaces (55). To determine whether SufC dimerization induces the exposure of hydrophobic regions in SufB-SufD protomers, we compared the fluorescence of the native and cross-linked complexes (described above). After adding ATP/Mg2+ and an oxidant to the purified mutant SufBCD complex (SufC-Y86C/C167A), we added ANS to the mixture and immediately measured its fluorescence. The results revealed a remarkable increase in fluorescence intensity, depending on the incubation time (Fig. 12A), indicating that a gross structural change of the SufBCD complex accompanied SufC dimerization (cross-link formation). No such fluorescence increase was observed when ATP/Mg2+ or oxidant was omitted from the reaction mixtures or in a control experiment using the single-mutant SufBCD complex where the dimer is not covalently stabilized (SufC-C167A) (Fig. 12A). Therefore, this conformational change was surely elicited by SufC dimerization.
FIGURE 12.
Fluorescent labeling analyses of the mutated SufBCD complex. A, ANS detects exposure of hydrophobic regions in SufB-SufD protomers. B, exposure of Cys-405 of SufB located inside the heterodimer interface between the SufB and SufD protomers under the cross-linked conditions detected by the DACM assays.
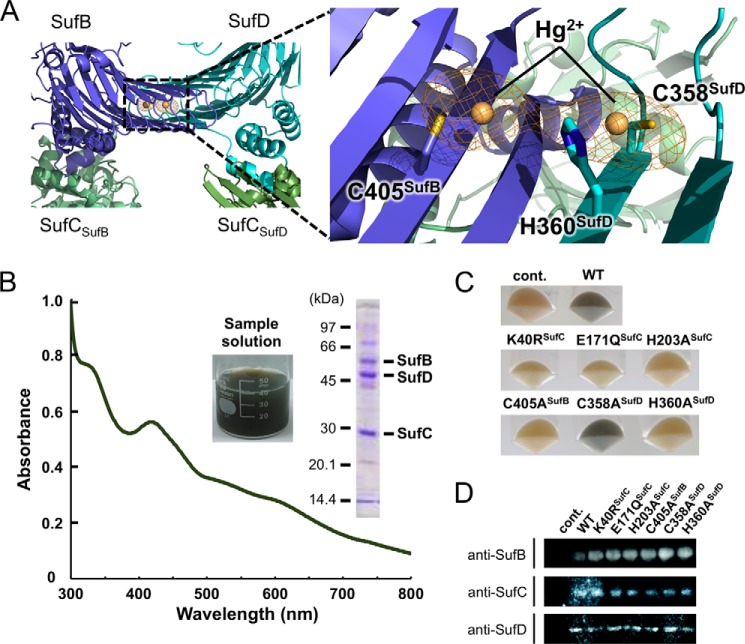
Next, we used another fluorescent reagent to determine whether the interface between SufB and SufD protomers is exposed. To this end, we used the fluorescent thiol reagent DACM, which has a high quantum yield when it reacts with the free cysteine residues on the protein surface (56). The SufBCD complex has a large number of cysteine residues (13 cysteines in SufB, 3 cysteines in SufD, and 1 cysteine in SufC), most of which are buried inside the molecule. We focused on Cys-405 of SufB, which is located at the heterodimer interface between the SufB and SufD protomers (Fig. 13A). This cysteine residue, which is strictly conserved among SufB homologues, is a potential Fe-S cluster assembly site (20). To ascertain whether the Cys-405 could be exposed and detected by DACM, we replaced Cys-405 of SufB with an alanine, in combination with the SufC-Y86C/C167A mutation. Each mutant complex (Y86C/C167A/C405A and Y86C/C167A) was incubated under cross-linked conditions (in the presence of ATP/Mg2+ and an oxidant) and further incubated after addition of DACM. Fluorescence intensity increased following formation of the SufC cross-linked dimer in the complex, indicating that several cysteine residues in the complex were exposed, whereas a significant decrease in fluorescence intensity was observed upon introduction of the SufB C405A mutation (Fig. 12B). Control experiments, in which the incubation was performed under non-cross-linked conditions (i.e. in the absence of ATP/Mg2+ and oxidant), exhibited no difference between mutant complexes. These results demonstrated that SufC dimer formation leads to exposure of the heterodimer interface of the SufB-SufD protomers (at least of Cys-405 of SufB, which is otherwise buried inside the dimer interface). Notably, the invariant residue His-360 of SufD, another candidate for the cluster coordination residue (20), is located close to the Cys-405 of SufB (Fig. 13A), strongly implying that His-360 of SufD could also be exposed by the conformational change.
