Background: Get4/5 is required for the efficient transfer of tail-anchored proteins to Get3.
Results: The Get3·Get4/5 complex forms an intermediate mediated by electrostatic interactions.
Conclusion: The rapid association of the Get3·Get4/5 intermediate complex is followed by a conformational change to the stable inhibited structure dominated by hydrophobic interactions.
Significance: These results provide insight into the mechanism of tail-anchored protein targeting.
Keywords: kinetics, membrane protein, protein targeting, Saccharomyces cerevisiae, structural biology
Abstract
Tail-anchored (TA) proteins, defined as having a single transmembrane helix at their C terminus, are post-translationally targeted to the endoplasmic reticulum membrane by the guided entry of TA proteins (GET) pathway. In yeast, the handover of TA substrates is mediated by the heterotetrameric Get4/Get5 complex (Get4/5), which tethers the co-chaperone Sgt2 to the targeting factor, the Get3 ATPase. Binding of Get4/5 to Get3 is critical for efficient TA targeting; however, questions remain about the formation of the Get3·Get4/5 complex. Here we report crystal structures of a Get3·Get4/5 complex from Saccharomyces cerevisiae at 2.8 and 6.0 Å that reveal a novel interface between Get3 and Get4 dominated by electrostatic interactions. Kinetic and mutational analyses strongly suggest that these structures represent an on-pathway intermediate that rapidly assembles and then rearranges to the final Get3·Get4/5 complex. Furthermore, we provide evidence that the Get3·Get4/5 complex is dominated by a single Get4/5 heterotetramer bound to one monomer of a Get3 dimer, uncovering an intriguing asymmetry in the Get4/5 heterotetramer upon Get3 binding. Ultrafast diffusion-limited electrostatically driven Get3·Get4/5 association enables Get4/5 to rapidly sample and capture Get3 at different stages of the GET pathway.
Introduction
The targeting of membrane proteins to their correct location in the cell is a highly regulated process (1). The majority of membrane proteins are targeted via the signal recognition particle, which typically recognizes the initial hydrophobic transmembrane domain as it emerges from the ribosome (2). However, the ubiquitous TA4 proteins, defined topologically by a single transmembrane domain near the C terminus, are unable to access the signal recognition particle pathway as their targeting signal, the single transmembrane domain, emerges from the ribosome only after protein synthesis is complete and must be targeted to the endoplasmic reticulum post-translationally (3, 4). In eukaryotes, TA proteins account for 1–2% of the proteome and are involved in many essential cellular processes such as apoptosis, vesicle fusion, and protein trafficking (5, 6).
A series of genetic and biochemical experiments in yeast and mammalian cells identified members of a dedicated pathway for delivery of TA proteins to the endoplasmic reticulum. In yeast, these factors comprise the guided entry of TA proteins (GET) pathway that consists of six proteins, Get1–5 and Sgt2, all with homologs in higher eukaryotes (7, 8). The first committed step in TA targeting is the formation of an Sgt2·TA complex (9), which may be assisted by chaperones. TA substrate is transferred to Get3 in a Get4/5-dependent manner (9). Extensive structural characterization of Get3, the central TA targeting factor, demonstrated that it undergoes ATP-dependent conformational changes from an open to closed form required for capturing the TA substrate (10–15). Once formed, this Get3·TA complex is then localized to the endoplasmic reticulum by the membrane proteins Get1 and Get2, which stimulate release of the TA protein and subsequent insertion into the endoplasmic reticulum membrane (16–19).
Efficient delivery of a TA substrate to Get3 requires the heterotetrameric Get4/5 that provides the link between Sgt2 and Get3 (9, 20, 21). Get4/5 stabilizes the ATP-bound state of Get3, delaying ATP release and inhibiting ATP hydrolysis, thereby locking Get3 in a conformation competent for TA substrate binding (22). Recently, the first crystal structure of a Get3·Get4/5 complex provided insight into the role of nucleotide in complex formation where Get4 binds to two functionally distinct binding interfaces that govern anchoring and ATPase regulation on a closed Get3 (23). Importantly, mutations introduced at these interfaces demonstrated that Get4-mediated regulation of ATP hydrolysis by Get3 were critical for efficient TA targeting. A recent study has now shown that Get4/5 binding to Get3 occurs on rapid diffusion-limited time scales, suggesting an electrostatic interaction (24). However, the nature of this interaction cannot be explained by the aforementioned Get3·Get4/5 structure.
This report describes the crystal structure of a Get3·Get4/5 intermediate complex from Saccharomyces cerevisiae in two crystal forms, at 2.8 and 6.0 Å, respectively. The structure represents an initial binding interaction mediated by electrostatics that facilitates the rate of subsequent complex formation. This is supported by kinetic analysis of Get3·Get4/5 complex formation that provides direct evidence for the two-step complex formation mechanism. Finally, mass spectrometry and multiangle light scattering were used to demonstrate that, under physiological steady-state conditions, a single Get4/5 heterotetramer is bound to one monomer of a Get3 dimer. This work allowed us to generate a refined model for Get3·Get4/5 complex formation.
Experimental Procedures
Protein Cloning, Expression, and Purification
The sequences of Get4 and Get5 were cloned as described previously (25). To generate the Get4/5N used in this study, this construct was further modified by truncating the C terminus of Get4 (residues 291–312) and by the addition of a stop codon after residue 54 within Get5 and verified by DNA sequencing. All Get4/5 proteins were overexpressed in BL21-Gold(DE3) (Novagen) grown in 2× YT (16 g/liter tryptone, 10 g/liter yeast extract, and 5 g/liter NaCl) medium at 37 °C and induced for 3 h by the addition of 0.5 mm isopropyl β-d-1-thiogalactopyranoside. Cells were lysed using a Microfluidizer (Microfluidics) and purified as a complex by nickel affinity chromatography (Qiagen). The affinity tag was removed by an overnight tobacco etch virus protease digest at room temperature while dialyzing against 20 mm Tris, pH 7.5, 30 mm NaCl, and 5 mm β-mercaptoethanol (BME). A second nickel-nitrilotriacetic acid column was used to remove any remaining His-tagged protein, and then the sample was loaded onto a 6-ml Resource Q anion exchange column (GE Healthcare). The peak containing the Get4/5N complex was collected and concentrated to 15–20 mg/ml. Initial purifications of the Get4/5N complex were verified to be a single monodispersed species by size exclusion chromatography (SEC) using a Superdex 200 16/60 column (GE Healthcare) equilibrated with 20 mm Tris, pH 7.5, 100 mm NaCl, and 5 mm BME. Full-length Get4/5 used in biochemical studies was further purified using a Superdex 200 16/60 column (GE Healthcare) equilibrated with GET buffer (50 mm K-HEPES, pH 7.5, 150 mm KOAc, 10 mm Mg(OAc)2, 10% (v/v) glycerol, and 5 mm BME). Fractions containing Get4/5 were pooled and concentrated to ∼5 mg/ml.
The S. cerevisiae Get3 coding region was cloned as described previously (10). A His6 tag followed by a tobacco etch virus protease site was fused to the N terminus, and a stop codon was placed in front of the C-terminal His6 tag. All S. cerevisiae Get3 mutants were generated using the QuikChange method. All Get3 proteins were expressed in BL21-Gold(DE3), grown in 2× YT (16 g/liter tryptone, 10 g/liter yeast extract, 5 g/liter NaCl) medium, and induced with 0.5 mm isopropyl β-d-1-thiogalactopyranoside for 16 h at 22 °C. Cells were lysed using a Microfluidizer and purified by nickel affinity chromatography. The affinity tag was removed by an overnight tobacco etch virus protease digest at room temperature while dialyzing against 20 mm Tris, pH 7.5, 100 mm NaCl, and 5 mm BME. A second nickel-nitrilotriacetic acid column was used to remove any remaining His-tagged protein, and then the sample was run on a Superdex 200 16/60 column (GE Healthcare) equilibrated with dialysis buffer (crystallization trials) or GET buffer (biochemical studies). Fractions corresponding to a dimer of Get3 were pooled and concentrated to 15–20 mg/ml.
