Background: Hypoxia inhibits myogenic differentiation, but the underlying mechanisms are not well understood.
Results: We used microarray analysis to identify genes and pathways affected by hypoxia. Gain- and loss-of-function studies indicate that Bhlhe40 inhibits myogenic differentiation and that hypoxia up-regulates Bhlhe40 through an HIF1α-independent, p53-dependent mechanism.
Conclusion: Hypoxia inhibits myogenesis through p53-dependent induction of Bhlhe40.
Significance: Inhibition of Bhlhe40 or p53 may facilitate muscle repair after ischemic injuries.
Keywords: basic helix-loop-helix transcription factor (bHLH), differentiation, hypoxia, muscle regeneration, p53, skeletal muscle
Abstract
Satellite cells are muscle-resident stem cells capable of self-renewal and differentiation to repair injured muscles. However, muscle injury often leads to an ischemic hypoxia environment that impedes satellite cell differentiation and reduces the efficiency of muscle regeneration. Here we performed microarray analyses and identified the basic helix-loop-helix family transcription factor Bhlhe40 as a candidate mediator of the myogenic inhibitory effect of hypoxia. Bhlhe40 is strongly induced by hypoxia in satellite cell-derived primary myoblasts. Overexpression of Bhlhe40 inhibits Myog expression and mimics the effect of hypoxia on myogenesis. Inhibition of Bhlhe40, conversely, up-regulates Myog expression and promotes myogenic differentiation. Importantly, Bhlhe40 knockdown rescues myogenic differentiation under hypoxia. Mechanistically, Bhlhe40 binds to the proximal E-boxes of the Myog promoter and reduces the binding affinity and transcriptional activity of MyoD on Myog. Interestingly, hypoxia induces Bhlhe40 expression independent of HIF1α but through a novel p53-dependent signaling pathway. Our study establishes a crucial role of Bhlhe40 in mediating the repressive effect of hypoxia on myogenic differentiation and suggests that inhibition of Bhlhe40 or p53 may facilitate muscle regeneration after ischemic injuries.
Introduction
Satellite cells reside between the sarcolemma and basement membrane of myofibers (1) and are responsible for postnatal muscle growth and regeneration of damaged muscles (2). Postnatal myogenesis involves activation of quiescent satellite cells, proliferation of the activated satellite cells (called myoblasts), and differentiation and fusion of myoblasts into multinuclear myofibers. This process is orchestrated by several basic helix-loop-helix family myogenic regulatory factors (MRFs),2 including Myf5, Myf6, MyoD, and Myog (2). Each MRF alone can transform some types of nonmyogenic cells into myoblasts (3–6). By contrast, knocking out any of these factors impedes postnatal myogenesis (7–9). Because of the crucial role of MRFs in myogenesis, physiological stresses that inhibit their expression also impede myogenesis (10–12).
Low oxygen tension (hypoxia) is one of the physiological stresses repressing myogenesis. A hypoxic microenvironment could result from ischemic insults, cachexia, or acute exposure to high altitude (10, 13). The physiological oxygen (O2) volume is 2–10% in muscle, but it can drop to 1% after an ischemic insult (14, 15). A hypoxic culture condition (1% O2) has been shown to repress myogenesis through various mechanisms, such as activating the Notch signaling pathway, inhibiting the PI3K pathway, or accelerating MyoD degradation (16–19). Although hypoxia has been reported to trigger multiple signaling pathways to repress myogenesis, ectopic expression of MyoD or Myog only partly rescues myogenesis (10). Therefore, additional factors that modulate the activity of MyoD, Myog, and other MRFs may also have been involved in repressing myogenesis under hypoxia.
Bhlhe40 (also called DEC1, STRA13, SHARP2, and Bhlhe2) belongs to the basic helix-loop-helix family and acts as a transcriptional repressor involved in cell proliferation, apoptosis, adipogenesis, fatty acid oxidation, and circadian rhythms (20–26). Although Bhlhe40 is recognized as a transcriptional repressor in multiple biological processes, its role in myogenesis is controversial. It has been reported that Bhlhe40 is required for postnatal myogenesis through promoting myogenic differentiation (27). However, overexpression of Bhlhe40 has been reported to inhibit myogenic differentiation in other studies (10, 28).
Here we report Bhlhe40 as a hypoxic responsive transcription repressor that mediates the repression of Myog expression through decreasing the transcriptional activity of MyoD, leading to inhibition of myogenic differentiation. Moreover, we demonstrate that hypoxia induces Bhlhe40 through the p53 pathway independent of HIF1α. These data together suggest that, other than the classical MRFs, Bhlhe40 also plays a key role in myogenesis, especially under hypoxic conditions.
Experimental Procedures
Animals
Mice were from The Jackson Laboratory (HIF1αfl/fl, stock no. 007561; Myf5Cre, stock no. 007893; MyoDiCre, stock no. 014140), and PCR genotyping was carried out using protocols described by the supplier. Mice were housed in the animal facility with free access to water and standard rodent chow. All procedures involving the use of animals were performed in accordance with the guidelines presented by the Purdue University Animal Care and Use Committee.
Primary Myoblast Isolation, Culture, and Differentiation
Primary myoblasts were isolated from hind limb skeletal muscles that were minced and digested in type I collagenase and dispase B mixture (Roche Applied Science). The digestion was stopped with F-10 Ham's medium containing 17% FBS, and the cells were filtered from debris, centrifuged, and cultured in growth medium (F-10 Ham's medium supplemented with 17% FBS, 4 ng/ml basic fibroblast growth factor, and 1% penicillin-streptomycin) on uncoated dishes for 3 days, and 5 ml of growth medium was added each day. Then the supernatant was collected, centrifuged, and trypsinized with 0.25% trypsin. After washing off the trypsin using growth medium and centrifugation, primary myoblasts were seeded on collagen-coated dishes, and the growth medium was changed every 2 days. Myoblasts were induced to differentiate on Matrigel-coated dishes and cultured in differentiation medium (DMEM supplemented with 2% horse serum and 1% penicillin-streptomycin). Unless indicated otherwise, primary myoblasts were cultured in normal humidified tissue culture incubators with 5% CO2.
