Abstract
PUF proteins are a conserved group of sequence specific RNA-binding proteins that bind to RNA in a modular fashion. The RNA-binding domain of PUF proteins typically consists of eight clustered Puf repeats. Plant genomes code for large families of PUF proteins that show significant variability in their predicted Puf repeat number, organization, and amino acid sequence. Here we sought to determine whether the observed variability in the RNA-binding domains of four plant PUFs results in a preference for nonclassical PUF RNA target sequences. We report the identification of a novel RNA binding sequence for a nucleolar Arabidopsis PUF protein that contains an atypical RNA-binding domain. The Arabidopsis PUM23 (APUM23) binding sequence was 10 nucleotides in length, contained a centrally located UUGA core element, and had a preferred cytosine at nucleotide position 8. These RNA sequence characteristics differ from those of other PUF proteins, because all natural PUFs studied to date bind to RNAs that contain a conserved UGU sequence at their 5′ end and lack specificity for cytosine. Gel mobility shift assays validated the identity of the APUM23 binding sequence and supported the location of 3 of the 10 predicted Puf repeats in APUM23, including the cytosine-binding repeat. The preferred 10-nucleotide sequence bound by APUM23 is present within the 18S rRNA sequence, supporting the known role of APUM23 in 18S rRNA maturation. This work also reveals that APUM23, an ortholog of yeast Nop9, could provide an advanced structural backbone for Puf repeat engineering and target-specific regulation of cellular RNAs.
Keywords: Arabidopsis, post-transcriptional regulation, RNA, RNA binding protein, RNA metabolism, APUM23, PUF protein, Protein-RNA interaction
Introduction
PUF (Pumilio) proteins are a conserved family of eukaryotic RNA-binding proteins that have important roles in post-transcriptional regulation of gene expression. PUF proteins bind to specific nucleotide sequences that are typically located in the 3′-untranslated region (UTR) of mRNAs, and function at the molecular level to regulate the stability and translation of mRNAs (1). The RNA-binding domain of classical PUF proteins, the Pumilio homology domain (PUM-HD),2 folds into a crescent-shaped structure that contains eight tandem Puf repeats each consisting of three α-helices. The second α-helix of each Puf repeat binds in a modular fashion to a single nucleotide on the inner concave surface of the PUM-HD. PUF proteins typically recognize eight nucleotides in their target RNA sequence, where the first four nucleotides are UGUR (where R is a purine) (2). The binding of the PUM-HD to RNAs is anti-parallel, with RNA bases 1 through 8 sequentially interacting with Puf repeats 8 through 1. There are, however, some atypical interactions between PUF proteins and their RNA targets. For example, the eight Puf repeats of Caenorhabditis elegans FBF-2 and yeast Puf4 bind to nine-nucleotide RNA targets and do so by flipping away one nucleotide from the RNA binding surface (3, 4). Yeast Puf3 possesses a binding pocket between Puf repeats 8 and 8′ (a pseudo-Puf repeat) that accommodates a cytosine located upstream of the UGUR core at the −2 position (5).
Three key amino acids (the tripartite recognition motif (TRM)) lie within a five-amino acid motif in the second α-helix of each Puf repeat (12XX5, where X is any hydrophobic amino acid). The TRM (positions 1, 2, and 5) provides nucleotide binding specificity toward the target RNA (6). Amino acids at positions 1 and 5 can contact the edge of the base through hydrogen bonding and/or van der Waals forces, whereas the amino acid at position 2 is involved in a stacking interaction with two adjacent bases. In the original RNA recognition code for Puf repeats, cysteine and glutamine at positions 1 and 5 bind adenine, whereas asparagine and glutamine at these positions bind uracil, and serine and glutamate recognize guanine (7). Puf repeats that naturally recognize cytosine have not yet been identified. However, cytosine-binding repeats have been engineered using a yeast three-hybrid approach and demonstrated that Puf repeats with serine and arginine at positions 1 and 5, respectively, bind specifically to cytosine (8, 9). Sequence database searches of annotated PUF sequences that contain arginine and serine at these positions indicate that natural cytosine binding Puf repeats likely exist (9). This TRM specificity scheme is not entirely simplistic, because positional and PUF-specific nucleotide binding preferences complicate the prediction of binding sites. A detailed study of PUF binding characteristics has provided an expanded repertoire of binding specificities that will be valuable for future engineering of PUF proteins customized for RNA target-specific regulation (6, 10). These engineered PUFs have already been shown to regulate the stability, localization, translation, and processing of their target mRNAs in vivo (11).
Only a small number of studies have been reported on the characterization of plant PUF proteins. The Arabidopsis thaliana PUF family is extensive, consisting of up to 26 members (12–14). These proteins demonstrate considerable variability in their number of Puf repeats, the position of these repeats in the primary sequence, and the identity of their putative TRMs. Over half of the Arabidopsis PUFs are predicted to possess eight tandemly arranged Puf repeats. Six of these proteins (APUM1 through APUM6) are conserved in the amino acids that comprise the TRM in each Puf repeat when compared with the TRMs in human PUMILIO1 (PUM1). The remaining PUF proteins are predicted to contain 3–10 Puf repeats, some of which contain large gaps between individual repeats (12–14). APUM1 through APUM6 were shown to bind the typical UGUR-containing Nanos response element that is bound by Drosophila PUMILIO (12, 13). Functional data for only four Arabidopsis PUF proteins have been reported (14–18). A mutated form of one of these proteins (APUM23) displayed shoot and root developmental abnormalities, defects in preribosomal RNA processing, and disrupted auxin homeostasis (14, 18). In vivo expression of APUM23 and APUM24 (orthologs of yeast Nop9 and Puf6p, respectively) as fusions to fluorescent proteins indicated that these proteins localize to nucleoli (13, 14). APUM23 deviates from the typical tandem eight Puf repeat arrangement in that it is predicted to possess 10 Puf repeats that are spread throughout the central region of the protein rather than clustered at the C-terminal region (13).
Here we report the RNA consensus target sequences of four APUM proteins using systematic evolution of ligands by exponential enrichment (SELEX) (19). Three of the APUM proteins preferred classical PUF RNA targets that were eight or nine nucleotides in length. The fourth, APUM23, bound specifically to a novel RNA target that was 10 nucleotides in length and possessed a noncanonical nucleotide sequence core, and it had a binding preference for cytosine at the eighth nucleotide position. This 10-nucleotide APUM23 target sequence is present in the 18S rRNA sequence, thereby supporting the known functional role of APUM23 in 18S rRNA processing. These results indicate that APUM23 provides a unique sequence-specific RNA-binding protein scaffold that could be engineered to modulate the processing, stability, translation, and localization of cellular target mRNAs.