FIGURE 13.
In vivo Fe-S cluster formation on the SufBCD complex. A, anomalous difference map for Hg2+ ion (contoured at 6.0 σ) in the Hg derivative of the SufBCD complex. A square highlights the binding site of Hg2+, of which the right panel shows a close-up view. Two Hg2+ ions bound to Cys-405 in SufB and Cys-358 in SufD are adjacent to His-360. These residues are depicted with a stick model, and Hg2+ ions are shown as orange balls. B, UV-visible absorption spectrum of the SufBCD complex at an early purification stage. The inset shows the sample solution and SDS-PAGE analysis of the partially purified SufBCD complex used in this measurement. C, colors of the harvested host cells overproducing the SufBCD complex and its variants. The blackish-green color represents the in vivo Fe-S cluster formation on the SufBCD complex. D, comparison of the expression level of SuB/SufC/SufD among the cells harboring the wild-type plasmid or the various mutant plasmids by immunoblot analyses using antibodies against SufB, SufC, and SufD. cont., control.
In Vivo Iron-Sulfur Cluster Formation
During the course of phase determination using the heavy atoms, we noticed that two clear electron densities derived from Hg2+ ion appeared inside the heterodimer interface between the SufB and SufD protomers (Fig. 13A): one Hg2+ ion bound to Cys-405 in SufB and the other bound to Cys-358 in SufD. One coordinated cysteine residue from SufB was the invariant Cys-405, which is presumably one of the residues composing the assembly site for the nascent Fe-S cluster (20). Interestingly, Cys-358 in SufD is located adjacent to His-360 of SufD, another candidate for cluster binding. This observation raises the possibility that Hg2+ ion binds to the authentic iron-binding site involved in Fe-S cluster assembly. Hence, we performed mutation analyses to ascertain whether these residues function as a cluster assembly site.
Because in vitro reconstitution experiments always run the risk that artificial Fe-S clusters will be formed at promiscuous sites (57), we assessed cluster assembly using the color of host cells overproducing the SufBCD complex as well as the SufA, SufS, and SufE components of the pathway. At an early stage of the purification, the fraction containing the SufBCD complex exhibited a blackish-green color; the UV-visible absorption spectrum indicated the presence of a nascent Fe-S cluster with absorption maxima at 340 and 420 nm and a broad shoulder at ∼500–650 nm (Fig. 13B). The color was lost gradually during purification, because the Fe-S cluster is intrinsically fragile with respect to oxygen. Hence, we speculated that cluster formation ability could be evaluated based on the color of the harvested cells prior to exposure of the nascent Fe-S cluster to air by disruption. Harvested cells expressing the wild-type SufBCD complex had a blackish-green color (Fig. 13C), quite similar to that of the partially purified SufBCD complex (Fig. 13B, inset). By contrast, control cells harboring only a vector plasmid were an unremarkable white (Fig. 13C). We thus reasoned that the color of the cells reflects in vivo cluster formation ability, at least for the SufBCD complex, even though the cells included other Fe-S proteins.
