Abstract
Neuronal electrical impulse propagation is facilitated by the myelin sheath, a compact membrane surrounding the axon. The myelin sheath is highly enriched in galactosylceramide (GalCer) and its sulfated derivative sulfatide. Over 50% of GalCer and sulfatide in myelin is hydroxylated by the integral membrane enzyme fatty acid 2-hydroxylase (FA2H). GalCer hydroxylation contributes to the compact nature of the myelin membrane, and mutations in FA2H result in debilitating leukodystrophies and spastic paraparesis. We report here the 2.6 Å crystal structure of sphingolipid α-hydroxylase (Scs7p), a yeast homolog of FA2H. The Scs7p core is composed of a helical catalytic cap domain that sits atop four transmembrane helices that anchor the enzyme in the endoplasmic reticulum. The structure contains two zinc atoms coordinated by the side chains of 10 highly conserved histidines within a dimetal center located near the plane of the cytosolic membrane. We used a yeast genetic approach to confirm the important role of the dimetal-binding histidines in catalysis and identified Tyr-322 and Asp-323 as critical determinants involved in the hydroxylase reaction. Examination of the Scs7p structure, coupled with molecular dynamics simulations, allowed for the generation of a model of ceramide binding to Scs7p. Comparison of the Scs7p structure and substrate-binding model to the structure of steroyl-CoA desaturase revealed significant differences in the architecture of the catalytic cap domain and location of the dimetal centers with respect to the membrane. These observations provide insight into the different mechanisms of substrate binding and recognition of substrates by the hydroxylase and desaturase enzymes.
Keywords: ceramide, crystal structure, endoplasmic reticulum (ER), membrane enzyme, metalloenzyme, myelin, sphingolipid, structural biology
Introduction
Electrical impulse propagation in the central and peripheral nervous system is facilitated by the myelin sheath, a highly organized multilamellar membrane that wraps around the neuronal axon (1). The myelin sheath is highly enriched in glycosphingolipids, with galactosylceramide (GalCer)2 and its sulfated derivative sulfatide representing the major glycolipid components. GalCer is generated from a ceramide-based precursor composed of a long chain base (LCB) moiety that is derived from palmitoyl-CoA and a very long chain fatty acid (VLCFA) moiety that is typically 26 carbons long (2). Over 50% of GalCer and sulfatide within the myelin sheath are hydroxylated at C-2 of the VLCFA moiety by the enzyme FA2H (3, 4). No other mammalian tissues contain such high levels of hydroxylated lipid. The hydroxylation of the VLCFA moiety, together with the amide and hydroxyl groups associated with the GalCer and sulfatide headgroups, leads to the formation of extensive hydrogen bonding networks that significantly contribute to the tightly organized and compact nature of the myelin membrane (1, 5). Mutations in FA2H cause hereditary leukodystrophy and spastic paraparesis in humans underscoring this enzyme's importance in the nervous system (6–9).
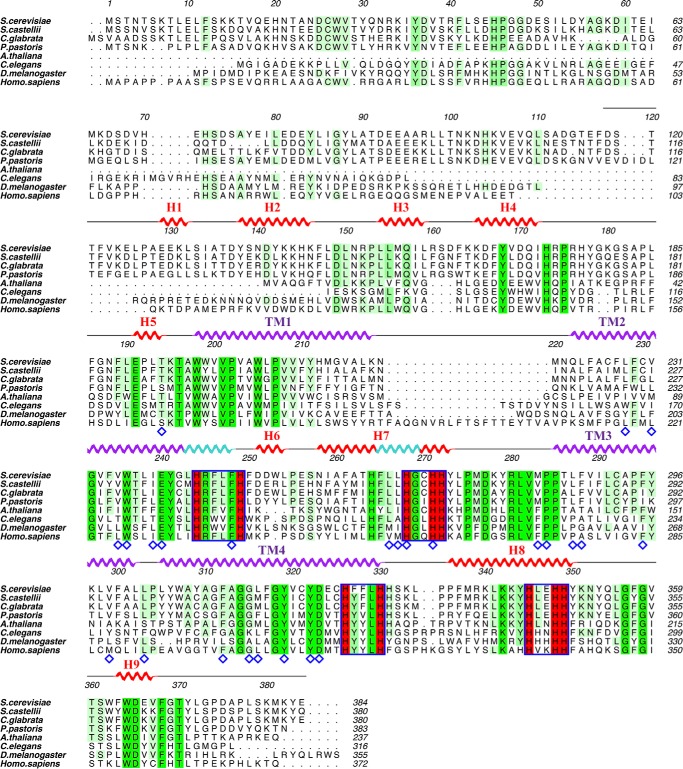
FA2H belongs to a superfamily of integral membrane di-iron-containing enzymes that hydroxylate or desaturate lipid-based substrates via a reaction mechanism that is dependent on NADH and oxygen (10). Members of this superfamily participate in an electron transport chain that moves electrons from NADH through cytochrome b5 reductase and cytochrome b5, with termination at the di-iron center of the enzyme for utilization in subsequent catalysis. Although enzymes within the membrane-bound arm of the superfamily do not share extensive sequence similarity, particular features of their sequences are highly conserved. For example, distinct sequence motifs, termed “histidine boxes,” contain histidine residues that are thought to coordinate two iron atoms at the active center of the enzyme (11, 12). Site-directed mutagenesis of these histidines has shown that they are essential for catalysis (12). In addition, hydropathy plots suggest that the membrane-bound family members contain four transmembrane helices that orient the enzymes such that their N and C termini are located on the cytosolic side of the membrane (13). Many of the integral membrane enzymes of the superfamily, including FA2H, have the intermediate cytochrome b5 electron donor directly fused to the catalytic domain at the N terminus (14).
The Saccharomyces cerevisiae endoplasmic reticulum (ER) contains five members of the membrane-bound hydroxylase/desaturase family, including sphingolipid α-hydroxylase (Scs7p), the yeast homolog of FA2H (11, 15). In S. cerevisiae, Scs7p is responsible for adding a hydroxyl group to the α-carbon of the VLCFA moiety of the ceramide substrate to generate inositol phosphoceramide, one of three major sphingolipids in yeast (15). Scs7p and human FA2H share 36% sequence identity. The other four members of the enzyme family in yeast are C-5 sterol desaturase (Erg3p) (16), methyl sterol oxidase (Erg25p) (17), Δ9 desaturase (Ole1p) (18), and sphingolipid C-4 hydroxylase (Sur2p) (15). When compared with other eukaryotic members of the hydroxylase/desaturase family, all five enzymes exhibit sequence conservation of the “histidine box” motifs and similar hydropathy plots indicative of the presence of four putative transmembrane helices. Scs7p and Ole1p are the only family members in yeast that contain a cytochrome b5 domain fused to the catalytic domain. The cytochrome b5 domain of Scs7p is fused at the N terminus of the enzyme, whereas the b5 domain of Ole1p is fused at the C terminus.
Only one integral membrane representative of the hydroxylase/desaturase superfamily, steroyl-CoA desaturase (SCD1) (19, 20), has been successfully crystallized due in large part to the difficulty of producing suitable quantities of stable protein for structural studies. We report here the 2.6-Å crystal structure of scScs7p, which is the first hydroxylase enzyme within the hydroxylase/desaturase superfamily to be characterized at the molecular level. The core structure is composed of a helical catalytic cap domain that sits atop four transmembrane helices. The dimetal-binding site is located within the catalytic cap domain and is occupied by zinc atoms coordinated by the highly conserved histidine side chains from four histidine box motifs. A phenotypic assay was used in conjunction with site-directed mutations to confirm the role of the dimetal-binding histidine residues. This approach revealed that Tyr-322 and Asp-323 are critical determinants involved in the hydroxylase reaction. Although global folds of the scScs7p and SCD1 enzymes are similar, the two structures exhibit significant differences in the architecture of the cytosolic cap domain, the location of the dimetal center, and the modes of substrate access and binding. Comparison of the scScs7p and SCD1 structures, coupled with molecular dynamics simulations, allows modeling of a ceramide substrate below the dimetal center of scScs7p.
Experimental Procedures
Construct Design and Mutagenesis
For protein expression, the SCS7 gene, encoding sphingolipid α-hydroxylase (Scs7p), was PCR-amplified from S. cerevisiae genomic DNA and subsequently inserted into vector pSGP46, containing both C-terminal ZZ and His10 affinity tags along with a 3C protease site for tag removal, via ligation-independent cloning as described previously (21). A deletion construct, with N-terminal residues 2–95 removed (Δ95scScs7p), was also engineered using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA) with the wild-type scScs7p in vector pSGP46 as the template. All constructs were confirmed by DNA sequencing.
Protein Expression and Purification
The overproduction of scScs7p and Δ95scScs7p for structural studies followed the general procedures outlined previously (21), with some modifications. Both scScs7p and Δ95scScs7p in pSGP46 were transformed into S. cerevisiae strain BJ5460 (ATCC 208285; MATa ura3-52 trp1 lys2-801 leu2 Δ1 his3-Δ200 pep4::HIS3 prb1 Δ1.6R can1 GAL+). A 1-ml culture of cells expressing either scScs7p or Δ95scScs7p in BJ5460 was utilized to inoculate 100 ml of SD-Ura media. After overnight growth, the 100-ml culture was added to 8 liters of SD-Ura media in a BioFlo 3000 fermentor (New Brunswick Scientific) and grown at 30 °C with agitation at 500 rpm to an A600 of 1.0. At this time, an additional 2 liters of 5× YP media (1% yeast extract; 2% peptone) was added to the fermentor. Expression of scScs7p and Δ95scScs7p from pSGP46 was controlled by the ADH2 promoter, which is induced when glucose is completely depleted from the growth medium. Thus, the glucose concentration of the medium was monitored utilizing a glucose monitor. Upon depletion of glucose, the cells were grown for 16–18 h and harvested via centrifugation.