Experimental Procedures
Protein Expression and Purification
The PUM-HDs of APUM2 (At2g29190, amino acids 614–963), APUM6 (At4g25880, amino acids 496–861), and APUM12 (At5g56510, amino acids 258–596), and full-length APUM23 (At1g72320, 731 amino acids using the second translation start site) were PCR-amplified from Arabidopsis thaliana leaf cDNA using primers listed in supplemental Table S1. Nop9 (NM_001181444.3, 666 amino acids) was PCR-amplified from Saccharomyces cerevisiae genomic DNA using primers listed in supplemental Table S1. PCR products were cloned into the BamHI/SalI sites of pGEX-6p-1 (GE Healthcare), resulting in a translational fusion to the C terminus of the GST tag. Escherichia coli (BL21(DE3)) expression of APUM and Nop9 proteins was induced by 0.05 mm isopropyl β-d-thiogalactopyranoside at 21 °C. Cells were pelleted and resuspended in lysis buffer (25 mm Tris-HCl, pH 8.0, 350 mm NaCl and 3 mm DTT) with the addition of 1 mg/ml lysozyme, 0.17 mg/ml PMSF, and protease inhibitor (Complete EDTA-free; Roche). The supernatant fraction was passed through a gravity column containing glutathione-Sepharose 4B resin (GE Healthcare). The resin was immediately washed four times with 10 column volumes of wash buffer (25 mm Tris-HCl, pH 8.0, 500 mm NaCl and 3 mm DTT) before elution with elution buffer (25 mm Tris-HCl, pH 8.0, 250 mm NaCl, 30 mm reduced l-glutathione, and 3 mm DTT). These conditions were effective for the purification of sufficient protein quantity and quality (greater than 90% purity) for use in the SELEX analysis. Recombinant protein was further purified for use in most of the mobility shift assays by size exclusion chromatography using a Superdex 200 Increase 10/300 GL size exclusion column (GE Healthcare) attached to an FPLC (AKTA; GE Healthcare). The running buffer contained 10 mm Tris-HCl, pH 8.0, and 250 mm NaCl. Purified protein fractions were pooled and used fresh or stored at −80 °C after the addition of glycerol to 20% (v/v).
The five-amino acid motif substitutions in APUM23 were constructed by removing the appropriate segment of the wild-type coding region and replacing it with a mutated coding region (synthesized by GenScript) in the GST-APUM23 translational fusion construct. Recombinant protein purification was performed as described for the wild-type proteins.
SELEX
SELEX was performed similarly to that described previously for the murine PUM2 protein (20) with some modifications. A degenerate DNA oligonucleotide (5′-GGGATCCGAATTCCCGACT(N)20GGAAGCTTACTCGAGCGC-3′) was used to produce the starting random pool of RNA that had a complexity of 1 × 1012. 20 pmol of this oligonucleotide provided ∼12-fold coverage of each unique sequence combination. PCR amplification of this oligonucleotide was achieved using SELEX primers. The forward SELEX primer (5′-GGACCTAATACGACTCACTATAGGGATCCGAATTCCCGACT-3′) contained the T7 RNA polymerase binding site (bold) and was paired with the reverse SELEX primer (5′-GCGCTCGAGTAAGCTTCC-3′). After seven cycles of amplification (94 °C for 35 s, 55 °C for 35 s, and 68 °C for 45 s) using Platinum Pfx polymerase (Invitrogen) and a final extension of 2 min at 72 °C, the dsDNA was electrophoresed on 3% agarose gel, and the 79 bp fragment was eluted. RNA pools were transcribed from 1 μg of purified dsDNA template using 100 units of T7 RNA Polymerase-Plus (Ambion) in a reaction containing 2 mm NTPs and 33 nm [α-32P]UTP (3000 Ci/mmol; PerkinElmer Life Sciences) for 2 h at 37 °C, followed by DNase (TURBO; Ambion) digestion at 37 °C for 1 h. RNA was isolated (TRIzol; Invitrogen), dissolved in reaction buffer (10 mm HEPES, pH 7.4, 1 mm EDTA, 50 mm KCl, 1 mm DTT, 0.01% (w/v) BSA, and 0.01% (v/v) Tween 20), and then passed through an equilibrated spin column (Bio-Gel P-6; Bio-Rad). A parallel nonradioactive trial was performed to provide an RNA concentration reference, which was quantified by spectrophotometry (model ND-1000; NanoDrop).
For each round of selection, RNAs were precleared in 20 μl of GST-coupled glutathione-Sepharose 4B resin to reduce nonspecific RNA binding prior to RNA enrichment. 2 μl of precleared RNA was subjected to isotope counting using a scintillation counter (Beckman). 2 nmol of GST-tagged PUM protein and 0.4 nmol of precleared RNA were mixed in 100 μl of reaction buffer containing 50 units of RNase Inhibitor (SUPERase·In; Ambion). This 5:1 molar ratio of protein:RNA was necessary to initiate enrichment of the RNA in the first two rounds of SELEX, because the APUM proteins did not possess full activity. In the third and fourth rounds, the protein:RNA molar ratio added was equal, and in the fifth and subsequent rounds, the ratio was 1:5. The RNA enrichment profiles were normalized according to these ratios. After a 30-min incubation at room temperature, the RNA-protein complexes were mixed with 20 μl of glutathione-Sepharose 4B resin for 15 min at room temperature, with gentle resuspension of the matrix every 5 min. The resin was washed three times in 400 μl of reaction buffer followed by elution of the protein-RNA complexes with 200 μl of elution buffer (25 mm Tris-HCl, pH 8.0, 200 mm NaCl, 30 mm reduced glutathione, and 3 mm DTT). Bound RNA was released using 300 μl of TRIzol followed by chloroform extraction and ethanol precipitation. The RNA pellet was dissolved in 19 μl of DEPC-treated water. 2 μl was used for scintillation counting to calculate the percentage of RNA bound. The remaining RNA was reverse-transcribed using SuperScript III (Invitrogen) and 1.2 μm SELEX reverse primer in a 40-μl reaction, and the cDNA product was precipitated by ethanol. The PCR amplifications from cDNA templates for each SELEX cycle were performed in 200-μl reactions with 1.875 μm SELEX forward and reverse primers, 1 mm dNTPs, 1 mm MgCl2, and 4 units of Platinum Pfx polymerase for 10–12 thermal cycles (94 °C for 35 s, 55 °C for 35 s, and 68 °C for 30 s) and a 3-min final extension at 68 °C. The resulting dsDNA was recovered by gel elution and used for the subsequent transcription. This SELEX cycle was repeated 9–12 times until no further enrichment was observed. The final dsDNA was digested with EcoRI and HindIII and cloned into pBlueScript for Sanger sequencing. Only unique and reliable sequences from independent transformants were used for analysis using MEME (21) to obtain the consensus sequence, logo graph, and the E value. The number of sequenced clones that were used for MEME analysis were: 52 for APUM2, 60 for APUM6, 38 for APUM12, 112 for APUM23, and 49 for Nop9. Default MEME settings were used, with the width of the consensus sequence set at 4–20 nucleotides. The refined logo graphs were generated by WebLogo graph tool using the populations validated by MEME.