Get3·Get4/5N complex was formed by equilibrating 105 μmol of Get4/5N with 100 μmol of Get3 at room temperature in 500 μl of 20 mm Tris, pH 7.5, 10 mm NaCl, 5 mm BME, 1 mm MgCl2, and either 1 mm ADP, AMP-PNP, or ATP. Prior to complex formation, Get3 had been pre-equilibrated with 1 mm MgCl2 and either 1 mm ADP or 1 mm AMP-PNP for 5 min at room temperature. Get3·Get4/5N complex was further separated from free Get4/5N using Superdex 200 10/300 (GE Healthcare) equilibrated with 20 mm Tris, pH 7.5, 10 mm NaCl, and 5 mm BME. Get3·Get4/5N complex using full-length Get4 was formed as above but in the absence of nucleotide and MgCl2. This complex was further separated from free Get4/5N using Superdex 200 16/60 (GE Healthcare) equilibrated with 20 mm Tris, pH 7.5, 10 mm NaCl, and 5 mm BME. All complexes were concentrated to 10–12 mg/ml before use in crystallization experiments.
Crystallization
Get3·Get4/5N crystal trials were carried out using the sitting drop vapor diffusion method at room temperature by equilibrating equal volumes of the protein complex solution and reservoir solution using a TTP LabTech Mosquito robot and commercially purchased kits (Hampton Research, Qiagen, and Molecular Dimensions Ltd.). The 2.8-Å Get3·Get4/5N intermediate complex crystals grew in the presence of 17% PEG 3350, 0.24 m sodium citrate, and 30 mm tris(2-carboxyethyl)phosphine. Crystals were cryoprotected by transferring directly to 10 μl of a reservoir solution supplemented with 20% glycerol, 1 mm ADP or AMP-PNP, and 1 mm MgCl2 and incubated for ≥10 min before being flash frozen in liquid nitrogen. The 6.0-Å Get3·Get4/5N intermediate complex crystals grew in the presence of 12% PEG 3350 and 0.1 m sodium malonate, pH 5.0, and were cryoprotected by transferring directly to 10 μl of a reservoir solution supplemented with 20% glycerol before being flash frozen in liquid nitrogen.
Data Collection, Structure Solution, and Refinement
All structures were solved using data sets collected on Beamline 12-2 at the Stanford Synchrotron Radiation Lightsource. Each structure was solved from a single data set that was integrated using MOSFLM (26) or XDS (27) and scaled and merged using SCALA (28, 29). The crystal of Get3·Get4/5N in the intermediate conformation diffracted to 2.8 Å, and phases were obtained by molecular replacement with Phaser (30) as implemented in PHENIX (31) using a monomer of the open (apo) form of Get3 (Protein Data Bank code 3A37) and one Get4/5N heterodimer (Protein Data Bank code 3LKU) as search models. Coordinates were refined using REFMAC v6.3 (28, 29) with NCS and B-factor restraints. Manual rebuilding was performed using Coot (32). Translation/libration/screw groups were included in the later refinement stages and were determined using the TLSMD web server (33). These associated regions within the structure were allowed a certain degree of variation based on predicted motions they might have during data collection. The final model refined to an R-factor of 22.4% (Rfree = 26.1%). The second structure in the intermediate conformation was solved to 6.0 Å using methods similar to those described above. This structure lacked the Get4 C-terminal truncation engineered for the other structures and refined to an R-factor of 27.4% (Rfree = 30.3%). Full crystallographic statistics are in Table 1.
TABLE 1.
Summary of crystallographic data
r.m.s.d., root mean square deviation.
| Intermediate complex |
||
|---|---|---|
| 2.8 Å | 6.0 Å | |
| Data set | ||
| Wavelength (Å) | 1.0000 | 0.9795 |
| Resolution range (Å) | 29.8–2.8 (2.9–2.8)a | 30.0–6.0 (6.6–6.0) |
| Space group | P 21212 | P 21 |
| Cell parameters | ||
| a, b, c (Å) | 112.0, 238.0, 52.2 | 152.4, 127.3, 210.3 |
| α, β, γ (°) | 90, 90, 90 | 90, 110.2, 90 |
| Unique reflections | 34,114 (4,406) | 18,388 (4,452) |
| Completeness (%) | 97.0 (95.7) | 96.3 (98.7) |
| Redundancy | 3.3 (3.1) | 3.2 (3.4) |
| Rsymb | 0.06 (0.58) | 0.06 (0.78) |
| Mean I/σ(I) | 8.3 (1.6) | 8.9 (1.4) |
| Refinement | ||
| Reflections: work/free | 32,332/1,710 | 17,462/917 |
| Rwork/Rfree | 0.224/0.261 | 0.274/0.303 |
| No. protein atoms | 7,750 | 41,984 |
| No. ligand atoms | 1 | 4 |
| Protein B-factors (Å2) | 111.0 | 361.8 |
| Water/ligand B-factors (Å2) | 111.1 | 357.5 |
| r.m.s.d. of bond lengths (Å) | 0.0148 | 0.0031 |
| r.m.s.d. of bond angles (°) | 1.70 | 0.74 |
| Ramachandran preferred (%) | 96.0 | 91.0 |
| Ramachandran allowed (%) | 3.3 | 6.7 |
| Ramachandran outliers (%) | 0.7 | 2.3 |
a Values in parentheses are for the highest resolution shell.
b Rsym = ΣhklΣi|Ii(hkl) − <I(hkl)>|/ΣhklΣiIi(hkl) where Ii(hkl) is ith observation of reflection hkl and I(hkl) is the weighted average intensity for all observations i of reflection hkl.
Structure Analysis and Figures
Schematic representations of protein structures were prepared using PyMOL (Schrödinger, LLC), whereas surface representations were prepared using UCSF Chimera (34). Surface figures were made in Chimera. Electrostatic surface potentials were calculated using Adaptive Poisson-Boltzmann Solver with default values as implemented in the PDB2PQR web server (35, 36).
Isothermal Titration Calorimetry
Get3·Get4/5N binding experiments were carried out using the MicroCal iTC200 system (GE Healthcare). Binding affinities were measured by titrating 350 μm Get4/5N into a sample cell containing 50 μm Get3 D57V. All samples were diluted in GET buffer containing 1 mm ATP. For the first titration, 0.4 μl of Get4/5N was injected. For all following titrations, 2 μl of Get4/5N was injected for 20 intervals spaced 120 s apart at 25 °C. The stirring speed and reference power were 1000 rpm and 5 μcal/s. Affinity constants were calculated from the raw data using Origin v7.4 software (MicroCal).
Multiangle Light Scattering
100 μl of 35 μm Get3 and 100 μl of 35 μm Get4/5 were prepared in GET buffer containing 1 mm ATP and either loaded separately or together onto a Superdex 200 10/300 (GE Healthcare) column equilibrated in GET buffer. Samples were analyzed using a Dawn HELEOS II multiangle light scattering unit (Wyatt Technology).
Alkylation and Mass Spectrometry
Get4/5 S48C/C177T was reduced with 2.5 mm tris(2-carboxyethyl)phosphine in GET buffer (without reducing agent) at room temperature for 2 h. For each alkylation reaction, 1 μm Get4/5 alone or in the presence of 2 μm Get3 was preincubated in 2 mm ATP for 10 min to allow for complex formation followed by the addition of 30 μm N-ethylmaleimide. The reaction was quenched with 9.0 mm DTT at the indicated time points, concentrated under vacuum, and redissolved in 0.2% formic acid, and 50 pmol of protein was analyzed on an LC-MSD SL 1100 series (Agilent) mass spectrometer. The samples were chromatographed on a 2.1 × 150-mm Zorbax 300SB-C3 column (Agilent) using a gradient consisting of 0.2% formic acid and 0.2% formic acid in 89.8% acetonitrile and 10% methanol. The m/z values of the intact proteins were measured in the single quadrupole, deconvoluted, and quantified using the ChemStation software (Agilent). Control experiments in which different ratios of unalkylated and alkylated proteins were mixed and subjected to MS analysis showed the quantification of the ratio of alkylated species to be reliable (37).