For purification of scScs7p and Δ95scScs7p, the cell pellets from a 10-liter fermentor growth was resuspended in 50 mm Tris, pH 8.0, 150 mm NaCl at a ratio of 1 ml of resuspension buffer per 0.35 g wet cell pellet weight. The cells were lysed by four passes at ∼24,000 p.s.i. through a Microfluidics M110 Microfluidizer. PMSF was added to the cell lysate at a ratio of 1.7 mg of PMSF/liter of cell pellet utilized. The lysed cells were centrifuged at 4000 × g for 10 min at 4 °C to remove cell debris. The supernatant was subsequently ultracentrifuged at 125,000 × g for 120 min at 4 °C to isolate the membrane fraction. The membrane fraction was resuspended in 50 mm Tris, pH 8.0, 150 mm NaCl at a ratio of 10 ml of buffer/g of membrane, along with the addition of 2 tablets of Complete Protease Inhibitor mixture (Roche Applied Science), using a homogenizer. For solubilization, β-octyl glucoside (βOG) was added to a final concentration of 2% (w/v). The mixture was stirred at room temperature for 90 min, followed by ultracentrifugation at 125,000 × g for 40 min at 4 °C to remove insoluble debris. Affinity purification was carried out in batch utilizing the C-terminal ZZ tag in conjunction with IgG-Sepharose 6 Fast Flow resin (GE Healthcare) pre-equilibrated in 50 mm Tris, pH 8.0, 150 mm NaCl, 1% βOG (w/v). A total of 10 ml of pre-equilibrated IgG-Sepharose 6 Fast Flow resin was added to the solubilized membrane fraction, followed by incubation with rocking for 2.5 h at 4 °C. The resin was then poured into a 13 × 2.5-cm column and washed with 100 ml of 50 mm Tris, pH 8.0, 150 mm NaCl, 1% βOG (w/v). The protein was subsequently cleaved from the IgG-Sepharose resin by adding 1 mg of rhinovirus 3C protease, engineered with N-terminal His6 and maltose-binding protein affinity tags (22), in 10 ml of 50 mm Tris, pH 8.0, 150 mm NaCl, 1 mm DTT, 1% βOG (w/v) and incubating for 16 h at 4 °C. The cleaved protein was eluted from the IgG-Sepharose resin by washing with 100 ml of 50 mm Tris, pH 8.0, 150 mm NaCl, 1% βOG (w/v). 3C protease was removed by adding 1 ml of His-Select cobalt resin (Sigma) equilibrated in 50 mm Tris, pH 8.0, 150 mm NaCl, 1% βOG (w/v) to the combined elution and wash fractions and incubating for 1 h with rocking at 4 °C. The slurry was poured into a 13 × 1-cm column and washed with 10 column volumes of 50 mm Tris, pH 8.0, 150 mm NaCl, 1% βOG (w/v). The wash fractions were pooled, concentrated to ∼2 ml using an Amicon Ultra-15 concentrator with a 50-kDa molecular mass cutoff (30 kDa for Δ95scScs7p; Millipore), and subjected to size-exclusion chromatography using a Superdex 200 HiLoad 16/60 column (GE Healthcare) equilibrated in 20 mm Hepes, pH 7.0, 150 mm NaCl, 1% βOG (w/v). Peak fractions were pooled and concentrated to ∼3 mg/ml for crystallization trials. 10-Liter fermenter growths consistently yielded ∼2.5 mg of pure scScs7p and Δ95scScs7p.
Crystallization
Initial crystallization screening was conducted using the 1536 condition tailored membrane protein screen (23) in the High-Throughput Crystallization Laboratory at the Hauptman-Woodward Institute (24). Crystals of scScs7p were grown at 14 °C using the microbatch-under-oil method by combining 2 μl of protein solution at 3.3 mg/ml with 2 μl of 20% polyethylene glycol (PEG) 3350 (w/v), 100 mm Hepes, pH 7.0, 300 mm ammonium sulfate and then covering the drop with 50 μl of mineral oil. Crystals appeared within 2 days and grew to ∼100 μm in a week. Crystals of Δ95scScs7p were also grown at 14 °C using the microbatch-under-oil method by combining 2 μl of protein solution at 2.5 mg/ml with 2 μl of 10% PEG 2000, 100 mm sodium citrate, pH 5.6, 50 mm ammonium sulfate, followed by covering the drop with 50 μl of mineral oil. Δ95scScs7p crystals grew to a size of ∼300 μm within a week. We noted that scScs7p and Δ95scScs7p crystals deteriorated over time in the crystallization trays. Thus, crystals were harvested after 1 week of growth and frozen in liquid nitrogen for diffraction analysis. For cryopreservation, crystals were harvested into the crystallization mixtures described above that also included 20% ethylene glycol. To generate the heavy atom derivative of scScs7p, crystals were harvested from drops and soaked in a solution of 11% PEG 3350, 50 mm Hepes, pH 7.0, 150 mm ammonium sulfate, 75 mm NaCl, and 1% βOG (w/v), which was saturated with K2HgI4 for 25 min. The crystals were then back-soaked into 11% PEG 3350, 50 mm Hepes, pH 7.0, 150 mm ammonium sulfate, 75 mm NaCl, 1% βOG (w/v), and 20% ethylene glycol for 2 min, followed by freezing in liquid nitrogen.
Structure Solution and Refinement
X-ray diffraction data were collected on beamline 17-ID at the Advanced Photon Source (Argonne National Laboratory) using a Pilatus 6 M detector and processed with HKL2000 (25). Data collection statistics are reported in Table 1. Single isomorphous replacement with anomalous scattering (SIRAS) phasing was utilized to solve the structure of scScs7p. PHENIX.autosol (26) located the positions of eight mercury atoms within the asymmetric unit using data to 6 Å resolution. These initial phases were then used in conjunction with the native scScs7p dataset to further improve the phases and extend the data to 3.8 Å in PHENIX. At this stage, the experimental map exhibited clear molecular boundaries for the two molecules of scScs7p in the asymmetric unit. Manual model building was carried out using COOT (27). The mercury atoms were found to interact with the cysteine residues in the protein, which aided in building a model with proper amino acid register. An x-ray fluorescence scan revealed a 2:1 ratio of zinc to iron in the crystals (data not shown). We therefore modeled two zinc atoms in the structure instead of the iron atoms that were expected to be present based on previous biochemical studies on enzymes of the hydroxylase/desaturase family. No electron density was observed for the N-terminal 116 residues corresponding to the cytochrome b5 domain and linker. The cytochrome b5 domain is present in the crystals based both on their brown color and on a characteristic signal present at the iron-edge in an x-ray fluorescence scan. We subsequently carried out molecular replacement with PHASER (28) utilizing a cytochrome b5 search model, but we were unsuccessful in locating this domain. Thus, we conclude that the N-terminal cytochrome b5 domain is disordered within scScs7p crystals. The final model was refined in PHENIX.refine to R and Rfree values of 24.6 and 27.1%, respectively, utilizing NCS restraints, individual B-factor, and TLS refinement. The structure of Δ95scScs7p was solved to 2.6 Å resolution using single-wavelength anomalous dispersion methods at the zinc edge to generate initial phases. Two molecules were found in the asymmetric unit, corresponding to scScs7p monomers composed of residues 117–384 and residues SGLEVL from the C-terminal affinity tag. The final model was refined to R and Rfree values of 19.7 and 23.9%, respectively utilizing individual B-factor and TLS refinement (Table 1). The two monomers were virtually identical, with a root mean square deviation between monomers of 0.33 Å (268 Cα atoms).
TABLE 1.