Electrophoretic Mobility Shift Assays
EMSAs were performed as described previously (13), with minor modifications. Recombinant APUM proteins were reconstituted in binding buffer (10 mm HEPES, pH 7.4, 1 mm EDTA, 50 mm KCl, 1 mm DTT, 0.01% Tween 20). Synthetic RNAs (Dharmacon) were radiolabeled using [γ-32P]ATP (3000 Ci/mmol; PerkinElmer Life Sciences) and T4 polynucleotide kinase (Thermo Scientific). Binding reactions were in a 20-μl volume containing 0.01 or 0.05 nm labeled RNA and 0.03–1024 nm range of gradient protein concentrations. The reactions were incubated at room temperature for 30 min and electrophoresed on a 6% nondenaturing acrylamide gel at 96 V for 20 min at 4 °C. The gels were dried and exposed to a storage phosphor screen (Kodak), and the screen was scanned using a PhosphorImager (Molecular Imager FX; Bio-Rad). Densitometry was performed using Quantity One software (version 4.5.1; Bio-Rad) and ImageJ (1.47v; Wayne Rasband, National Institutes of Health), and the data were analyzed using Prism 5 (version 5.03, GraphPad). To determine the apparent dissociation constant and binding curve, the fractions of RNA bound to protein (the relative pixel intensity in the complex band divided by the sum of the pixel intensities in the complex band plus the free RNA band) were plotted against protein concentrations in a semi-log graph format. The curves were fitted in the equation of “One site - specific binding” defined by the Prism 5 software. The average apparent dissociation constant values (± S.E.) were derived from three or four trials, depending on the particular experiment.
The activity of the recombinant proteins was determined by EMSA using an excess amount of RNA to saturate the protein binding capacity. 50 nm of the cognate radiolabeled RNA was incubated with 5, 2, and 1 nm of protein in a 20-μl volume, and electrophoresed alongside 4, 0.6, and 0.1 nm free RNA as a concentration standard. The absolute amount of RNA bound by protein of different concentrations was determined by comparing the pixel intensity of the complex band with the free RNA bands in the concentration standard. RNA concentration was plotted on a graph against protein concentration, and the slope of the line was measured to determine the percentage of active protein molecules.
Structural Modeling of APUM23
The SWISS-MODEL protein structure homology server (22) was used to produce APUM23 models that guided the mapping of predicted Puf repeat locations and TRM identity, as described in the text. The classical PUF protein structure consisting of α-helical repeats and precisely positioned TRM amino acids provided the template for these models. Partial models were produced by analyzing two overlapping regions of the APUM23 polypeptide: amino acids 1–382 and 221–731. 94 and 67 templates were identified for these two sequences, respectively. The top 20 templates that were most similar in sequence to APUM23 were modeled for each region. Models that demonstrated the typical PUF concave structure, Puf repeat α-helical structure, and TRM positioning were used for repeat and TRM predictions.
Results
Prediction of Puf Repeats in the Selected APUM Proteins
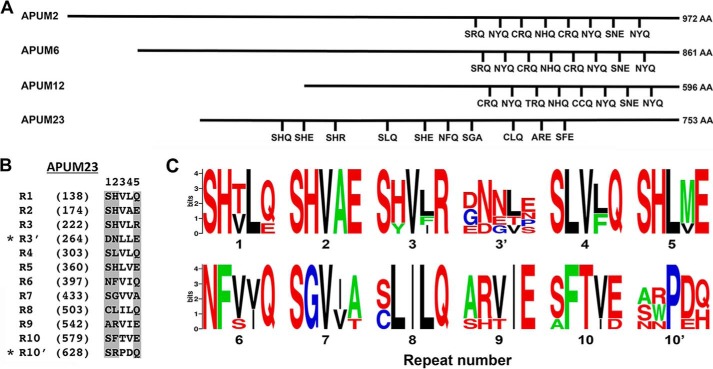
Selection of the APUM proteins that were analyzed in this study (APUM2, 6, 12, and 23) was based on the conservation or variability in the number and position of their predicted Puf repeats and on the TRM composition of their repeats (Fig. 1A). APUM2 and APUM6 are predicted to possess a PUM-HD with the characteristic eight tandem Puf repeats that are tightly clustered at the C-terminal region of the proteins, and they share identical TRMs with human PUM1. APUM12 also appears to contain eight Puf repeats, although the TRMs in repeats 1, 3, and 5 differ from those in human PUM1. We predict that APUM23 possesses 10 Puf repeats that are not clustered within the C-terminal region but rather are unevenly positioned throughout the central region of the protein (Fig. 1A). Our prediction for the number and location of the APUM23 Puf repeats and the identity of their TRMs was based on three lines of evidence. First was their sequence identity to TRMs that are common in Puf repeats from other PUF proteins. Second was the positioning of the predicted TRMs in α-helical regions in a three-dimensional model of APUM23. Third was the level of conservation of the five-amino acid motif that contains the TRM sequence in plant orthologs of APUM23. Modeling the three-dimensional structure of the entire APUM23 protein sequence (see “Experimental Procedures”), when referenced against the crystal structure of human PUM1, resulted in a distorted structure in a portion of the model. Therefore, two overlapping partial models were obtained that together modeled the entire protein. Twelve putative Puf repeats were identified in these models (Figs. 1B and 2), many of which possessed previously identified TRMs located within the α-helix on the inner, concave face of the protein that contacts RNA (6, 7). The position of these TRMs on each of the modeled repeats were oriented in a typical Puf repeat fashion, with the TRMs (amino acids at positions 1, 2, and 5) exposed to the outer portion of the second α-helix and the hydrophobic amino acids (positions 3 and 4) hidden on the inside of the helix. Repeat 3′ was considered an unlikely Puf repeat candidate because it possesses an unusual predicted TRM (DNE) that is not conserved at this position in other species, including other members of the dicotyledonous class of plants (Figs. 1C and supplemental Fig. S1). This repeat may be positioned away from the RNA binding surface in vivo, allowing repeats 3 and 4 to bind to their adjacent target bases. Putative Puf repeat 10′ possesses a TRM (SRQ) (Fig. 1B) that is present in other PUF proteins (6). However, the presence of the 10′ repeat at this position is not highly conserved within other dicotyledonous plants (Figs. 1C and supplemental Fig. S1), and the combination of proline and aspartic acid at the third and fourth position (Fig. 1B) of its predicted five-amino acid motif is atypical. Aside from repeats 3′ and 10′, the remaining repeats displayed high conservation in APUM23 orthologs from other species (Figs. 1C and supplemental Fig. S1).
FIGURE 1.