For hypoxia treatment (∼1% O2), primary myoblasts were put into gas-tight modular incubator chambers (Billups-Rothenberg, Del Mar, CA) that were flushed with a custom gas mixture containing 5% air, 5% CO2, and 90% N2, 30 p.s.i./min, for 2.5 min each day. The p53 inhibitor Pifithrin-α was purchased from Sigma and used to treat satellite cell-derived primary myoblasts at 5 μm concentration.
Microarray Analysis of Myoblasts under Hypoxic Conditions
Three replicates of total RNA extracted from myoblasts cultured under hypoxic conditions (1% O2) or normoxic conditions (21% O2) for 2 days were shipped to Miltenyi Biotec, where the quality of RNA was checked via the Agilent 2100 Bioanalyzer platform (Agilent Technologies). Two replicates of high-quality (RNA integrity number > 8.2) RNA from the normoxic control group or hypoxia-treated group were labeled with Cy3 or Cy5 and hybridized to Agilent whole mouse genome oligo microarrays 8 × 60,000 (two-color). Fluorescence signals of the hybridized Agilent oligo microarrays were detected using a DNA microarray scanner (Agilent Technologies). Agilent feature extraction software was used to read out and process the microarray image files and to determine feature intensities and ratios. Only spots on both replicates that had the same trend (both up or down) were selected for further analysis using DAVID 6.7 (29).
Adenovirus Production to Overexpress Bhlhe40
The adenovirus with Bhlhe40 insertion was generated using the AdEasy system (30). Briefly, the Bhlhe40 ORF was cloned using a pair of primers (5′-CAGCGGCCGCATGGAACGGATCCCCAG and 5′-TTCTCGTGTTAGTCTTTGGTTTCTAAGTTT) and inserted into the pAdTrack-CMV plasmid. The formed pAdTrack-CMV-Bhlhe40 (pAdTrack-CMV as the control) plasmid was digested by PmeI, and then we transfected the DH5a-competent cell with pAdEasy-1. The positive recombinant plasmid was detected by PacI digestion. Then 293A cells (60–70% confluent) in 10-cm culture dishes were transfected with 4 μg of PacI-digested recombinant plasmid using Lipofectamine 2000 (Life Technologies) according to the protocol of the manufacturer. After 14 days of incubation, the recombinant adenovirus was released by four freeze-thaw-vortex cycles. Two more rounds of infection were adapted to amplify the recombinant virus, and the titers were determined by the expression of GFP. To analyze the influence of Bhlhe40 on myoblasts, myoblasts were treated with adenovirus for 1 day and cultured in virus-free medium for at least one more day.
Immunostaining and Image Acquisition
Myoblasts or myotubes were fixed with 4% paraformaldehyde and then blocked with blocking buffer (5% goat serum, 2% BSA, 0.2% Triton X-100, and 0.1% sodium azide in PBS) for at least 1 h. Then the samples were incubated with primary antibodies (MF20 was from the Developmental Studies Hybridoma Bank) in blocking buffer overnight. After washing with PBS, the samples were incubated with secondary antibodies and DAPI for 45 min at room temperature. Fluorescent images were captured using a Leica DM 6000B fluorescence microscope. As the adenoviral vector contains the green fluorescence protein gene, cells transduced with adenoviral vectors show green fluorescence. We calculated the number of different stainings only in cells with green fluorescence.
Total RNA Extraction, cDNA Synthesis, and Real-time PCR
Total RNA was extracted from cells using TRIzol reagent according to the protocols of the manufacturer. RNA was treated with RNase-free DNase I to remove contaminating genomic DNA. The purity and concentration of total RNA were measured by Nanodrop 3000 (Thermo Fisher). Random primers and Moloney murine leukemia virus reverse transcriptase were used to convert RNA into cDNA. Real-time PCR was performed using a Roche Lightcycler 480 PCR system with SYBR Green Master Mix and gene-specific primers (18S, 5′-AACGGTCTAGACAACAAGCTG and 5′-AGTGGTCTTGGTGTGCTGAC; MyoD, 5′-GGCTACGACACCGCCTACTA and 5′-CGACTCTGGTGGTGCATCTG; Myog, 5′-TGCCCAGTGAATGCAACTCC and 5′-TTGGGCATGGTTTCGTCTGG; p53, 5′-ACTCAGACTGACTGCCTCTG and 5′-TCTCAGCCCTGAAGTCATAA; and Bhlhe40, 5′-CCCACATGTACCAAGTGTAC and 5′-CCTTCAGCTGGGCAATGCAC). The Ct value of 18S rRNA was used as an internal control, and the 2−ΔΔCT method was used to analyze the relative mRNA expression of various genes (31).
Protein Extraction and Western Blot Analysis
Cultured myoblasts were washed with PBS and homogenized with radioimmune precipitation assay buffer (50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS). Protein concentrations were determined using Pierce BCA protein assay reagent. Proteins were separated by 10% SDS-PAGE, electrotransferred onto a PVDF membrane (Millipore Corp., Billerica, MA) and incubated with specific primary antibodies. MyoD (catalog no. M318), Myog (catalog no. F5D), and GAPDH (catalog no. 6C5) antibodies were from Santa Cruz Biotechnology, Bhlhe40/Sharp2 (catalog no. ab23794) was from Abcam, FLAG (catalog no. 2368) was from Cell Signaling Technology, and p53 (catalog no. bs-0033R) was from Bioss. Immunodetection was performed using ECL Western blotting substrate (Pierce) and detected with a FluoChem R imaging system (ProteinSimple).