Get4/5 PEGylation Assay
Get4/5 Q34C/C177T containing a His6 tag was reduced with 2.5 mm tris(2-carboxyethyl)phosphine in GET buffer (without reducing agent) at room temperature for 2 h. For each PEGylation reaction, 0.45 μm Get4/5 alone or in the presence of 2 μm Get3 was preincubated in 2 mm ATP for 10 min to allow complex formation followed by the addition of 60 μm PEG maleimide (10-kDa conjugates; Sigma). The reaction was quenched with 9.0 mm DTT at the indicated time points, and the extent of Get4/5 PEGylation was followed by Western blotting with an anti-His antibody (Qiagen). The ratio of unmodified Get4/5 to PEGylated Get4/5 was determined using ImageQuant software (GE Healthcare).
Fluorescent Labeling
Following purification over an UnoQ column (GE Healthcare), Get4/5 S48C/C177T and Get4/5N S48C/C177T were labeled with thiol-reactive acrylodan. Protein was dialyzed in labeling buffer (50 mm K-HEPES, pH 7.0, and 300 mm NaCl) and treated with 2 mm tris(2-carboxyethyl)phosphine to reduce the disulfide bonds. The labeling reaction was carried out using a 10–30-fold excess of dye over protein. The reaction was incubated overnight at 4 °C and stopped by adding 2 mm DTT. Excess dye was removed by gel filtration using Sephadex G-25 (Sigma) equilibrated with GET buffer.
Association and Dissociation Rate Measurements
All rate measurements were performed on a KinTek Corp. stopped-flow apparatus using sample prepared in GET buffer. For association rate measurements, acrylodan-labeled Get4/5 was held constant at 0.2 μm, Get3 concentration was varied as indicated, and ATP was present at 2 mm. Observed rate constants (kobsd) were plotted as a function of Get3 concentration and fit to the following equation (Equation 1).
in which kon is the association rate constant and koff is the dissociation rate constant.
For dissociation rate measurements, a pulse-chase experiment was used. A complex between acrylodan-labeled Get4/5 (at 0.15 μm) and Get3 (at 0.3 μm) was preformed by incubation in 2 mm ATP for 10 min followed by addition of unlabeled Get4/5 at 6 μm as the chase to initiate Get3·Get4/5 dissociation. The time course for change in fluorescence (Fobsd) was fit to the following double exponential function (Equation 2).
in which Fe is the fluorescence when the reaction reaches equilibrium, ΔF1 and kfast are the magnitude and rate constant of the fluorescence change in the fast phase, and ΔF2 and kslow are the magnitude and rate constant of the fluorescence change in the slow phase.
Kinetic Modeling
The kinetic model was derived using KinTek Explorer Professional Software (KinTek Corp.) (38, 39). Kinetic and equilibrium simulations were carried out using the rate constants listed in Fig. 1H, and values for the experimental data were taken from Rome et al. (24).
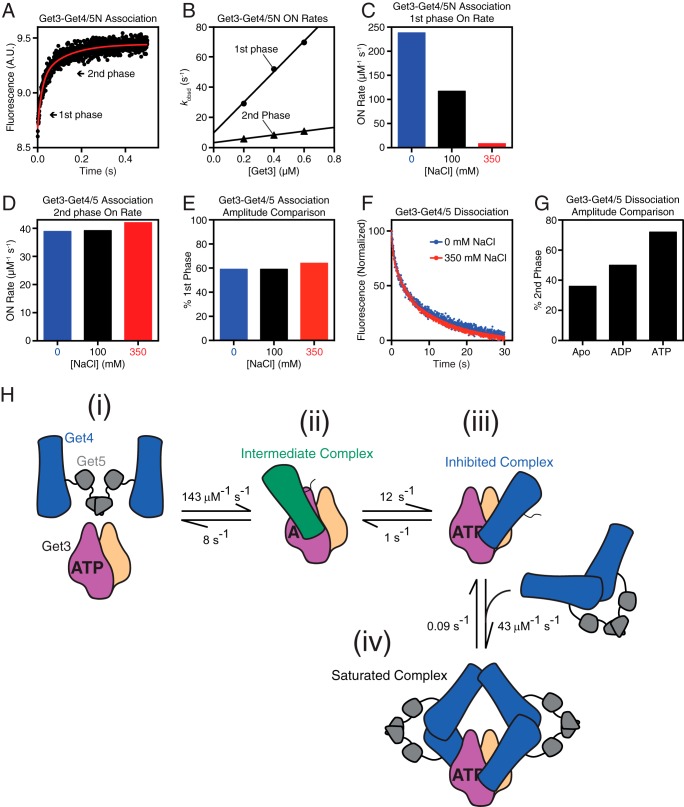
FIGURE 1.
Get4/5 undergoes a conformational change upon binding Get3. A, time course of Get4/5N binding to ATP-bound Get3. Arrows indicate the two kinetic phases. B, observed rate constants during binding (kobsd) are analyzed as a function of Get3 concentration to determine the association rate constant kon for both the first (circles) and second (triangles) kinetic phases. C and D, summary of association rate constants in different salt concentrations for the first (C) and second (D) kinetic phases during Get3·Get4/5 binding. E, the relative amplitude of the kinetic phases for Get3·Get4/5 association is invariant to buffer ionic strength. F, dissociation rate measurement of Get3 from Get4/5 in 0 (blue) and 350 mm (red) NaCl. G, relative amplitude of the second kinetic phase during Get3 dissociation from Get4/5N is dependent on nucleotide. H, proposed model for Get3·Get4/5 complex association based on kinetic data as detailed in the text. In steps ii and iii, Get4/5 is presented as the Get4/5N in our crystal structures with Get4 either green (intermediate complex) or blue (inhibited complex). A.U., absorbance units.
Kinetic Simulations
Simulation of Get3·Get4/5 association was carried out using the KinTek Explorer association function. Equations used to model the associations are listed in Table 2 where G3 denotes Get3, G45 denotes fluorescently labeled Get4/5, Get345 denotes the intermediate Get3·Get4/5 complex, G345* denotes the inhibited Get3·Get4/5 complex after rearrangement, and G345*G45 denotes the saturated complex with two Get4/5 bound to the Get3 dimer (refer to Fig. 1H). Modeling used the following initial conditions: [G45]0 = 0.2 μm, [G3]0 = 0.1, 0.18, and 0.26 μm. The following fluorescence values (F in arbitrary units) were assigned to each species: F = 5 for G45, F = 10 for G345 and G345*, F = 20 for G345*45. For the equation G345 ⇔ G345*, the forward and reverse rate constants were varied until the simulation results matched experimental data.
TABLE 2.
Equations used to model Get3·Get4/5 association kinetics
| Equation | kfor | krev |
|---|---|---|
| μm−1 s−1 | s−1 | |
| G3 + G45 ⇔ G345 | 144 | 8 |
| G345 ⇔ G345* | Varied | Varied |
| G345* + G45 ⇔ G345*G45 | 43.5 | 0.09 |
Simulation of the dissociation kinetics of the Get3·Get4/5 complexes was carried out using the KinTek Explorer two-step mixing function, which accurately reflects the experimental pulse-chase setup. In the first time period (T1), 0.3 μm G3 and 0.15 μm G45 were mixed for 3 s allowing for equilibrium to be reached. During the second time period (T2), 6 μm unlabeled Get4/5 (chase; designated C) was added to this mixture. Dissociation of the Get3·Get4/5 complexes during T2 involves a collection of exchange reactions between G45 and the chase (unlabeled Get4/5) represented by equations in Table 3.