Predicted location and sequence of TRMs from the APUM proteins analyzed by SELEX. A, location map of the TRMs within each of the predicted Puf repeats in the four APUM proteins used in this study. The three-letter TRMs are shown. The length of each protein (in amino acids, AA) is shown at the right of each line. B, the five-amino acid sequence motifs from each of the predicted APUM23 Puf repeats are shown. The numbers in parentheses refer to the position of the first amino acid from each motif in the APUM23 amino acid sequence. Shaded letters identify the amino acids at motif positions 1, 2, and 5 that are predicted to provide nucleotide contacts. Repeats 3′ and 10′ (*) are repeats that were identified in the APUM23 model as possible Puf repeats (Fig. 2) but do not possess TRMs that are conserved within dicotyledonous plants and thus are not considered to be nucleotide-interacting Puf repeats. C, conservation of the five-amino acid motifs in the predicted Puf repeats. Logo graphs of the five-amino acid motifs involved in nucleotide binding from the predicted Puf repeats of APUM23 orthologs consisting of five dicotyledonous plant species shown in the alignment in supplemental Fig. S1 (Arabidopsis lyrata, Brassica rapa, Populus euphratica, Vitus venifera, and Phaseolus vulgaris). The first, second, and fifth amino acids in each logo graph correspond to the predicted TRMs.
FIGURE 2.

Models of APUM23 referenced against the structure of human PUM1. The APUM23 amino acid sequence was split into two overlapping parts, and each part was modeled independently using the Swiss-Model server. A and B, ribbon diagrams of the N-terminal region of APUM23 (amino acids 1–382) comprising putative Puf repeats 1, 2, 3, 3′, 4, and 5 (A) and the C-terminal region (amino acids 221–753) putative repeats 4, 5, 6, 7, 8, 9, 10, and 10′ (B) are shown. Based on their location in the second helix, the amino acids in the putative TRM (i.e. amino acids 1, 2, and 5 of the five-amino acid RNA binding motif on the inner facing helix) are identified. The putative repeats are numbered with their corresponding TRM (red). The three-digit numbers (black) refer to the linear position of the specific amino acid (single-letter amino acid code) within the protein. Blue lines point to individual amino acids on the inner facing α-helix.
SELEX Identifies a Novel APUM RNA Target Sequence
SELEX was performed to determine a consensus RNA binding sequence for the four APUM proteins selected for this study. Between nine and twelve rounds of in vitro selection were carried out for each recombinant APUM protein (Fig. 3). The round of selection that demonstrated the highest percentage of RNA binding was used for PCR amplification, cloning, and sequencing. The consensus RNA binding sequences that were obtained for each APUM protein were assembled into logo graphs (Fig. 4; see “Experimental Procedures” for details on consensus sequence determination and logo graph assembly). The APUM2 and APUM6 consensus sequences consisted of eight nucleotides, whereas the APUM12 and APUM23 sequences consisted of nine and ten nucleotides, respectively. A 5′ UGUA core was present in the APUM2, 6, and 12 RNA sequences. Subtle degeneracy was observed at nucleotide position 4 in the sequences of APUM2 and APUM6, whereas a higher degree of degeneracy was observed at position 5 in these sequences. Degeneracy at position 5 is common for many other PUF protein targets (23). APUM12 deviated from the classical one repeat-one nucleotide pattern for PUF binding, because it showed an insertion of an additional uracil near the 3′ end of its consensus sequence when compared with the target sequences of APUM2 and APUM6 (Fig. 4). The APUM23 consensus sequence of 10 nucleotides matched the predicted number of TRMs in this protein (Fig. 4D and supplemental Table S2). This RNA target lacked the typical 5′ UGUA core, but rather contained a conserved four nucleotide “core” (UUGA) that was located centrally in the consensus sequence. It also preferred cytosine at nucleotide position 8 and showed strong binding to guanine at positions 1, 9, and 10. All four APUM proteins showed high affinity binding to their cognate RNA probes in EMSAs, with apparent dissociation constants ranging from 0.113 to 2.16 nm (Fig. 4).
FIGURE 3.

RNA enrichment profiles from the SELEX experiments. SELEX assays were performed using the PUM-HDs of APUM 2, 6, and 12, and the complete APUM23 protein. The percentage of RNA that bound to the APUM-coupled matrix was determined after each cycle of enrichment. DNA resulting from the cycle that had the highest percentage of bound RNA was used for cloning and sequencing.
FIGURE 4.
SELEX consensus RNA target sequences for the APUM proteins. Logo graphs of the RNA SELEX products for APUM2 (A), APUM6 (B), APUM12 (C), and APUM23 (D) are shown. The numbers below the letters in the logo graph refer to the nucleotide position in the consensus sequence (5′ to 3′ direction). The E value of the consensus sequence is indicated in parentheses above the logo graph. The TRMs of the predicted Puf repeats are shown below their corresponding nucleotide. The uracil at nucleotide position 7 in the APUM12 logo graph (C) is not aligned with a Puf repeat, because this nucleotide is likely extruded from the RNA binding surface of the protein (see text). The Puf repeats are numbered in reverse, because the binding of Puf proteins to RNA is anti-parallel. Representative mobility shift assays of each APUM protein are shown below the corresponding logo graph. A GST sample served as a negative control. The lowest and highest concentration of protein in the exponential dilution series is indicated above each gel. Protein concentrations are corrected so that they reflect the concentration of active protein in each sample. The average apparent dissociation constant value (Kd) for the protein bound to the RNA consensus sequence is shown below each gel, as is the sequence of the cognate RNA used. The bottom right panel in D shows Coomassie Blue-stained lanes from SDS-PAGE gels that show GST-APUM23 protein that was purified using consecutive glutathione affinity chromatography alone (G) or both glutathione affinity chromatography and size exclusion chromatography steps (S). Gel marker lines are in kilodaltons.
Nucleotides in the RNA consensus sequences of each APUM protein were aligned with a predicted Puf repeat that was represented by its TRM (Fig. 4). The alignment of nucleotides with TRMs from APUM2 and APUM6 was of high confidence, because their TRMs and RNA consensus sequences are conserved with those of human PUM1. APUM12 is also quite conserved. However, the additional ninth nucleotide in its consensus RNA target could be attributable to the extrusion of one of the bases. Yeast Puf4 and APUM12 have identical TRMs and identical nine-nucleotide RNA consensus sequence. In the Puf4-RNA complex, the uracil at nucleotide position 7 is flipped away from the binding surface (3, 4). The conserved TRMs and target sequences of Puf4p suggest that a similar extrusion of uracil from the RNA-binding surface exists for APUM12, as is reflected in the TRM alignment shown in Fig. 4C. The alignment of the APUM23 Puf repeats with the nucleotides in its cognate RNA target is supported, in part, by the observation that cytosine at position 8 aligns with Puf repeat 3, a repeat that contains a predicted cytosine binding TRM (SHR; Fig. 4D) (8, 9). TRMs that are predicted to be natural cytosine binders are rare, and the cytosine binding preference by this TRM has only been demonstrated in an engineered PUF protein (9). When considering the base-specifying amino acids at positions 1 and 5 in each TRM, eight of the ten TRM:nucleotide pairing assignments for APUM23 (TRMs 1–6, 8, and 10; Fig. 4D) are conserved with previously observed nucleotide interactions for TRMs (6, 8, 9). The nucleotide targets of the remaining two Puf repeats (repeat 7, SGA; repeat 9, ARE) have not been characterized elsewhere. Consistent with its role in 18S rRNA processing, the preferred APUM23 RNA binding sequence (GAAUUGACGG) is present at nucleotide position 1142 in the 18S rRNA sequence, indicating a direct binding of APUM23 to this RNA.
