Abstract
Intralumenal vesicle formation of the multivesicular body is a critical step in the delivery of endocytic cargoes to the lysosome for degradation. Endosomal sorting complex required for transport III (ESCRT-III) subunits polymerize on endosomal membranes to facilitate membrane budding away from the cytoplasm to generate these intralumenal vesicles. The ATPase Vps4 remodels and disassembles ESCRT-III, but the manner in which Vps4 activity is coordinated with ESCRT-III function remains unclear. Ist1 is structurally homologous to ESCRT-III subunits and has been reported to inhibit Vps4 function despite the presence of a microtubule-interacting and trafficking domain-interacting motif (MIM) capable of stimulating Vps4 in the context of other ESCRT-III subunits. Here we report that Ist1 inhibition of Vps4 ATPase activity involves two elements in Ist1: the MIM itself and a surface containing a conserved ELYC sequence. In contrast, the MIM interaction, in concert with a more open conformation of the Ist1 core, resulted in stimulation of Vps4. Addition of the ESCRT-III subunit binding partner of Ist1, Did2, also converted Ist1 from an inhibitor to a stimulator of Vps4 ATPase activity. Finally, distinct regulation of Vps4 by Ist1 corresponded with altered ESCRT-III disassembly in vitro. Together, these data support a model in which Ist1-Did2 interactions during ESCRT-III polymerization coordinate Vps4 activity with the timing of ESCRT-III disassembly.
Keywords: ATPase, endosomal sorting complexes required for transport (ESCRT), protein conformation, protein sorting, protein stability, Did2, Ist1, Vps4, endosomal sorting
Introduction
The endosomal sorting complexes required for transport (ESCRTs)2 mediate the sorting of ubiquitinated cargoes into multivesicular bodies (MVBs) (Refs. 1, 2 and reviewed in Ref. 3), the budding of enveloped viruses (4–7), and abscission of the cellular bridge during cytokinesis (4, 5, 8–11). Common to these cellular activities is the requirement for membrane remodeling, which is mediated by ESCRT-III (reviewed in Ref. 12). During this process, monomeric ESCRT-III subunits in the cytoplasm undergo dynamic polymerization into a spiral-like filament on membranes to induce membrane budding and/or scission (13–19). Physiological ESCRT-III polymerization is a highly ordered process involving the sequential recruitment of the seven ESCRT-III subunits: the core subunits Vps20/CHMP6, Snf7/CHMP4, Vps24/CHMP3, and Vps2/CHMP2 as well as the accessory subunits Did2/Fti1/CHMP1, Ist1/hIst1, and Vps60/CHMP5 (17, 20–27). Subsequently, the AAA-ATPase Vps4 interacts with the ESCRT-III polymer to remodel and ultimately disassemble the ESCRT-III polymer using energy derived from ATP hydrolysis, thereby recycling individual ESCRT-III subunits to their cytoplasmic pool (17, 24, 28–31). In this manner, Vps4-mediated disassembly of ESCRT-III is the only ATP-dependent step in the ESCRT pathway and is essential for all ESCRT-dependent cellular processes.
Although Vps4 regulates ESCRT-III function, ESCRT-III subunits themselves also regulate Vps4 activity, suggesting direct coordination between these activities. The Vps4 microtubule-interacting and trafficking (MIT) domain binds to MIT-interacting motifs (MIMs) present in the C termini of some ESCRT-III subunits, including Vps2, Did2, and Ist1 (Refs. 9, 32–36 and reviewed in Ref. 37). This interaction serves to recruit Vps4 to the site of ESCRT action as well as to stimulate Vps4 ATPase activity through an unknown mechanism (20, 24, 38, 39). Vps4 stimulation also occurs through additional interactions between acidic residues in ESCRT-III subunits and the Vps4 linker region (38, 39) and the Vps4 pore (38, 40), which is formed upon Vps4 oligomerization into the functional AAA-ATPase ring structure (41–44). The presumption is that these ESCRT-III MIM-dependent and MIM-independent Vps4 regulatory activities are revealed as ESCRT-III subunits polymerize on membranes and undergo conformational changes from closed to open states (45–49). Stimulation of Vps4 ATPase activity, either directly by ESCRT-III or via the co-factor Vta1 (20, 50–53), subsequently promotes disassembly of ESCRT-III (17, 30, 31). Disassembly occurs as ESCRT-III subunits are fed through the Vps4 pore (38, 40, 43, 54), similar to other AAA-ATPases such as ClpX (Refs. 55–57 and reviewed in Refs. 58, 59). How Vps4 activity is held in check to permit ESCRT-III assembly and function prior to disassembly by Vps4 is unresolved.
The ESCRT-III subunit Ist1 may play a special role in coordinating Vps4 and ESCRT-III functions by exerting both positive and negative regulation of Vps4 activity. Similar to Did2 and Vps2, Ist1 has been implicated as a positive regulator of Vps4. Ist1 is essential for cytokinesis in mammalian cells (8, 9), synthetic genetic defects in MVB sorting are observed in ist1Δ vta1Δ and ist1Δ vps60Δ yeast strains (23, 26), and a C-terminal fragment of Ist1 containing its MIM element stimulates Vps4 ATPase activity in vitro (23). By contrast, overexpression of Ist1 disrupts MVB sorting in yeast, and full-length Ist1 inhibits Vps4 ATPase activity in vitro (23), indicating that Ist1 can also negatively regulate ESCRT function. These dual activities, inhibition and stimulation of Vps4, make Ist1 unique among the ESCRT-III subunits. However, the mechanism that mediates switching between Ist1 positive and negative regulation of Vps4 are unclear.
To examine the relationships between Ist1 conformation and Vps4 regulation, a structure-function study of Ist1 was conducted. Here we report that alterations in the conformation of the Ist1 core domain altered regulation of Vps4 function. Both negative and positive regulation of Vps4 by Ist1 required MIM-MIT interactions, whereas a highly conserved ELYC region located in the Ist1 core region was required for negative regulation. These structure-function studies suggested that Ist1 MIM-Vps4 MIT domain interactions represent the primary mode of interaction between Vps4 and Ist1, whereas secondary interactions dependent upon changes in ESCRT-III core conformation modulate Vps4 function. Conversion of Ist1 from an inhibitor to a stimulator of Vps4 ATPase activity in vitro has also been observed upon addition of Did2, the ESCRT-III subunit to which Ist1 binds specifically (20, 23, 26, 45, 60). We propose that Ist1 binding to Did2 during ESCRT-III polymerization induces conformational changes in Ist1 that alter regulation of Vps4 to coordinate Vps4 and ESCRT-III functions.
Experimental Procedures
Plasmids and Strains
Yeast IST1 was amplified from Saccharomyces cerevisiae genomic DNA with the 5′ oligomeric primer designed to remove the intron of Ist1 and cloned into the BamHI and XhoI sites of pET28b (Novagen), generating pET28-Ist1. Mutagenesis of Ist1 was performed using the Gene Tailor site-directed mutagenesis system (Invitrogen) with a pBS-Ist1 template. The pET28a Ist1(L168A,Y172A) construct was supplied by Dr. Zhaohui Xu (University of Michigan) (60). All cloned PCR products and mutant plasmids were sequenced to exclude unexpected mutations. The Ist1 promoter was amplified from yeast genomic DNA and subcloned into the NotI and BamHI sites of pRS415 (61), yielding the pRS415 Promoter(Ist1). The Ist1 coding sequences for the WT and mutants were subcloned from pET28b bacterial expression vectors into pRS415 Promoter(Ist1) via the BamHI and SalI sites. Alternatively, the BamH1 and Xho1 sites were used for pET28a Ist1(L168A,Y172A). The yeast strains used in this study included SEY6210 (62); TVY1, MATα ura3–52 leu2–3,112 his3-Δ200 trp 1-Δ901 lys2–801 suc2-Δ9 pep4Δ::LEU2 (63); JPY193, pep4Δ::LEU2 ist1Δ::HIS3 (this study); JPY275, pep4Δ::LEU2 vta1Δ::HIS3 (this study); JPY194, pep4Δ::LEU2 ist1Δ::HIS3 vta1Δ::HIS3 (this study); and JPY283, pep4Δ::LEU2 vps4Δ::TRP1 (this study). GST-Vps4 (pMB54, Ref. 20) and GST-Vps4Δ31–87 (pMB40, Ref. 30) were supplied by Dr. Marcus Babst (University of Utah).
Ist1 Antibody Generation
Purified full-length Ist1 (pET28-Ist1) lacking the His6 tag was used for antiserum production (Covance). A New Zealand rabbit was immunized with Ist1, and test bleeds were obtained. Bleeds were tested for detection of Ist1 in WT (SEY6210) and ist1Δ yeast strains and recombinantly expressed and purified Ist1. This polyclonal antibody detected purified WT Ist1 and Ist1 MIM mutants equivalently (data not shown).
Protein Expression and Purification
Protein expression for GST, GST-Vps4, Did2-His6, and His6-Ist1 was performed in the BL21-DE3 bacterial strain at 16 °C for 16 h with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. His6 fusion proteins were purified by Ni2+-affinity chromatography (5 ml HiTrap chelating FF column; GE Healthcare Life Sciences), treated with thrombin to remove the His6 tag from Ist1, incubated with ATP to dissociate chaperones, and subjected to size exclusion chromatography (SEC) (Superdex200 16/60 HiLoad for Ist1 or Superdex75 16/60 HiLoad for Did2, GE Healthcare Life Sciences) in 25 mm HEPES, 200 mm KCl (pH 7.5). Purification of Ist1 and Did2 included treatment with ATP to minimize contaminating ATPase activities, and the final purified Ist1 and Did2 proteins were analyzed to confirm the absence of significant contaminating ATPase activity (Fig. 2C and data not shown). Purified Ist1 was run on SDS-PAGE with Benchmark Ladder (GE Healthcare Life Sciences) to confirm sample purity (Figs. 2C, 4D, 5A, 7A, and 8B). GST-Vps4 fusion protein was purified as described previously (30).
FIGURE 2.