Knockdown of Bhlhe40 Using Lentivirus
To deplete endogenous Bhlhe40, the shRNA target 5′-GCGAGGTTACAGTGTTTATA corresponding to the 3′ UTR of Bhlhe40 was chosen according to the description in Ref. 32. The Plko.1-Bhlhe40 shRNA was purchased from Thermo Scientific (catalog no. TRCN0000081853). The protocol to generate lentivirus was modified on the basis of Ref. 33. Briefly, 293T cells (80–90% confluent) in collagen-coated 10-cm culture dishes were co-transfected with 4 μg of pLKO.1-Bhlhe40shRNA or pLKO.1 (as the control), 4 μg of pHR′-CMV-ΔR8.20vpr, and 2 μg of pHR′-CMV-VSV-G using Lipofectamine 2000 (Life Technologies) according to the protocol of the manufacturer. Viruses were harvested 4 times every 12 h from 24 h post-transfection and filtered through a 0.45-μm pore size filter. Then the viruses were concentrated at 22,000 rpm for 2 h in a Sorvall T 647.5 rotor. The viral pellets were suspended in 100 μl of TNE buffer (50 mm Tris-HCl (pH 7.8), 130 mm NaCl, and 1 mm EDTA) and incubated overnight at 4 °C. Along with transduction of Bhlhe40shRNA to myoblast, Polybrene (10 μg/ml) and HEPES (10 mm) were added to increase the transduction efficiency. Puromycin (1 μg/ml) was used to select the infected myoblasts 1 day post-infection.
Bhlhe40 KD Cell Line Selection
C2C12 cells were electroporated with pLKO.1-Bhlhe40shRNA or pLKO.1 (1 μg/106 cells). Two days after electroporation, puromycin (1 μg/ml) was added to the culture medium. Seven to ten days after puromycin selection, a single cell clone was detached by trypsin (0.25×) and transferred to 12-well plates. When the cell confluency was over 70%, cells were passaged, and some of them were used to do RNA extraction and Bhlhe40 expression detection.
Chromatin Immunoprecipitation
The commercial Bhlhe40 antibody was not suitable for chromatin immunoprecipitation, so we generated the Bhlhe40-FLAG plasmid by cloning the Bhlhe40 gene into the pCMV-FLAG-2b plasmid and electroporated Bhlhe40-FLAG into primary myoblasts. As a control, 1 day after electroporation, we added Bhlhe40 shRNA lentivirus to the electroporated primary myoblasts. The treated primary myoblasts were cross-linked with 1% formaldehyde in Ham's F-10 medium for 10 min at room temperature, followed by the addition of 125 mm glycine for 5 min at room temperature, after which cells were scraped into SDS lysis buffer. The cells were further sonicated and diluted for immunoprecipitation with the indicated antibodies. The immunoprecipitates were eluted and reverse-cross-linked overnight at 65 °C. DNA fragments were purified using the Cycle Pure kit (Omega), and the Myog promoter E1/E2 region (5′-GAATCACATGTAATCCACTGGA and 5′-ACGCCAACTGCTGGGTGCCA) was quantified by qPCR.
Statistical Analysis
The data are presented as mean ± S.E. p values were calculated using unpaired two-tailed Student's t test. p <0.05 was considered to be statistically significant.
Results
Microarray Analysis to Identify Hypoxia-induced Genes in Primary Myoblasts
To identify hypoxia-regulated genes in an unbiased manner, we performed a microarray analysis of primary myoblasts cultured under hypoxia (1% O2) or normoxia. This analysis identified 865 genes that were up- or down-regulated at least 2-fold (-fold change ≥ 2 and p ≤ 0.01) by hypoxia in both replicates (Fig. 1A). Of these, 641 genes (515 protein-coding genes and 126 lncRNAs or non-annotated genes) were up-regulated, and 224 genes (173 protein-coding genes and 51 long non-coding RNAs or non-annotated genes) were down-regulated (Fig. 1A). Gene ontology analysis shows that the top two categories of up-regulated genes are related to the cell cycle and metabolic process and that the top two categories of down-regulated genes are related to the protein catabolic process and muscle organ development (data not shown). Selective cohorts of genes that were up- or down-regulated by hypoxia are listed in the heat map in Fig. 1B. Particularly, many genes related to glycolysis and HIF1α signaling were up-regulated, whereas many genes related to oxidative phosphorylation and muscle organ development were down-regulated (Fig. 1B). These results are consistent with the important role of O2 in oxidative phosphorylation and the known function of hypoxia in activating HIF1α signaling and inhibiting myogenesis. Interestingly, we found that hypoxia also activated the p53 signaling pathway (Fig. 1B), presumably to protect myoblasts from oxidative stress-induced cell death. These results demonstrate the validity of our microarray analysis.
FIGURE 1.
Microarray analysis to identify hypoxia-regulated genes in primary myoblasts. A, genes affected by hypoxia (-fold change ≥ 2 and p ≤ 0.01) in two biological replicates were merged and filtered, including 641 up-regulated genes and 224 down-regulated genes. B, heat map showing selective pathways and genes regulated by hypoxia. Green indicates down-regulated genes and pathways, and red (orange) shows up-regulated genes and pathways. C, -fold increases of Bhlhe40 under hypoxia compared with normoxia. D, relative protein levels of Bhlhe40 under normoxia and hypoxia.
Overexpression of Bhlhe40 Inhibits the Differentiation of Myoblasts
We next focused on transcriptional factors that are activated by hypoxia to mediate its effect on myogenesis. Among the top 50 hypoxia up-regulated genes, we identified Bhlhe40, a member of the basic helix-loop-helix family transcriptional factors, whose expression was up-regulated 7.7-fold (Fig. 1C). Western blotting confirmed that the protein level of Bhlhe40 was also increased by hypoxia (Fig. 1D). Interestingly, another closely related member of this family, Bhlhe41, was also up-regulated 5.3-fold (data not shown). Because basic helix-loop-helix transcriptional factors such as Myf5, MyoD, and Myog are important regulators of myogenesis, we decided to focus the rest of our study on Bhlhe40, whose expression is induced more robustly than Bhlhe41 by hypoxia (Fig. 1C).