TABLE 3.
Equations used to model Get3·Get4/5 dissociation kinetics
| Equation | kfor | krev |
|---|---|---|
| μm−1 s−1 | s−1 | |
| G3 + C ⇔ G3C | 144 | 8 |
| G345 + C ⇔ G345C | 144 | 8 |
| G345* + C ⇔ G345*C | 43.5 | 0.09 |
| G45 + G3C ⇔ G345C | 144 | 8 |
| G45 + G3C* ⇔ G345*C | 43.5 | 0.09 |
| C + G3C* ⇔ G3C*C | 43.5 | 0.09 |
| G345C ⇔ G345*C | 12 | 1 |
| G3C + C ⇔ G3CC | 144 | 8 |
| G3C ⇔ G3C* | 12 | 1 |
To simulate the equilibrium of complex formation between Get3 and Get4/5, the KinTek Explorer equilibrium titration function was used. For these simulations, 0.05 μm G45 was held constant, and the fluorescence at equilibrium was simulated with increments of 0.005 μm G3 until a final concentration of 0.5 μm G3 was reached.
Results and Discussion
Formation of the Get3·Get4/5 Complex Follows an Electrostatically Driven Intermediate
To understand the mechanism of assembly for the Get3·Get4/5 complex, we characterized the assembly pathway using pre-steady-state kinetics. Complex formation was monitored using acrylodan-labeled Get4/5 whose fluorescence is enhanced upon binding Get3 (24). The Get3·Get4/5 complex has been demonstrated to be highly stable but dynamic, and its formation follows rapid biphasic association at near diffusion-limited time scales (24). The observed association rate constants in both kinetic phases are linearly dependent on Get3 concentration (24), indicating that both phases represent bimolecular association of Get3 with Get4/5. Although the initial report used full-length Get4/5, the same biphasic association was observed with a truncated Get4/5N that lacks the homodimerization domain of Get5 and hence forms a heterodimer (Fig. 1, A and B) (23, 25). This rules out the possibility that the second kinetic phase arises from binding of Get3 to the second Get4 molecule in a Get4/5 heterotetramer.
Additional characterizations strongly suggest the presence of an additional rearrangement step in the assembly pathway. First, the rate of the initial “burst” phase is strongly dependent on ionic strength, consistent with an electrostatically driven association (Fig. 1C) (24), whereas the second association phase is independent of ionic strength (Fig. 1D) (24). This suggests a structural rearrangement to adopt a more stable interface prior to the second association phase. Second, despite the large changes in the relative association rates of the salt-sensitive and -insensitive phases, the amplitudes of these two phases are unaltered by changes in ionic strength (Fig. 1E). This argues against the possibility that the biphasic association arises from parallel pathways, i.e. that the second association phase arises from a subpopulation of Get3 (or Get4/5) that binds the interaction partner with slower kinetics. Third, the dissociation rate constant extrapolated from the fast association phase (Fig. 1B) is 8–10 s−1 (kobsd = kon[Get3] + koff); this koff value is 10–100-fold faster than those of the final Get3·Get4/5 complex determined directly in pulse-chase experiments (Fig. 1F) (24), suggesting that the initial association gives rise to an unstable intermediate that must subsequently rearrange to adopt more stable interfaces. Further in support of this model are results from the dissociation rate measurements. Although dissociation of the Get3·Get4/5 complex also exhibits biphasic kinetics, neither the rates nor the magnitude of the phases is dependent on ionic strength (Fig. 1F). Finally, the magnitude of the slow dissociation phase increases successively when apo-Get3 is compared with ADP- and ATP-bound Get3 (Fig. 1G), consistent with previous data showing that ATP strengthens binding between Get3 and Get4/5 by inducing Get3 into a more closed conformation (22–25, 31).
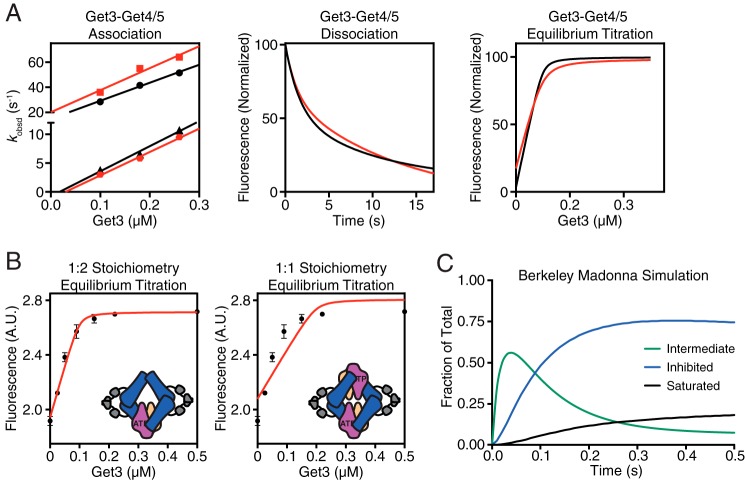
These new data, coupled with kinetic simulation, support a multistep mechanism for the Get3·Get4/5 interaction as the simplest model to explain all the available observations (Fig. 1H) (24). In this model, association of Get3 with Get4/5 involves the following steps. (i) Resting Get3 bound to ATP is biased toward a “closed” conformation. (ii) Initial binding of Get4/5 to Get3 is rapid and generates an intermediate dominated by electrostatic interactions. (iii) This is followed by a conformational change to the final stable structure dominated by hydrophobic interactions. (iv) Under the experimental conditions, the second subunit in the Get3 dimer can bind another Get4/5 to give rise to the second phase in association kinetics. Using the experimentally observed kon and koff values from both the salt-sensitive and salt-insensitive phases, the equilibrium binding data, and kinetic modeling, we were able to completely assign the rate constant for individual microscopic steps in this model. Analytical simulations based on this model reproduced the experimentally observed association/dissociation kinetics and equilibrium titrations (Fig. 2A). Furthermore, the equilibrium titration data with full-length Get4/5 (24) could only be fit to a 1:2 stoichiometry in which a Get3 dimer is bound to two different Get4/5 heterotetramers (Fig. 2B), consistent with the biphasic nature of complex formation.
FIGURE 2.
Modeling of experimental kinetic data. A, comparison of experimental (black) and simulated (red) binding data for the Get3·Get4/5 complex. Association rate measurements (left), dissociation rate measurements (center), and equilibrium titrations (right) are shown. B, left, model depicting a 1:2 binding stoichiometry of Get3 with Get4/5. This configuration was used to fit an equilibrium titration of acrylodan-labeled Get4/5 full length (100 nm) with Get3 in 2 mm ATP. Right, same as on left but fit with a model assuming a 1:1 binding stoichiometry. Error bars represent S.D. C, Berkeley Madonna simulation of the time course of Get3·Get4/5 complex formation. A.U., absorbance units.
Although binding of a second Get4/5 to Get3 was observed (Fig. 1H), multiple observations suggest that the stable inhibited Get3·Get4/5 complex (species iii) dominates under most conditions, and accumulation of the saturated Get3·Get4/5 complex (species iv) is modest. First, previous assays of Get3 activity show that binding of one full-length Get4/5 is sufficient to inhibit the ATPase activity of Get3 (22). Second, kinetic simulations show that during complex assembly with relatively stoichiometric amounts of proteins the saturated Get3·Get4/5 complex accumulates to <20% of all the complexes formed (Fig. 2C). Coupled with the different Get3·Get4/5N complexes in various crystal structures (23), it appears that binding of Get4/5 to Get3 has a preferred but not obligatory stoichiometry.