As expected, both SufB C405A and SufD H360A mutants had white cells, indicating that these residues are indispensable for cluster assembly, whereas Cys-358 of SufD, the other binding site for Hg2+ ions, was not involved in cluster formation (Fig. 13C). These results suggest that Cys-405 of SufB and His-360 of SufD could serve as the in vivo cluster binding sites. Furthermore, mutants in residues essential for SufC ATP hydrolysis (K40R, E171Q, and H203A; described above) also had white cells (Fig. 13C). In combination with the findings described above, SufC dimerization and conformational changes are indispensable for nascent Fe-S cluster formation (discussed below). The wild-type and mutant SufBCD proteins were present at equal levels in the cells as confirmed by immunoblot analyses (Fig. 13D).
Discussion
Structural Features
In this study, we determined the crystal structure of the SufBCD complex for the first time, revealing its unusual architecture. The core domains of the SufB-SufD protomers consisted of many long strands (8–13 residues in each strand) arranged in a helical architecture, a so-called β-helix. Indeed, the SufD fold (and probably the SufB fold as well) has been categorized as a novel folding superfamily (superhelix turns made of two very long strands each) in the SCOP2 classification database. Moreover, the respective β-helix core domains in SufB and SufD are associated by anti-parallel β-strands to form a novel heterodimeric structure. In the ABC transporters in general, the transmembrane domains consist of a total of 12 helices with 6 helices per monomer, but they have structural diversity appropriate for their respective substrates and functions. In SMC proteins, the function-specific domain consists of a long coiled-coil arm that forms a V-shaped dimeric molecule by interacting with the hinge region for DNA binding. Therefore, the novel β-helix architecture of the SufB-SufD protomers appears to be specialized for the Fe-S cluster biogenesis systems.
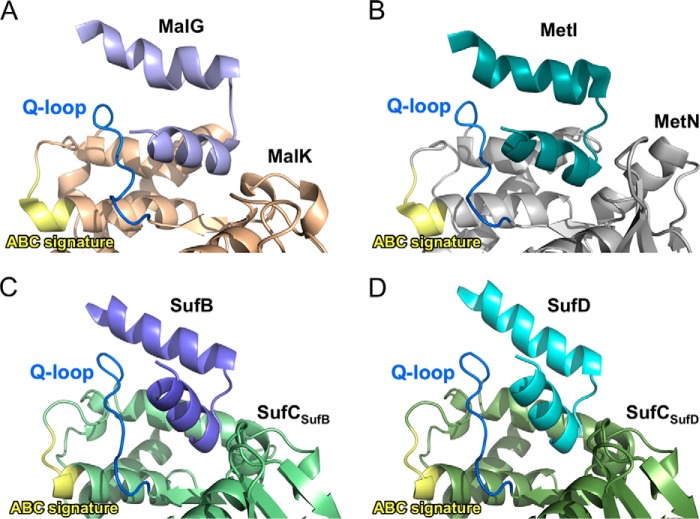
Despite the structural variations in their substrate- and function-specific domains or subunits, ABC proteins share a common mechanism for transmitting the driving force for their respective functions. Recent extensive structural analyses in ABC transporters revealed that the so-called “transmission interface” transmits the dynamic motion of the ABC ATPase to the transmembrane domain during ATP binding and hydrolysis (49), where the Q-loop in the ABC ATPase domain associates with the two short helix-turn-helix motifs in the transmembrane domains. Structural motifs involved in the interaction between SufC and SufB/SufD bear striking similarity to the corresponding configurations in other structurally characterized ABC proteins (Fig. 14). From a structural standpoint, the SufBCD complex also shares the mode of the transmission of the driving force with other ABC proteins: the ATPase activity of SufC drives the conformational change of SufB-SufD protomers for Fe-S cluster biogenesis.
FIGURE 14.
Comparison of the transmission interface in the SufBCD complex and ABC proteins from E. coli. A–D, close-up views of the interface between the substrate/function-specific subunit and the ABC ATPase subunit in the resting (inward-facing) state of MalFGK maltose transporter (Protein Data Bank code 3FH6) (A), the inward-facing state of the MetNI methionine transporter (Protein Data Bank code 3DHW) (B), and the resting state of the SufBCD complex at the SufB-SufC interface (C) and the SufD-SufC interface (D). Each subunit in the complex is depicted in a different color. Substrate/function-specific subunit is displayed only for the two short helix-turn-helix involved in the interaction. Color coding for the conserved motifs in ABC ATPases is the same as in Fig. 5.