Crystallographic statistics
| Crystallographic parameter | scScs7p | scScs7p mercury derivative | ΔN95 scScs7p |
|---|---|---|---|
| Space group | I213 | I213 | I213 |
| No. in asymmetric unit | 2 | 2 | 2 |
| Unit cell length (Å) | |||
| a | 202.99 | 203.20 | 202.76 |
| b | 202.99 | 203.20 | 202.76 |
| c | 202.99 | 203.20 | 202.76 |
| α = β = γ (°) | 90.00 | 90.00 | 90.00 |
| Wavelength (Å) | 1.74 | 1.74 | 1.25 |
| Resolution (Å) | 50-3.80 | 50-6.0 | 50-2.6 |
| Highest resolution shell (Å) | 3.94-3.80 | 6.1-6.0 | 2.65-2.60 |
| Rmergea | 12.2 (159.2)b | 11.7 (56.9) | 7.1 (164.6) |
| Rmeas | 13.8 (182.4) | 7.8 (193.3) | |
| Rpim | 6.5 (87.4) | 3.3 (99.4) | |
| CC*c | 1.00 (0.68) | 1.00 (0.52) | |
| Total observations | 60,376 | 29,274 | 227,550 |
| Total unique | 13,804 | 3594 | 42,246 |
| I/σ(I) | 8.5 (1.1) | 13.5 (4.0) | 12.1 (0.8) |
| Completeness (%) | 99.5 (99.7) | 100 (100) | 99.2 (87.9) |
| Redundancy | 4.4 (4.3) | 8.1 (8.5) | 5.4 (3.4) |
| No. of reflections in refinement | 13,852 | 42,170 | |
| No. of reflections in Rfree set | 699 | 2133 | |
| No. of atoms in refinement | 4470 | 4859 | |
| R | 24.6 (35.4) | 19.7 (34.1) | |
| Rfree | 27.1 (37.7) | 23.9 (37.6) | |
| Average B-factor, protein (Å2) | 65.8 | 83.6 | |
| Average B-factor, ligand (Å2) | 59.3 | 106.2 | |
| Mean positional error (Å) | 0.58 | 0.40 | |
| r.m.s.d.d bond length (Å) | 0.003 | 0.007 | |
| r.m.s.d. bond angle (°) | 0.723 | 1.102 | |
| Ramachandran plot | |||
| Allowed (%) | 94.0 | 94.7 | |
| Generous (%) | 6.0 | 4.9 | |
| Disallowed (%) | 0.0 | 0.4 | |
Yeast Viability Assay
S. cerevisiae exhibits sensitivity toward the antifungal cyclic lipodepsinonapeptide syringomycin E, produced by Pseudomonas syringae pv. syringae (29). We replaced SCS7 with the hygromycin B resistance marker to generate an scs7-Δ strain in BY4742 (MATα, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0). The scs7-Δ strain was used to assay the function of constructs used for crystallization and to analyze the effect of point mutations on Scs7p activity. Genes encoding both the scScs7p-ZZ-His10 and Δ95scScs7p-ZZ-His10 crystallization constructs were cloned into the BamHI1 and EcoRI sites of vector pRS316-ADH1 (30). The variants H173A, H244A, H249A, H264A, H268A, H271A, H272A, Y319A, Y322A, D323A, H326A, H330A, H331A, H345A, H348A, and H349A were created using the QuikChange mutagenesis kit with wild-type scScs7p in pRS316-ADH1 as the template. All constructs were confirmed by DNA sequencing. Both wild-type and mutant scScs7p constructs were transformed into the scs7-Δ strain. To test for growth on syringomycin E, yeast cells were grown in SD-Ura media overnight at 30 °C to an A600 of ∼1.0. The cells were subsequently diluted to 0.3 A600 units, and 2 μl of each dilution was spotted onto SD-Ura plates and SD-Ura plates containing 0.5 μg/ml syringomycin E. The plates were incubated at 30 °C for 2 days and analyzed for growth. For each mutant, at least two colonies from independent transformations were chosen to ensure that they behaved similarly on plates containing syringomycin E. The expression of all mutant constructs was confirmed via Western blot analysis using the protocol outlined previously (31) (data not shown). A total of 15 μl of each mutant construct was run out on a 4–20% SDS-polyacrylamide gel, followed by transfer to a PVDF membrane and probing with anti-mouse IgG. Western blot analysis was performed three independent times to confirm expression of all the mutant constructs.
Modeling of the Enzyme-Substrate Complex
To gain insight into how substrate binds to scScs7p, molecular dynamics was performed with GROMACS (32) utilizing the CHARMM36 force field (33) and SPCE water model. Topology and parameter files for the phytoceramide substrate were generated with SwissParam (34). The model of Δ95scScs7p was inserted into a pre-equilibrated lipid bilayer comprised of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine using the CHARMM-GUI (35) and solvated in an 11 × 11 × 10-nm3 rectangular box. Charge was neutralized by replacing seven water molecules with seven chloride ions, followed by energy minimization utilizing a steepest decent protocol. The energy-minimized system was then subjected to a 100-ps NVT equilibration using a modified Berendsen thermostat at a temperature of 310 K, followed by a 1-ns NPT equilibration using the Parrinello-Rahman barostat. Molecular dynamics simulations for Δ95scScs7p within the 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine bilayer were performed until a stable structure with the proper metal coordination was identified. The stabilized Δ95scScs7p structure was then utilized to generate the enzyme-substrate complex. Phytoceramide, containing a 26-carbon VLCFA moiety, was used as the model substrate based on the findings of Beeler and co-workers (11). The substrate was modeled in two different orientations, with the VLCFA moiety placed in the upper channel and the LCB moiety in the lower channel and vice versa. Both ligand-bound models were subsequently equilibrated at 310 K prior to running a final 100-ns MD simulation. All simulations were carried out on the cluster at the Center for Computational Resources at the University at Buffalo.
Structural Analysis
Model validation was conducted using MOLPROBITY (36), and figures were generated using CCP4_MG (37). The positioning of scScs7p within the membrane was generated using the PPM server (38). Measurements of the distances between zinc atoms and the cytosolic membrane were carried out using GEOMCALC in the CCP4 suite of programs (39). Atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession numbers 4ZR0 (full-length scScs7p) and 4ZR1 (Δ95scScs7p).
Results
Structure of scScs7p
We cloned Scs7p orthologs from nine different yeast species and subsequently expressed these constructs in S. cerevisiae to identify protein constructs that exhibited enhanced stability and suitability for crystallization (21). Purified protein from the S. cerevisiae Scs7p (scScs7p) ortholog was readily crystallized. The sequence of scScs7p shares 36% sequence identity with Homo sapiens FA2H (Fig. 1). Crystallization utilizing the microbatch-under-oil method generated brown-colored crystals of scScs7p that diffracted to 3.8 Å resolution. Although residues 117–384, corresponding to the hydroxylase domain, were successfully built into the electron density map, there was no interpretable electron density for the N-terminal 116 residues. This region, corresponding to the heme-containing cytochrome b5 domain and linker peptide, is disordered within the crystals. To improve resolution, we engineered a hydroxylase domain construct with the N-terminal cytochrome b5 domain removed (Δ95scScs7p). Crystals of the Δ95scScs7p construct were colorless, confirming the absence of the heme-containing cytochrome b5 domain, and diffracted to 2.6 Å resolution. The resulting electron density map was significantly higher in quality than that of the full-length construct, allowing for a more accurate model of the hydroxylase domain to be built.
FIGURE 1.
Sequence alignment of Scs7p orthologs. The sequences of Scs7p family members were aligned using PROMALS3D (49) and displayed using ALINE (50). The NCBI database ID for each sequence is as follows: S. cerevisiae (NP_013999); S. castellii (XP_003675406); C. glabrata (XP_446117); Pichia pastoris (CABH01000051.1); Arabidopsis thaliana (NP_181023); Caenorhabditis elegans (NP_492678); Drosophila melanogaster (NP_610279); and H. sapiens (NP_077282). The helical secondary structure for the hydroxylase domain is displayed above the sequence alignment. Helices from the catalytic cap domain are colored red, and transmembrane helices are colored purple. π-Helix segments within helices are depicted in light blue. Completely conserved residues within the alignment are shaded green, and highly conserved residues are shaded in lime. Histidine box motifs are outlined in blue with the conserved histidines shaded in red. Residues lining the substrate binding channels are denoted with a blue diamond.
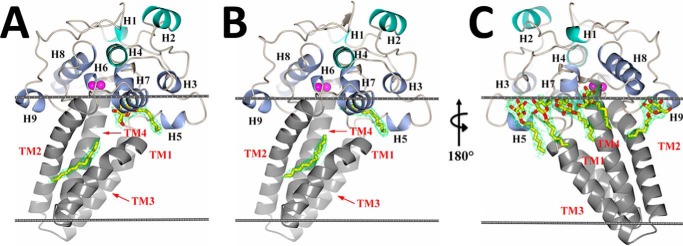
The structure of the hydroxylase domain of scScs7p is composed of 13 helical secondary structural elements resembling a mushroom in overall shape (Fig. 2). The four predicted transmembrane helices (TM1–TM4) form the stalk, and the other nine helices (H1–H9) form the catalytic cap of the mushroom. TM2 and TM4 contain 27 and 26 residues, respectively, whereas TM1 and TM3 are shorter and comprise 21 and 18 residues. Short 3-residue loops connect TM1/TM2 and TM3/TM4 on the luminal side of the ER membrane. Consistent with previous structural predictions for the membrane-bound members of the hydroxylase/desaturase family, the N and C termini are located on the cytosolic side of the ER membrane (13).
FIGURE 2.
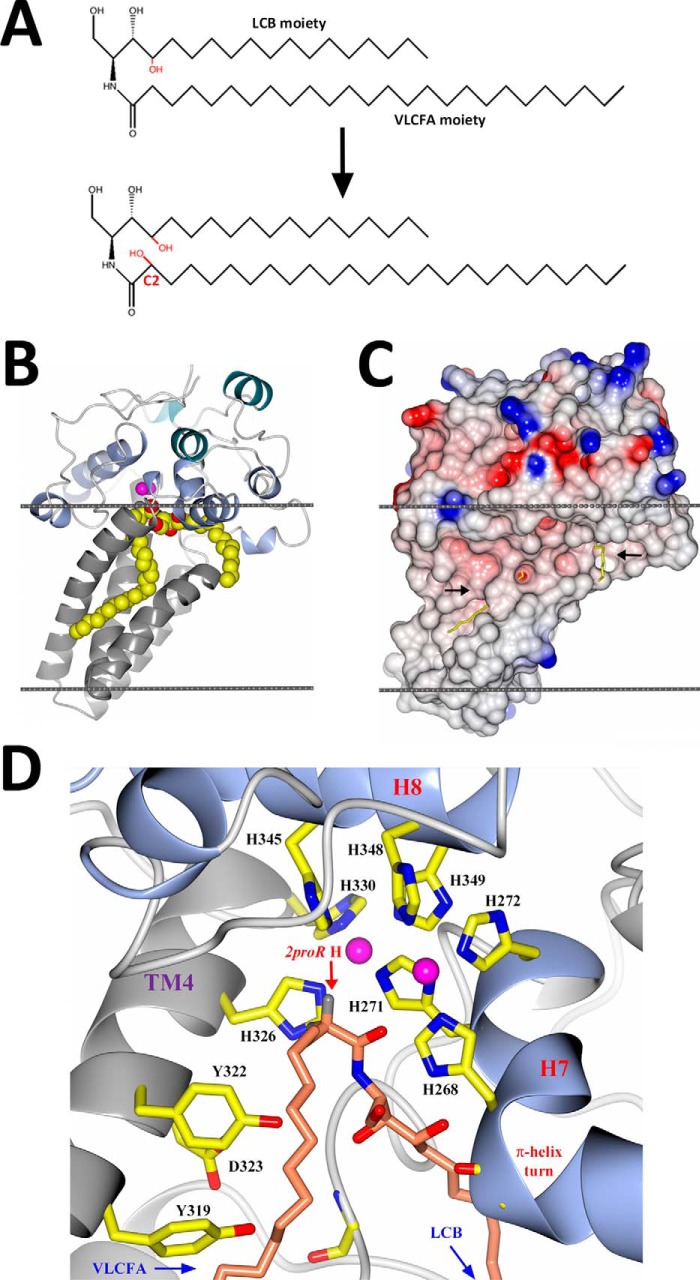
Structure of scScs7p. Schematic representations of the secondary structural elements of the scScs7p hydroxylase domain placed within a model lipid bilayer (gray lines) are depicted. Transmembrane helices TM1–TM4 in the stalk domain are colored in gray, and the helices at the apex of the catalytic cap domain (H1, H2, and H4) are colored green. The remaining helices (H3, H5, H6, H7, H8, and H9) are colored blue. The two zinc atoms within the dimetal center are colored magenta. 2Fo − Fc electron density (green) for unidentified lipid (or βOG), carried along from the expression and purification process, is shown bound in the lower and upper channels of the enzyme for monomer A (A) and monomer B (B). C, view of A and B rotated 180° depicting βOG detergent molecules bound near the amphipathic helices at the proposed membrane interface. 2Fo − Fc electron density contoured at 1.5σ for βOG is shown in green. Unidentified lipid and βOG molecules are depicted as sticks, with carbon and oxygen atoms colored yellow and red, respectively.