The identification of an atypical RNA consensus sequence for APUM23 led us to determine whether an ortholog of APUM23 also showed specificity for the APUM23 RNA sequence. APUM23 and its S. cerevisiae ortholog (Nop9) are nucleolar proteins that have a conserved role in 18S rRNA maturation (14, 24). However, Nop9 shares only 20% amino acid identity (39% similarity) with APUM23 (supplemental Fig. S1), and only three of the predicted TRMs present in APUM23 are conserved in Nop9 (Fig. 5A). Nop9 showed very low binding affinity to the preferred 10-nucleotide APUM23 consensus sequence (Kd = 605 nm) (Fig. 5B). We then performed a Nop9 SELEX experiment to determine its sequence preference. SELEX enrichment of RNA binding achieved maximal levels after only the third round of selection (Fig. 5C). Sequence analysis did not identify any RNA consensus sequence motif for Nop9 (supplemental Table S3). This indicates that Nop9 lacks RNA sequence specificity in vitro, as was suggested previously (24).
FIGURE 5.

A, comparison of the S. cerevisiae Nop9 and APUM23 five-amino acid RNA-binding motifs from their predicted Puf repeats. The position of the first amino acid in each motif within the corresponding Puf repeat is indicated in parentheses. B, representative mobility shift gel for the Nop9 interaction with the preferred APUM23 RNA target sequence (23-WT RNA). The details are as described in Fig. 4. C, RNA enrichment profiles from the Nop9 SELEX experiment. The percentage of RNA that bound to the Nop9-coupled matrix was determined after each cycle of enrichment.
Validation of the APUM RNA Consensus Sequences
To validate the APUM RNA consensus sequences that were derived from the SELEX experiments, EMSAs using nucleotide-substituted RNAs were performed. RNAs consisting of 10 nucleotides were used in these assays, because this was the length of the longest consensus sequence. The sequence UGUAUAUA was used as the wild-type cognate RNA sequence (6-WT) for APUM2 and APUM6 (Fig. 4, A and B) and was flanked on either end by uracil (Table 1). The central eight nucleotides matched the preferred consensus RNA sequence for APUM6 (Fig. 4B). This sequence is present in the Nanos response element (NRE1) RNA that is bound by Drosophila PUMILIO (25). The cognate wild-type RNA for APUM12 (12-WT; Table 1) differed from the APUM6 wild-type RNA in that the last two nucleotides were swapped, matching the nine-nucleotide preferred consensus sequence for APUM12 (Fig. 4C).
TABLE 1.
Actual and relative apparent dissociation constants (Kd) of APUM2, APUM6, and APUM12 when bound to wild-type and nucleotide substituted RNA
| Protein | RNA | RNA sequence | Kd | Krela |
|---|---|---|---|---|
| nm | ||||
| APUM2 | 6-WT | UUGUAUAUAUb | 0.11 ± 0.01 | 1 |
| 6-U5A | UUGUAAAUAUc | 0.07 ± 0.01 | 0.64 | |
| 12-WT | UUGUAUAUUA | 0.39 ± 0.05 | 3.4 | |
| 6-G2U | UUUUAUAUAU | >74.0d | >654 | |
| APUM6 | 6-WT | UUGUAUAUAU | 2.16 ± 0.17 | 1 |
| 6-U5A | UUGUAAAUAU | 1.29 ± 0.23 | 0.60 | |
| 12-WT | UUGUAUAUUA | 23.4 ± 3.60 | 11 | |
| 6-G2U | UUUUAUAUAU | >155 | >71 | |
| APUM12 | 12-WT | UUGUAUAUUA | 1.49 ± 0.08 | 1 |
| 6-WT | UUGUAUAUAU | 6.30 ± 0.69 | 4.2 | |
| 6-U5A | UUGUAAAUAU | 13.8 ± 2.45 | 9.3 | |
| 6-G2U | UUUUAUAUAU | >318 | >213 |
a Krel values represent the relative affinity of the specific APUM protein to wild-type or mutant RNAs.
b The UGUA core of the wild-type RNA is highlighted with bold type. 6-WT and 12-WT refer to the SELEX consensus sequences for APUM6 and APUM12, respectively.
c Substituted nucleotides are underlined.
d The Kd value could not be determined because RNA binding was not observed for the maximal protein concentration used for this RNA-protein combination. Thus, the Kd value is shown as being greater than the highest concentration of protein used.
A substitution in the conserved UGUA core sequence (6-G2U) resulted in a large decrease in the affinity of APUM2, APUM6, and APUM 12 to this RNA, to the point that the apparent dissociation constant could not be determined with the amount of protein used in our assays (Table 1). Substitutions at positions outside of the core (positions 5 and 8) showed smaller changes in binding affinity. A substitution at the variable nucleotide position 5 (6-U5A) resulted in a small increase in affinity to APUM2 and APUM6 (Table 1), perhaps because of subtle differences in binding in the SELEX versus EMSA assays. The APUM12 consensus sequence showed a strong preference for UA at its final two positions (Fig. 4C). When these nucleotides were substituted with AU (i.e. the APUM6 wild-type probe, 6-WT) to conform to the typical 8-nucleotide PUF consensus sequence, there was a greater than 4-fold drop in the binding affinity of APUM12 to the modified RNA (Table 1). An additional substitution at position 5 (6-U5A) in the RNA reduced the binding affinity of APUM12 another 2.2-fold. Overall, APUM 2, 6, and 12 are typical PUF proteins in that there are eight tandem Puf repeats predicted from its amino acid sequence, and their target RNA possesses a conserved UGUA core. APUM12 differs in that it prefers 9 nucleotides rather than the typical 8 nucleotides.
Thirteen nucleotide-substituted RNAs were used in mobility shift experiments to validate the SELEX-derived consensus target sequence for APUM23. The binding affinity of APUM23 to these base-substituted RNAs showed a high correlation with the nucleotide composition of its RNA consensus sequence shown in Fig. 4D. Single-base substitutions within the UUGA core showed 14–31-fold decreases in APUM23 binding affinity compared with the wild-type RNA (Table 2). A double substitution in the UUGA core (UG56GU) that mimicked the classical UGUA core showed close to 200-fold reduction in binding affinity to APUM23. RNAs with substitutions at positions outside the core also validated the APUM23 consensus RNA target sequence. For instance, the consensus sequence showed a greater preference for cytosine than uracil at position 8 (Fig. 4D), and a uracil substitution at this position (C8U) resulted in a 2.5-fold reduction in affinity of the protein to this RNA (Table 2). Adenine and guanine substitutions at this position (C8A and C8G) showed 6.7- and 12-fold reductions in affinity, supporting the lack of adenine or guanine at this position in the consensus sequence (Fig. 4D). These data indicate that cytosine is the preferred nucleotide at position 8. Changes at nucleotide position 10 resulted in relatively high reductions in affinity (18- and 29-fold reduction), whereas changes at nucleotide positions 1, 2, and 3 showed a lower reduction in affinity (1.1–10-fold decreases). Finally, APUM23 did not possess a measurable apparent dissociation constant with the classical eight-nucleotide PUF target (UGUAUAUA, human PUM1) because of its low affinity binding (Table 2). Coupled with the UG56GU results discussed above, this indicates that the UGU core, a component of the RNA targets of all other characterized Puf proteins, is not a preferred core in the RNA target of APUM23. Overall, these APUM23 mobility shift data validate the identification of a newly described, 10-nucleotide PUF consensus RNA target sequence that contains a unique central UUGA core and a preference for cytosine at nucleotide position 8.