Insight into Iron-Sulfur Cluster Biogenesis
In vitro biochemical experiments and in vivo functional analyses based on the crystal structure of the SufBCD complex provided unprecedented insights into the molecular mechanism of Fe-S cluster biogenesis. The main findings are summarized as follows: 1) SufC of ABC-type ATPase forms a transient head to tail dimer within the SufBCD complex during the catalytic step of ATP binding and hydrolysis; 2) SufC dimerization drives gross structural changes of the SufB-SufD protomers, leading to the exposure of Cys-405 of SufB (and probably also His-360 of SufD) inside the heterodimer interface; 3) the conformational changes are directly related to nascent Fe-S cluster formation on the SufBCD complex; and 4) Cys-405 of SufB and His-360 of SufD are most likely to work in concert, possibly serving as the site of in vivo cluster synthesis.
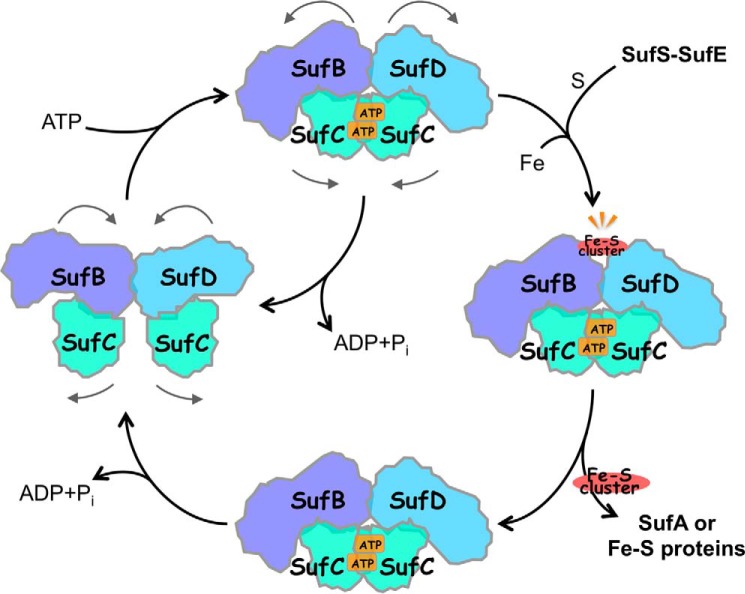
Based on these findings, we propose a mechanism for Fe-S cluster biogenesis for the SufBCD complex (Fig. 15). In the resting state, the SufC ABC-type ATPase in the complex is ready for ATP binding, and the nascent cluster assembly site at the SufB and SufD interface is buried inside the complex. When SufC forms the head to tail dimer upon ATP binding, its dynamic motion is transmitted to the SufB-SufD protomers of the function-specific subunits via the transmission interface. Consequently, the invariant residues involved in Fe-S cluster assembly, Cys-405 in SufB and His-360 in SufD, are exposed to the surface to construct and transfer the nascent Fe-S cluster.
FIGURE 15.
Proposed mechanism of Fe-S cluster biogenesis for the SufBCD complex. Biogenesis cycle starts (at left) in the resting state in which SufC is ready for ATP binding. Upon binding of ATP, SufC forms a head to tail dimer. Consequently, the Fe-S cluster binding site between the SufB and SufD interface is exposed to the surface. Nascent Fe-S cluster is built/transferred and ATP is hydrolyzed, restoring the SufBCD complex to its resting state.
New Perspective on the Suf System
The Suf machinery is thought to represent the ancestral system for Fe-S cluster biogenesis in all kingdoms of life (16). Genes homologous to SufB and SufC are present in a wide range of bacteria, Archaea, and plastids, suggesting that the Suf system is almost ubiquitous in nature. In this study, we revealed that SufB and SufD share novel structural features. Therefore, the dynamic motion of the SufB1-SufC2-SufD1 complex, experimentally demonstrated here, is universally applicable to all Suf systems, even the SufB2-SufC2 complex in the Archaeal Suf system.