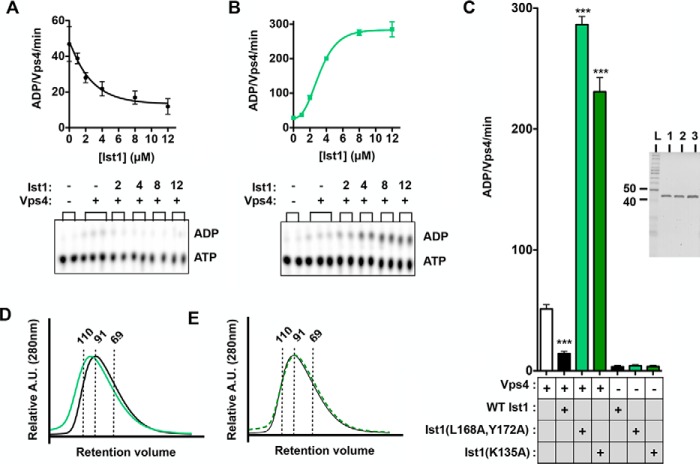
Ist1(L168A,Y172A) and Ist1(K135A) hyperstimulate Vps4 ATPase activity. A and B, ATPase activities for titrations of 500 nm-12 μm WT Ist1 (A) or Ist1(L168A,Y172A) (B) in the presence of 500 nm Vps4. Vps4-specific activity is expressed as ADP generated per Vps4 molecule per minute. Raw data of TLC plates for a single time point (16′) are shown, which were used to calculate Vps4-specific activities. C, ATPase activities of Ist1(L168A,Y172A) and Ist1(K135A) alone or in the presence of 500 nm Vps4. Results are presented as mean + S.D. of triplicate experiments, with statistical differences from 500 nm Vps4 alone indicated (***, p < 0.001). The purity of recombinant Ist1 proteins as assessed by SDS-PAGE analysis and Coomassie staining is shown in the right panel (L, ladder; lane 1, WT Ist1; lane 2, Ist1(L168A,Y172A); lane 3, Ist1(K135A)). D and E, UV traces for SEC analyses of Ist1. D, WT Ist1 (black) and Ist1(L168A,Y172A) (green). E, WT Ist1 (black) versus Ist1(K135A) (dashed green). A.U., absorption units. Apparent molecular weights are indicated by dashed lines.
FIGURE 4.
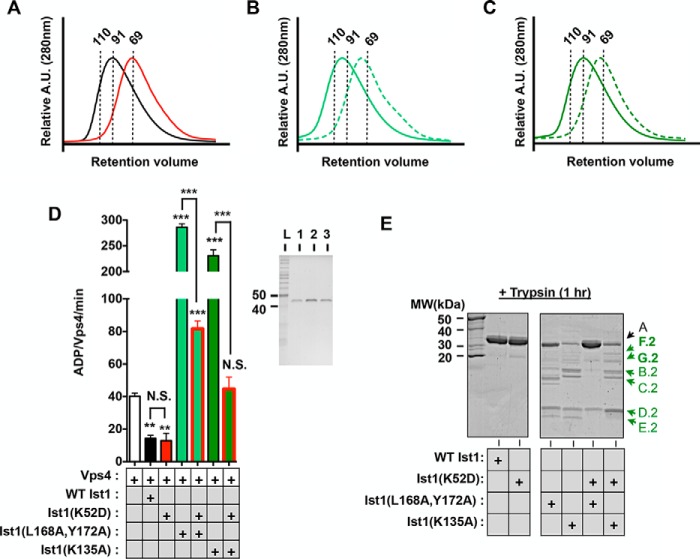
K52D stabilizes a more closed Ist1 conformation to prevent Vps4 hyperstimulation. A–C, UV traces for SEC analyses of Ist1: A, WT Ist1 (black) and Ist1(K52D) (red); B, Ist1(L168A,Y172A) (solid) and Ist1(L168A,Y172A,K52D) (dashed); C, Ist1(K135A) (solid) and Ist1(K135A,K52D) (dashed). A.U., absorption units. D, ATPase activities of 500 nm Vps4 in the presence of 8 μm Ist1 K52D mutants with or without L168A,Y172A and K135A. Results are presented as mean ± S.D. of triplicate experiments, with statistical differences from 500 nm Vps4 alone indicated (***, p < 0.001; **, p < 0.005). The purity of recombinant Ist1 proteins as assessed by SDS-PAGE analysis and Coomassie staining is shown in the right panel. Lane 1, Ist1(K52D); lane 2, Ist1(L168A,Y172A, K52D); lane 3, Ist1(K135A, K52D). L, ladder; N.S., not significant. E, limited proteolysis of Ist1 with trypsin. Aliquots were removed at the 1-h time point for SDS-PAGE analysis and Coomassie staining. Additional peptide fragments in F.2 and G.2 are highlighted in the Ist1(K135A,K52D) sample. MW, molecular weight.
FIGURE 5.
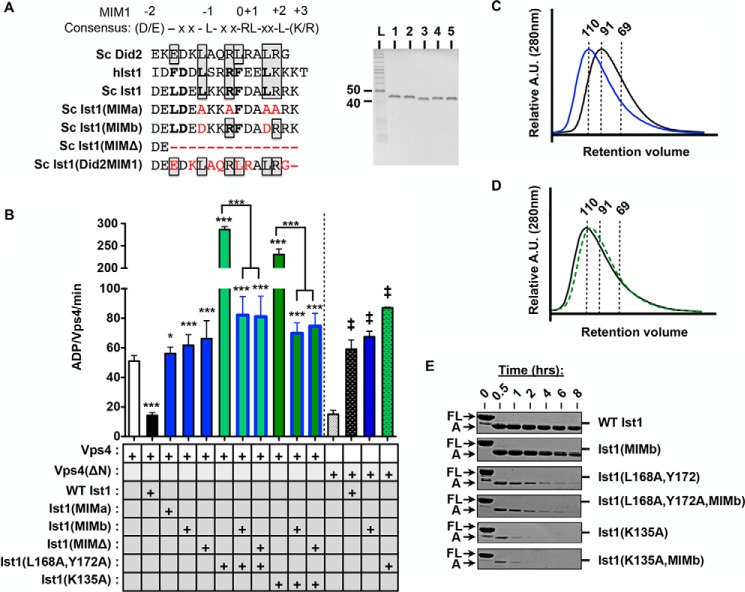
Disrupting the Ist1 MIM-Vps4 MIT interaction results in loss of Vps4 inhibition and hyperstimulation. A, sequence alignment of α6 of yeast (Sc) Ist1, human (h) Ist1, and Sc Did2. MIM1 residues are boxed, and MIM3 residues are bold. Some residues contribute to both MIM1 and MIM3 modes of interaction (33). Residues mutated in the Ist1 MIM mutants used in this study are highlighted in red. The purity of recombinant Ist1 proteins as assessed by SDS-PAGE analysis and Coomassie staining is shown in the right panel. Lane 1, Ist1(MIMa); lane 2, Ist1(MIMb); lane 3, Ist1(MIMΔ); lane 4, Ist1(L168A,Y172A,MIMb); lane 5, Ist1(K135A,MIMb). L, ladder. B, ATPase activity of 500 nm Vps4 or Vps4(ΔN) in the presence of 8 μm Ist1 MIM mutants. Results are presented as mean ± S.D. of triplicate experiments, with statistical differences from 500 nm Vps4 alone indicated (*, p < 0.05; ***, p < 0.001) of Vps4(ΔN) alone (‡, <0.001). C and D, UV traces for SEC analyses of Ist1: C, WT Ist1 (black) and Ist1(MIMb) (blue); D, Ist1(L168A,Y172A,MIMb (black) and Isg1(K135A,MIMb) (green, dashed). A.U., absorption units. E, limited proteolysis of Ist1 with trypsin. Aliquots were removed at various time points for SDS-PAGE analysis and Coomassie staining. Full-length (FL) Ist1 and the A fragments are indicated.
FIGURE 7.
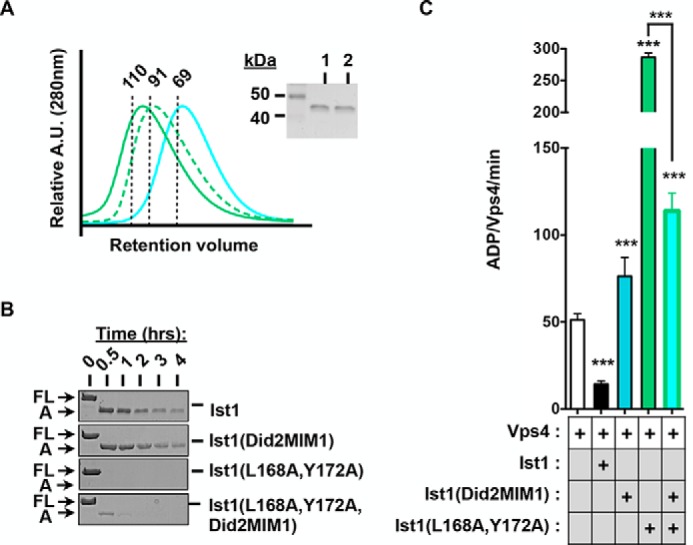
The Did2-MIM1 element cannot functionally replace the Ist1 MIM element. A, UV traces for SEC analyses of Ist1(Did2-MIM1) (cyan), Ist1(L168A,Y172A) (green), and Ist1(L168A,Y172A, Did2-MIM1) (green, dashed). The purity of recombinant Ist1 proteins as assessed by SDS-PAGE analysis and Coomassie staining is shown in the right panel. Lane 1, Ist1(Did2-MIM1); lane 2, Ist1(L168A,Y172A,Did2-MIM1). A.U., absorption units. B, limited proteolysis of Ist1 with trypsin. Aliquots were removed at various time points for SDS-PAGE analysis and Coomassie staining. Full-length (FL) Ist1 and the A fragments are indicated. C, ATPase activities of 500 nm Vps4 in the presence of 8 μm Ist1 Did2-MIM1 chimeras with or without L168A,Y172A. Results are presented as mean + S.D. of triplicate experiments, with statistical differences from 500 nm Vps4 alone indicated (***, p < 0.001).
FIGURE 8.