TABLE 2.
Actual and relative apparent dissociation constants of WT and mutant APUM23 proteins when bound to wild-type and nucleotide substituted RNA
| Protein | RNA | RNA sequence | Kd | Krela |
|---|---|---|---|---|
| nm | ||||
| WT | WT | GAAUUGACGGb | 0.41 ± 0.07 | 1 |
| G1U | UAAUUGACGGc | 2.18 ± 0.23 | 5.4 | |
| G1C | CAAUUGACGG | 4.19 ± 0.64 | 10 | |
| A2C | GCAUUGACGG | 0.91 ± 0.14 | 2.2 | |
| A3G | GAGUUGACGG | 0.43 ± 0.07 | 1.1 | |
| U4G | GAAGUGACGG | 7.68 ± 1.25 | 19 | |
| UG56GU | GAAUGUACGG | 77.4 ± 31.7 | 191 | |
| G6U | GAAUUUACGG | 12.4 ± 1.88 | 31 | |
| A7U | GAAUUGUCGG | 5.54 ± 0.97 | 14 | |
| C8U | GAAUUGAUGG | 1.03 ± 0.12 | 2.5 | |
| C8A | GAAUUGAAGG | 2.72 ± 0.26 | 6.7 | |
| C8G | GAAUUGAGGG | 4.94 ± 0.41 | 12 | |
| G10U | GAAUUGACGU | 7.16 ± 0.80 | 18 | |
| G10C | GAAUUGACGC | 11.9 ± 1.51 | 29 | |
| Human PUM1 | UGUAUAUA | >140d | >346 | |
| SHVLQ-1-NYVIQ | G10U | GAAUUGACGU | 2.55 ± 0.34 | 0.056 |
| WT | GAAUUGACGG | 45.3 ± 5.95 | 1 | |
| SHVLR-3-NYVIQ | C8U | GAAUUGAUGG | 6.95 ± 0.67 | 0.23 |
| WT | GAAUUGACGG | 30.4 ± 3.49 | 1 | |
| SHVLR-3-SNVVE | C8G | GAAUUGAGGG | 85.3 ± 5.81 | 0.30 |
| WT | GAAUUGACGG | 282 ± 30.0 | 1 | |
| SHLVE-5-NYVIQ | G6U | GAAUUUACGG | 62.2 ± 16.7 | 0.66 |
| WT | GAAUUGACGG | 94.5 ± 15.5 | 1 |
a Krel values represent the relative affinity of wild-type or mutant proteins to wild-type or mutant RNAs. Mutant protein Krel values that are lower for the mutant RNA compared to the wild-type RNA are underlined.
b The UUGA central core in the APUM23 wild-type RNA is highlighted with bold type.
c Modified nucleotides are underlined. The human PUM1 RNA is the human PUMILIO1 consensus sequence.
d Kd value could not be determined because RNA binding was not observed for the maximal protein concentration used for this RNA-protein combination. Thus, the Kd value is shown as being greater than the highest concentration of protein used.
Motif Swapping Supports the Identity Predictions of Three Puf Repeats
In an attempt to determine whether the APUM23 Puf repeats could be modified to alter their base specificity, we engineered the protein by swapping the five-amino acid RNA-binding motif in three of the predicted Puf repeats. Successfully altering base specificity would provide further support for the interaction of these repeats with their corresponding nucleotides shown in Fig. 4D. We modified Puf repeat 1 to determine whether it binds to the consensus sequence boundary at position 10, Puf repeat 3 to confirm binding to cytosine at nucleotide position 8, and Puf repeat 5 to confirm binding to the core guanine nucleotide at position 6. The five-amino acid motif sequence NYVIQ was chosen for substitution into each of these repeats so as to alter the binding preference of the substituted repeat to uracil. NYQ is a strong uracil binding TRM in other PUF proteins (6, 10), and its five-amino acid motif is present in two Puf repeats that are predicted to bind with uracil in the consensus RNAs of APUM2 and APUM6 (Fig. 4, A and B). A TRM with strong guanine binding preference (SNE) (6, 10) was also substituted into Puf repeat 3 to provide additional supporting evidence that this is the repeat that normally binds to cytosine at nucleotide position 8. SNVVE was used as the substituted motif, because this amino acid sequence was present at Puf repeat 7 in APUM2 and APUM12 and corresponded with guanine in the UGU core of their respective target RNAs.
Substitutions in predicted Puf repeat 1 (aligns with a guanine at consensus target sequence position 10; Fig. 4D) involved a switch from SHVLQ to NYVIQ (SHVLQ-1-NYVIQ) in an attempt to alter the binding specificity of this repeat to uracil (Fig. 6A). In the nucleotide substitution assays described earlier, wild-type APUM23 showed an 18-fold decrease in affinity to G10U RNA compared with its affinity to its cognate wild-type RNA (Table 2). In contrast, the SHVLQ-1-NYVIQ substitution in Puf repeat 1 showed a 17.9-fold increase in affinity to the G10U RNA compared with wild-type RNA (Krel = 0.056) (Table 2 and Fig. 6B). Two motif substitutions were made in predicted Puf repeat 3 (Fig. 6A), the repeat that aligns with the preferred cytosine base at nucleotide position 8. The SHVLR-3-NYVIQ substitution mutant demonstrated a 4.4-fold increase in binding affinity to C8U RNA (Krel = 0.23; Fig. 6C and Table 2) compared with wild-type RNA, whereas the wild-type protein was shown to have a corresponding decrease in affinity (2.5-fold) to the C8U RNA over its cognate RNA (Table 2). The SHVLR-3-SNVVE mutant bound more than 3-fold more tightly to its cognate RNA (C8G) than to wild-type RNA (Fig. 6D and Table 2), whereas the wild-type protein had a 12-fold decrease in binding to C8G compared with wild-type RNA (Table 2). A substitution in Puf repeat 5 (SHLVE-5-NYVIQ) showed enhanced binding to the G6U RNA over wild-type RNA (∼1.5-fold) (Fig. 6E and Table 2), whereas the wild-type protein bound 31-fold less tightly to G6U RNA compared with wild-type RNA (Table 2). The altered affinity of these motif-swapped proteins supports our predicted interactions between Puf repeats 1, 3, and 5 and nucleotides 10, 8, and 6, respectively. These motif-substituted proteins all had lower binding affinities for their cognate RNAs compared with the wild-type protein binding to its cognate RNA. This was especially noticeable for the SHVLR-3-SNVVE and SHLVE-5-NYVIQ substitutions. This indicates that although the substitutions altered the specificity of the repeat, they may have also altered the local structure at these positions, thereby leading to a reduced binding affinity.