Structural and mechanistic understanding of Suf systems may enable the development of new antibiotics that target the SufBCD complex. Indeed, a recent study demonstrated that the Suf system of malaria parasites is essential for survival and plays a fundamental role in maintaining the apicoplast organelle (58). In eukaryotes, including mammalian cells, the ISC system (59) and its dependent CIA system (60) are responsible for nascent Fe-S cluster biogenesis. The extensively studied scaffold protein of the ISC machinery, IscU, has a completely different sequence and tertiary/quaternary structure than the SufBCD complex (61). Therefore, the Suf system, especially the SufB-SufD protomers with its characteristic β-helix fold and dynamic motion, is an eligible target for drug design with minimal risk of harm to the human body.
Author Contributions
F. W. O., K. F., Y. T., and K. W. conceived the project; G. K., K. F., Y. T., and K. W. designed and conducted the experiments; K. H. performed protein purification and crystallization and determined the crystal structure; K. H., E. Y., and N. T. carried out functional analyses and genetic studies; S. K. and K. I. performed single-particle EM reconstructions; K. H. and T. M. performed SAXS experiments; and K. H., K. I., T. M., F. W. O., K. F., Y. T., and K. W. wrote the manuscript.
Acknowledgments
We thank Dr. E. Yamashita, Dr. A. Higashiura, Dr. K. Hirata, and Dr. K. Yamashita for assistance with data collection at the SPring-8 synchrotron radiation facility (Hyogo, Japan). We also thank N. Kaseda, N. Kamimura, H. Kouriki, and Y. Motoyama of the University of Miyazaki for technical assistance, Dr. T. Ishizuka and Dr. Y. Xu of the University of Miyazaki for assistance with fluorescence measurements, and Y. Erikawa for plasmid construction and complementation experiments.
This work was supported by Platform for Drug Discovery, Informatics, and Structural Life Science funded by the Ministry of Education, Culture, Sports, Science, and Technology of Japan. The synchrotron radiation experiments were performed on BL44XU and BL32XU at SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute under Proposals 2011B6535, 2012B6726, 2013AB6828, 2013B6863, 2014AB6963, 2015A6557, and PDIS1235. This work was also supported by the Program to Disseminate Tenure Tracking (to K. W.) from the Ministry of Education, Culture, Sports, Science, and Technology; Ministry of Education, Culture, Sports, Science, and Technology Grants-in-Aid for Scientific Research 25840023 and 21770112 (to K. W.), 15H04472 (to Y. T.), and 23370052 (to K.F.); a Grant-in-Aid for the Japan Society for the Promotion of Science Fellows 12J01292 (to K. H.); a grant from the Japan Foundation for Applied Enzymology (to K. W.); and National Institutes of Health Grants GM 81706 and GM 112919 (to F. W. O.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The atomic coordinates and structure factors (codes 5AWF and 5AWG) have been deposited in the Protein Data Bank (http://wwpdb.org/).
N. Tanaka and Y. Takahashi, manuscript in preparation.
- ABC
- ATP-binding cassette
- SMC
- structural maintenance of chromosome
- Fe-S
- iron-sulfur
- r.m.s.d.
- root mean square deviation
- RCT
- random conical tilt
- SAXS
- small angle x-ray scattering
- ANS
- 1-anilinonaphthalene-8-sulfonate
- DACM
- N-(7-dimethylamino-4-methylcoumarinyl)-maleimide.