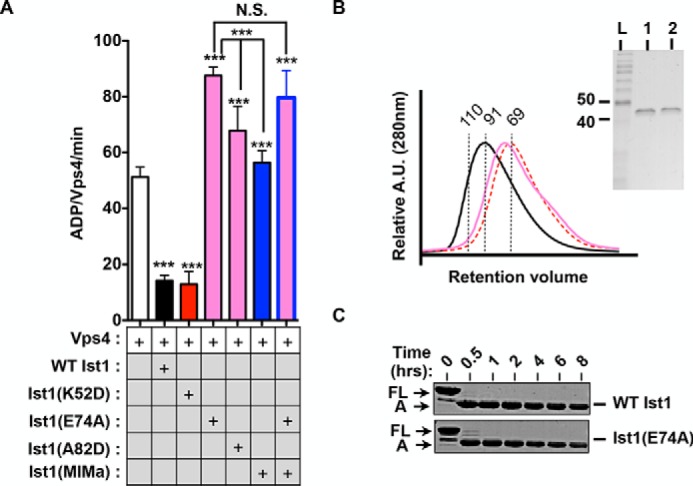
Mutations in the Ist1 ELYC region result in loss of Vps4 inhibition. A, ATPase activities of 500 nm Vps4 in the presence of 8 μm Ist1 ELYC regions with or without MIM mutants. Results are presented as mean + S.D. of triplicate experiments, with statistical differences from 500 nm Vps4 alone indicated (***, p < 0.001; N.S., not significant). B, UV traces for SEC analyses of WT Ist1 (black), Ist1(E74A) (pink), and Ist1(K52D) (red, dashed). A.U., absorption units. The purity of recombinant Ist1 proteins as assessed by SDS-PAGE analysis and Coomassie staining is shown in the right panel. Lane 1, Ist1(E74A); lane 2, Ist1(A82D). L, ladder. C, limited proteolysis of Ist1 with trypsin. Aliquots were removed at various time points for SDS-PAGE analysis and Coomassie staining. Full-length (FL) Ist1 and the A fragments are indicated.
ATPase Assay
Measurement of Vps4 ATPase activity was performed as described previously (20, 30, 31, 53), but reactions were initiated by addition of 6 mm ATP (compared with 1 or 2 mm ATP) to remain within a linear range of ATP hydrolysis for Vps4 hyperstimulated by Ist1 mutants. ATPase activities from three independent experiments are shown as mean ± S.D. The significance of difference in rates was assessed by t tests using Prism5 (GraphPad). An example of this analysis, including images of TLC plates and determination of rates, is presented in Ref. 53. 500 nm Vps4 was used in all ATPase assays because this concentration of Vps4 exhibits submaximal specific activity (41–50 ADP molecules/Vps4/min), making it amenable for observing stimulation or inhibition of Vps4 ATPase activity. Maximal inhibition or stimulation of 500 nm Vps4 by WT Ist1 or Ist1 mutants was achieved at [Ist1] ≤ 8 μm, as ascertained from titrations from 0.5–12 μm Ist1 in the presence of Vps4 (Fig. 2, A and B, and data not shown). Vps4 activity with 8 μm Ist1 mutants is therefore presented for comparison (Figs. 2C, 4D, 5B, 7C, and 8A).
Limited Proteolysis
Ist1 was diluted from >100 μm stocks in Ist1 purification buffer (25 mm HEPES and 200 mm KCl (pH 7.5)) to ∼13 μm in ATPase buffer (20 mm HEPES, 100 mm KOAc, and 5 mm MgOAc (pH 7.5)), incubated at 30 °C for 30 min prior to addition of trypsin (Sigma-Aldrich, catalog no. T8658) at a final ratio of 1:1000 (w/w) trypsin:Ist1. 10-μl samples were taken at the indicated time points (e.g. 5 min, 1 h, 2 h, and 4 h) and quenched by adding 12 μl of 5× Laemmli sample buffer (64) and heating at 100 °C for 10 min. The samples and protein ladder (Benchmark protein ladder or Benchmark prestained protein ladder, Invitrogen) were resolved by SDS-PAGE and stained with Coomassie Blue (Bio-Rad) (Figs. 3, A and B; 4E; 5E, 6D, and 7D). Alternatively, 10-μl samples were quenched by adding 12 μl of 2% trifluoroacetic acid, separated by HPLC (Agilent 1200 system, Agilent Zorbax SB C18 column), and analyzed by positive-mode electron spray ionization mass spectrometry (Agilent 6224 TOF system). These data were correlated to Ist1 amino acid sequences with an accuracy of 10 ppm using Agilent Mass Hunter Qualitative Analysis/BioConfirm software (version B.05.00).
FIGURE 3.
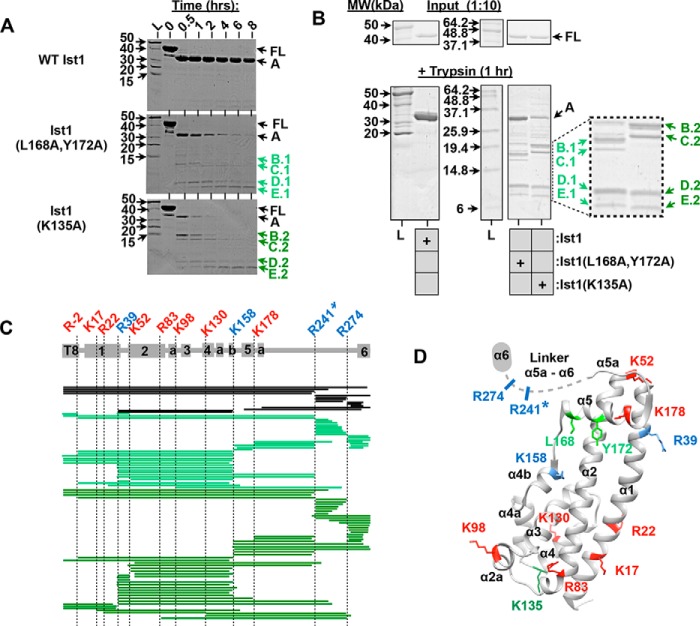
Ist1 hyperstimulatory mutants exhibit increased sensitivity to trypsin cleavage. A and B, limited proteolysis of Ist1 with trypsin. Aliquots were removed at various time points for SDS-PAGE analysis and Coomassie staining. Full-length (FL) Ist1 is cleaved into smaller peptide fragments (A–E). The ladder (L) is shown with corresponding molecular weights indicated (kilodalton). B, distinct cleavage patterns for Ist1(L168A,Y172A) versus Ist1(K135A) are highlighted using samples from a 1-h time point (B–E.1 and B–E.2, respectively). Coomassie staining of a 1:10 dilution of Ist1 samples before addition of trypsin (Input) was performed to confirm equal loading of Ist1. MW, molecular weight. C and D, summary of tryptic cleavage sites in Ist1 identified by mass spectrometry. C, tryptic peptides are shown for WT Ist1 (black), Ist1(L168A,Y172A) (light green), and Ist1(K135A) (dark green) in relation to Ist1 secondary structural domains (α1, α2, α2a, α3, α4, α4a, α4b, α5, α5a, and α6). The T8 tag (residues −17 through −1) remains following cleavage of His6 tag by thrombin. D, location of tryptic cleavage sites in the Ist1 NTD crystal structure (67). Fragment A, the predominant species apparent in cleavage of WT Ist1, is generated by cleavage at R241 (asterisk). Tryptic cleavage sites that are common to WT Ist1, Ist1(L168A,Y172A), and Ist1(K135A) are highlighted in blue, whereas those that are unique to Ist1(L168A,Y172A) and Ist1(K135A) are highlighted in red.
FIGURE 6.
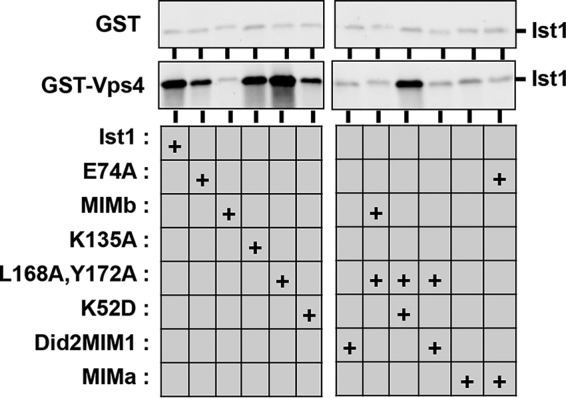
Ist1 MIM mutants are defective for binding to Vps4. GST pulldowns using GST alone or GST-Vps4 and purified Ist1 and Ist1 mutants. Isolated Ist1 was detected by Western blotting.
SEC
Analytical SEC was performed with Superdex200 GL 10/300 (GE Healthcare Life Sciences) in 25 mm HEPES, 200 mm KCl (pH 7.5). This buffer was used because SEC performed with ATPase buffer resulted in reduced recovery of Ist1 and an extended lagging shoulder (data not shown). Ist1 stocks were normalized to 200 μm in 25 mm HEPES, 200 mm KCl (pH 7.5), and 172 μg of sample was resolved (0.75 ml/min flow rate at 4 °C). The UV traces shown are representative of at least two runs. Fractions from SEC runs were collected, subjected to SDS-PAGE analysis, and visualized by Coomassie staining (data not shown). Indicated apparent molecular weights were deduced from gel filtration standards (Bio-Rad) (dashed black lines in Figs. 2, D and E; 4, A–C; 5, C and D; 7A; and 8B).
Protein-Protein Interaction
GST or GST-Vps4 was prebound to glutathione-Sepharose 4B (GE Healthcare Life Sciences) in PBS + 0.05% Tween 20, incubated at 4 °C for 1 h, washed with PBS + 0.05% Tween 20, and equilibrated with ATPase buffer + 0.05% Tween 20. Purified Ist1 proteins (2 μg) were added to GST or GST-Vps4 in the presence of 0.1 mg/ml BSA. Following extensive washing with ATPase buffer + 0.05% Tween 20, bound material was eluted with 20 mm Tris, 100 mm NaCl, 20 mm glutathione, and 1 mm DTT (pH 7.5). Samples were resolved by SDS-PAGE. Western blotting was performed using anti-Ist1 (this study) and Cy3-conjugated Goat anti-Rabbit IgG secondary (Life Technologies) and detected using the Typhoon FLA 7000 system.
Subcellular Fractionation
Subcellular fractionation of cells grown in minimal medium was performed as described previously (23), except that cell extracts were subjected to centrifugation at 13,000 × g for 10 min at 4 °C to separate the soluble and pellet fractions. Samples (0.2 A600 equivalents) were resolved by SDS-PAGE and Western-blotted for Snf7 (31) and Ist1 (this study). Phosphoglycerate kinase (Life Technologies) and Pep12 (Life Technologies) were used a markers for the soluble and pellet fractions, respectively. Western blots were developed using HRP-conjugated goat anti-rabbit IgG or goat anti-mouse (Life Technologies), along with SuperSignal West Femto and SuperSignal West Pico substrates (Thermo), and the Autochemi system (UVP, Upland, CA). Quantitation was performed with the ImageQuant software package. Subcellular fractionation data represent three independent experiments (with representative Western blots shown) and are graphed as mean ± S.D.