We and others have reported previously that hypoxia inhibits the expression of Myog and myogenic differentiation (10, 16). To investigate whether this effect is mediated by Bhlhe40, we first used adenovirus to overexpress Bhlhe40 in primary myoblasts. To examine whether Bhlhe40 affects myogenic differentiation, myoblasts transduced with GFP-expressing (as a control) or Bhlhe40-expressing adenovirus were induced to differentiate by serum withdrawal. Within 1 day after induced differentiation, the control myoblasts had already displayed an obviously elongated morphology, a hallmark of differentiation (Fig. 2A, top panels). In contrast, the Bhlhe40-overexpressing (BhlheOE) myoblasts were mostly still spherical (Fig. 2A, bottom panels), indicative of a differentiation defect. On day 3 after induced differentiation, more than 70% of the control myoblasts formed uniformly aligned multinucleated myotubes, whereas the BhlheOE myoblasts formed only few poorly aligned myotubes (Fig. 2B). We further quantified the differentiation index, which measures the fraction of myonuclei that are located in myosin heavy chain (MF20+)-expressing cells. Although more than 40% of nuclei in the control group were located in MF20+ cells, only 15% of nuclei in the BhlheOE group were MF20+ (Fig. 2C). At the molecular level, Bhlhe40 mRNA and its protein expression were both elevated in BhlheOE myoblasts compared with control myoblasts (Fig. 2, D and E). Importantly, BhlheOE led to a 30% reduction in Myog mRNA (Fig. 2D) and nearly abolished the expression of Myog at the protein level (Fig. 2E), consistent with the observed defects in myogenic differentiation. Interestingly, the mRNA and protein expression of MyoD was not affected by BhlheOE (Fig. 2, D and E). These results indicate that Bhlhe40 inhibits the Myog-driven terminal differentiation process without altering the expression of MyoD.
FIGURE 2.
Bhlhe40 overexpression down-regulates Myog and inhibits the differentiation of primary myoblasts. A and B, primary myoblasts were induced to differentiate (DF) for 1 day (A) and 3 days (B) after they were infected by adenovirus (control and Bhlhe40OE, both also expressed GFP) for 2 days. The differentiated myoblasts were stained with MF20 (red), and nuclei were counterstained with DAPI (blue). C, differentiation index of myoblasts 24 h after serum withdrawal. The differentiation index was calculated by dividing the number of nuclei in myotubes (MF20-positive elongated cells) by the total number of nuclei. Only adenovirus-infected cells (green cells) were used for quantification). n = 3 experiments using different batches of primary myoblasts, with five different areas analyzed in each experiment. D and E, relative expression of Bhlhe40, Myog, and MyoD in control and Bhlhe40OE myoblasts, as determined by qPCR (D) and Western blot analysis (E). Error bars represent mean ± S.D. *, p < 0.05.
Knockdown of Bhlhe40 Up-regulates Myog and Leads to Precocious Differentiation
We next performed a Bhlhe40 loss-of-function study using lentiviral shRNA-mediated knockdown. Primary myoblasts were treated with lentivirus for 2 days and selected in puromycin (1 μg/ml) for 2 additional days. The expression of Bhlhe40 in the shRNA knockdown (BhlheKD) group was only 40% of that in the GFP shRNA control group (Fig. 3A). Western blot analysis confirmed that the increased Myog expression was associated with a reduction in Bhlhe40 protein (Fig. 3B). We further selected two clones of single cell-derived C2C12 myoblasts (KD1 and KD2) after transfection with the pLKO.1-Bhlhe40 shRNA plasmid. The mRNA level of Bhlhe40 in the two clones of KD myoblasts was only 20–40% of that in control myoblasts (Fig. 3C). Consistent with the results obtained with a mixture of satellite cell-derived primary myoblasts, Myog expression was increased five to seven times in the Bhlhe40 KD cell clones (Fig. 3D). Myog protein levels were also increased robustly in the two clones of KD myoblasts compared with the control myoblasts (Fig. 3E). The abundance of Myog+ cells under proliferation conditions was 5–21 times higher in the two KD clones than in the control myoblasts (Fig. 3, F and G). In addition, MF20+ cells emerged in the two KD clones but were absent in the control myoblasts (Fig. 3F). We further induced the BhlheKD primary myoblasts to differentiate after treatment with lentivirus for 2 days and selection in puromycin (1 μg/ml) for 2 additional days. 24 h (day 1) after induced differentiation, BhlheKD myoblasts and control myoblasts showed a comparable morphology and differentiation index (Fig. 3, H, top panels, and I). 72 h (day 3) after induced differentiation, although 93% of nuclei in the control group were located in MF20+ cells, only 65% of nuclei in the BhlheKD group were in MF20+ cells (Fig. 3, H, top panels, and I). The fusion index was also lower in BhlheKD than in control myoblasts (Fig. 3J). Interestingly, the protein levels of Bhlhe40 increased gradually during normal differentiation (Fig. 3K). These data suggest that Bhlhe40 is dynamically regulated during myogenesis and that blockage of its expression leads to precocious myogenic differentiation, resulting in reduced proliferation and overall worsened myogenesis.
FIGURE 3.
Bhlhe40 knockdown up-regulates the expression of Myog and promotes myogenic differentiation. A and B, primary myoblasts infected by lentivirus (control and Bhlhe40KD) were collected for qPCR (A) and Western blot (B) analysis to determine the relative expression of Bhlhe40 and Myog. C—E, three clones of stably transduced C2C12 myoblasts infected with control or Bhlhe40KD lentivirus (KD1 and KD2) were analyzed. The relative expression of Bhlhe40 and Myog was determined by qPCR (C and D) and Western blots (E), respectively. F, immunostaining of Myog (red), myosin heavy chain (MF20, green) and DAPI (blue) in the three clones of proliferating cells. G, percentage of Myog-positive cells in the control and Bhlhe40KD clones. H, myoblasts infected by control or BhlheKD lentivirus were induced to differentiate (diff) for 1–3 days under normoxic conditions and stained with MF20 (red) and DAPI (cyan). I, differentiation index of myotubes formed by the treated myoblasts shown in H. J, fusion index of treated myoblasts shown in H, bottom panels. The fusion index measures the percentage of myonuclei present in multinucleated myofibers. D, day. K, protein levels of Bhlhe40 during differentiation. n = 3 different batches of myoblasts with two replicates using each batch of myoblasts. Error bars represent mean ± S.D. *, p < 0.05.