References
- 1.Higgins C. F. (1992) ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8, 67–113 [DOI] [PubMed] [Google Scholar]
- 2.Schmitt L., and Tampé R. (2002) Structure and mechanism of ABC transporters. Curr. Opin. Struct. Biol. 12, 754–760 [DOI] [PubMed] [Google Scholar]
- 3.Locher K. P., Lee A. T., and Rees D. C. (2002) The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science 296, 1091–1098 [DOI] [PubMed] [Google Scholar]
- 4.Hopfner K. P., and Tainer J. A. (2003) Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr. Opin. Struct. Biol. 13, 249–255 [DOI] [PubMed] [Google Scholar]
- 5.Holland I. B., and Blight M. A. (1999) ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J. Mol. Biol. 293, 381–399 [DOI] [PubMed] [Google Scholar]
- 6.Hirano T. (2005) SMC proteins and chromosome mechanics: from bacteria to humans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hopfner K. P., Karcher A., Shin D. S., Craig L., Arthur L. M., Carney J. P., and Tainer J. A. (2000) Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell 101, 789–800 [DOI] [PubMed] [Google Scholar]
- 8.Graumann P. L. (2001) SMC proteins in bacteria: condensation motors for chromosome segregation? Biochimie 83, 53–59 [DOI] [PubMed] [Google Scholar]
- 9.Balk J., and Lobréaux S. (2005) Biogenesis of iron-sulfur proteins in plants. Trends Plant Sci. 10, 324–331 [DOI] [PubMed] [Google Scholar]
- 10.Takahashi Y., and Tokumoto U. (2002) A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J. Biol. Chem. 277, 28380–28383 [DOI] [PubMed] [Google Scholar]
- 11.Beinert H., Holm R. H., and Münck E. (1997) Iron-sulfur clusters: nature's modular, multipurpose structures. Science 277, 653–659 [DOI] [PubMed] [Google Scholar]
- 12.Rees D. C., and Howard J. B. (2003) The interface between the biological and inorganic worlds: iron-sulfur metalloclusters. Science 300, 929–931 [DOI] [PubMed] [Google Scholar]
- 13.Loiseau L., Ollagnier-de-Choudens S., Nachin L., Fontecave M., and Barras F. (2003) Biogenesis of Fe-S cluster by the bacterial Suf system: SufS and SufE form a new type of cysteine desulfurase. J. Biol. Chem. 278, 38352–38359 [DOI] [PubMed] [Google Scholar]
- 14.Layer G., Gaddam S. A., Ayala-Castro C. N., Ollagnier-de Choudens S., Lascoux D., Fontecave M., and Outten F. W. (2007) SufE transfers sulfur from SufS to SufB for iron-sulfur cluster assembly. J. Biol. Chem. 282, 13342–13350 [DOI] [PubMed] [Google Scholar]
- 15.Gupta V., Sendra M., Naik S. G., Chahal H. K., Huynh B. H., Outten F. W., Fontecave M., and Ollagnier de Choudens S. (2009) Native Escherichia coli SufA, coexpressed with SufBCDSE, purifies as a [2Fe-2S] protein and acts as an Fe-S transporter to Fe-S target enzymes. J. Am. Chem. Soc. 131, 6149–6153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tokumoto U., Kitamura S., Fukuyama K., and Takahashi Y. (2004) Interchangeability and distinct properties of bacterial Fe-S cluster assembly systems: functional replacement of the isc and suf operons in Escherichia coli with the nifSU-like operon from Helicobacter pylori. J. Biochem. 136, 199–209 [DOI] [PubMed] [Google Scholar]
- 17.Outten F. W., Djaman O., and Storz G. (2004) A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol. Microbiol. 52, 861–872 [DOI] [PubMed] [Google Scholar]
- 18.Saini A., Mapolelo D. T., Chahal H. K., Johnson M. K., and Outten F. W. (2010) SufD and SufC ATPase activity are required for iron acquisition during in vivo Fe-S cluster formation on SufB. Biochemistry 49, 9402–9412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrovic A., Davis C. T., Rangachari K., Clough B., Wilson R. J., and Eccleston J. F. (2008) Hydrodynamic characterization of the SufBC and SufCD complexes and their interaction with fluorescent adenosine nucleotides. Protein Sci. 17, 1264–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wada K., Sumi N., Nagai