ESCRT-III Disassembly Assay
ESCRT-III disassembly assays were performed as described previously (31). Yeast (pep4Δ vps4Δ or pep4Δ ist1Δ vta1Δ) was lysed with a Dounce in 20 mm Pipes (pH 6.8), 100 mm KCl, 50 mm KOAc, 5 mm MgOAc, and 100 mm sorbitol with protease inhibitors. The lysate was cleared by a 5-min, 300 × g spin, and the cleared lysate was subjected to a 10-min spin at 10,000 × g. The pelleted membranes were washed twice in lysis buffer and resuspended in disassembly assay buffer (20 mm HEPES (pH 7.4), 100 mm KOAc, 5 mm MgOAc, and 200 mm sorbitol with protease inhibitors) at 50 A600 equivalents/ml. Reactions with various amounts of purified Vps4 and Ist1 were performed with 0.5 A600 equivalent membranes and an ATP regeneration system (1 mm ATP) at 30 °C for 10 min. Membranes were repelleted via a 10-min spin at 13,000 × g, and Western blotting was performed to assess levels of Snf7 in the pellet and soluble fractions. The UVP Autochemi system and ImageQuant software package were used to quantify Snf7 levels. Disassembly assay data represent two or three independent experiments with reactions performed in duplicate or triplicate within experiments and are graphed as the mean ± S.E. (representative Western blots are shown).
Results
Ist1 Core Folding Is a Critical Determinant of Vps4 Regulation
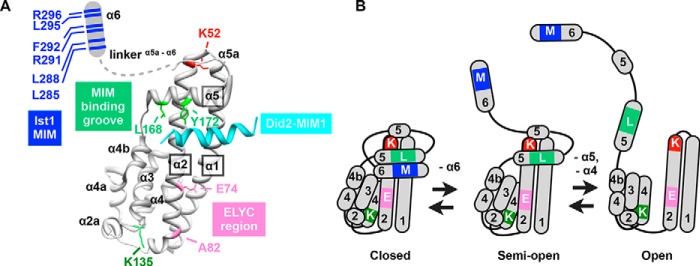
ESCRT-III subunits contain six characteristic α helices and share a common “ESCRT-III fold” that is characterized by a cleft formed by α5 binding to the α1/2 hairpin, forming the α1/2/5 groove, and packing of the α3/4 bundle against the opposite face of the α1/2 hairpin (Fig. 1) (21, 45–47, 60). Monomeric ESCRT-III subunits in the cytoplasm are autoinhibited by intramolecular binding of their C termini (α6) to the α1/2/5 groove (Fig. 1B, closed conformation), whereas recruitment into ESCRT-III polymers on membranes leads to displacement of α6 from the α1/2/5 groove (Fig. 1B, semi-open conformation) and additional conformational changes within the Ist1 core (α1–5) (Fig. 1B, open conformation) (45–49, 60). MIM elements are located in the α6 helices of Vps2, Did2, and Ist1 (Figs. 1 and 5A). Therefore, we predicted that Ist1 intramolecular interactions between the MIM-containing α6 and α1/2/5 groove in a closed conformation may contribute to Vps4 inhibition, whereas a more open Ist1 conformation resulting in displacement of α6 or additional conformational changes in the Ist1 core (α1-α5) may generate Vps4 stimulation in a MIM-dependent manner. To explore this model, site-directed mutagenesis of Ist1 was implemented with the goal of generating forms with altered Vps4 regulation and Ist1 conformation.
FIGURE 1.
Putative Vps4 regulatory elements in Ist1. A, site-directed mutagenesis of Ist1. Residues mutated in these studies are indicated in the Ist1 NTD-Did2 MIM1 co-crystal structure (PDB code 3GGZ (67)), including the Did2-MIM1 element (cyan), the Ist1 MIM element (blue, Leu285, Leu288, Arg291, Phe292, Leu295, and Arg296), the Did2-MIM1-binding motif (light green, Leu168 and Tyr172), a highly conserved lysine of unknown function (dark green, Lys135), the ELYC region (pink, Glu74 and Ala78), and a highly conserved lysine corresponding to hIST1 (Arg51) that was defective for Ist1 homopolymerization in vitro (45) (red, Lys52). The Ist1 α6 and adjacent linker region are drawn as a schematic in a theoretical semi-open Ist1 conformation. B, model for Ist1 conformational changes. In the closed conformation, α6 is buried within the α1/2/5 groove. In the semi-open conformation, α6 is displaced from the α1/2/5 binding groove. In the open conformation, the α1,2,5 groove is dissolved as α5 is displaced from the closed end of the α1/2 hairpin. Further conformational changes in the Ist1 core domain may occur, including unpacking of the α3/4 bundle from the open end of the α1/2 hairpin. Mutations are indicated in distinct Ist1 conformations: MIM residues, M (α6); MIM-binding motif, L (α5); K135, K (α4); ELYC region, E (α2); and K52, K (α2).
Consistent with previous observations (23), titration of WT Ist1 (500 nm-12 μm) resulted in concentration-dependent inhibition of Vps4 ATPase activity (Fig. 2, A and C). In striking contrast, two mutant forms of Ist1 hyperstimulated Vps4 ATPase activities (i.e. greater stimulation than observed in other ESCRT-III subunits (20)): Ist1(L168A,Y172A) (Fig. 2, B and C) and Ist1(K135A) (Fig. 2C). Ist1(L168A,Y172A) is altered in the α1/2/5 groove (Fig. 1A) and is defective for binding to the Did2 MIM1 (60), suggesting that L168A,Y172A may destabilize the closed conformation of full-length Ist1. By contrast, Lys135 is located in α4 and may contribute to packing against the opposite end of the α1/2 hairpin (Fig. 1A). Together, these mutants suggest that changes in Ist1 conformation contribute to Vps4 hyperstimulation.
To assess Ist1 conformation, SEC and limited proteolysis were performed. SEC analysis of WT Ist1 (35 kDa) revealed an apparent molecular mass of 94 kDa (Fig. 2, D and E), consistent with previous analyses indicating that Ist1 behaves as an elongated monomer (23). In addition, Ist1(L168A,Y172A) and Ist1(K135A) eluted with apparent masses of 104 and 94 kDa, respectively (Fig. 2, D and E). This similarity to WT Ist1 suggests that these mutants also behave as elongated monomers, with Ist1(L168A,Y172A) adopting a subtly more open conformation.
Transient conformational changes in Ist1 were assessed by limited proteolysis with trypsin (Fig. 3). Trypsin treatment of Ist1 resulted in the initial generation of a 30-kDa fragment (indicated as A in Fig. 3). However, the stability of this fragment varied between the WT and mutant Ist1 forms. Although the A fragment from WT Ist1 persisted for more than 8 h, the A fragments from Ist1(L168A,Y172A) and Ist1(K135A) were cleaved more rapidly into smaller species (Fig. 3, A and B, fragments B–D), suggesting more open conformations. Next, mass spectrometry analyses were conducted to identify the location of trypsin cleavage in wild-type versus mutant Ist1 forms (Fig. 3, C and D). These analyses revealed that the A fragment of WT Ist1, Ist1(L168A,Y172A), and Ist1(K135A) resulted from cleavage at Arg241 located within the linker region between α5a and α6 (Fig. 3C), suggesting that this region was equally susceptible to trypsin. Increased proteolysis of the Ist1(L168A,Y172A) and Ist1(K135A) core occurred through cleavage sites both common with WT Ist1 (Arg39 and Lys158, blue residues in Fig. 3, C and D) and sites unique to these mutants (Lys17, Arg22, Lys52, Arg83, Lys98, Lys130, and Lys178; red residues in Fig. 3, C and D). These data suggest that Ist1(L168A,Y172A) and Ist1(K135A) adopt more open conformations than WT Ist1 via unfolding of their core domains.
A third mutation (K52D), which is located near the tip of the α1,2 hairpin (Fig. 1A), resulted in a form of Ist1 that eluted later in SEC analyses (Fig. 4A). This elution profile is suggestive of a more closed Ist1 conformation. The K52D mutation converted Ist1(L168A,Y172A) and Ist1(K135A) to a more closed conformation, as revealed by the increased elution volumes of Ist1(L168A,Y172A,K52D) and Ist1(K135A,K52D) relative to Ist1(L168A,Y172A) and Ist1(K135A), respectively (Fig. 4, B and C). Similarly, the A fragments of Ist1(L168A,Y172A,K52D) and Ist1(K135A,K52D) were less sensitive to trypsin, although to a lesser extent in Ist1(K135A,K52D) (Fig. 4E). Therefore, the effect of the K52D mutation to stabilize a more closed Ist1 conformation was dominant to the L168A,Y172A and K135A mutations. Importantly, these behaviors corresponded to a loss of Vps4 hyperstimulation by Ist1(L168A,Y172A,K52D) and Ist1(K135A,K52D), further highlighting a requirement for an open Ist1 conformation to maximally stimulate Vps4 (Fig. 4D). By contrast, Ist1(K52D) inhibited Vps4 to a level similar as WT Ist1, suggesting that the closed conformation adopted by Ist1(K52D) did not prevent negative Vps4 regulation.