The Structure of a Get3·Get4/5 Intermediate Complex
The recent crystal structure of an ATP-bound Get3·Get4/5N complex demonstrated a stable interface that included conserved hydrophobic interactions (23). The structure allowed rationalization of how Get4 stabilized an ATP-bound Get3; however, it was unclear how the structure could account for the multistep Get3·Get4/5 assembly seen experimentally. To address this question, we set out to obtain a structure of Get4/5N bound to a Get3 dimer under conditions that would stabilize an initial intermediate complex. As before (23), Get3 and Get4/5 were expressed and purified separately, then combined, and purified as a 1:1 complex in low ionic strength conditions by SEC (23). Initial crystals were obtained using a Get4/5N construct similar to that used for solving the structure of the Get4/5N heterodimer (25). This resulted in a crystal that diffracted to 6.0 Å in space group P21. Phases for this crystal form were obtained by molecular replacement using an “open” form of Get3 (Protein Data Bank code 3A37) (14) and the Get4/5N heterodimer (Protein Data Bank code 3LKU) as search models. The final model was refined to an R/Rfree of 27.4/30.3% (Table 1). To improve the diffraction limit, purification and crystallization trials were performed in the presence of either ADP or the non-hydrolyzable ATP analogue AMP-PNP. In addition, 22 residues from the C terminus of Get4 were truncated as they are disordered in previous structures (12, 25). This resulted in a new crystal form that diffracted to 2.8 Å in space group P21212. The final model was refined to an R/Rfree of 22.4/26.1% (Table 1). Although ADP and AMP-PNP produced identical crystal forms, the highest resolution data set reported here was collected from a crystal that grew in the presence of AMP-PNP, although the nucleotide was not visible in the density.
The Get3·Get4/5 Intermediate Complex Is Dominated by an Electrostatic Interface
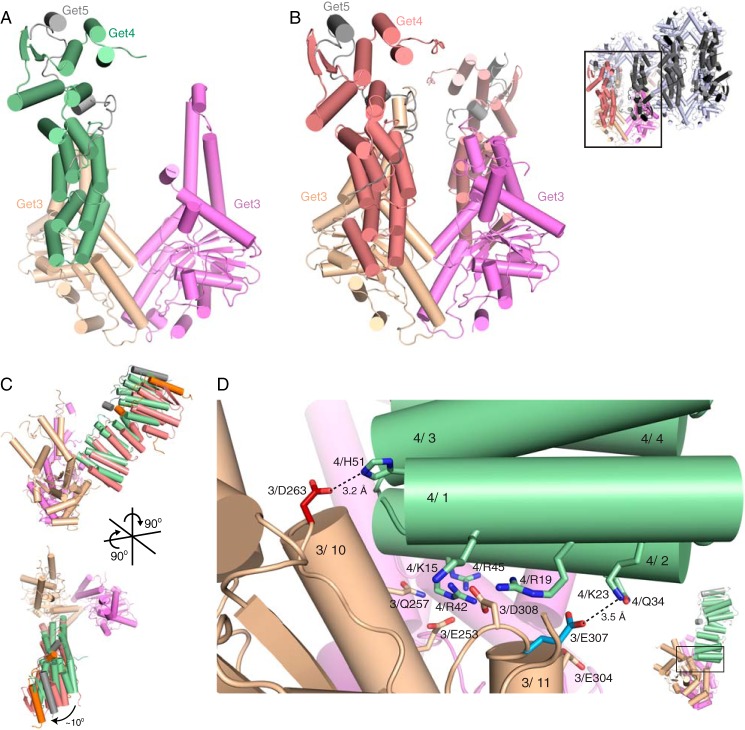
In both crystal forms, Get4 binds in a similar orientation on Get3 (Fig. 3, A and B). The 2.8-Å structure contains a 2:1 complex of a Get3 dimer bound to a single Get4/5N heterodimer (Fig. 3A). This crystal form is related to another Get3 crystal form (14) in which limited space in the crystal lattice allows for only one Get4/5N molecule despite the 1:1 stoichiometry of the complex put into crystallization trays (as measured by SEC). Although nucleotide was present throughout purification and crystallization, the nucleotide-binding pocket appears empty. For the 6.0-Å structure (Fig. 3B), the asymmetric unit contained a 1:1 complex with eight copies of Get3 bound to eight Get4/5N heterodimers (Fig. 3B, top right).
FIGURE 3.
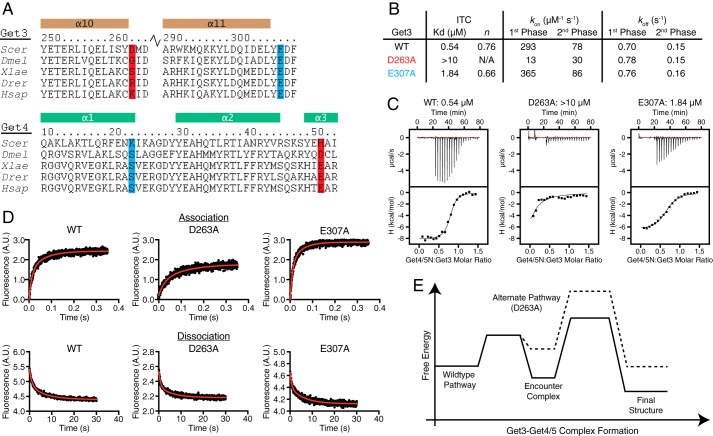
Crystal structures of a Get3/4/5 intermediate complex. A, the asymmetric unit of the 2.8-Å structure of Get4/5N (Get4, green; Get5N, gray) bound to Get3 (wheat and magenta). B, left, one subunit from the 6.0-Å structure containing two Get4/5N molecules (Get4, salmon; Get5N, gray) bound to a Get3 dimer (wheat and magenta). Top right, the asymmetric unit of the 6.0-Å structure of Get4/5N (Get4, red; Get5N, gray) bound to Get3 (wheat and magenta). The black box outlines one Get3 dimer (wheat and magenta) bound to two Get4/5N heterodimers (Get4, salmon; Get5N, gray). The rest of the asymmetric unit is colored as follows: Get3, light gray; Get4, dark gray; Get5N, black. C, two views showing the relative orientation of Get4 monomers from the 2.8- (green) and 6.0-Å (red) crystal forms. Alignment was based on helices α10 and α11 from the contacting Get3 monomer. D, view of the interface from the 2.8-Å structure showing interactions between Get4 (green) and Get3 (wheat and purple). Residues Asp263 (red) and Glu307 (blue) were tested for interaction and colored based on phenotype. These residues are unique to this interface and are not found in the ATP-bound structure (23). Dashed lines represent potential interactions and are labeled with their atomic distances.
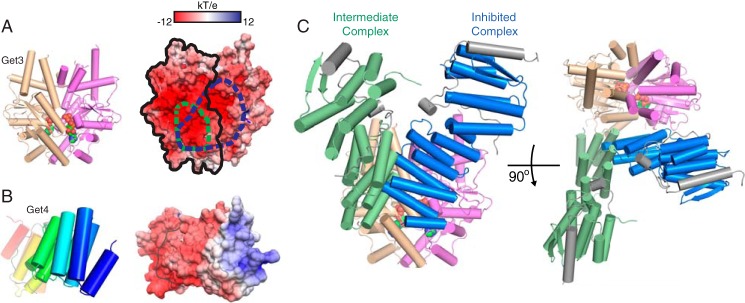
In both cases, Get3 is in an open conformation, and Get4/5N binds in the same interface and orientation, defined here as the intermediate complex. Despite the differences in crystallization and space group, there is little difference in the interface between the two structures, only a slight 10° rotation at the furthest point (Fig. 3C). The higher resolution structure will be used for reference for the rest of the text. In this intermediate complex, Get4 interacts with a single monomer of Get3 in an orientation compatible with both the open and closed structures, burying ∼970 Å2 of surface area (Figs. 3D and 4A). The interactions in this interface are electrostatic and involve the positively charged surface of Get4 binding to the negatively charged surface of Get3 (Fig. 4, A and B).