Bhlhe40 Regulates Myog Expression through Modulating MyoD Transcriptional Activity
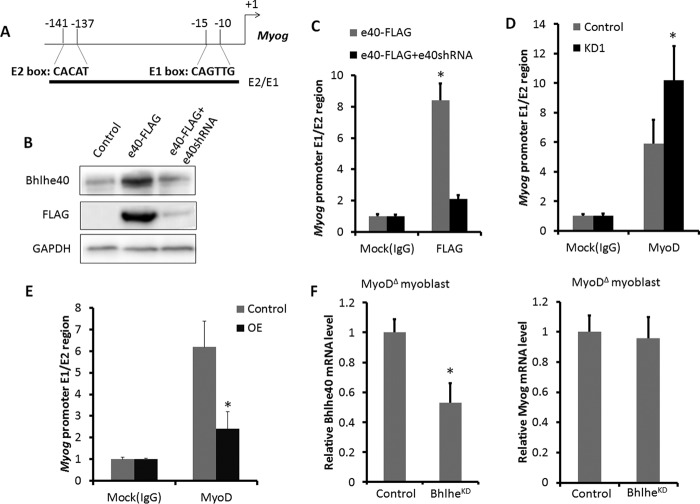
Bhlhe40 belongs to the E-protein family that binds to the E-box (CANNTG) DNA sequence located in the promotor region of target genes to regulate their expression. There are two E-boxes in the proximal promoter of Myog (Fig. 4A). We used ChIP to determine whether Bhlhe40 can bind directly to these E-box DNA sequences. Because of the lack of commercially available ChIP-grade antibodies, we generated the Bhlhe40-FLAG plasmid to overexpress FLAG-tagged Bhlhe40 in primary myoblasts (Fig. 4B). In addition, we used Bhlhe40 shRNA lentivirus, which efficiently blocked Bhlhe40-FLAG expression, as a negative control (Fig. 4B). As expected, the ChIP assay using FLAG antibody showed an 8-fold enrichment of E-box DNA sequences compared with the control group using IgG antibody, and this enrichment was abolished by shRNA knockdown of Bhlhe40 (Fig. 4C). These results demonstrate that Bhlhe40 binds directly to the Myog promoter but do not explain why Bhlhe40 inhibits Myog expression.
FIGURE 4.
Bhlhe40 regulates Myog expression through modulating the DNA binding and transcriptional activity of MyoD. A, schematic of the Myog promoter. E-boxes (E1 and E2) are depicted relative to the +1 transcriptional start site. The solid bar represents the region amplified by the indicated primer sets as described under “Experimental Procedures” and used in the PCR. B, relative levels of Bhlhe40 protein in myoblasts transfected with empty vector, Bhlhe40-FLAG plasmid, and Bhlhe40-FLAG plasmid plus Bhlhe40 shRNA lentivirus infection. C—E, -fold enrichment of the Myog E-box promoter region (shown in A) by Bhlhe40-FLAG (C) and MyoD (D and E), as determined by chromatin immunoprecipitation using antibodies against FLAG and MyoD, respectively. BhlheKD enhanced the binding activity of MyoD on the E-box region of the Myog promoter (D), and BhlheOE reduced the binding activity of MyoD on the E-box region of the Myog promoter (E). F, relative expression of Bhlhe40 and Myog in MyoDΔ myoblasts infected with control or Bhlhe40KD lentivirus. n = 3 different batches of primary myoblasts with two replicates using each batch of myoblasts. Error bars represent mean ± S.D. *, p < 0.05.
We hypothesized that Bhlhe40 disrupts the binding of MyoD to the E-boxes of Myog promoter. Indeed, ChIP assays using MyoD antibody showed that MyoD occupancy at the E-boxes of the proximal promoter of Myog was enhanced significantly by Bhlhe40 KD (Fig. 4D). In contrast, MyoD occupancy at the E-boxes of the proximal promoter of Myog was reduced significantly by BhlheOE (Fig. 4E). We further predicted that the occupancy at the Myog promoter by Bhlhe40 would reduce the transcriptional activity of MyoD, leading to inhibition of Myog expression. To investigate whether Bhlhe40 regulates Myog expression through MyoD, we treated MyoD knockout (MyoDΔ) myoblasts with control or Bhlhe40KD lentivirus. Strikingly, Bhlhe40 KD failed to up-regulate Myog expression in the absence of MyoD (MyoDΔ myoblasts) (Fig. 4F). These results support our hypothesis that Bhlhe40 inhibits Myog expression through reducing the binding of MyoD to the Myog promoter.
Knockdown of Bhlhe40 Alleviates Hypoxic Repression of Myoblast Differentiation
To examine whether the up-regulation of Bhlhe40 under hypoxia mediates the inhibitory effect of hypoxia on myogenic differentiation, we carried out rescue experiments. Primary myoblasts were infected with control or Bhlhe40KD lentivirus and induced to differentiate under hypoxia conditions. Bhlhe40KD robustly improved the differentiation of myoblasts, manifested by more obvious elongation of myotubes (Fig. 5A) and a higher abundance of myotubes expressing sarcomeric myosin heavy chain (MF20+, Fig. 5B). On average, the differentiation index was about three times higher in the Bhlhe40KD group than that in the control group (Fig. 5C). Myog protein expression was also up-regulated in Bhlhe40KD cells under hypoxia conditions (Fig. 5D). These data suggest that Bhlhe40 mediates the inhibitory effect of hypoxia on myogenic differentiation.
FIGURE 5.
Inhibition of Bhlhe40 rescues myogenic differentiation under hypoxia. A and B, myoblasts infected by control or BhlheKD lentivirus were induced to differentiate for 2 days under hypoxic conditions and stained with MF20 (red) and DAPI (cyan). C, differentiation index 2 days post-differentiation. C, control. D, relative levels of Myog and Bhlhe40 proteins in control and Bhlhe40 KD myoblasts grown under hypoxia for 48 h. n = 3 different batches of primary myoblasts, with five different areas analyzed using each batch of myoblasts. Error bars represent mean ± S.D. *, p < 0.05.
Hypoxia Induces Bhlhe40 through the p53 signaling pathway independent of HIF1α
We next sought to determine how hypoxia up-regulates Bhlhe40. Because hypoxia stabilizes HIF1α, we asked whether hypoxia activates Bhlhe40 through HIF1α. To examine this possibility, myoblasts extracted from WT mice and HIF1α conditional knockout (Myf5Cre/HIF1αf/f) mice were incubated under hypoxic conditions (1% O2) for 48 h. The expression of Bhlhe40 was subsequently determined. Hypoxia similarly up-regulated the expression of Bhlhe40 in WT and HIF1α KO myoblasts (8.2-fold versus 7.8-fold up-regulation, respectively; Fig. 6A). This observation indicates that the induction of Bhlhe40 by hypoxia is independent of HIF1α.