The catalytic cap domain is defined by three stretches of residues according to their location with respect to the transmembrane helices. The N-terminal stretch comprises 91 residues (Phe-117 through Ala-197) prior to TM1. There is a short stretch of 38 residues (Phe-248 through Pro-284) between TM2 and TM3, followed by a 54-residue stretch (His-331 through Glu-384) beyond TM4 at the C terminus. The helices within the catalytic cap domain can be divided into two layers. Helices H1, H2, and H4 are located the apex of the catalytic cap domain, whereas helices H3, H5, H6, H7, H8, and H9 lie closer to cytosolic face of the membrane. Moreover, helices H3, H5, H7, and H9 are amphipathic in nature, and their observed orientation suggests that they provide additional stabilization of the catalytic cap domain on the surface of the membrane. Indeed, residues within helices H5, H7, and H9 form stabilizing interactions with βOG detergent molecules utilized to stabilize the protein during purification and crystallization (Fig. 2C).
Dimetal Center
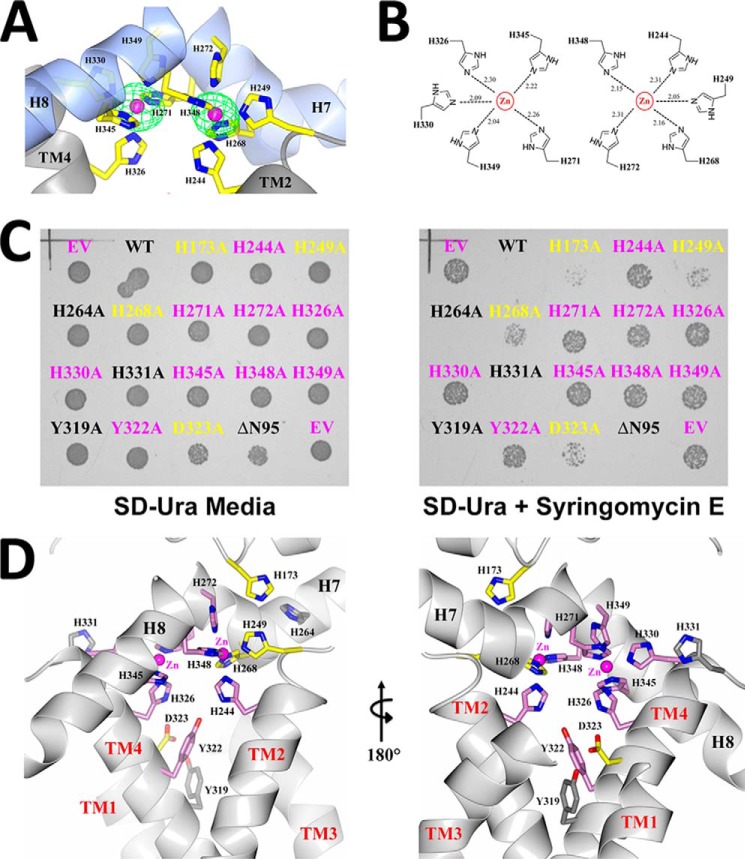
Despite the fact that Scs7p and FA2H are classified as di-iron enzymes within the hydroxylase/desaturase superfamily (13), we routinely observed the presence of zinc atoms in purified enzyme preparations and within crystals of scScs7p. X-ray fluorescence measurements, coupled with diffraction data collection at the zinc absorption edge yielded anomalous difference peaks that unambiguously identified the metal atoms in the crystal as zinc. Two zinc atoms were modeled within the core of the catalytic cap domain (Fig. 3A). These reside 6.25 Å apart and are coordinated by the side chains of 10 highly conserved histidine residues (Fig. 1). Based on its depth and spatial location with respect to the resolved detergent molecules in the structure, the dimetal-binding site is proposed to lie at the surface of the cytosolic side of the ER membrane. The zinc-coordinating histidine residues are located in four motifs, defined as Ia, Ib, IIa, and IIb (11). Motifs Ia and IIa are located at the C-terminal ends of TM2 and TM4, respectively, whereas motifs Ib and IIb are located in H7 and H8 within the catalytic cap domain (Fig. 1). Each zinc atom is coordinated to the Nϵ2 atoms of five histidine side chains, with coordination exhibiting square pyramidal geometry as follows: zinc1 with His-244 (TM2), His-249 (TM2), His-268 (H7), His-272 (H7), and His-348 (H8); and zinc2 with His-271 (H7), His-326 (TM4), His-330 (TM4), His 345 (H8), and His-349 (H8). The average Zn-Nϵ2 distance is 2.20 Å (Fig. 3B).
FIGURE 3.
scScs7p dimetal center. A, schematic depiction of the dimetal center and coordinating histidine side chains. Anomalous electron density (green) contoured at 10σ is shown for the zinc atoms (pink). B, chemical structure rendering of the histidine side chains, along with distances, from motifs Ia, Ib, IIa, and IIb that participate in the coordination of the zinc atoms in the dimetal center of scScs7p. C, colonies of wild-type and mutant constructs of scScs7p grown on SD-Ura plates in the absence (left) or presence (right) of syringomycin E. Constructs that maintain wild-type activity do not grow on plates containing syringomycin E, i.e. syringomycin E sensitivity is observed. Mutant constructs that affect hydroxylase activity lose their sensitivity to syringomycin E and thus grow on plates containing syringomycin E. Mutants that do not alter hydroxylase activity are labeled in black. Mutants that significantly alter or somewhat alter enzyme activity are labeled pink and yellow, respectively. D, schematic depiction of the proposed binding site for the substrate headgroup, with side chain atoms of mutated residues colored and labeled as above. The abbreviations used are as follows: WT, wild type; EV, empty vector; ΔN95, N-terminal deletion construct utilized for crystallization.
In vitro assay of the hydroxylase activity of scScs7p was not pursued because of the difficulties associated with reconstituting the necessary electron transport chain and the extremely hydrophobic nature of the ceramide substrate. We therefore turned to a phenotypic assay of mutant SCS7 alleles in whole yeast cells as a means to evaluate the roles of various residues in scScs7p function. S. cerevisiae exhibits sensitivity toward the antifungal cyclic lipodepsinonapeptide syringomycin E, produced by P. syringae pv. syringae (29). We generated an scs7-Δ strain and evaluated point mutations by transforming the deletion strain with a multicopy plasmid containing a normal or mutated copy of SCS7 expressed under control of the ADH1 promoter. Cells expressing Scs7p with normal activity from this plasmid do not grow on plates containing syringomycin E, i.e. syringomycin E sensitivity is observed. Mutant constructs that affect hydroxylase activity lose their sensitivity to syringomycin E and thus grow on plates containing the lipopeptide. Mutation of each of the 10 zinc-coordinating histidines to alanine resulted in the reduction or complete loss of this sensitivity on plates containing syringomycin E (Fig. 3, C and D), reinforcing the important role that these histidine residues play with respect to the catalytic activity of the enzymes of the hydroxylase/desaturase superfamily (12).
Putative Substrate Channels
Electron density corresponding to the hydrocarbon tail of an unidentified lipid molecule or a βOG detergent molecule is observed within two orthogonal channels that intersect just below the dimetal center (Fig. 2). The lower channel runs perpendicular to the lipid bilayer and is formed by transmembrane helices TM2, TM3, and TM4. The upper channel runs parallel to the lipid bilayer and is formed by amphipathic helices H5 and H7, along with the N-terminal portion of TM3. The lower channel is lined with hydrophobic residues, including Trp-236 (TM2), Phe-248 (TM2), Phe-288 (TM3), Leu-291 (TM3), Phe-295 (TM3), Leu-298 (TM3), Val-299 (TM3), and Phe-312 (TM4). Surprisingly, there are a significant number of residues that are hydrophilic or have side chains with the potential to form ionic or hydrogen bonds within the lower channel. These include His-244 (TM2), Tyr-319 (TM4), Tyr-322 (TM4), and Asp-323 (TM4), which are all highly conserved in membrane-bound hydroxylase enzymes across species (Fig. 1). His-244 and Phe-248 are part of histidine motif Ia. Conversely, the upper channel is lined solely by hydrophobic residues Leu-191 (H5), Leu-266 (H7), Leu-267 (H7), Met-283 (TM3), Pro-284 (TM3), and Leu-287 (TM3).