The Ist1 MIM Element Is Essential for Inhibition and Hyperstimulation of Vps4 Activity
Previous studies have highlighted interactions between other ESCRT-III MIM elements and the Vps4 MIT domain as being critical for ESCRT function (9, 20, 32–36). To determine the role of the Ist1 MIM element in positive and negative Vps4 regulation, mutant forms of Ist1 were generated in which the MIM-containing α6 was deleted (residues 1–287, Ist1(MIMΔ)) or altered by site-directed mutagenesis (Ist1(L288A, R291A, L295A, R296A) or Ist1(MIMa) and Ist1(L288D, L295D) or Ist1(MIMb)) (Fig. 5A) and tested for their effects on Vps4 ATPase activity (Fig. 5B). In contrast to our initial predictions, Ist1(MIMa), Ist1(MIMb), and Ist1(MIMΔ) were unable to inhibit Vps4, suggesting that accessibility of α6 in a semi-open conformation was required for negative regulation of Vps4 (Fig. 1B). In addition, all Ist1 MIM mutants exhibited a low level of Vps4 stimulation, suggesting a MIM-MIT-independent mode of Vps4 stimulation. Combining the MIM mutants with L168A,Y172A or K135A resulted in a loss of Vps4 hyperstimulation, and a form of Vps4 lacking the MIT domain (Vps4(ΔN)) was defective for both inhibition by WT Ist1 as well as hyperstimulation by Ist1(L168A,Y172A). In SEC, combining Ist1(MIMb) with L168A,Y172A or K135A resulted in an earlier elution volume, suggesting that aspects of their conformation were more open (Fig. 5, C and D; apparent molecular weight, 110 kDa). In addition, the A fragments of Ist1(L168A,Y172A,MIMb) and Ist1(K135A,MIMb) were not stabilized relative to Ist1(L168A,Y172A) and Ist1(K135A), respectively (Fig. 5E). Therefore, in contrast to Ist1(L168A,Y172A,K52D) and Ist1(K135A,K52D), defects in Vps4 hyperstimulation by Ist1(L168A,Y172A,MIMb) and Ist1(K135A,MIMb) were not related to rescue of a more closed conformation. Instead, loss of Vps4 regulation in the context of the Ist1 MIM mutants was related to reduced Vps4 binding, as revealed in a GST-Vps4 pulldown assay (Fig. 6). These findings suggest that MIM-MIT interactions are required for robust binding between Ist1 and Vps4 to permit both positive and negative regulation.
Next we sought to test whether a form of Ist1 with its α6 bound more strongly to the α1,2,5 groove would reduce Vps4 inhibition or stimulation (Fig. 1B, closed conformation). The Did2-MIM1 element binds with high affinity to the Ist1 α1,2,5 groove (60). Therefore, we replaced the Ist1 MIM element with the Did2 MIM1 element to yield the Ist1(Did2-MIM1) chimera (Fig. 5A) and tested its biochemical activities (Fig. 7). Compared with WT Ist1, Ist1(Did2-MIM1) eluted later in SEC analyses (Fig. 7A, apparent molecular mass of 69 kDa), whereas the A fragment was equally susceptible to proteolysis (Fig. 7B). These observations are consistent with Ist1(Did2-MIM1) adopting a more closed conformation and further support that cleavage at Arg241 to generate the A fragment can occur in the closed conformation. Ist1(Did2-MIM1) failed to bind or inhibit Vps4 activity (Fig. 7C), consistent with inaccessibility of the Did2-MIM1 for interactions with the Vps4 MIT domain.
To displace the Did2-MIM1 element from the Ist1 α1,2,5 groove, we introduced the L168A,Y172A double mutation (60), resulting in Ist1(L168A,Y172A,Did2-MIM1). This molecule eluted earlier in SEC analyses relative to Ist1(Did2-MIM1), and its A fragment was more sensitive to trypsin, suggesting displacement of the Did2-MIM1 as well as unfolding of the Ist1(L168A,Y172A,Did2-MIM1) core domain in the open conformation (Fig. 1B). However, Ist1(L168A,Y172A, Did2-MIM1) exhibited reduced binding to Vps4 and was unable to maximally stimulate Vps4 (Figs. 6 and 7C). Therefore, the Did2 MIM1 element cannot substitute the Ist1 MIM to mediate Vps4 hyperstimulation, apparently because of an inability to equivalently interact with the Vps4 MIT domain.
The Ist1 ELYC Region Is Essential for Inhibition of Vps4 Activity
Ist1 contains a highly conserved ELYC sequence located in α2 of the Ist1 core that has been suggested previously to play a role in Vps4 inhibition (Fig. 1A) (23). Several additional highly conserved, surface-exposed residues are located in this region, including alanine 82 (74ELYCELLLA82). To test the role of this ELYC region in Vps4 regulation, we generated Ist1(E74A) and Ist1(A82D). Ist1(E74A) and Ist1(A82D) were unable to inhibit Vps4 activity but, instead, stimulated Vps4 to greater levels than Ist1 MIM mutants (Fig. 8A). Combining the E74A and MIMa mutations led to Vps4 stimulation similar to E74A alone, suggesting that additional Vps4 stimulation was MIM-MIT-independent. Ist1(E74A) eluted later in SEC analyses, suggesting a more closed conformation (Fig. 8B). However, the MIM element was still accessible, as indicated by robust binding of Ist1(E74A) to Vps4 (Fig. 6). In limited proteolysis experiments, sensitivity of the A fragment of Ist1(E74A) was not increased relative to WT Ist1, indicating that folding of the core domain was not altered (Fig. 8C). Taken together, these data suggested that the Ist1 ELYC region is involved in a secondary, weaker affinity interaction in conjunction with the Ist1 MIM element to inhibit Vps4 activity in a semi-open conformation.
Did2 Converts Ist1 from an Inhibitor to a Stimulator of Vps4 ATPase Activity
Binding of Did2 to Ist1 has been suggested to alter the conformation of the Ist1 core as Ist1 incorporates into ESCRT-III polymers (45, 60). To test whether Did2 binding altered Ist1 regulation of Vps4, the effect of Ist1 on Vps4 ATPase activity in the presence of Did2 was assessed (Fig. 9). Addition of 2 or 4 μm Did2 alone stimulated Vps4, consistent with previous observations (20, 39). Although WT Ist1 alone inhibited Vps4 ATPase activity, mixing WT Ist1 and Did2 led to Vps4 stimulation that was greater than that observed by addition of Did2 alone. This suggests that Did2 binding to Ist1 induces a more open Ist1 conformation, thereby switching Ist1 from an inhibitor to a stimulator of Vps4 activity.
FIGURE 9.
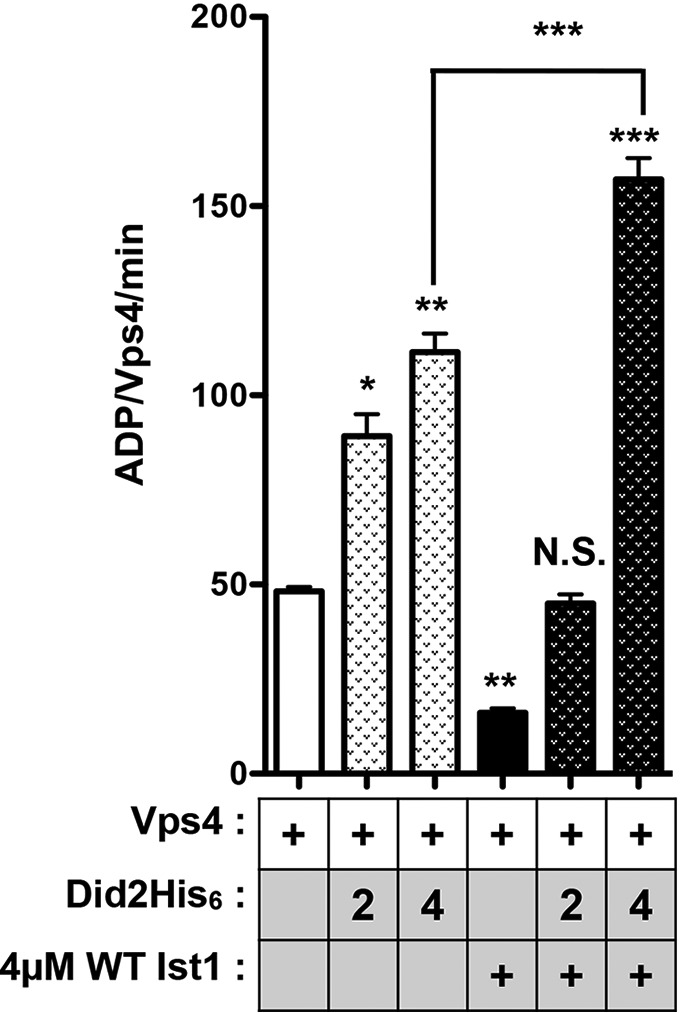
Did2 converts WT Ist1 from an inhibitor to a stimulator of Vps4. Shown is ATPase activity of 500 nm Vps4 in the presence of Did2His6 (2 or 4 μm) with or without 4 μm WT Ist1. Results are presented as mean + S.D. of triplicate experiments, with statistical differences from 500 nm Vps4 alone indicated (**, p < 0.01; ***, p < 0.001; N.S., not significant).
Ist1 Conformation Affects Vps4 Function
Vps4 disassembly of ESCRT-III is critical for ESCRT function (17, 24, 28–31). Therefore, the effects of altered Ist1 conformation on Vps4 function were examined in two contexts: subcellular fractionation of ESCRT-III subunits in vivo and ESCRT-III disassembly in vitro.
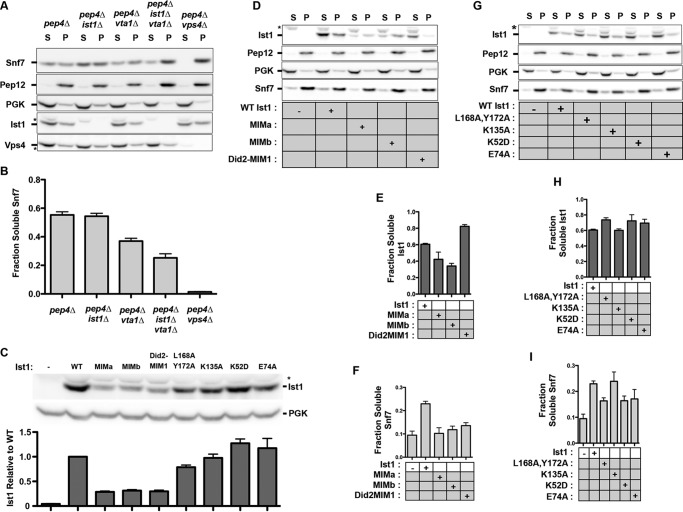
Although loss of Ist1 alone does not affect MVB sorting, synthetic genetics defects have been observed in an ist1Δ vta1Δ background (23, 26, 60). Consistent with these studies, we observed a defect in Snf7 recycling in the ist1Δ vta1Δ genetic background (Fig. 10, A and B). This sensitized genetic background was utilized to assess mutant Ist1 function by their re-expression from the endogenous promoter. Ist1 mutants with defective MIM function (Ist1(MIMa), Ist1(MIMb), and Ist1(Did2-MIM1)) displayed decreased protein levels in vivo (Fig. 10C), thereby precluding interpretations pertaining to complementation of Snf7 disassembly (Fig. 10, D and F). Relative to WT Ist1, Ist1(MIMa) and Ist1(MIMb) were more membrane-associated, whereas Ist1(Did2-MIM1) was less membrane-associated (Fig. 10, D and E). These effects correlate with the more open and closed Ist1 conformations observed via SEC analyses, respectively (Figs. 5C and 7A), and are consistent with previous reports showing that displacement of C-terminal portions of other ESCRT-III subunits (α5 and/or α6) promotes membrane association in vivo (9, 45, 48, 49).