FIGURE 4.
Comparing the intermediate and inhibited interfaces. A, left, orientation of Get3 used in subsequent panels to highlight binding interface. For surface representations, one monomer is outlined in black, and the interaction surface of Get4 on Get3 is highlighted by dashed lines for encounter complex (green) and inhibited complex (blue). Right, accessible surface color-ramped based on electrostatic potential from −12 (red) to +12 kT/e (blue). B, surface properties of Get4 as in A except that Get4 is color-ramped from the N terminus (blue) to the C terminus (red), and Get5N is colored gray. C, two views of the overlay of Get4 from the intermediate complex (green) and inhibited complex (blue) on the closed Get3 dimer from Protein Data Bank code 4PWX (23). The Get4 from the intermediate complex was orientated by aligning helices α10 and α11 on one monomer of Get3.
In this orientation, Get4 α1 sits on top of a groove formed by the loop before Get3 α1 and the loop following Get3 α11 such that the N terminus of Get4 α1 packs against the loops following Get3 α9 and α10, resulting in an interaction between Lys23 on Get4 α1 and Glu307 on Get3 (Fig. 3D). In addition, Get4 α2 packs roughly perpendicularly to Get3 α10 and α11 such that the N terminus contacts Get3 α11 and the C terminus contacts Get3 α10. This results in interactions of the invariant residues Arg42 on Get4 α2 with Asp308 on Get3 and Arg45 on Get4 α2 with both Glu253 and Gln257 on Get3 α10 (Fig. 3D). The N terminus of Get4 α3, which is oriented between α1 and α2, contacts the top of Get3 α10, allowing His51 on Get4 α3 to interact with Asp263 on Get3 α10 (Fig. 3D).
Comparison of the Two Distinct Get3·Get4/5 Complexes
In the previously reported ATP-bound Get3·Get4/5N complex, Get4/5N is bound across the dimer interface of a closed Get3, forming interactions with both monomers, which we define as the inhibited complex (Fig. 4C). This is in contrast to the current structure, which is bound to an open Get3 and interacts with only one monomer (Fig. 4C). Because the Get4-binding surfaces do not undergo a significant conformational change between the open and closed Get3 structures, we aligned the helices (α10 and α11) involved in Get4 binding from our intermediate complex onto the inhibited complex. Both structures have overlapping binding sites and predominantly utilize the N terminus of Get4 for the interaction (Fig. 4B). The difference in orientation involves a rotation centered on α2 of Get4. This rotation allows Get4 α2 to form the majority of the interactions in the inhibited complex, whereas Get4 α1 is responsible for many interactions in the intermediate complex. Based on the multistep assembly observed from kinetic analysis, we believe that the new structure dominated by electrostatics represents the intermediate Get3·Get4/5 complex (Fig. 1H, species ii) prior to rearrangement to the final structure stabilized by hydrophobic interactions.
Mutational Analysis of the Intermediate Complex
The intermediate complex interface involves numerous electrostatic residues on both Get3 and Get4. Of these interactions, only Get3 Asp263-Get4 His51 and Get3 Glu307-Get4 Lys23 are unique to the intermediate interface and are not found in the inhibited complex (Fig. 5A). The first interaction (Asp263-His51) is not conserved; however, in higher eukaryotes, there is conservation of a likely salt bridge (Fig. 5A). The conserved Get3 Glu307 also interacts with the cytoplasmic domain of Get2 (16, 17), whereas Get4 Lys23 is not conserved (Fig. 5A). Substitution of Get3 Asp263 leads to a loss of observable binding to Get4/5N in isothermal titration calorimetry measurements (Fig. 5, B and C), whereas substitution of Get3 Glu307 had a more modest loss in affinity (Fig. 5, B and C).
FIGURE 5.
Get3 D263A is defective for complex formation. A, sequence alignments of regions involved in contacts in the Get3-Get4 interface using ClustalW (43). Scer, S. cerevisiae; Dmel, Drosophila melanogaster; Xlae, Xenopus laevis; Drer, Danio rerio; Hsap, Homo sapiens. Helices are indicated above the sequence and labeled. Highlighted residues are unique to the intermediate interface and were tested for binding: red, S. cerevisiae Get3 Asp263 and S. cerevisiae Get4 His51; blue, S. cerevisiae Get3 Glu307 and S. cerevisiae Get4 Lys23. B, summary of the data obtained from isothermal titration calorimetry (ITC) and kinetic experiments. C, representative isotherms from ITC experiments. D, top, representative association rate measurements of Get4/5 binding to wild type and mutant Get3 at 260 nm. Each plot represents the average of 10 experiments. Bottom, representative dissociation rate measurements with wild type and mutant Get3. Each plot represents the average of three experiments. E, free energy diagram of the effects of the Get3 D263A mutation on Get3·Get4/5 complex formation. A.U., absorbance units.
If the intermediate complex is on-pathway to the final Get3·Get4/5 complex, one would predict that disrupting the interactions unique to this intermediate would slow down Get3·Get4/5 association. Disrupting the putative Get3 Glu307-Get4 Lys23 interaction (E307A) resulted in similar association rates compared with wild type Get3 and, together with the isothermal titration calorimetry results, suggests that the interaction is not essential for complex formation (Fig. 5, B–D). In contrast, significantly reduced rates of Get3·Get4/5 association were observed with Get3 D263A (Fig. 5, B–D). Although the association rates are slightly higher than previously reported, the differences are small (∼2-fold) and do not change the interpretation. Both mutants had virtually identical dissociation rates compared with wild type Get3; therefore, the difference in affinity for D263A is specific for complex formation kinetics (Fig. 5, B–D).
If Get3 D263A follows the same assembly pathway as wild type Get3, one would expect that once the Get3 D263A·Get4/5 complex is formed it has the same equilibrium stability as the wild type complex because Asp263 does not participate in interface interactions in the final complex (Fig. 5E). However, the weakened equilibrium stability of the Get3 D263A·Get4/5 complex indicates that the changes induced by D263A are more complex, and this mutant likely follows a different assembly pathway leading to a non-native complex. Because a reaction follows the fastest pathway, this observation in turn suggests that the D263A mutation slows the native assembly pathway more extensively than was experimentally measured. Collectively, mutational analysis of the intermediate complex provides independent evidence that the structure observed here represents an on-pathway intermediate during Get3·Get4/5 assembly.