A pocket is formed at the intersection of the two channels, directly below the dimetal center. The unique secondary structure observed for helix H7, which harbors histidine motif Ib, plays a vital role in the formation of this pocket, which could accommodate the hydrophilic headgroup of a ceramide substrate. Helix H7 is kinked by 70° at Leu-267, due to the insertion of one turn of the π-helix. As a result, the histidine side chains in motif Ib are properly aligned near the dimetal center in an appropriate configuration for coordination. This 70° kink presumably provides access for the substrate headgroup to the dimetal center for initiation of the hydroxylase reaction. Interestingly, there is no significant sequence identity in the region of H7 outside of the Scs7p/FA2H family of enzymes, suggesting that the observed secondary structure for H7 is unique to both the substrate specificity and function of hydroxylation (Fig. 1).
N-terminal Cytochrome b5 Domain Interaction
The hydroxylase reaction proceeds upon the transfer of an electron to the dimetal center via the fused N-terminal cytochrome b5 domain. Electron transfer is thought to occur through an interaction that involves an anionic surface on cytochrome b5 composed of the carboxylate side chains of conserved glutamic acid residues and the heme moiety (40). The interaction surface for the cytochrome b5 domain on the hydroxylase domain of scScs7p is unknown. Although the N-terminal cytochrome b5 domain is present in crystals of full-length scScs7p, this domain is disordered in the structure, suggesting that the interaction between the two domains is transient in nature. We constructed point mutants containing the substitutions H173A, H264A, and H331A at sites that are located outside of the histidine box motifs to gain insight into which surface of the hydroxylase domain of Scs7p might interact with cytochrome b5. The side chains of all three histidines are surface accessible, with His-173 and His-264 located on one side of the dimetal center and His-331 on the opposite side (Fig. 3D). Only the H173A mutation resulted in the loss of syringomycin E sensitivity, suggesting its involvement in electron transfer from cytochrome b5 (Fig. 3C). His-173 is highly conserved among the hydroxylase enzymes within this family (Fig. 1).
Comparison of Hydroxylase and Desaturase Scaffolds
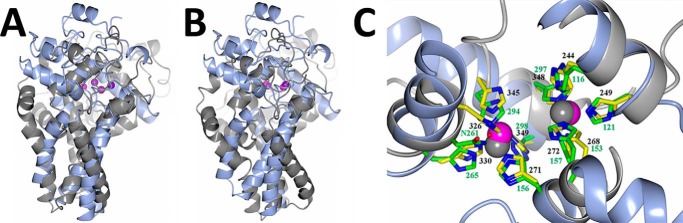
There are only 21 residues within the 268-residue hydroxylase domain of scScs7p that are identical compared with SCD1, a desaturase representative within the hydroxylase/desaturase superfamily (19, 20). Despite the less than 8% sequence identity, scScs7p and SCD1 exhibit a similar tertiary scaffold, predominantly consisting of helical secondary structure. Both enzymes are composed of a cytosolic catalytic cap domain that sits atop a stalk of four transmembrane helices, with the N- and C-terminal ends of the enzymes located on the cytosolic side of the ER membrane (Fig. 4). The residues involved in metal coordination at the dimetal centers of scScs7p and SCD1 share the highest sequence and structural conservation between enzymes (Fig. 5). Both structures have zinc atoms bound within their dimetal centers, despite the utilization of different expression systems in the generation of the proteins used for structural studies (19, 20). The zinc atoms are similarly spaced ∼6.5 Å apart from each other. Nine of the 21 residues that are conserved between the enzymes are zinc-coordinating histidine residues located in the histidine box motifs. Zinc is coordinated in virtually the same manner in both structures, exhibiting square pyramidal geometry.
FIGURE 4.
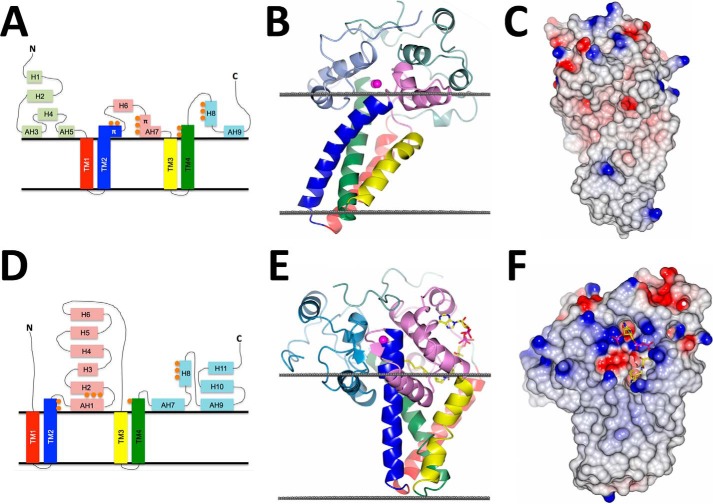
Comparison of scScs7p and SCD1 crystal structures. Topology diagram, schematic representation, and space-filling models of scScs7p (A–C) and mSCD1 (D–F) (Protein Data Bank code 4YMK (19)) depicting the differences associated with the following: 1) the cytoplasmic catalytic domains; 2) transmembrane helical arrangement; 3) location of the dimetal center with respect to the cytosolic membrane of the ER; and 4) lack of a polar electrostatic surface equivalent to that observed for SCD1 in scScs7p. Views in C and F are rotated ∼90° compared with B and E, with an additional ∼30° rotation of scScs7p along the y axis in to compensate for the observed tilt within the membrane helices. The surfaces in C and F are colored according to the electrostatic potential, from −0.5 V (red) to +0.5 V (blue). Steroyl-CoA is shown bound to SCD1 in E and F.
FIGURE 5.
Superposition between scScs7p and SCD1. Schematic rendering of the superposition between scScs7p (light blue) and mouse SCD1 (gray). Although the overall fold is conserved between the two enzymes, there are significant differences between the helices in the transmembrane stalk and the catalytic cap domain, regardless of whether the superposition is carried out based on secondary structural elements (A) or when the zinc atoms and histidine box residues are utilized for the superposition (B). C, dimetal center is the most conserved region between the two enzymes. Histidine side chains from scScs7p are colored with yellow carbons and numbered in black font, and those from mouse SCD1 are colored with green carbons and numbered in green font. Note that SCD1 coordinates the two zinc atoms within the dimetal center with nine histidine residues and Asn-261 via a bridging water molecule (red). Nitrogen atoms are colored dark blue. The zinc atoms from scScs7p are colored magenta, and those from SCD1 are colored gray.
Although the global architecture is similar between enzymes, the root mean square deviation between scScs7p and SCD1 is strikingly poor (3.9 Å for 138 equivalent Cα atoms). In the SCD1 structure, the transmembrane helices are splayed apart at the cytosolic face of the ER membrane, with the catalytic cap domain of the enzyme nestled within. In contrast, the transmembrane helices of scScs7p exhibit a closed conformation, attributable primarily to differences in helical geometry observed in TM2 and TM3 between the structures. There is a single turn of π-helix in the last turn of TM2 in scScs7p that is lacking in the SCD1 structure, resulting in a kink that positions TM2 in closer proximity to the N-terminal end of TM3. TM3 in SCD1 is kinked due to the presence of Pro-226, whereas there is no kink in TM3 in the scScs7p structure (Fig. 4). A hydrogen bond formed between the carbonyl oxygen of Pro-226 and the side chain of Arg-249 in TM4 is hypothesized to stabilize the kink within the membrane interior of SCD1 (19). Neither Pro-226 nor Arg-249 are conserved in the membrane-bound hydroxylase enzymes (Fig. 1).
There are also significant differences between scScs7p and SCD1 in the catalytic cap domain. The catalytic cap domain of scScs7p is composed of nine helices within three stretches of 91, 38, and 54 residues (Fig. 4). The N-terminal stretch contains five helices, whereas the central and C-terminal stretches each contain two helices. Four of the nine helices in the catalytic cap domain, H3, H5, H7, and H9 are amphipathic in nature. In contrast, only the 93-residue central and 90-residue C-terminal stretches of SCD1 contain helices, of which three, H1, H7, and H9, are amphipathic (Fig. 4). The different locations of the helices within the catalytic caps result in vastly different domain architectures for each enzyme. For SCD1, the two domains, C1 and C2, lie on opposite sides of the dimetal center. It is hypothesized that the cytochrome b5 domain accesses the dimetal center for subsequent electron transfer via a groove between C1 and C2 (19). In contrast, the helices in the N- and C-terminal stretches of scScs7p serve to bury helices H6 and H7 in the central stretch, suggesting an alternative binding mode for cytochrome b5 and mechanism for electron transfer (Fig. 4).
The major difference between the dimetal centers of scScs7p and SCD1 is the location of these groups with respect to the cytosolic membrane. In SCD1, TM2 and TM4 extend three helical turns out of the membrane and form interactions with residues within the catalytic cap domain (19, 20). Zinc-coordinating histidine residues His-120, His-125, and His-269 in SCD1 are contained within the portions of TM2 and TM4 that extend out of the membrane. As a result, the dimetal center is located ∼8.5 Å above the plane of the cytosolic membrane. In stark contrast, the dimetal center of scScs7p resides only ∼2.8 Å above the plane of the cytosolic membrane (Fig. 4). The two factors responsible for the difference in location of the dimetal center in scScs7p are the presence of the π-helix turn in TM2 that bends the end of the helix closer to the membrane and the increased number of amphipathic helices in the catalytic cap that stabilize this domain closer to the membrane interface. The observed distance differences in the location of the dimetal center with respect to the membrane may play a role in the different catalytic activities carried out by each enzyme as well as their substrate specificities.