FIGURE 10.
Ist1 conformation leads to altered membrane association in vivo. A, subcellular fractionation of the ESCRT-III subunits Ist1and Snf7 in pepΔ, pepΔ ist1Δ, pepΔ vta1Δ, pep4Δ ist1Δ vta1Δ and pep4Δ vps4Δ yeast strains. Blots are representative of experiments performed four times. Phosphoglycerate kinase and Pep12 served as markers for the 13,000 × g soluble (S) and pellet (P) fractions. Subcellular fractionation of the ATPase Vps4 is also shown. Nonspecific species detected by Ist1 and Vps4 antibodies are indicated (asterisks). B, quantitation of soluble Snf7 in the four experiments is indicated. Error bars indicate mean ± S.D. C, whole cell lysates showing total Ist1 protein levels in pep4Δ ist1Δ vta1Δ, with phosphoglycerate kinase (PGK) shown as a loading control. D and G, subcellular fractionation of ESCRT-III subunits Ist1 and Snf7 in ist1Δ vta1Δ pepΔ yeast with plasmids expressing WT Ist1 and Ist1 mutants. Blots are representative of experiments performed three times. Phosphoglycerate kinase and Pep12 served as markers for the 13,000 × g soluble and pellet fractions. A nonspecific species detected by Ist1 antibody is indicated (asterisks). E—I, quantitation of three experiments. E and H, relative amount of Ist1 in soluble fraction. H, relative amount of Snf7 in soluble fraction. Error bars indicate mean ± S.D.
In contrast to the Ist1 MIM mutants, the protein levels of Ist1(L168A,Y172A), Ist1(K135A), Ist1(K52D), and Ist1(E74A) were similar to WT Ist1 (Fig. 10C). Ist1(L168A,Y172A) exhibited decreased membrane association (Fig. 8, G and H) and decreased Snf7 recycling (Fig. 8, G and I). This is consistent with a previous report showing that defects in Ist1 binding to Did2 correlate with defects in MVB sorting (60). Ist1(K52D) and Ist1(E74A) exhibited decreased membrane association (Fig. 10, G and H), which correlated with reduced Snf7 disassembly (Fig. 10, G and I). Ist1(K135A) exhibited levels and membrane association equivalent to WT Ist1, and these behaviors corresponded to comparable Snf7 recycling. Therefore, altered Ist1 conformation in vitro correlated with Ist1 subcellular localization in vivo, presumably contributing to altered Snf7 disassembly.
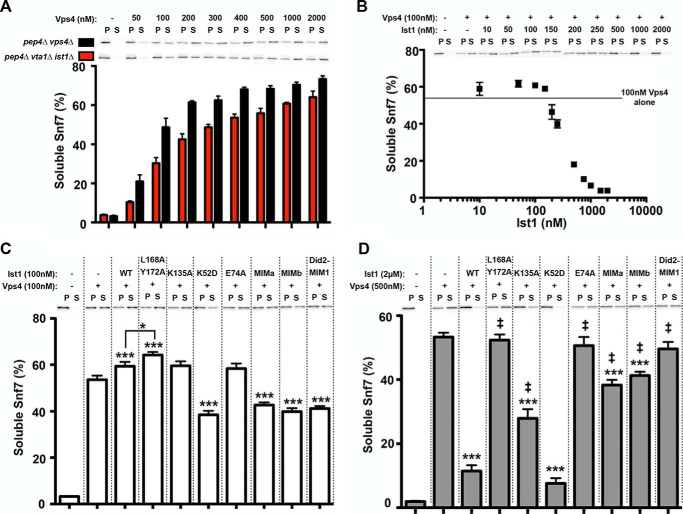
To circumvent differences in Ist1 protein levels observed in vivo, a previously established in vitro ESCRT-III disassembly assay was performed in the presence of equal amounts of purified Ist1 (Fig. 11). ESCRT-III-containing membranes were isolated from pep4Δ vta1Δ ist1Δ yeast and examined for Vps4-dependent release of Snf7. ESCRT-III substrate from pep4Δ vta1Δ ist1Δ yeast was less responsive to disassembly by Vps4 than ESCRT-III substrate from pep4Δ vps4Δ (Fig. 11A), consistent with reduced recycling of Snf7 to the soluble pool in subcellular fractionation experiments (Fig. 10, A and B). Intriguingly, titration of WT Ist1 (10 nm-2 μm) with 100 nm Vps4 generated a biphasic response (Fig. 10B). Low concentrations of Ist1 (10–150 nm) stimulated Vps4-mediated Snf7 release, whereas higher concentrations (200 nm-2 μm) inhibited Vps4-mediated Snf7 release. These behaviors are consistent with the ability of Ist1 to both positively or negatively affect ESCRT function in vitro and in vivo (Figs. 2–7 and Refs. 23, 26, 60). Subsequently, the effects of Ist1 mutants on Vps4-mediated ESCRT-III disassembly were examined at 100 nm or 2 μm Ist1 to address both modes of regulation.
FIGURE 11.
Ist1 exerts positive and negative regulation of ESCRT-III disassembly in vitro. A, Vps4-mediated Snf7 disassembly from pep4Δ vps4Δ (black columns) and pep4Δ vta1Δ ist1Δ (red columns). Vps4 was titrated from 50 nm-2 μm. Representative blots of Snf7 localization to the soluble (S) and pellet (P) fractions are shown. Quantitation of two experiments with reactions performed in duplicate is indicated as percent Snf7 in soluble fraction. Error bars indicate mean ± S.E. B, effects of WT Ist1 on Vps4-mediated Snf7 disassembly from pep4Δ vta1Δ ist1Δ. WT Ist1 was titrated from 10 nm-2 μm in the presence of 100 nm Vps4. Basal Snf7 disassembly by 100 nm Vps4 is indicated by the line to highlight enhanced Snf7 disassembly in the presence of WT Ist1 at low [Ist1], followed by inhibition at high [Ist1]. Representative blots of Snf7 localization to the soluble and pellet fractions are shown. Quantitation of three experiments with reactions performed in duplicate is indicated as percent Snf7 in soluble fraction. Error bars indicate mean ± S.E. C, effects of 100 nm Ist1 mutants on Vps4-mediated Snf7 disassembly at 100 nm Vps4. Representative blots of Snf7 localization to the soluble and pellet fractions are shown. Quantitation of three experiments with reactions performed in triplicate is indicated as percent Snf7 in soluble fraction. Statistical differences from addition of Vps4 alone (***, p < 0.001) or from addition of Vps4 and WT Ist1 are indicated (*, p < 0.05). D, effects of excess (2 μm) Ist1 mutants on Vps4-mediated Snf7 disassembly in the presence of 500 nm Vps4. Representative blots of Snf7 localization to the soluble and pellet fractions are shown. Quantitation of three experiments with reactions performed in triplicate is indicated as percent Snf7 in soluble fraction. Error bars indicate mean ± S.E. Statistical differences from addition of Vps4 alone (***, p < 0.001) or from addition of Vps4 and WT Ist1 are indicated (‡, p < 0.001).
Positive regulation of ESCRT-III disassembly by Ist1 in the presence of 100 nm Vps4 was examined first (Fig. 11C). Relative to WT Ist1, ESCRT-III disassembly was enhanced further by the hyperstimulatory Ist1(L168A, Y172A) mutant (p < 0.05), revealing that the increased ATPase activity observed correlates with increased Vps4 function. Two mutants (Ist1(K135A) and Ist1(E74A)) that were both defective for inhibition of Vps4 ATPase activity and retained functional MIM elements exhibited a similar enhancement of Snf7 disassembly compared with WT Ist1. In contrast, mutants that were defective for binding to the Vps4 MIT domain (Ist1(MIMa), Ist1(MIMb), and Ist1(Did2MIM)) and/or possessed a more folded Ist1 core domain (Ist1(K52D)) exhibited partial inhibition of disassembly activity. These results further highlight the importance of the MIM element in Ist1 as well as the conformational state of the Ist1 core region to permit positive regulation of ESCRT function.
Negative regulation of ESCRT-III disassembly was subsequently examined with 500 nm Vps4 and excess Ist1 (2 μm) (Fig. 11D). Addition of WT Ist1 and Ist1(K52D) inhibited Snf7 disassembly, consistent with their abilities to inhibit Vps4 ATPase activity (Fig. 4D). By contrast, all of the other mutants examined (which either adopted a more open core conformation or contained mutations in the Ist1 ELYC region or MIM element) exhibited partial or complete deficits in inhibiting Vps4-mediated ESCRT-III disassembly. Therefore, Ist1 mutants that were defective for inhibition of Vps4 ATPase activity were similarly unable to inhibit Vps4-mediated ESCRT-III disassembly.
Discussion
In recent years, considerable progress has been made toward elucidating distinct modes of Vps4 regulation by ESCRT-III subunits (20, 23, 24, 38, 39, 65). The studies presented here provide additional insights into the bimodal regulation of Vps4 by Ist1 and its effects on ESCRT function via conformational changes in Ist1.
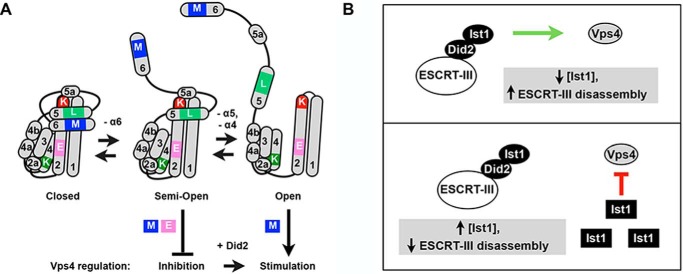
Negative regulation of Vps4 by Ist1 results from the combination of Ist1 MIM and ELYC interactions with Vps4. How does the Ist1 MIM element contribute to inhibition of Vps4 activity whereas the MIM1 elements of Did2 and Vps2 stimulate Vps4 (20)? The Ist1 MIM element itself is functionally distinct from the Did2 MIM1 because the Did2 MIM1 could not replace the Ist1 MIM element in Vps4 binding and/or regulation (Figs. 6 and 7). Although the effect on Ist1 conformation may have contributed to these effects, the Ist1 MIM has also been demonstrated to bind to MIT domains in unique ways. hIst1 α6 binds to the Vps4A MIT domain in a manner similar to Vps2 and Did2 MIM1 elements (9, 32), hIst1 residues upstream of α6 bind to a distinct surface of the Vps4A MIT domain in a MIM2 mode (9, 33), and hIst1 α6 can bind to the MIT domain of Spartin via a different orientation, referred to as the MIM3 mode (32). These MIM2 and MIM3 modes of association may enhance the strength of Ist1-Vps4 interactions or alter presentation of the Ist1 core in a manner that permits ELYC interaction to inhibit Vps4. The ELYC motif is not found in other ESCRT-III subunits, potentially explaining why Vps4 inhibition is unique to Ist1. Interactions between Vps4 and the ELYC region are weaker compared with the MIM-MIT interaction (Fig. 6), suggesting that the MIM acts as the primary binding interaction that subsequently permits additional interactions to mediate distinct Vps4 regulation (Fig. 12A).