Stoichiometry between Get3 and Get4/5
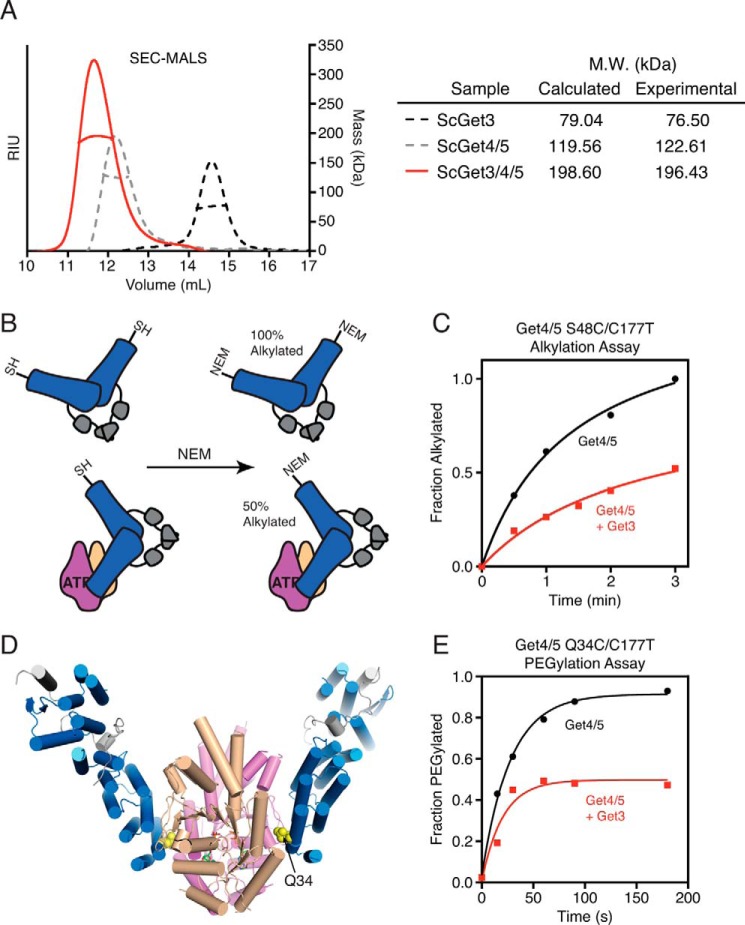
The stoichiometry of the Get3·Get4/5 interaction is not fully determined. A recent SEC-multiangle light scattering analysis using an ATPase-deficient Get3 (D57N) suggested that the complex contains one Get4/5 heterotetramer bound to one Get3 dimer (40). Using wild type Get3, SEC-multiangle light scattering analysis with equimolar concentrations of Get3 and Get4/5 confirmed that the size of the complex is consistent with a single Get4/5 heterotetramer bound to a single Get3 dimer (Fig. 6A) (23).
FIGURE 6.
Get3 binds to one-half of the Get4/5 heterotetramer. A, SEC-multiangle light scattering (MALS) analysis of the Get3·Get4/5 complex (ScGet/3/4/5). B, model depicting the N-ethylmaleimide (NEM) accessibility of a solvent-exposed Cys residue (S48C) on Get4/5 alone and in complex with Get3. C, results of the alkylation assay shown in B. D, ATP-bound structure of Get3·Get4/5N (Protein Data Bank code 4PWX) demonstrating the location of Q34C within the Get3·Get4/5 interface. E, results of the PEGylation assay with Get4 Q34C/5 alone or in complex with Get3. RIU, refractive index units. Sc, S. cerevisiae.
One puzzling observation arises from these data: despite the presence of two Get4 molecules in full-length Get4/5, there has been no evidence for binding of Get3 to the second Get4 in the Get4/5 heterotetramer. To directly test this asymmetry of Get4/5 during complex formation, we developed an alkylation protection assay. An engineered cysteine at the Get3·Get4/5 interface (Get4 S48C (24)) is allowed to react with N-ethylmaleimide in the absence and presence of various factors (Fig. 6B). In the free Get4/5, Get4 Cys48 is solvent-exposed (25) and rapidly alkylates to completion (Fig. 6, B and C). Consistent with binding at a single Get4 interface, only 50% of Get4 Cys48 was protected from alkylation by Get3 (Fig. 6C). This protection pattern was observed at Get3 concentrations nearly 1000-fold above the dissociation constant for the Get3·Get4/5 complex, indicating that the 50% protection did not arise from incomplete Get3·Get4/5 binding. These results strongly suggest that only one site of a Get4/5 heterotetramer is able to bind Get3.
To further support these results, accessibility of Get4/5 was probed using a 10-kDa PEG maleimide. A second site (Get4 Q34C) that is also occluded in the Get3·Get4/5N structure (Fig. 6D) was chosen to further validate the interface. Importantly, substitution of Gln34 to alanine does not impair binding to Get3 (23). In this reaction, Get4 Q34C with PEG maleimide forms a covalent adduct that gives a 10-kDa increase in mass that can be detected by SDS-PAGE and Coomassie staining. In the absence of Get3, this reaction resulted in 100% PEGylation of Get4/5 over a 2-min time course, whereas addition of saturating Get3 resulted in 50% PEGylation (Fig. 6E). This provides corroborating evidence that only one Get4 molecule in a heterotetramer binds Get3. Together, these data indicate that once Get3 binds to Get4/5 the other Get4 molecule in the Get4/5 heterotetramer is inhibited from further interaction with another Get3 dimer.
A Model for Get3·Get4/5 Complex Formation
Multiple lines of evidence support a model in which Get3 and Get4/5 rapidly form an electrostatic intermediate complex and then undergo a structural rearrangement to a more stable complex (24) (Fig. 2A). This conformational change is consistent with previous results showing that Get4/5 binding induces Get3 into an “occluded” state, leading to Get3 ATPase inhibition and delayed ATP dissociation kinetics (22). Based on this model, once Get3 has transitioned to the occluded conformation, complex dissociation would be insensitive to buffer ionic strength (Fig. 1F) but dependent on nucleotide (Fig. 1G).
The initial Get3·Get4/5 complex presented here (Fig. 3, A and B) reveals a novel binding interface composed of electrostatic interactions (Fig. 3D). This structure provides the molecular basis for the initial salt-dependent association seen in our kinetic description of the Get3·Get4/5 interaction (Fig. 1). Although Get3 is in an open conformation, these structures likely represent the true on-pathway intermediate because the Get3-Get4 interface does not change between the open and closed forms. This is corroborated by the D263A mutation introduced within this interface that drastically slows down complex formation (Fig. 5). These data coupled with the kinetic experiments argue that these structures represent the initial intermediate complex during Get3 and Get4/5 binding.
As we reported previously, Get4/5 is able to precisely discriminate between nucleotide states of Get3, enabling Get4/5 to regulate Get3 activity and prime it for efficient capture of the TA substrate (22–24). The structure described here provides evidence for an additional interaction dominated by electrostatic interactions and characterized by fast association rates. This ultrafast diffusion-limited association of Get4/5 to Get3 (24) may function as an additional mechanism for Get4/5 to select for the appropriate conformational state of Get3 or inversely enables Get3 to constantly sample numerous Get4/5 until transfer of TA substrate occurs. Interestingly, this may also function to ensure that Get3 is quickly recruited following dissociation from the membrane to prevent rebinding to Get1 or Get2 (24). This type of interaction has been seen in other systems including barnase-barstar (41) and ribosome-interacting proteins (42) and likely represents a recurring theme in protein-protein interactions. This work provides further evidence for the importance of Get4/5 in TA targeting.
Author Contributions
H. B. G., M. E. R., S. S., and W. M. C. designed the study and wrote the paper. H. B. G. determined the 2.8-Å crystal structure and characterized the intermediate complex. M. E. R. performed experiments to define stoichiometry and kinetic analysis. J. W. C. determined the structure of the 6.0-Å complex. M. R. helped conceptually with the study. S. H. assisted in the mass spectrometry analysis. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Graeme Card, Ana Gonzalez, and Michael Soltis for help with data collection at Stanford Synchrotron Radiation Lightsource Beamline 12-2. We are grateful to Gordon and Betty Moore for support of the Molecular Observatory at Caltech. Operations at Stanford Synchrotron Radiation Lightsource are supported by the United States Department of Energy and National Institutes of Health.
This work was supported by National Science Foundation Graduate Research Fellowship DGE-1144469 (to M. E. R.) and National Institutes of Health Training Grant 5T32GM007616-33 (to H. B. G. and M. R.) and Research Grants R01GM097572 (to W. M. C.) and R01GM107368 (to S. S.). The Proteome Exploration Laboratory was supported by Gordon and Betty Moore Foundation Grant GBMF775 and the Beckman Institute (to S. H.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The atomic coordinates and structure factors (codes 5BW8 and 5BWK) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- TA
- tail-anchored
- GET
- guided entry of TA proteins
- BME
- β-mercaptoethanol
- SEC
- size exclusion chromatography
- AMP-PNP
- 5′-adenylyl-β,γ-imidodiphosphate.