The serendipitous capture of the steroyl-CoA substrate in SCD1 crystal structures provides insight into the similarities and differences in substrate binding between the hydroxylase and desaturase enzymes. In SCD1, the CoA moiety binds to the catalytic cap domain, between helices H2 and H4 and near the end of TM4 (19, 20). The adenosine, diphosphate, and pantothenate groups of the CoA moiety form electrostatic and hydrogen bonds with polar residues that include Asn-75, Asn-148, Arg-155, Asp-156, Arg-188, Lys-189, and Lys-194. Collectively, this highly conserved cluster of residues forms an electrostatic surface within the catalytic cap domain of SCD1 that aids in substrate recognition and binding (19, 20). There is no equivalent electrostatic patch on the surface of the catalytic cap in scScs7p, as the corresponding residues in scScs7p for Asn-148, Arg-155, and Asp-156 are hydrophobic residues Ile-259, Leu-266, and Leu-267 (Fig. 4). The lack of conservation of these residues, coupled with the significantly different architectures observed for the catalytic cap domains of SCD1 and scScs7p, suggests that fatty acyl-CoA substrates are not hydroxylated by scScs7p.
The fatty acyl tail of steroyl-CoA binds within a kinked hydrophobic tunnel in SCD1 that is composed of residues from TM2 and TM4. The kink in the tunnel is generated by the positioning of the side chains of Trp-149 and Thr-257, two highly conserved residues in the desaturase branch of the hydroxylase/desaturase family. In addition to forming a kink, these residues cap the top of the tunnel. As a consequence, the shape of the tunnel forces the acyl chain to bind in a conformation poised for desaturation at the carbon 9–10 bond. Tyr-104, also highly conserved among desaturase enzymes, is located at the base of the tunnel, where it serves as a plug that restricts fatty acyl chains with more than 18 carbons from binding within the tunnel (19, 20). Although we do not observe bound substrate in scScs7p, there is an equivalent channel lined by residues located within TM2, TM3, and TM4 that is occupied by a lipid molecule. The channel in scScs7p is more open below the dimetal center as there are no residues equivalent to Trp-149 and Thr-257. Moreover, the equivalent residue to Tyr-104 in scScs7p is Gly-232, which would not be expected to impose any restriction on the length of fatty acyl chain that can bind within the channel. The implications that these differences between SCD1 and scScs7p have for substrate specificity, recognition, and binding are discussed below.
Modeling Ceramide Binding to scScs7p and Implications for Catalysis
We carried out molecular dynamics simulations to gain further insight into how a ceramide substrate could bind to scScs7p. Prior to the simulations, models were created that placed the VLCFA moiety in the lower channel bound by TM2, TM3, and TM4 and LCB moiety of the ceramide in the upper channel bound by H5, H7, and TM3 and vice versa. Three different starting positions, each of which differed by a shift of the substrate headgroup by a few angstroms with respect to the dimetal center in a direction perpendicular to the membrane, were utilized for both binding modes. The molecular dynamics simulations resulted in ending positions for the ceramide substrate that were very similar to the starting positions. In each case, the VLCFA and LCB moieties remained in their respective channels, and the headgroup was located below the dimetal center. There were no significant changes observed in the scScs7p structure over the course of the simulations. Models that placed the VLCFA moiety in the upper channel resulted in the positioning of C-2 of the VLCFA far away from the dimetal center, with distances from C-2 to the zinc atoms of 9.0 and 9.2 Å. These long distances do not favor hydroxylation and thus the binding pose that places the VLCFA moiety in the upper channel is unlikely to be the catalytically competent conformation for the substrate. When the VLCFA moiety was placed in the lower channel and the LCB moiety in the upper channel, the simulations placed C-2 much closer to the dimetal center, with distances of 5.2 and 5.9 Å (Fig. 6). In this orientation, the pro-(R) hydrogen of C-2 lies 4.9 and 5.3 Å away from the zinc atoms, consistent with the C-9/C-10 distances to the dimetal center for steroyl-CoA bound to SCD1 (4.7 and 5.4 Å) (19, 20) and with the production of (R)-enantiomers by FA2H (41).
FIGURE 6.
Proposed model for ceramide binding to scScs7p. A, chemical drawing of the 2-hydroxylase reaction depicting the substrate, ceramide B, and resulting hydroxylated product, ceramide C. The enzyme hydroxylates C-2 on the VLCFA moiety of the ceramide. B, schematic depiction of the hydroxylase domain of scScs7p, colored as in Fig. 2, showing ceramide B bound in the active site channels below the dimetal center. Carbon, nitrogen, and oxygen atoms are depicted as spheres and colored yellow, blue, and red, respectively. The positioning of scScs7p within the membrane (gray) was generated using the PPM server (38). C, electrostatic surface rendering of the view in B. Black arrows point to openings between TM2 and TM3, which lead to the active site channels. D, schematic depiction of the junction of the upper and lower channels below the dimetal center. The turn of π-helix in H7 forms a pocket for the substrate headgroup to bind. Histidine residues derived from the histidine box motifs in H7, H8, and TM4 are labeled accordingly, along with the side chains of Tyr-319, Tyr-322, and Asp-323. Zinc atoms are colored pink. The carbon, nitrogen, and oxygen atoms of the substrate are colored salmon, blue, and red, respectively. The 2pro(R) hydrogen is also labeled.
The highly conserved residues Tyr-319, Tyr-322, and Asp-323 are located at the C-terminal end of TM4 (Fig. 1), where they introduce functional groups that could participate in the stabilization of the ceramide headgroup through interaction with the hydroxyl groups and amine group. In our model, the majority of the interactions made between the ceramide headgroup and scScs7p are hydrogen bonds to protein main chain nitrogen and carbonyl atoms. Interestingly, the phenolic oxygen of Tyr-322 is positioned 4 Å away from the nitrogen atom in the ceramide headgroup. Mutation of Tyr-322 to alanine results in the loss of syringomycin E sensitivity, suggesting that this residue plays a role in binding ceramide or optimally positioning the substrate for catalysis (Fig. 3). Indeed, the equivalent residue within TM4 in the SCD1 structure is Trp-262, which interacts with the carbonyl oxygen of the fatty acid (19, 20). The side chains of Tyr-319 and Asp-323 in scScs7p are greater than 5 Å away from the nearest hydrogen-bonding partner in the ceramide headgroup. The side chain of Asp-323 interacts with the phenolic oxygen of Tyr-319 within the helix register of TM4. The observed Tyr-319/Asp-323 interaction may aid in the positioning of the side chain of Tyr-322 for its interaction with the substrate. Based on maintenance of sensitivity to syringomycin E, substitution of alanine for Tyr-319 does not diminish scScs7p activity, suggesting that it does not directly interact with the substrate (Fig. 3).
During our molecular dynamics simulations, we observed the movement of water molecules from the aqueous exterior to the dimetal center via two pathways along the plane of the membrane. One route involved interactions with the main chain carbonyl oxygen atoms of Gly-179 and Gly-181, along with the side chains of His-271, Asp-277, and Arg-280. His-271 is located in histidine box motif Ib within H7, whereas Asp-277 and Arg-280 are located in the loop connecting H7 to TM3. The other route involved interactions with the side chain atoms of Asp-323 and Glu-324, located in the C-terminal portion of TM4. His-271, Asp-277, Arg-280, and Asp-323 are highly conserved residues within the hydroxylase family of enzymes (Fig. 1). As water is generated as a by-product of the hydroxylation of the ceramide substrate, one or both of these routes would provide water egress to the aqueous exterior. Interestingly, mutation of Asp-323 to alanine results in the loss of syringomycin E sensitivity (Fig. 3). Because the side chain of Asp-323 is not predicted to interact with the ceramide substrate, it may be that this residue is involved in water egress during the hydroxylase reaction.
Discussion
We report here the x-ray crystal structure of the integral membrane enzyme sphingolipid α-hydroxylase Scs7p, a representative of the hydroxylase branch of the hydroxylase/desaturase superfamily. Scs7p is the yeast ortholog of human FA2H, which is responsible for hydroxylating GalCer and sulfatide within the myelin sheath. The enzyme adopts a mushroom-like shape similar to the overall fold of the related steroyl CoA desaturase, but with significant differences from these structures relevant to the differing substrate specificities and catalytic action of the two enzymes.
We routinely observed the presence of zinc atoms in purified enzyme preparations and within crystals of scScs7p, despite the fact that scScs7p is classified as a di-iron enzyme. We made significant attempts to increase the iron content within the enzyme, including reduction of the zinc content in the expression media, stripping out zinc with subsequent addition of iron during purification, and manipulation of the yeast expression conditions to favor iron incorporation over zinc (42). Although these attempts led to the production of highly purified enzyme with iron incorporated at the dimetal center up to ∼50%, we were unsuccessful at achieving 100% replacement with iron. Moreover, the incorporation of zinc into scScs7p is not a result of the use of S. cerevisiae as an overexpression system, as zinc atoms were also observed bound to scScs7p when we utilized an insect cell expression system. Our results are consistent with the observation of zinc bound within the dimetal centers of the structures of human and mouse SCD1, both of which were determined utilizing protein derived from an insect cell expression system (19, 20). It is quite possible that zinc simply serves as a suitable surrogate for iron during overexpression, given their similar size and charge properties (19). In addition to the structures of SCD1 and that of scScs7p presented here, there are other examples of zinc replacement at iron-binding sites, including the structure of quinol-dependent nitric-oxide reductase (43).