FIGURE 12.
Model for bimodal regulation of Vps4 and ESCRT function by Ist1. A, model of Ist1 conformations and their effects on Vps4 regulation. In the semi-open Ist1 conformation, the Ist1 MIM element (M, blue box) and ELYC region (E, pink box) interact with Vps4 to inhibit ATPase activity. Unfolding of the Ist1 core domain in a more open conformation permits Vps4 hyperstimulation in a MIM-dependent manner. Did2 induces the conversion between Ist1 inhibitory and stimulatory activities, presumably through generating the open conformation of Ist1. B, effects of Ist1 concentration on Vps4 regulation. Ist1 incorporates into ESCRT-III via Did2, leading to an open Ist1 conformation (circle) and stimulation of Vps4-mediated ESCRT-III disassembly (green arrow). Saturation of ESCRT-III with Ist1 leads to the accumulation of soluble Ist1 in a closed conformation (square), resulting in inhibition of Vps4-mediated ESCRT-III disassembly (red line).
Vps4 stimulation by Ist1 is also MIM-MIT-dependent but additionally requires a more open Ist1 core conformation (Fig. 12A). Unfolding of the Ist1 core domain in the contexts of Ist1(L168A,Y172A) and Ist1(K135A) resulted in Vps4 hyperstimulation (Figs. 2 and 3), and this effect was blocked by introducing the K52D mutation, which stabilized the Ist1 core in a more closed conformation (Fig. 4). Ist1(K52D) was examined on the basis of its similarity to hIST1(R51D), which has been demonstrated previously to be defective for homopolymerization in vitro (45). Altered Ist1 core conformation may affect Vps4 regulation in different ways. First, conformational changes may alter the presentation of secondary Vps4 regulatory elements in Ist1. For example, the Vps4 inhibitory ELYC region, which is located in α2 of the Ist1 core domain, may be unfolded in an open state, leading to loss of Vps4 inhibition. Alternatively, the ELYC region itself may be involved in interactions with Did2 (for instance during ESCRT-III polymerization). This notion is supported by the hIST1 ELYC region being implicated in crystal packing interactions correlated with hIST1 homopolymerization in vitro (45), which may mimic aspects of ESCRT-III heteropolymerization in vivo. Finally, the open conformation may reveal a secondary surface that acts in concert with the Ist1 MIM element to synergistically stimulate Vps4 activity. An obvious candidate for a secondary stimulatory surface is a cluster of acidic residues located in the linker region that connects α5–6 or α5, which interact with the Vps4 linker and/or pore regions to stimulate ATPase activity in other ESCRT-III subunits (38–40). We speculate that the Ist1 conformational changes associated with hyperstimulation of Vps4 resemble changes that occur with Ist1 binding to Did2 (Fig. 12A). This model is supported by the result that addition of Did2 converted Ist1 from an inhibitor to a stimulator of Vps4 ATPase activity.
These studies also highlight a physiological link between Ist1 regulation of Vps4 ATPase activity and ESCRT-III disassembly through interactions with Did2. Addition of low concentrations of Ist1 stimulated ESCRT-III disassembly from membranes in vitro, whereas high concentrations of Ist1 resulted in potent inhibition of ESCRT-III disassembly (Fig. 11). This pattern is consistent with the model in which Ist1 association with Did2 within ESCRT-III polymers reveals positive regulation of ESCRT function, whereas saturation of this association and subsequent accumulation of soluble Ist1 leads to negative regulation of Vps4 function (Fig. 12B). This behavior may explain how reduced Ist1 protein levels in vivo, which occurs during starvation (66), promotes MVB sorting by lowering the ratio of Ist1:Did2 to minimize the soluble, inhibitory pool of Ist1.
Bimodal regulation of ESCRT function by Ist1 may be important for other cellular processes, including cytokinesis in mammalian cells (8, 9). Intriguingly, hIst1 and CHMP1/hDid2 bind specifically to another MIT domain-containing AAA-ATPase, Spastin. Spastin alters microtubule-severing activities during cytokinesis and has also been implicated in mitotic spindle disassembly, nuclear envelope sealing, and/or neuron function (Refs. 67–71 and reviewed in Ref. 72). It is tempting to speculate that hIst1 may regulate Spastin ATPase activity in a manner analogous to Ist1 regulation of Vps4 in these studies, thereby positioning hIst1 as a coordinator of membrane trafficking and cytoskeletal dynamics. In both contexts, we predict that ATPase regulation by Ist1 is altered in response to binding to Did2 or ESCRT-III polymerization, which, in turn, would be dependent on physiological cues such as receptor signaling, protein degradation, nutrient composition, and cellular proliferation.
Author Contributions
J. T., B. A. D., and D. J. K. conceived the study and wrote the paper. J. T. and B. A. D. designed and conducted the experiments along with J. A. P. J. T. and L. M. B. conceived, performed, and analyzed the mass spectrometry analyses. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Bruce Horazdovsky and Shirley Dean for discussions and Benjamin Madden for technical advice pertaining to mass spectrometry analyses.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM73024 (to D. J. K.) and National Institutes of Health F31 Predoctoral Fellowship F31GM106693 (to J. A. T.). This work was also supported by a Fraternal Order of the Eagles postdoctoral fellowship (to B. A. D.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- ESCRT
- endosomal sorting complexes required for transport
- MVB
- multivesicular body
- AAA-ATPase
- ATPase associated with diverse cellular activities
- MIT
- microtubule-interacting and trafficking
- MIM
- MIT-interacting motif
- SEC
- size exclusion chromatography.
References
- 1.Katzmann D. J., Babst M., and Emr S. D. (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145–155 [DOI] [PubMed] [Google Scholar]
- 2.Stringer D. K., and Piper R. C. (2011) A single ubiquitin is sufficient for cargo protein entry into MVBs in the absence of ESCRT ubiquitination. J. Cell Biol. 192, 229–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piper R. C., and Katzmann D. J. (2007) Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 23, 519–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlton J. G., Agromayor M., and Martin-Serrano J. (2008) Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc. Natl. Acad. Sci. U.S.A. 105, 10541–10546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlton J. G., and Martin-Serrano J. (2007) Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316, 1908–1912 [DOI] [PubMed] [Google Scholar]
- 6.Morita E., Sandrin V., McCullough J., Katsuyama A., Baci Hamilton I., and Sundquist W. I. (2011) ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe 9, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stuchell M. D., Garrus J. E., Müller B., Stray K. M., Ghaffarian S., McKinnon R., Krusslich H. G., Morham S. G., and Sundquist W. I. (2004) The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. J. Biol. Chem. 279, 36059–36071 [DOI] [PubMed] [Google Scholar]
- 8.Agromayor M., Carlton J. G., Phelan J. P., Matthews D. R., Carlin L. M., Ameer-Beg S., Bowers K., and Martin-Serrano J. (2009) Essential role of hIST1 in cytokinesis. Mol. Biol. Cell 20, 1374–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bajorek M., Morita E., Skalicky J. J., Morham S. G., Babst M., and Sundquist W. I. (2009) Biochemical analyses of human IST1 and its function in cytokinesis. Mol. Biol. Cell 20, 1360–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guizetti J., Schermelleh L., Mäntler J., Maar S., Poser I., Leonhardt H., Müller-Reichert T., and Gerlich D. W. (2011) Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science 331, 1616–1620 [DOI] [PubMed] [Google Scholar]
- 11.Morita E., Sandrin V., Chung H. Y., Morham S. G., Gygi S. P., Rodesch C. K., and Sundquist W. I. (2007) Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26, 4215–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCullough J., Colf L. A., and Sundquist W. I. (2013) Membrane fission reactions of the mammalian ESCRT pathway. Annu. Rev. Biochem. 82, 663–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cashikar A. G., Shim S., Roth R., Maldazys M. R., Heuser J. E., and Hanson P. I. (2014) Structure of cellular ESCRT-III spirals and their relationship to HIV budding. eLife 3, e02184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabrikant G., Lata S., Riches J. D., Briggs J. A., Weissenhorn W., and Kozlov M. M. (2009) Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput. Biol. 5, e1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanson P. I., Roth R., Lin Y., and Heuser J. E. (2008) Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 180, 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Im Y. J., Wollert T., Boura E., and Hurley J. H. (2009) Structure and function of the ESCRT-II-III interface in multivesicular body biogenesis. Dev. Cell 17, 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saksena S., Wahlman J., Teis D., Johnson A. E., and Emr S. D. (2009) Functional reconstitution of ESCRT-III assembly and disassembly. Cell 136, 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wollert T., and Hurley J. H. (2010) Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464, 864–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wollert T., Wunder C., Lippincott-Schwartz J., and Hurley J. H. (2009) Membrane scission by the ESCRT-III complex. Nature 458, 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azmi I. F., Davies B. A., Xiao J., Babst M., Xu Z., and Katzmann D. J. (2008) ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev. Cell 14, 50–61 [DOI] [PubMed] [Google Scholar]
- 21.Babst M., Katzmann D. J., Estepa-Sabal E. J., Meerloo T., and Emr S. D. (2002) ESCRT-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev. Cell 3, 271–282 [DOI] [PubMed] [Google Scholar]
- 22.Babst M., Katzmann D. J., Snyder W. B., Wendland B., and Emr S. D. (2002) Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3, 283–289 [DOI] [PubMed] [Google Scholar]
- 23.Dimaano C., Jones C. B., Hanono A., Curtiss M., and Babst M. (2008) Ist1 regulates Vps4 localization and assembly. Mol. Biol. Cell 19, 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nickerson D. P., West M., Henry R., and Odorizzi G. (2010) Regulators of Vps4 ATPase activity at endosomes differentially influence the size and rate of formation of intralumenal vesicles. Mol. Biol. Cell 21, 1023–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nickerson D. P., West M., and Odorizzi G. (2006) Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J. Cell Biol. 175, 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rue S. M., Mattei S., Saksena S., and Emr S. D. (2008) Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol. Biol. Cell 19, 475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teis D., Saksena S., and Emr S. D. (2008) Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell 15, 578–589 [DOI] [PubMed] [Google Scholar]