FIGURE 6.
Hypoxia induces Bhlhe40 through p53 and independent of HIF1α. A, qPCR analysis of Bhlhe40 expression in myoblasts from Myf5-HIF1αf/f mice or WT mice cultured under hypoxia (1% O2) or normoxia (21% O2) for 48 h. Different letters (a and b) indicate significant difference, p < 0.05. B and C, myoblasts incubated in hypoxia and normoxia chambers were collected for qPCR (B) and Western blot analysis (C) to determine the expression of p53. D, qPCR analysis of Bhlhe40 expression in myoblasts treated with 5 μm PFT (an inhibitor of p53) for 4 h. DMSO, dimethyl sulfoxide. E, Western blot analysis of Bhlhe40 and Myog protein expression in myoblasts treated with 5 μm PFT treated with hypoxia (H) or normoxia (N) for 48 h. C, control. F, immunostaining of MF20 (red) showing differentiation (for 2 days) of myoblasts under normoxia and hypoxia conditions with or without addition of the p53 inhibitor Pifithrin-α. G, differentiation index of cells treated as shown in F. Data were from three different batches of primary myoblasts with two replicates using each batch of myoblasts. Error bars represent mean ± S.D. *, p < 0.05.
Because our microarray analysis indicates that hypoxia activates the p53 signaling pathway in myoblasts, we asked whether p53 mediates the induction of Bhlhe40 by hypoxia. We first confirmed that p53 was up-regulated by hypoxia at both mRNA and protein levels (Fig. 6, B and C). To examine whether p53 is necessary for the induction of Bhlhe40, we used Pifithrin-α (PFT, 5 μm) to inhibit p53. The Bhlhe40 mRNA level was reduced by more than 60% by PFT treatment (4 h) under normoxic conditions (Fig. 6D). The protein level of Bhlhe40 was also decreased by PFT under both normoxia and hypoxia conditions, with a concomitant increase in Myog expression under hypoxia (Fig. 6E). Lastly, we examined whether inhibition of p53 could rescue the differentiation defects of myoblasts under hypoxia. Under normoxic conditions, PFT treatment had no effect on myoblast differentiation (Fig. 6, F, top panels, and G). Under hypoxic conditions, however, the abundance of MF20+ myotubes was increased strikingly in the presence of PFT (Fig. 6F, bottom panels). On average, PFT treatment improved the differentiation index about three times under hypoxia conditions (Fig. 6G). Taken together, these data demonstrate that hypoxia up-regulates Bhlhe40 through the p53 signaling pathway independent of HIF1α.
Discussion
In this study, we identified Bhlhe40 as a hypoxia-regulated factor that inhibits myogenic differentiation through modulating the DNA binding and transcriptional activity of MyoD. Previously, Bhlhe40 has been recognized as a factor regulated by hypoxia in tumor cells and adipocytes (25, 35, 36). Here we report that Bhlhe40 is induced by hypoxia in myoblasts. We found Bhlhe40 from the microarray analysis of myoblasts treated by hypoxia. Bhlhe40 has been identified as a transcriptional repressor in multiple biological processes (20–26). Because Bhlhe40 is induced by hypoxia, and hypoxia inhibits myogenic differentiation, we hypothesize that Bhlhe40 may mediate the inhibitory effect of hypoxia on myogenic differentiation. To test our hypothesis, we used gain- and loss-of-function approaches to demonstrate a direct role of Bhlhe40 in regulating the expression of Myog, a master factor controlling myogenic differentiation. Importantly, Bhlhe40 knockdown rescued the myogenic differentiation defects under hypoxic conditions.
Bhlhe40 has been reported to repress MRFs and the differentiation of human myoblasts (28). However, contradictory roles of Bhlhe40 in myogenesis have been reported in two other studies (10, 27). We found that Bhlhe40OE inhibited Myog expression and myogenic differentiation. In contrast, Bhlhe KD up-regulated Myog expression and promoted precocious differentiation at the expense of proliferation. The differentiation defects shown in BhlheKD myoblasts indicates that a proper level of Bhlhe40 is necessary for not only preventing the precocious differentiation of myoblasts but also for inhibiting apoptosis and inactivating the Notch signaling pathway (27).
Bhlhe40 has been shown to inhibit the transcriptional activity of MyoD through disrupting the interaction between P300/CBP-associated factor (P/CAF) and MyoD (34). Because MyoD is an important transcription activator of Myog, it is speculated that Bhlhe40 regulates Myog through modulating the transcriptional activity of MyoD. To examine this possibility, we verified, by ChIP assay, that Bhlhe40 binds directly to the Myog promoter. This result is consistent with a previous report (34). However, we found that this binding does not alter Myog expression in the absence of MyoD, assuring that Bhlhe40 acts through MyoD to regulate Myog expression. Indeed, MyoD binding to the Myog promoter was enhanced when Bhlhe40 was knocked down and disrupted when Bhlhe40 was overexpressed, suggesting that Bhlhe40 interferes with MyoD on Myog promoter binding. Previous work has indicated that formation of the MyoD-P/CAF complex stabilizes the binding of MyoD to the promoter of Myog (37) and that Bhlhe40 disrupts the formation of the MyoD-P/CAF complex (34). Our results suggest that Bhlhe40 destabilizes the binding of MyoD to the promoter of Myog through disrupting the formation of the MyoD-P/CAF complex. Because Bhlhe40 and MyoD both belong to E-proteins that are known to form heterodimers (34), the Bhlhe40/MyoD heterodimer would prevent the formation of the transcriptionally active MyoD-P/CAF complex. These results together demonstrate that Bhlhe40 regulates Myog expression through modulating the transcriptional activity of MyoD.