References
- 1.Shao S., and Hegde R. S. (2011) Membrane protein insertion at the endoplasmic reticulum. Annu. Rev. Cell Dev. Biol. 27, 25–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shan S. O., and Walter P. (2005) Co-translational protein targeting by the signal recognition particle. FEBS Lett. 579, 921–926 [DOI] [PubMed] [Google Scholar]
- 3.Kutay U., Hartmann E., and Rapoport T. A. (1993) A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 3, 72–75 [DOI] [PubMed] [Google Scholar]
- 4.Kutay U., Ahnert-Hilger G., Hartmann E., Wiedenmann B., and Rapoport T. A. (1995) Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 14, 217–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borgese N., Brambillasca S., and Colombo S. (2007) How tails guide tail-anchored proteins to their destinations. Curr. Opin. Cell Biol. 19, 368–375 [DOI] [PubMed] [Google Scholar]
- 6.Borgese N., Gazzoni I., Barberi M., Colombo S., and Pedrazzini E. (2001) Targeting of a tail-anchored protein to endoplasmic reticulum and mitochondrial outer membrane by independent but competing pathways. Mol. Biol. Cell 12, 2482–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hegde R. S., and Keenan R. J. (2011) Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 12, 787–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chartron J. W., Clemons W. M. Jr., and Suloway C. J. (2012) The complex process of GETting tail-anchored membrane proteins to the ER. Curr. Opin. Struct. Biol. 22, 217–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang F., Brown E. C., Mak G., Zhuang J., and Denic V. (2010) A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol. Cell 40, 159–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suloway C. J., Chartron J. W., Zaslaver M., and Clemons W. M. Jr. (2009) Model for eukaryotic tail-anchored protein binding based on the structure of Get3. Proc. Natl. Acad. Sci. U.S.A. 106, 14849–14854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mateja A., Szlachcic A., Downing M. E., Dobosz M., Mariappan M., Hegde R. S., and Keenan R. J. (2009) The structural basis of tail-anchored membrane protein recognition by Get3. Nature 461, 361–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bozkurt G., Stjepanovic G., Vilardi F., Amlacher S., Wild K., Bange G., Favaloro V., Rippe K., Hurt E., Dobberstein B., and Sinning I. (2009) Structural insights into tail-anchored protein binding and membrane insertion by Get3. Proc. Natl. Acad. Sci. U.S.A. 106, 21131–21136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu J., Li J., Qian X., Denic V., and Sha B. (2009) The crystal structures of yeast Get3 suggest a mechanism for tail-anchored protein membrane insertion. PLoS One 4, e8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamagata A., Mimura H., Sato Y., Yamashita M., Yoshikawa A., and Fukai S. (2010) Structural insight into the membrane insertion of tail-anchored proteins by Get3. Genes Cells 15, 29–41 [DOI] [PubMed] [Google Scholar]
- 15.Suloway C. J., Rome M. E., and Clemons W. M. Jr. (2012) Tail-anchor targeting by a Get3 tetramer: the structure of an archaeal homologue. EMBO J. 31, 707–71922124326 [Google Scholar]
- 16.Mariappan M., Mateja A., Dobosz M., Bove E., Hegde R. S., and Keenan R. J. (2011) The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature 477, 61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stefer S., Reitz S., Wang F., Wild K., Pang Y. Y., Schwarz D., Bomke J., Hein C., Löhr F., Bernhard F., Denic V., Dötsch V., and Sinning I. (2011) Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex. Science 333, 758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F., Whynot A., Tung M., and Denic V. (2011) The mechanism of tail-anchored protein insertion into the ER membrane. Mol. Cell 43, 738–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto Y., and Sakisaka T. (2012) Molecular machinery for insertion of tail-anchored membrane proteins into the endoplasmic reticulum membrane in mammalian cells. Mol. Cell 48, 387–397 [DOI] [PubMed] [Google Scholar]
- 20.Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., Sevier C. S., Ding H., Koh J. L., Toufighi K., Mostafavi S., Prinz J., St Onge R. P., VanderSluis B., Makhnevych T., Vizeacoumar F. J., Alizadeh S., Bahr S., Brost R. L., Chen Y., Cokol M., Deshpande R., Li Z., Lin Z. Y., Liang W., Marback M., Paw J., San Luis B. J., Shuteriqi E., Tong A. H., van Dyk N., Wallace I. M., Whitney J. A., Weirauch M. T., Zhong G., Zhu H., Houry W. A., Brudno M., Ragibizadeh S., Papp B., Pál C., Roth F. P., Giaever G., Nislow C., Troyanskaya O. G., Bussey H., Bader G. D., Gingras A. C., Morris Q. D., Kim P. M., Kaiser C. A., Myers C. L., Andrews B. J., and Boone C. (2010) The genetic landscape of a cell. Science 327, 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Battle A., Jonikas M. C., Walter P., Weissman J. S., and Koller D. (2010) Automated identification of pathways from quantitative genetic interaction data. Mol. Syst. Biol. 6, 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rome M. E., Rao M., Clemons W. M., and Shan S. O. (2013) Precise timing of ATPase activation drives targeting of tail-anchored proteins. Proc. Natl. Acad. Sci. U.S.A. 110, 7666–7671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gristick H. B., Rao M., Chartron J. W., Rome M. E., Shan S. O., and Clemons W. M. Jr. (2014) Crystal structure of ATP-bound Get3-Get4-Get5 complex reveals regulation of Get3 by Get4. Nat. Struct. Mol. Biol. 21, 437–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rome M. E., Chio U. S., Rao M., Gristick H., and Shan S. O. (2014) Differential gradients of interaction affinities drive efficient targeting and recycling in the GET pathway. Proc. Natl. Acad. Sci. U.S.A. 111, E4929–E4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chartron J. W., Suloway C. J., Zaslaver M., and Clemons W. M. Jr. (2010) Structural characterization of the Get4/Get5 complex and its interaction with Get3. Proc. Natl. Acad. Sci. U.S.A. 107, 12127–12132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Battye T. G., Kontogiannis L., Johnson O., Powell H. R., and Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 29.Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., and Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Painter J., and Merritt E. A. (2006) TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 39, 109–111 [Google Scholar]
- 34.Pettersen E. F, Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 35.Baker N. A., Sept D., Joseph S., Holst M. J., and McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dolinsky T. J., Nielsen J. E., McCammon J. A., and Baker N. A. (2004) PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen T. X., Jaru-Ampornpan P., Lam V. Q., Cao P., Piszkiewicz S., Hess S., and Shan S. O. (2013) Mechanism of an ATP-independent protein disaggregase: I. structure of a membrane protein aggregate reveals a mechanism of recognition by its chaperone. J. Biol. Chem. 288, 13420–13430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson K. A. (2009) Fitting enzyme kinetic data with KinTek Global Kinetic Explorer. Methods Enzymol. 467, 601–626 [DOI] [PubMed] [Google Scholar]
- 39.Johnson K. A., Simpson Z. B., and Blom T. (2009) Global kinetic explorer: a new computer program for dynamic simulation and fitting of kinetic data. Anal. Biochem. 387, 20–29 [DOI] [PubMed] [Google Scholar]
- 40.Mateja A., Paduch M., Chang H. Y., Szydlowska A., Kossiakoff A. A., Hegde R. S., and Keenan R. J. (2015) Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo. Science 347, 1152–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schreiber G., and Fersht A. R. (1996) Rapid, electrostatically assisted association of proteins. Nat. Struct. Biol. 3, 427–431 [DOI] [PubMed] [Google Scholar]
- 42.Sandikci A., Gloge F., Martinez M., Mayer M. P., Wade R., Bukau B., and Kramer G. (2013) Dynamic enzyme docking to the ribosome coordinates N-terminal processing with polypeptide folding. Nat. Struct. Mol. Biol. 20, 843–850 [DOI] [PubMed] [Google Scholar]
- 43.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., and Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]