A surprising aspect of the studies reported here is that the Δ95scScs7p crystallization construct exhibits syringomycin E sensitivity, indicative of hydroxylase function. This indicates that the electron transfer to the truncation mutant must be provided by some endogenous source of cytochrome b5 (Fig. 3) (14). Indeed, the Saccharomyces castellii and Candida glabrata Scs7p orthologs do not contain the His-72 residue in their cytochrome b5 domains, which is required for heme binding (Fig. 1) (44). In contrast to these observations for scScs7p, the fused cytochrome b5 domain is required for activity of other family members such as yeast Δ9 desaturase (Ole1p), which when similarly truncated is not functional in cells, implying that it cannot be rescued by an alternative cytochrome b5 (18), Similarly, in humans, FA2H containing point mutations in the cytochrome b5 domain causes mild forms of hereditary leukodystrophies (45). It is not clear what evolutionary advantage is attained through the fusing of a cytochrome b5 domain to a hydroxylase or desaturase domain. One hypothesis is that the fusion provides a kinetic advantage for enzyme function (44).
Although it is clear that FA2H and Scs7p hydroxylate C-2 of a VLCFA moiety, the breadth of the substrate specificity of these enzymes has not been definitively established. Candidate substrates include free fatty acids, fatty acyl-CoA derived substrates, and ceramide. Establishing substrate specificities is complicated by the difficulties associated with reconstituting the electron transport chain for in vitro assays and by the hydrophobic nature of the substrates. Lipid analysis of gene deletions in S. cerevisiae suggests that phytoceramide is the preferred substrate for Scs7p (15), and these findings are further supported by experiments carried out with rat brain microsomes, which indicate that free fatty acid substrates and fatty acyl-CoA-derived substrates are not effectively modified (46). The structural architecture observed in scScs7p provides strong evidence that ceramide is an optimal substrate for the enzyme. The elongated shape and open pocket below the dimetal center provide suitable space for the binding of a substrate that contains both an LCB and VLCFA moiety. The lack of a polar electrostatic patch in scScs7p and the different structural architectures observed between the catalytic cap domains of scScs7p and SCD1 would seem to rule out the binding of a fatty acyl-CoA derived substrate.
Molecular dynamics simulations suggest that ceramide binds optimally to scScs7p with the VLCFA moiety located in the lower channel and the LCB moiety in the upper channel. Detergent or lipid molecules are bound in both of these channels in the scScs7p crystal structure, further indicating that these channels bind the VLCFA and LCB moieties of ceramide. Additional support for the location of VLCFA in the lower channel comes from inspection of the equivalent hydrophobic channel in SCD1, responsible for binding the fatty acyl tail of steroyl-CoA. The SCD1 hydrophobic channel is plugged at its base by Tyr-104. Interestingly, Tyr-104 is known to be responsible for limiting the substrate specificity for SCD1 to substrates containing fatty acyl chain lengths between 14 and 19 carbons (19). Although Tyr-104 is highly conserved in the desaturase branch of the hydroxylase/desaturase family, the marine copepod desaturase from Calanus hyperboreus has a threonine at this position, which allows it to desaturate substrates containing fatty acyl chains with lengths between 22 and 26 carbons (47). No such plug is present in the lower channel in the scScs7p structure, where the equivalent residue to Tyr-104 is Gly-232, suggesting that a VLCFA moiety can be accommodated. Moreover, Gly-232 is highly conserved in the hydroxylase branch of the hydroxylase/desaturase superfamily (Fig. 1).
Occupation of the lower and upper channels by the VLCFA and LCB moieties, respectively, places the C-2 of the VLCFA below the dimetal center in an orientation that is poised for hydroxylation, with distances to the zinc atoms of 5.2 and 5.9 Å. These distances are consistent with the spacing observed between the metal atoms and C-9 and C-10 for steroyl-CoA bound to SCD1 (19, 20). Tyr-319, Tyr-322, and Asp-323, highly conserved residues within the hydroxylase branch of the hydroxylase/desaturase family, are located near the substrate headgroup. Both Y322A and D323A mutant constructs resulted in the loss of syringomycin E sensitivity in the cellular activity assay, suggesting a role for each in catalysis. Tyr-322 is positioned 4 Å away from the nitrogen atom in the ceramide headgroup. The equivalent residue in SCD1 is Trp-262, which forms a hydrogen bond with the carbonyl oxygen of the fatty acyl chain within the steroyl-CoA substrate. Given both the observed loss of syringomycin E sensitivity and its location with respect to the substrate, we hypothesize that Tyr-322 forms a similar interaction with the ceramide headgroup. Tyr-319 and Asp-323 interact as part of the helical geometry of TM4. The observed interaction between Tyr-319 and Asp-323 could play a role in stabilizing the turn of helix in TM4 and subsequently the position of Tyr-322 for optimal interaction with the substrate headgroup. We hypothesize that the side chain of Asp-323 plays a role in the egress of water, generated during the hydroxylase reaction, to the aqueous cytosol.
The differences in substrate specificities between SCD1 and scScs7p provide insight into the entry and egress of ceramide into the enzyme for subsequent catalysis. In SCD1, binding of the CoA moiety of the substrate to the polar surface within the catalytic cap domain drives insertion of the fatty acyl tail into the kinked hydrophobic pocket that is capped by residues Trp-149 and Thr-257 (19, 20). The side chains of Trp-149 and Thr-257 are stabilized by hydrogen bonds with Gln-143. Bai et al. (19) propose that breaking the hydrogen bond between Gln-143 and Thr-257 provides an opening between TM4 and the loop between H1 and H2 that would allow the desaturated product to be released from the pocket. The area below the dimetal center in the scScs7p is not capped as it is in the SCD1 structure and thus is more open. We propose that the predominantly hydrophobic ceramide substrate would access and depart the active site directly from within the confines of the lipid bilayer through an opening between transmembrane helices TM2 and TM3. Direct entry from the lipid bilayer would orient the LCB and VLCFA moieties in the appropriate channels and the headgroup below the dimetal center, analogous to the positioning of the ligand sphingosine 1-phosphate upon binding to its cognate G protein-coupled receptor (48). The observed difference in proximity of the dimetal centers of scScs7p and SCD1 to the plane of the cytosolic membrane (2.8 Å versus 8. Å) provides additional support for different modes of substrate access and egress for the enzymes.
In summary, the crystal structure of scScs7p presented here provides the first structural snapshot of a hydroxylase enzyme representative of the membrane-bound hydroxylase/desaturase superfamily. The location of the metal-binding histidine boxes with respect to the positions of the four transmembrane helices and the placement of the metal center at the surface of the membrane facing the cytosol confirms previous structural predictions made for scScs7p. Examination of the structure, along with molecular dynamics simulations, suggests that ceramide binds below the dimetal center, with the VLCFA placed in the lower channel and the LCB moiety placed in the upper channel. Furthermore, results of cell-based activity assays suggest roles for highly conserved residues Tyr-322 and Asp-323 of TM4 in substrate binding. Coupled with the structural determination of SCD1, a desaturase enzyme within the same membrane-bound hydroxylase/desaturase superfamily, a detailed analysis can now be made of the molecular determinants of substrate specificity and hydroxylation or desaturation of lipid substrates by these enzymes.
Author Contributions
M. G. M. conceived and coordinated the study and wrote the paper. M. E. D., S. M. C., and G. Z. were responsible for cloning and construction of yeast deletion strains. G. Z. was responsible for expression, solubilization, purification, and functional characterization. M. K.-R. and G. Z. were responsible for crystallization, and G. Z. carried out data collection and crystallographic analyses. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
X-ray diffraction experiments and fluorescence scans were conducted at the Advanced Photon Source. Use of IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by companies of the Industrial Macromolecular Crystallography Association through a contract with the Hauptman-Woodward Medical Research Institute. GM/CA CAT has been funded in whole or in part with federal funds from the National Institutes of Health Grants Y1-CO-1020 from NCI and Y1-GM-1104 from NIGMS. Use of the Advanced Photon Source was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
This work was supported by National Institutes of Health Grant U54GM094611 from NIGMS. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The atomic coordinates and structure factors (codes 4ZR0 and 4ZR1) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- GalCer
- galactosylceramide
- βOG
- n-octyl-β-d-glucopyranoside
- ER
- endoplasmic reticulum
- FA2H
- fatty acid 2-hydroxylase
- LCB
- long chain base
- m
- mouse
- PEG
- polyethylene glycol
- sc
- Saccharomyces cerevisiae
- VLCFA
- very long chain fatty acid
- scScs7p
- S. cerevisiae Scs7p.