It has been reported that ectopic expression of Myog only partly rescues the myogenic differentiation under hypoxic conditions (10). Because Bhlhe40 is up-regulated by hypoxia and negatively regulates Myog expression, we examined whether knockdown of Bhlhe40 rescues the myogenic differentiation defects under hypoxia conditions. We found that Bhlhe40 knockdown only partly rescues myogenic differentiation. This may be due to the relatively low efficiency of knockdown, which is about 40% in our study. Another possibility is that multiple factors are involved in mediating the effect of hypoxia. Hypoxia triggers global changes in gene expression, as indicated by our microarray results. The Notch signaling pathways, PI3K pathways, and stability of MyoD mRNA have all been reported to regulate myogenesis under hypoxia conditions (16–19). Although both MyoD and Myog are key factors in myogenic differentiation, their overexpression could only partly rescue myogenesis under hypoxia (10), again suggesting that multiple factors are involved in mediating the effect of hypoxia. Nevertheless, our study provides strong evidence that Bhlhe40 is the key factor mediating the effect of hypoxia on myogenesis.
Defining the intrinsic signaling pathways regulating the expression of Bhlhe40 in myoblasts is key to understanding how hypoxia regulates myogenesis. It has been widely accepted that hypoxia regulates gene expression through stabilizing the transcriptional factor HIF1α. HIF1α has been reported as an inducer of Bhlhe40 in adipocytes (25, 35, 36). Interestingly, we found that hypoxia induces Bhlhe40 in the absence of HIF1α, therefore excluding HIF1α as a regulator of Bhlhe40. Because our microarray data indicate that hypoxia activates the p53 signaling pathway, we sought to determine whether p53 is involved in regulating Bhlhe40. p53 has been shown to target Bhlhe40 in the breast cancer cell line MCF7 and the colon carcinoma cell line RKO (38). We found that pharmacological inhibition of p53 blocks the induction of Bhlhe40 by hypoxia, demonstrating that p53 is an inducer of Bhlhe40 in hypoxia-treated myoblasts. Importantly, inhibition of p53 partially rescued the differentiation defects of hypoxia-treated myoblasts, indicating that hypoxia regulates Bhlhe40 at least partially through p53. In a recent study, p53 has been reported to inhibit the transcription of Myog directly by binding to the promoter of Myog under genotoxic stress (39). Our study sheds light on an alternative avenue through which p53 regulates Myog. Our results further raise the possibility of promoting muscle regeneration under ischemic hypoxia conditions through temporal blockage of p53.
Author Contributions
C. W. and S. K. conceived the study and wrote the paper. C. W., W. L., and Z. L. performed the experiments. L. C. and X. L. provided technical assistance. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Jun Wu for laboratory management and maintaining mouse colonies and other members of the Kuang laboratory for technical assistance and discussions.
This work was partially sponsored by the Muscular Dystrophy Association and the National Institutes of Health Grant R01 AR060652 (to S. K.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- MRF
- myogenic regulatory factor
- qPCR
- quantitative PCR
- KD
- knockdown
- PFT
- Pifithrin-α.
References
- 1.Mauro A. (1961) Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 9, 493–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuang S., and Rudnicki M. A. (2008) The emerging biology of satellite cells and their therapeutic potential. Trends Mol. Med. 14, 82–91 [DOI] [PubMed] [Google Scholar]
- 3.Edmondson D. G., and Olson E. N. (1989) A gene with homology to the myc similarity region of myod1 is expressed during myogenesis and is sufficient to activate the muscle differentiation program. Genes Dev. 3, 628–640 [DOI] [PubMed] [Google Scholar]
- 4.Davis R. L., Weintraub H., and Lassar A. B. (1987) Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987–1000 [DOI] [PubMed] [Google Scholar]
- 5.Braun T., Buschhausen-Denker G., Bober E., Tannich E., and Arnold H. H. (1989) A novel human-muscle factor related to but distinct from myod1 induces myogenic conversion in 10t1/2 fibroblasts. EMBO J. 8, 701–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun T., Bober E., Winter B., Rosenthal N., and Arnold H. H. (1990) Myf-6, a new member of the human gene family of myogenic determination factors: evidence for a gene-cluster on chromosome-12. EMBO J. 9, 821–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Megeney L. A., Kablar B., Garrett K., Anderson J. E., and Rudnicki M. A. (1996) MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 10, 1173–1183 [DOI] [PubMed] [Google Scholar]
- 8.Parker M. H., Seale P., and Rudnicki M. A. (2003) Looking back to the embryo: defining transcriptional networks in adult myogenesis. Nat. Rev. Genet. 4, 497–507 [DOI] [PubMed] [Google Scholar]
- 9.Hasty P., Bradley A., Morris J. H., Edmondson D. G., Venuti J. M., Olson E. N., and Klein W. H. (1993) Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 364, 501–506 [DOI] [PubMed] [Google Scholar]
- 10.Yun Z., Lin Q., and Giaccia A. J. (2005) Adaptive myogenesis under hypoxia. Mol. Cell Biol. 25, 3040–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong J. F., Yan X., Zhu M. J., Ford S. P., Nathanielsz P. W., and Du M. (2009) Maternal obesity downregulates myogenesis and β-catenin signaling in fetal skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 296, E917-E924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma J. F., Hall D. T., and Gallouzi I.-E. (2012) The impact of mRNA turnover and translation on age-related muscle loss. Ageing Res. Rev. 11, 432–441 [DOI] [PubMed] [Google Scholar]
- 13.Hoppeler H., and Vogt M. (2001) Muscle tissue adaptations to hypoxia. J. Exp. Biol. 204, 3133–3139 [DOI] [PubMed] [Google Scholar]
- 14.Sair M., Etherington P. J., Peter Winlove C., and Evans T. W. (2001) Tissue oxygenation and perfusion in patients with systemic sepsis. Crit. Care Med. 29, 1343–1349 [DOI] [PubMed] [Google Scholar]