R., Iwasaki K., Sato T., Suzuki K., Hasegawa Y., Kitaoka S., Minami Y., Outten F. W., Takahashi Y., and Fukuyama K. (2009) Molecular dynamism of Fe-S cluster biosynthesis implicated by the structure of the SufC2-SufD2 complex. J. Mol. Biol. 387, 245–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nachin L., Loiseau L., Expert D., and Barras F. (2003) SufC: an unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 22, 427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Outten F. W., Wood M. J., Munoz F. M., and Storz G. (2003) The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe-S cluster assembly in Escherichia coli. J. Biol. Chem. 278, 45713–45719 [DOI] [PubMed] [Google Scholar]
- 23.Chahal H. K., Dai Y., Saini A., Ayala-Castro C., and Outten F. W. (2009) The SufBCD Fe-S scaffold complex interacts with SufA for Fe-S cluster transfer. Biochemistry 48, 10644–10653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wollers S., Layer G., Garcia-Serres R., Signor L., Clemancey M., Latour J. M., Fontecave M., and Ollagnier de Choudens S. (2010) Iron-sulfur (Fe-S) cluster assembly: the SufBCD complex is a new type of Fe-S scaffold with a flavin redox cofactor. J. Biol. Chem. 285, 23331–23341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyde S. C., Emsley P., Hartshorn M. J., Mimmack M. M., Gileadi U., Pearce S. R., Gallagher M. P., Gill D. R., Hubbard R. E., and Higgins C. F. (1990) Structural model of ATP-binding proteins associated with cystic fibrosis, multidrug resistance and bacterial transport. Nature 346, 362–365 [DOI] [PubMed] [Google Scholar]
- 26.Hirano T. (2002) The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 16, 399–414 [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 28.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutchinson E. G., and Thornton J. M. (1996) PROMOTIF: a program to identify and analyze structural motifs in proteins. Protein Sci. 5, 212–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laskowski R. A., MacArthur M. W., Moss D. S., and Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 32.Kleywegt G. (1996) Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr. D 52, 842–857 [DOI] [PubMed] [Google Scholar]
- 33.Andreeva A., Howorth D., Chothia C., Kulesha E., and Murzin A. G. (2014) SCOP2 prototype: a new approach to protein structure mining. Nucleic Acids Res. 42, D310–D314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeLano W. L. (2012) The PyMOL Molecular Graphics System, version 1.5.0.1, Schroedinger, LLC, New York [Google Scholar]
- 35.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 36.Shaikh T. R., Gao H., Baxter W. T., Asturias F. J., Boisset N., Leith A., and Frank J. (2008) SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc. 3, 1941–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ludtke S. J., Baldwin P. R., and Chiu W. (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 38.Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D., Konarev P. V., and Svergun D. I. (2012) New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svergun D., Barberato C., and Koch M. H. J. (1995) CRYSOL: a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 40.Franke D., and Svergun D. I. (2009) DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Volkov V. V., and Svergun D. I. (2003) Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakamura M., Saeki K., and Takahashi Y. (1999) Hyperproduction of recombinant ferredoxins in Escherichia coli by coexpression of the ORF1-ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster. J. Biochem. 126, 10–18 [DOI] [PubMed] [Google Scholar]
- 43.Kovach M. E., Elzer P. H., Hill D. S., Robertson G. T., Farris M. A., Roop R. M. 2nd, and Peterson K. M. (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176 [DOI] [PubMed] [Google Scholar]