FIGURE 6.
RNA binding analysis for wild-type and mutated APUM23 bound to wild-type and nucleotide-substituted RNAs. A, logo map and predicted nucleotide preference for the five-amino acid motif swapping experiments. The labeling of nucleotide and TRM positions are as in Fig. 4. The predicted Puf repeats that were selected for mutagenesis are identified below the three-letter TRM of the specific Puf repeat. The expected nucleotide bound by the mutated Puf repeat (*) and the substituted five-amino acid motif in the Puf repeat (**) are shown. B–E, representative equilibrium binding data of substituted APUM23 Puf repeat motifs to their cognate (red lines) and wild-type (black lines) sequences. The fraction of RNA bound is shown on the vertical axis, and the concentration of mutant protein is shown on the horizontal axis.
Discussion
The SELEX-derived consensus RNA binding sequences of the four APUM proteins studied here provide insight into the diversity of RNA sequences bound by plant PUF-like proteins. In particular, a novel PUF consensus sequence for APUM23 was identified. This sequence was 10 nucleotides in length, contained an atypical core element (UUGA) that was located centrally in the sequence, and preferred cytosine at nucleotide position 8. All PUF RNA target sequences studied to date contained a UGU core located at the 5′ end of the sequence (1). The evidence for a central UUGA core in the APUM23 consensus sequence and for the presence of a cytosine binding Puf repeat is supported by EMSA analysis that used nucleotide substituted RNAs as well as swapped RNA binding motifs. Natural Puf repeats that are cytosine binders have not previously been identified. However, a yeast three-hybrid screen that was used to engineer cytosine recognition codes identified a cytosine-binding TRM code of S(Y/H/R)R (8, 9, 26). In addition to showing a preference for cytosine, Puf repeat 3 (contains a TRM code of SHR) had a weaker preference for uracil (Fig. 4D and Table 2). This weaker preference for uracil was also observed for an engineered cytosine-binder that had a TRM code of SYR (9). Thus, our SELEX and EMSA results that showed that Puf repeat 3 prefers cytosine, coupled with the reports of an engineered cytosine binding repeat with the same TRM, indicate that repeat 3 in APUM23 has natural cytosine binding characteristics.
The novel RNA consensus sequence identified for APUM23, as well as its atypical arrangement of Puf repeats, suggests that it has a unique three-dimensional structure when bound to RNA. The modeled structures of APUM23 (Fig. 2) assisted in predicting the identity of the Puf repeats in this protein. However, the structure of the protein in vivo is likely quite complex and differs from the classical PUF protein structure because of the presence of relatively large gaps between some of the predicted repeats (Fig. 1A). The resolved crystal structure of human Puf-A (27), an ortholog of Arabidopsis APUM24, shows that it has a unique PUF folding pattern. Puf-A folds into an L-shaped structure with 11 Puf repeats in two subdomains and binds structured RNA and DNA in a non-sequence-specific manner (27). Puf-A and APUM24 are also nucleolar targeted PUF proteins (13, 28), and Puf-A is known to function in rRNA processing, although at a different step from yeast Nop9 and APUM23 (14, 24, 27). The non-sequence-specific binding of Puf-A to nucleic acid is at least partially attributable to the presence of hydrophobic residues located within the TRMs of four consecutive repeats and the presence of basic surface patches on the protein (27). In contrast, the predicted repeats in APUM23 largely possess typical Puf-like TRMs, and we have shown that APUM23 binds to RNA with sequence specificity. Although APUM23 and Puf-A have different RNA binding characteristics and low conservation in their amino acid sequences, the Puf-A structure demonstrates that a nonclassical PUF protein can form a unique structure.
The identification of an RNA consensus target sequence for APUM23 provides insight into its functional roles in rRNA processing. Knock-out mutants of APUM23 have a partial defect in 35S ribosomal RNA processing that results in a small accumulation of a nonprocessed version of 18S rRNA on top of normal levels of fully processed 18S rRNA (14). Apum23 mutants displayed slower growth, shorter roots, and smaller, serrated and pointed leaves (14, 18). The S. cerevisiae homolog (Nop9) has a similar 18S rRNA processing role; however, nop9 mutants do not survive (24). Interestingly, the 18S rRNA sequence of Arabidopsis contains a single APUM23 consensus sequence (GAAUUGACGG) at nucleotide position 1142, as does the corresponding region in S. cerevisiae 18S rRNA. The frequency that this 10-nucleotide sequence is expected to appear randomly in RNA is approximately once per 1000 kb. This provides supporting evidence that APUM23 binds to the 10-nucleotide sequence in 18S rRNA. The nucleolar localization of APUM23 and Nop9 proteins (13, 14, 24) also supports a direct binding to 18S rRNA in its rRNA processing role. However, SELEX analysis of the yeast ortholog of APUM23 (Nop9) showed that this protein has no recognizable binding specificity toward RNA, and it binds to the 10-nucleotide APUM23 target with much lower affinity than APUM23. Despite having limited amino acid sequence and predicted TRM conservation, APUM23 partially complements the lethal yeast nop9 mutant (18), suggesting that these proteins recognize similar RNA targets in their conserved rRNA processing role. Perhaps APUM23 and Nop9 both interact with 18S rRNA, but using different binding mechanisms. Nop9 might require interactions with binding partners in vivo, in a similar manner to that proposed for another atypical nucleolar PUF protein, yeast Puf6 (27). Future studies will identify APUM23 binding partners to determine whether this protein assists in recruiting RNA processing machinery, similar to the roles of some other PUF proteins (29).
APUM23, a protein with uniquely dispersed Puf repeats, provides a newly identified backbone for a sequence-specific RNA-binding protein. Co-crystal structures of APUM23 bound to its cognate RNA will identify the nucleotide-amino acid interactions that occur, identify the precise number of functional Puf repeats, and determine whether extrusion of bases is a component of its binding to RNA. The RNA-binding motif swapping experiments provided evidence for specific binding of predicted Puf repeats 1, 3, and 5 and demonstrated that the APUM23 Puf repeats could be engineered to recognize different RNA targets. Also, the 10-nucleotide RNA-binding consensus sequence that is preferred by APUM23 may provide a greater range of specificity than the classical eight-repeat PUFs. These RNA binding characteristics demonstrate the potential for engineering APUM23 so that it can bind to specific cellular RNA targets to modulate the physiology and metabolism of these RNAs, as has been achieved for other PUF proteins (11, 30, 31).