- 15.Greenbaum A. R., Etherington P. J., Manek S., O'Hare D., Parker K. H., Green C. J., Pepper J. R., and Winlove C. P. (1997) Measurements of oxygenation and perfusion in skeletal muscle using multiple microelectrodes. J. Muscle Res. Cell Motil. 18, 149–159 [DOI] [PubMed] [Google Scholar]
- 16.Liu W., Wen Y., Bi P., Lai X., Liu X. S., Liu X., and Kuang S. (2012) Hypoxia promotes satellite cell self-renewal and enhances the efficiency of myoblast transplantation. Development 139, 2857–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Carlo A., De Mori R., Martelli F., Pompilio G., Capogrossi M. C., and Germani A. (2004) Hypoxia inhibits myogenic differentiation through accelerated myod degradation. J. Biol. Chem. 279, 16332–16338 [DOI] [PubMed] [Google Scholar]
- 18.Majmundar A. J., Skuli N., Mesquita R. C., Kim M. N., Yodh A. G., Nguyen-McCarty M., and Simon M. C. (2012) O-2 regulates skeletal muscle progenitor differentiation through phosphatidylinositol 3-kinase/AKT signaling. Mol. Cell Biol. 32, 36–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gustafsson M. V., Zheng X., Pereira T., Gradin K., Jin S., Lundkvist J., Ruas J. L., Poellinger L., Lendahl U., and Bondesson M. (2005) Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev. Cell 9, 617–628 [DOI] [PubMed] [Google Scholar]
- 20.Liu Y., Wang L., Lin X.-Y., Wang J., Yu J.-H., Miao Y., and Wang E.-H. (2013) The transcription factor DEC1 (BHLHE40/STRA13/SHARP-2) is negatively associated with tnm stage in non-small-cell lung cancer and inhibits the proliferation through cyclin D1 in A549 and BE1 cells. Tumor Biol. 34, 1641–1650 [DOI] [PubMed] [Google Scholar]
- 21.Honma S., Kawamoto T., Takagi Y., Fujimoto K., Sato F., Noshiro M., Kato Y., and Honma K. (2002) Dec1 and dec2 are regulators of the mammalian molecular clock. Nature 419, 841–844 [DOI] [PubMed] [Google Scholar]
- 22.Li Y., Zhang H., Xie M., Hu M., Ge S., Yang D., Wan Y., and Yan B. (2002) Abundant expression of DEC1/STRA13/SHARP2 in colon carcinoma: its antagonizing role in serum deprivation-induced apoptosis and selective inhibition of procaspase activation. Biochem. J. 367, 413–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zawel L., Yu J., Torrance C. J., Markowitz S., Kinzler K. W., Vogelstein B., and Zhou S. (2002) Dec1 is a downstream target of TGF-β with sequence-specific transcriptional repressor activities. Proc. Natl. Acad. Sci. U.S.A. 99, 2848–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ehata S., Hanyu A., Hayashi M., Aburatani H., Kato Y., Fujime M., Saitoh M., Miyazawa K., Imamura T., and Miyazono K. (2007) Transforming growth factor-β promotes survival of mammary carcinoma cells through induction of antiapoptotic transcription factor dec1. Cancer Res. 67, 9694–9703 [DOI] [PubMed] [Google Scholar]
- 25.Yun Z., Maecker H. L., Johnson R. S., and Giaccia A. J. (2002) Inhibition of PPAR γ 2 gene expression by the HIF-1-regulated gene dec1/stra13: a mechanism for regulation of adipogenesis by hypoxia. Dev. Cell 2, 331–341 [DOI] [PubMed] [Google Scholar]
- 26.Yamada K., and Miyamoto K. (2005) Basic helix-loop-helix transcription factors, BHLHB2 and BHLHB3: their gene expressions are regulated by multiple extracellular stimuli. Front. Biosci. 10, 3151–3171 [DOI] [PubMed] [Google Scholar]
- 27.Sun H., Li L., Vercherat C., Gulbagci N. T., Acharjee S., Li J., Chung T.-K., Thin T. H., and Taneja R. (2007) Stra13 regulates satellite cell activation by antagonizing notch signaling. J. Cell Biol. 177, 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lecomte V., Meugnier E., Euthine V., Durand C., Freyssenet D., Nemoz G., Rome S., Vidal H., and Lefai E. (2010) A new role for sterol regulatory element binding protein 1 transcription factors in the regulation of muscle mass and muscle cell differentiation. Mol. Cell Biol. 30, 1182–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 30.Luo J., Deng Z.-L., Luo X., Tang N., Song W.-X., Chen J., Sharff K. A., Luu H. H., Haydon R. C., Kinzler K. W., Vogelstein B., and He T.-C. (2007) A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat. Protoc. 2, 1236–1247 [DOI] [PubMed] [Google Scholar]
- 31.Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 (t) (−ΔΔ c) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 32.Hu G., and Chen J. (2013) A genome-wide regulatory network identifies key transcription factors for memory CD8(+) T-cell development. Nat. Comms. 4, 2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X., Lei M., and Erikson R. L. (2006) Normal cells, but not cancer cells, survive severe PLK1 depletion. Mol. Cell Biol. 26, 2093–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsiao S. P., Huang K. M., Chang H. Y., and Chen S. L. (2009) P/CAF rescues the BHLHE40-mediated repression of myod transactivation. Biochem. J. 422, 343–352 [DOI] [PubMed] [Google Scholar]
- 35.Chakrabarti J., Turley H., Campo L., Han C., Harris A. L., Gatter K. C., and Fox S. B. (2004) The transcription factor DEC1 (STRA13, SHARP2) is associated with the hypoxic response and high tumour grade in human breast cancers. Br. J. Cancer 91, 954–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giatromanolaki A., Koukourakis M. I., Sivridis E., Turley H., Wykoff C. C., Gatter K. C., and Harris A. L. (2003) DEC1 (STRA13) protein expression relates to hypoxia-inducible factor 1-α and carbonic anhydrase-9 overexpression in non-small cell lung cancer. J. Pathol. 200, 222–228 [DOI] [PubMed] [Google Scholar]
- 37.Mal A., and Harter M. L. (2003) Myod is functionally linked to the silencing of a muscle-specific regulatory gene prior to skeletal myogenesis. Proc. Natl. Acad. Sci. U. S. A. 100, 1735–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qian Y., Zhang J., Yan B., and Chen X. (2008) Dec1, a basic helix-loop-helix transcription factor and a novel target gene of the p53 family, mediates p53-dependent premature senescence. J. Biol. Chem. 283, 2896–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Z. J., Broz D. K., Noderer W. L., Ferreira J. P., Overton K. W., Spencer S. L., Meyer T., Tapscott S. J., Attardi L. D., and Wang C. L. (2015) P53 suppresses muscle differentiation at the myogenin step in response to genotoxic stress. Cell Death Differ. 22, 560–573 [DOI] [PMC free article] [PubMed] [Google Scholar]