- 28.Adell M. A., and Teis D. (2011) Assembly and disassembly of the ESCRT-III membrane scission complex. FEBS Lett. 585, 3191–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Babst M., Sato T. K., Banta L. M., and Emr S. D. (1997) Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16, 1820–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Babst M., Wendland B., Estepa E. J., and Emr S. D. (1998) The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17, 2982–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies B. A., Azmi I. F., Payne J., Shestakova A., Horazdovsky B. F., Babst M., and Katzmann D. J. (2010) Coordination of substrate binding and ATP hydrolysis in Vps4-mediated ESCRT-III disassembly. Mol. Biol. Cell 21, 3396–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo E. Z., and Xu Z. (2015) Distinct mechanisms of recognizing endosomal sorting complex required for transport III (ESCRT-III) protein IST1 by different microtubule interacting and trafficking (MIT) domains. J. Biol. Chem. 290, 8396–8408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kieffer C., Skalicky J. J., Morita E., De Domenico I., Ward D. M., Kaplan J., and Sundquist W. I. (2008) Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev. Cell 15, 62–73 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 34.Obita T., Saksena S., Ghazi-Tabatabai S., Gill D. J., Perisic O., Emr S. D., and Williams R. L. (2007) Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature 449, 735–739 [DOI] [PubMed] [Google Scholar]
- 35.Scott A., Gaspar J., Stuchell-Brereton M. D., Alam S. L., Skalicky J. J., and Sundquist W. I. (2005) Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc. Natl. Acad. Sci. U.S.A. 102, 13813–13818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stuchell-Brereton M. D., Skalicky J. J., Kieffer C., Karren M. A., Ghaffarian S., and Sundquist W. I. (2007) ESCRT-III recognition by VPS4 ATPases. Nature 449, 740–744 [DOI] [PubMed] [Google Scholar]
- 37.Hurley J. H., and Yang D. (2008) MIT domainia. Dev. Cell 14, 6–8 [DOI] [PubMed] [Google Scholar]
- 38.Merrill S. A., and Hanson P. I. (2010) Activation of human VPS4A by ESCRT-III proteins reveals ability of substrates to relieve enzyme autoinhibition. J. Biol. Chem. 285, 35428–35438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shestakova A., Curtiss M., Davies B. A., Katzmann D. J., and Babst M. (2013) The linker region plays a regulatory role in assembly and activity of the Vps4 AAA ATPase. J. Biol. Chem. 288, 26810–26819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han H., Monroe N., Votteler J., Shakya B., Sundquist W. I., and Hill C. P. (2015) Binding of substrates to the central pore of the Vps4 ATPase is autoinhibited by the microtubule interacting and trafficking (MIT) domain and activated by MIT interacting motifs (MIMs). J. Biol. Chem. 290, 13490–13499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landsberg M. J., Vajjhala P. R., Rothnagel R., Munn A. L., and Hankamer B. (2009) Three-dimensional structure of AAA ATPase Vps4: advancing structural insights into the mechanisms of endosomal sorting and enveloped virus budding. Structure 17, 427–437 [DOI] [PubMed] [Google Scholar]
- 42.Monroe N., Han H., Gonciarz M. D., Eckert D. M., Karren M. A., Whitby F. G., Sundquist W. I., and Hill C. P. (2014) The oligomeric state of the active Vps4 AAA ATPase. J. Mol. Biol. 426, 510–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott A., Chung H. Y., Gonciarz-Swiatek M., Hill G. C., Whitby F. G., Gaspar J., Holton J. M., Viswanathan R., Ghaffarian S., Hill C. P., and Sundquist W. I. (2005) Structural and mechanistic studies of VPS4 proteins. EMBO J. 24, 3658–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao J., Xia H., Yoshino-Koh K., Zhou J., and Xu Z. (2007) Structural characterization of the ATPase reaction cycle of endosomal AAA protein Vps4. J. Mol. Biol. 374, 655–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bajorek M., Schubert H. L., McCullough J., Langelier C., Eckert D. M., Stubblefield W. M., Uter N. T., Myszka D. G., Hill C. P., and Sundquist W. I. (2009) Structural basis for ESCRT-III protein autoinhibition. Nat. Struct. Mol. Biol. 16, 754–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lata S., Roessle M., Solomons J., Jamin M., Gottlinger H. G., Svergun D. I., and Weissenhorn W. (2008) Structural basis for autoinhibition of ESCRT-III CHMP3. J. Mol. Biol. 378, 818–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muzioł T., Pineda-Molina E., Ravelli R. B., Zamborlini A., Usami Y., Göttlinger H., and Weissenhorn W. (2006) Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell 10, 821–830 [DOI] [PubMed] [Google Scholar]
- 48.Shim S., Kimpler L. A., and Hanson P. I. (2007) Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic 8, 1068–1079 [DOI] [PubMed] [Google Scholar]
- 49.Zamborlini A., Usami Y., Radoshitzky S. R., Popova E., Palu G., and Göttlinger H. (2006) Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc. Natl. Acad. Sci. U.S.A. 103, 19140–19145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Azmi I., Davies B., Dimaano C., Payne J., Eckert D., Babst M., and Katzmann D. J. (2006) Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J. Cell Biol. 172, 705–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davies B. A., Norgan A. P., Payne J. A., Schulz M. E., Nichols M. D., Tan J. A., Xu Z., and Katzmann D. J. (2014) Vps4 stimulatory element of the cofactor Vta1 contacts the ATPase Vps4 α7 and α9 to stimulate ATP hydrolysis. J. Biol. Chem. 289, 28707–28718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lottridge J. M., Flannery A. R., Vincelli J. L., and Stevens T. H. (2006) Vta1p and Vps46p regulate the membrane association and ATPase activity of Vps4p at the yeast multivesicular body. Proc. Natl. Acad. Sci. U.S.A. 103, 6202–6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Norgan A. P., Davies B. A., Azmi I. F., Schroeder A. S., Payne J. A., Lynch G. M., Xu Z., and Katzmann D. J. (2013) Relief of autoinhibition enhances Vta1 activation of Vps4 via the Vps4 stimulatory element. J. Biol. Chem. 288, 26147–26156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang B., Stjepanovic G., Shen Q., Martin A., and Hurley J. H. (2015) Vps4 disassembles an ESCRT-III filament by global unfolding and processive translocation. Nat. Struct. Mol. Biol. 22, 492–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cordova J. C., Olivares A. O., Shin Y., Stinson B. M., Calmat S., Schmitz K. R., Aubin-Tam M. E., Baker T. A., Lang M. J., and Sauer R. T. (2014) Stochastic but highly coordinated protein unfolding and translocation by the ClpXP proteolytic machine. Cell 158, 647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin A., Baker T. A., and Sauer R. T. (2008) Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat. Struct. Mol. Biol. 15, 1147–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weber-Ban E. U., Reid B. G., Miranker A. D., and Horwich A. L. (1999) Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature 401, 90–93 [DOI] [PubMed] [Google Scholar]
- 58.Baker T. A., and Sauer R. T. (2012) ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim. Biophys. Acta 1823, 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maurizi M. R., and Stan G. (2013) ClpX shifts into high gear to unfold stable proteins. Cell 155, 502–504 [DOI] [PubMed] [Google Scholar]
- 60.Xiao J., Chen X. W., Davies B. A., Saltiel A. R., Katzmann D. J., and Xu Z. (2009) Structural basis of Ist1 function and Ist1-Did2 interaction in the multivesicular body pathway and cytokinesis. Mol. Biol. Cell 20, 3514–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simons R. W., Houman F., and Kleckner N. (1987) Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53, 85–96 [DOI] [PubMed] [Google Scholar]
- 62.Robinson J. S., Klionsky D. J., Banta L. M., and Emr S. D. (1988) Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell Biol. 8, 4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gerhardt B., Kordas T. J., Thompson C. M., Patel P., and Vida T. (1998) The vesicle transport protein Vps33p is an ATP-binding protein that localizes to the cytosol in an energy-dependent manner. J. Biol. Chem. 273, 15818–15829 [DOI] [PubMed] [Google Scholar]
- 64.Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 65.Shestakova A., Hanono A., Drosner S., Curtiss M., Davies B. A., Katzmann D. J., and Babst M. (2010) Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol. Biol. Cell 21, 1059–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jones C. B., Ott E. M., Keener J. M., Curtiss M., Sandrin V., and Babst M. (2012) Regulation of membrane protein degradation by starvation-response pathways. Traffic 13, 468–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Allison R., Lumb J. H., Fassier C., Connell J. W., Ten Martin D., Seaman M. N., Hazan J., and Reid E. (2013) An ESCRT-spastin interaction promotes fission of recycling tubules from the endosome. J. Cell Biol. 202, 527–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reid E., Connell J., Edwards T. L., Duley S., Brown S. E., and Sanderson C. M. (2005) The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B. Hum. Mol. Genet. 14, 19–38 [DOI] [PubMed] [Google Scholar]
- 69.Renvoisé B., Parker R. L., Yang D., Bakowska J. C., Hurley J. H., and Blackstone C. (2010) SPG20 protein spartin is recruited to midbodies by ESCRT-III protein Ist1 and participates in cytokinesis. Mol. Biol. Cell 21, 3293–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vietri M., Schink K. O., Campsteijn C., Wegner C. S., Schultz S. W., Christ L., Thoresen S. B., Brech A., Raiborg C., and Stenmark H. (2015) Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature 522, 231–235 [DOI] [PubMed] [Google Scholar]
- 71.Yang D., Rismanchi N., Renvoisé B., Lippincott-Schwartz J., Blackstone C., and Hurley J. H. (2008) Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat. Struct. Mol. Biol. 15, 1278–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lumb J. H., Connell J. W., Allison R., and Reid E. (2012) The AAA ATPase spastin links microtubule severing to membrane modelling. Biochim. Biophys. Acta 1823, 192–197 [DOI] [PubMed] [Google Scholar]