References
- 1.Aggarwal S., Yurlova L., and Simons M. (2011) Central nervous system myelin: structure, synthesis and assembly. Trends Cell Biol. 21, 585–593 [DOI] [PubMed] [Google Scholar]
- 2.Pruett S. T., Bushnev A., Hagedorn K., Adiga M., Haynes C. A., Sullards M. C., Liotta D. C., and Merrill A. H. Jr. (2008) Biodiversity of sphingoid bases (“sphingosines”) and related amino alcohols. J. Lipid Res. 49, 1621–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alderson N. L., Rembiesa B. M., Walla M. D., Bielawska A., Bielawski J., and Hama H. (2004) The human FA2H gene encodes a fatty acid 2-hydroxylase. J. Biol. Chem. 279, 48562–48568 [DOI] [PubMed] [Google Scholar]
- 4.Eckhardt M., Yaghootfam A., Fewou S. N., Zöller I., and Gieselmann V. (2005) A mammalian fatty acid hydroxylase responsible for the formation of α-hydroxylated galactosylceramide in myelin. Biochem. J. 388, 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coetzee T., Suzuki K., and Popko B. (1998) New perspectives on the function of myelin galactolipids. Trends Neurosci. 21, 126–130 [DOI] [PubMed] [Google Scholar]
- 6.Blackstone C. (2012) Cellular pathways of hereditary spastic paraplegia. Annu. Rev. Neurosci. 35, 25–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dick K. J., Eckhardt M., Paisán-Ruiz C., Alshehhi A. A., Proukakis C., Sibtain N. A., Maier H., Sharifi R., Patton M. A., Bashir W., Koul R., Raeburn S., Gieselmann V., Houlden H., and Crosby A. H. (2010) Mutation of FA2H underlies a complicated form of hereditary spastic paraplegia (SPG35). Hum. Mutat. 31, E1251–E1260 [DOI] [PubMed] [Google Scholar]
- 8.Edvardson S., Hama H., Shaag A., Gomori J. M., Berger I., Soffer D., Korman S. H., Taustein I., Saada A., and Elpeleg O. (2008) Mutations in the fatty acid 2-hydroxylase gene are associated with leukodystrophy with spastic paraparesis and dystonia. Am. J. Hum. Genet. 83, 643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pierson T. M., Simeonov D. R., Sincan M., Adams D. A., Markello T., Golas G., Fuentes-Fajardo K., Hansen N. F., Cherukuri P. F., Cruz P., Mullikin J. C., Blackstone C., Tifft C., Boerkoel C. F., Gahl W. A., et al. (2012) Exome sequencing and SNP analysis detect novel compound heterozygosity in fatty acid hydroxylase-associated neurodegeneration. Eur. J. Hum. Genet. 20, 476–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shanklin J., and Cahoon E. B. (1998) Desaturation and related modifications of fatty acids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 611–641 [DOI] [PubMed] [Google Scholar]
- 11.Dunn T. M., Haak D., Monaghan E., and Beeler T. J. (1998) Synthesis of monohydroxylated inositol phosphorylceramide (IPC-C) in Saccharomyces cerevisiae requires Scs7p, a protein with both a cytochrome b5-like domain and a hydroxylase/desaturase domain. Yeast 14, 311–321 [DOI] [PubMed] [Google Scholar]
- 12.Shanklin J., Whittle E., and Fox B. G. (1994) Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry 33, 12787–12794 [DOI] [PubMed] [Google Scholar]
- 13.Sperling P., Ternes P., Zank T. K., and Heinz E. (2003) The evolution of desaturases. Prostaglandins Leukot. Essent. Fatty Acids 68, 73–95 [DOI] [PubMed] [Google Scholar]
- 14.Mitchell A. G., and Martin C. E. (1997) Fah1p, a Saccharomyces cerevisiae cytochrome b5 fusion protein, and its Arabidopsis thaliana homolog that lacks the cytochrome b5 domain both function in the α-hydroxylation of sphingolipid-associated very long chain fatty acids. J. Biol. Chem. 272, 28281–28288 [DOI] [PubMed] [Google Scholar]
- 15.Haak D., Gable K., Beeler T., and Dunn T. (1997) Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J. Biol. Chem. 272, 29704–29710 [DOI] [PubMed] [Google Scholar]
- 16.Arthington B. A., Bennett L. G., Skatrud P. L., Guynn C. J., Barbuch R. J., Ulbright C. E., and Bard M. (1991) Cloning, disruption and sequence of the gene encoding yeast C-5 sterol desaturase. Gene 102, 39–44 [DOI] [PubMed] [Google Scholar]
- 17.Li L., and Kaplan J. (1996) Characterization of yeast methyl sterol oxidase (ERG25) and identification of a human homologue. J. Biol. Chem. 271, 16927–16933 [DOI] [PubMed] [Google Scholar]
- 18.Mitchell A. G., and Martin C. E. (1995) A novel cytochrome b5-like domain is linked to the carboxyl terminus of the Saccharomyces cerevisiae Δ-9 fatty acid desaturase. J. Biol. Chem. 270, 29766–29772 [DOI] [PubMed] [Google Scholar]
- 19.Bai Y., McCoy J. G., Levin E. J., Sobrado P., Rajashankar K. R., Fox B. G., and Zhou M. (2015) X-ray structure of a mammalian stearoyl-CoA desaturase. Nature 524, 252–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H., Klein M. G., Zou H., Lane W., Snell G., Levin I., Li K., and Sang B. C. (2015) Crystal structure of human stearoyl-coenzyme A desaturase in complex with substrate. Nat. Struct. Mol. Biol. 22, 581–585 [DOI] [PubMed] [Google Scholar]
- 21.Clark K. M., Fedoriw N., Robinson K., Connelly S. M., Randles J., Malkowski M. G., DeTitta G. T., and Dumont M. E. (2010) Purification of transmembrane proteins from Saccharomyces cerevisiae for x-ray crystallography. Protein Exp. Purif. 71, 207–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexandrov A., Dutta K., and Pascal S. M. (2001) MBP fusion protein with a viral protease cleavage site: one-step cleavage/purification of insoluble proteins. BioTechniques 30, 1194–1198 [DOI] [PubMed] [Google Scholar]
- 23.Koszelak-Rosenblum M., Krol A., Mozumdar N., Wunsch K., Ferin A., Cook E., Veatch C. K., Nagel R., Luft J. R., Detitta G. T., and Malkowski M. G. (2009) Determination and application of empirically derived detergent phase boundaries to effectively crystallize membrane proteins. Protein Sci. 18, 1828–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luft J. R., Snell E. H., and Detitta G. T. (2011) Lessons from high-throughput protein crystallization screening: 10 years of practical experience. Expert Opin. Drug Discov. 6, 465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otwinowski Z., and Minor W. (1997) Macromolecular crystallography, part A. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 26.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 28.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hama H., Young D. A., Radding J. A., Ma D., Tang J., Stock S. D., and Takemoto J. Y. (2000) Requirement of sphingolipid α-hydroxylation for fungicidal action of syringomycin E. FEBS Lett. 478, 26–28 [DOI] [PubMed] [Google Scholar]
- 30.Hoffman G. A., Garrison T. R., and Dohlman H. G. (2000) Endoproteolytic processing of Sst2, a multidomain regulator of G protein signaling in yeast. J. Biol. Chem. 275, 37533–37541 [DOI] [PubMed] [Google Scholar]
- 31.Kushnirov V. V. (2000) Rapid and reliable protein extraction from yeast. Yeast 16, 857–860 [DOI] [PubMed] [Google Scholar]
- 32.Pronk S., Páll S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., Shirts M. R., Smith J. C., Kasson P. M., van der Spoel D., Hess B., and Lindahl E. (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Best R. B., Zhu X., Shim J., Lopes P. E., Mittal J., Feig M., and Mackerell A. D. Jr. (2012) Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zoete V., Cuendet M. A., Grosdidier A., and Michielin O. (2011) SwissParam: a fast force field generation tool for small organic molecules. J. Comput. Chem. 32, 2359–2368 [DOI] [PubMed] [Google Scholar]
- 35.Jo S., Kim T., Iyer V. G., and Im W. (2008) CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 [DOI] [PubMed] [Google Scholar]
- 36.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B. 3rd, Snoeyink J., Richardson J. S., and Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., and Noble M. (2004) Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 38.Lomize M. A., Pogozheva I. D., Joo H., Mosberg H. I., and Lomize A. L. (2012) OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Potterton E., Briggs P., Turkenburg M., and Dodson E. (2003) A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 40.Dailey H. A., and Strittmatter P. (1979) Modification and identification of cytochrome b5 carboxyl groups involved in protein-protein interaction with cytochrome b5 reductase. J. Biol. Chem. 254, 5388–5396 [PubMed] [Google Scholar]
- 41.Guo L., Zhang X., Zhou D., Okunade A. L., and Su X. (2012) Stereospecificity of fatty acid 2-hydroxylase and differential functions of 2-hydroxy fatty acid enantiomers. J. Lipid Res. 53, 1327–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shakoury-Elizeh M., Protchenko O., Berger A., Cox J., Gable K., Dunn T. M., Prinz W. A., Bard M., and Philpott C. C. (2010) Metabolic response to iron deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 285, 14823–14833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hino T., Matsumoto Y., Nagano S., Sugimoto H., Fukumori Y., Murata T., Iwata S., and Shiro Y. (2010) Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science 330, 1666–1670 [DOI] [PubMed] [Google Scholar]
- 44.Napier J. A., Sayanova O., Stobart A. K., and Shewry P. R. (1997) A new class of cytochrome b5 fusion proteins. Biochem. J. 328, 717–718 [PMC free article] [PubMed] [Google Scholar]
- 45.Hama H. (2010) Fatty acid 2-hydroxylation in mammalian sphingolipid biology. Biochim. Biophys. Acta 1801, 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akanuma H., and Kishimoto Y. (1979) Synthesis of ceramides and cerebrosides containing both α-hydroxy and nonhydroxy fatty acids from lignoceroyl-CoA by rat brain microsomes. J. Biol. Chem. 254, 1050–1060 [PubMed] [Google Scholar]
- 47.Meesapyodsuk D., and Qiu X. (2014) Structure determinants for the substrate specificity of acyl-CoA Δ9 desaturases from a marine copepod. ACS Chem. Biol. 9, 922–934 [DOI] [PubMed] [Google Scholar]
- 48.Hanson M. A., Roth C. B., Jo E., Griffith M. T., Scott F. L., Reinhart G., Desale H., Clemons B., Cahalan S. M., Schuerer S. C., Sanna M. G., Han G. W., Kuhn P., Rosen H., and Stevens R. C. (2012) Crystal structure of a lipid G protein-coupled receptor. Science 335, 851–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pei J., Kim B. H., and Grishin N. V. (2008) PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bond C. S., and Schüttelkopf A. W. (2009) ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr. D Biol. Crystallogr. 65, 510–512 [DOI] [PubMed] [Google Scholar]
- 51.Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 52.Karplus P. A., and Diederichs K. (2012) Linking crystallographic model and data quality. Science 336, 1030–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]