- 44.Rangachari K., Davis C. T., Eccleston J. F., Hirst E. M., Saldanha J. W., Strath M., and Wilson R. J. (2002) SufC hydrolyzes ATP and interacts with SufB from Thermotoga maritima. FEBS Lett. 514, 225–228 [DOI] [PubMed] [Google Scholar]
- 45.Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perrière G., and Gouy M. (1996) WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78, 364–369 [DOI] [PubMed] [Google Scholar]
- 47.Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou J., and Rudd K. E. (2013) EcoGene 3.0. Nucleic Acids Res. 41, D613–D624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hollenstein K., Dawson R. J., and Locher K. P. (2007) Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 17, 412–418 [DOI] [PubMed] [Google Scholar]
- 50.Badger J., Sauder J. M., Adams J. M., Antonysamy S., Bain K., Bergseid M. G., Buchanan S. G., Buchanan M. D., Batiyenko Y., Christopher J. A., Emtage S., Eroshkina A., Feil I., Furlong E. B., Gajiwala K. S., Gao X., He D., Hendle J., Huber A., Hoda K., Kearins P., Kissinger C., Laubert B., Lewis H. A., Lin J., Loomis K., Lorimer D., Louie G., Maletic M., Marsh C. D., Miller I., Molinari J., Muller-Dieckmann H. J., Newman J. M., Noland B. W., Pagarigan B., Park F., Peat T. S., Post K. W., Radojicic S., Ramos A., Romero R., Rutter M. E., Sanderson W. E., Schwinn K. D., Tresser J., Winhoven J., Wright T. A., Wu L., Xu J., and Harris T. J. (2005) Structural analysis of a set of proteins resulting from a bacterial genomics project. Proteins 60, 787–796 [DOI] [PubMed] [Google Scholar]
- 51.Kitaoka S., Wada K., Hasegawa Y., Minami Y., Fukuyama K., and Takahashi Y. (2006) Crystal structure of Escherichia coli SufC, an ABC-type ATPase component of the SUF iron-sulfur cluster assembly machinery. FEBS Lett. 580, 137–143 [DOI] [PubMed] [Google Scholar]
- 52.Smith P. C., Karpowich N., Millen L., Moody J. E., Rosen J., Thomas P. J., and Hunt J. F. (2002) ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell 10, 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaitseva J., Jenewein S., Jumpertz T., Holland I. B., and Schmitt L. (2005) H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J. 24, 1901–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hung L. W., Wang I. X., Nikaido K., Liu P. Q., Ames G. F., and Kim S. H. (1998) Crystal structure of the ATP-binding subunit of an ABC transporter. Nature 396, 703–707 [DOI] [PubMed] [Google Scholar]
- 55.Hawe A., Sutter M., and Jiskoot W. (2008) Extrinsic fluorescent dyes as tools for protein characterization. Pharm. Res. 25, 1487–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Namihisa T., Saifuku K., Ishii H., Watanabe S., and Sekine T. (1983) N-(7-Dimethylamino-4-methylcoumarinyl)-maleimide (DACM): an alternative label for fluorescence tracing. J. Immunol. Methods 56, 125–134 [DOI] [PubMed] [Google Scholar]
- 57.Arakawa S., and Kimura T. (1979) Preparation and partial characterization of iron-sulfur, iron-selenium, and iron-tellurium complexes of bovine serum albumin. Biochim. Biophys. Acta 580, 382–391 [DOI] [PubMed] [Google Scholar]
- 58.Gisselberg J. E., Dellibovi-Ragheb T. A., Matthews K. A., Bosch G., and Prigge S. T. (2013) The suf iron-sulfur cluster synthesis pathway is required for apicoplast maintenance in malaria parasites. PLoS Pathog. 9, e1003655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson D. C., Dean D. R., Smith A. D., and Johnson M. K. (2005) Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74, 247–281 [DOI] [PubMed] [Google Scholar]
- 60.Lill R. (2009) Function and biogenesis of iron-sulphur proteins. Nature 460, 831–838 [DOI] [PubMed] [Google Scholar]
- 61.Shimomura Y., Wada K., Fukuyama K., and Takahashi Y. (2008) The asymmetric trimeric architecture of [2Fe-2S] IscU: implications for its scaffolding during iron-sulfur cluster biosynthesis. J. Mol. Biol. 383, 133–143 [DOI] [PubMed] [Google Scholar]