Author Contributions
C. Z. performed the research, and C. Z. and D. G. M. designed the research, analyzed the data, and wrote the article.
Supplementary Material
Acknowledgments
We are grateful to Isabelle Barrette-Ng and Ken Ng for helpful discussions on structural modeling, and we thank Dave Hansen for critical reading of the manuscript.
This work was supported by a Natural Sciences and Engineering Research Council of Canada Discovery Grant (to D. G. M.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Tables S1–S3 and Fig. S1.
- PUM-HD
- Pumilio homology domain
- TRM
- tripartite recognition motif
- SELEX
- systematic evolution of ligands by exponential enrichment.
References
- 1.Quenault T., Lithgow T., and Traven A. (2011) PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 21, 104–112 [DOI] [PubMed] [Google Scholar]
- 2.Lu G., Dolgner S. J., and Hall T. M. (2009) Understanding and engineering RNA sequence specificity of PUF proteins. Curr. Opin. Struct. Biol. 19, 110–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y., Opperman L., Wickens M., and Hall T. M. (2009) Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein. Proc. Natl. Acad. Sci. U.S.A. 106, 20186–20191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller M. T., Higgin J. J., and Hall T. M. (2008) Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p. Nat. Struct. Mol. Biol. 15, 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu D., Stumpf C. R., Krahn J. M., Wickens M., and Hall T. M. (2009) A 5′ cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs. Proc. Natl. Acad. Sci. U.S.A. 106, 20192–20197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell Z. T., Valley C. T., and Wickens M. (2014) A protein-RNA specificity code enables targeted activation of an endogenous human transcript. Nat. Struct. Mol. Biol. 21, 732–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X., McLachlan J., Zamore P. D., and Hall T. M. (2002) Modular recognition of RNA by a human pumilio-homology domain. Cell 110, 501–512 [DOI] [PubMed] [Google Scholar]
- 8.Filipovska A., Razif M. F., Nygård K. K., and Rackham O. (2011) A universal code for RNA recognition by PUF proteins. Nat. Chem. Biol. 7, 425–427 [DOI] [PubMed] [Google Scholar]
- 9.Dong S., Wang Y., Cassidy-Amstutz C., Lu G., Bigler R., Jezyk M. R., Li C., Hall T. M., and Wang Z. (2011) Specific and modular binding code for cytosine recognition in Pumilio/FBF (PUF) RNA-binding domains. J. Biol. Chem. 286, 26732–26742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheong C. G., and Hall T. M. (2006) Engineering RNA sequence specificity of Pumilio repeats. Proc. Natl. Acad. Sci. U.S.A. 103, 13635–13639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., Wang Z., and Tanaka Hall T. M. (2013) Engineered proteins with Pumilio/fem-3 mRNA binding factor scaffold to manipulate RNA metabolism. FEBS J. 280, 3755–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francischini C. W., and Quaggio R. B. (2009) Molecular characterization of Arabidopsis thaliana PUF proteins: binding specificity and target candidates. FEBS J. 276, 5456–5470 [DOI] [PubMed] [Google Scholar]
- 13.Tam P. P., Barrette-Ng I. H., Simon D. M., Tam M. W., Ang A. L., and Muench D. G. (2010) The Puf family of RNA-binding proteins in plants: phylogeny, structural modeling, activity and subcellular localization. BMC Plant Biol. 10, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbasi N., Kim H. B., Park N. I., Kim H. S., Kim Y. K., Park Y. I., and Choi S. B. (2010) APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 64, 960–976 [DOI] [PubMed] [Google Scholar]
- 15.Huh S. U., Kim M. J., and Paek K. H. (2013) Arabidopsis Pumilio protein APUM5 suppresses cucumber mosaic virus infection via direct binding of viral RNAs. Proc. Natl. Acad. Sci. U.S.A. 110, 779–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huh S. U., and Paek K. H. (2014) APUM5, encoding a Pumilio RNA binding protein, negatively regulates abiotic stress responsive gene expression. BMC Plant Biol. 14, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiang Y., Nakabayashi K., Ding J., He F., Bentsink L., and Soppe W. J. (2014) Reduced Dormancy5 encodes a protein phosphatase 2C that is required for seed dormancy in Arabidopsis. Plant Cell 26, 4362–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang T., Kerstetter R. A., and Irish V. F. (2014) APUM23, a PUF family protein, functions in leaf development and organ polarity in Arabidopsis. J. Exp. Bot. 65, 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellington A. D., and Szostak J. W. (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 [DOI] [PubMed] [Google Scholar]
- 20.White E. K., Moore-Jarrett T., and Ruley H. E. (2001) PUM2, a novel murine puf protein, and its consensus RNA-binding site. RNA 7, 1855–1866 [PMC free article] [PubMed] [Google Scholar]
- 21.Bailey T. L., and Elkan C. (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, AAAI Press, Menlo Park, CA: [PubMed] [Google Scholar]
- 22.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Cassarino T. G., Bertoni M., Bordoli L., and Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller M. A., and Olivas W. M. (2011) Roles of Puf proteins in mRNA degradation and translation. WIREs RNA 2, 471–492 [DOI] [PubMed] [Google Scholar]
- 24.Thomson E., Rappsilber J., and Tollervey D. (2007) Nop9 is an RNA binding protein present in pre-40S ribosomes and required for 18S rRNA synthesis in yeast. RNA 13, 2165–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta Y. K., Lee T. H., Edwards T. A., Escalante C. R., Kadyrova L. Y., Wharton R. P., and Aggarwal A. K. (2009) Co-occupancy of two Pumilio molecules on a single hunchback NRE. RNA 15, 1029–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Filipovska A., and Rackham O. (2012) Modular recognition of nucleic acids by PUF, TALE and PPR proteins. Mol. Biosyst. 8, 699–708 [DOI] [PubMed] [Google Scholar]
- 27.Qiu C., McCann K. L., Wine R. N., Baserga S. J., and Hall T. M. (2014) A divergent Pumilio repeat protein family for pre-rRNA processing and mRNA localization. Proc. Natl. Acad. Sci. U.S.A. 111, 18554–18559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang H. Y., Fan C. C., Chu P. C., Hong B. E., Lee H. J., and Chang M. S. (2011) hPuf-A/KIAA0020 modulates PARP-1 cleavage upon genotoxic stress. Cancer Res. 71, 1126–1134 [DOI] [PubMed] [Google Scholar]
- 29.Goldstrohm A. C., Hook B. A., Seay D. J., and Wickens M. (2006) PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 13, 533–539 [DOI] [PubMed] [Google Scholar]
- 30.Filipovska A., and Rackham O. (2011) Designer RNA-binding proteins: new tools for manipulating the transcriptome. RNA Biol. 8, 978–983 [DOI] [PubMed] [Google Scholar]
- 31.Abil Z., and Zhao H. (2015) Engineering reprogrammable RNA-binding proteins for study and manipulation of the transcriptome. Mol. Biosyst. 11, 2658–2665 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.