Abstract
Introduction
The study of xenobiotic metabolism and toxicity has been greatly aided by the use of genetically-modified mouse models and metabolomics.
Areas covered
Gene knockout mice can be used to determine the enzymes responsible for the metabolism of xenobiotics in vivo and to examine the mechanisms of xenobiotic-induced toxicity. Humanized mouse models are especially important since there exist marked species differences in the xenobiotic-metabolizing enzymes and the nuclear receptors that regulate these enzymes. Humanized mice expressing cytochromes P450 (CYPs) and nuclear receptors including the pregnane X receptor (PXR), the major regulator of xenobiotic metabolism and transport were produced. With genetically-modified mouse models, metabolomics can determine the molecular map of many xenobiotics with a level of sensitivity that allows the discovery of even minor metabolites. This technology can be used for determining the mechanism of xenobiotic toxicity and to find early biomarkers for toxicity.
Expert opinion
Metabolomics and genetically-modified mouse models can be used for the study of xenobiotic metabolism and toxicity by: 1) Comparison of the metabolomics profiles between wild-type and genetically-modified mice, and searching for genotype-dependent endogenous metabolites; 2) Searching for and elucidating metabolites derived from xenobiotics; 3) Discovery of specific alteration of endogenous compounds induced by xenobiotics-induced toxicity.
Keywords: genetically-modified mice, CYP, CYP3A4, metabolomics, pregnane X receptor, UPLC-ESI-QTOFMS, xenobiotics
1. Introduction
Drug-induced toxicity remains among the top reasons limiting the clinical use of drugs and the development of new chemical entities (NCEs) [1]. For the purpose of this review, drugs will be lumped with other foreign chemicals as xenobiotics since the technologies discussed cab apply to drugs, environmental and industrial chemicals and dietary compounds. Toxicity can be due to metabolism, which while important for the elimination of drugs, can also produce electrophilic metabolites that can cause cell death. For example, acetaminophen at high doses, is metabolized by cytochromes P450 (CYP) to a toxic quinone metabolite that depletes cellular glutathione and can covalently bind macromolecules resulting in elevated oxidative stress, mitochondrial damage and cell death [2, 3]. For decades, acetaminophen has served as the model to explore the mechanisms of drug-induced hepatotoxicities in order to understand idiosyncratic drug reactions involving increased liver enzymes, noted with many clinically-used drugs and drugs under development. In the past decade, these studies have been greatly aided by the use of genetically-modified mouse models and, more recently, by use of LCMS-based metabolomics.
1.1. Study of xenobiotic metabolism and toxicity
Many strategies have been used to study drug metabolism and to determine the mechanisms of drug- or xenobiotic-mediated toxicities, particularly liver toxicity, which is first diagnosed by the elevation of liver enzymes aspartate aminotransferase (AST) and alanine aminotransferase (ALT). In the pharmaceutical industry, rats have historically been used as the first line to investigate the metabolism and potential toxicity of drugs in whole animals [4]. Later studies can involve the use of dogs. Human hepatocytes have also been found to be of great value in the study of drug metabolism, but their utility in the prediction of drug toxicity is limited as hepatic toxicity, in particular subtle toxicity requires a whole liver context for complete manifestation [5]. Studies with human hepatocytes have been greatly aided by cryopreservation techniques but also limited due to the large interindividual variability in human livers. New engineered human hepatocytes from inducible stem cells could be used to standardize experimental human liver and hepatocytes [6, 7].
In vitro evaluation of the metabolic activation of xenobiotics through the use of liver subcellular fractions such as microsomes (fractions from endoplasmic reticulum) and trapping reagents (glutathione, N-acetylcysteine, m-anisaldehyde) have been employed to determine the production of electrophilic metabolites that result from xenobiotic metabolism [8–10]. As noted above, the use of hepatocyte cutures and liver slices have also been of great value for studying xenobiotic metabolism and the mechanisms of xenobiotic toxicity [11]. However, in vivo metabolism is the most relevant means to determine the fate of xenobiotics in mammals and to predict human responses to foreign compounds. In vivo animal models have wide utility for the investigation of toxicity mechanisms, but are limited by their predictive value for humans because mice can metabolize many xenobiotics quite differently from humans, due in part to the marked species differences in the metabolic enzymes, notably the CYPs. For example, the most abundant CYP in humans is CYP3A4, a CYP that metabolizes more than 50% of clinically-used drugs [12–14]. The corresponding mouse homologues are expressed at lower levels than that found for CYP3A4 in liver. In particular, CYP3A4 is expressed in high levels in intestine, incrontrast to low expression of the mouse CYP3As in this tissue [15–17]. There is less species differences in the expression and catalytic activities in the phase 2 conjugating enzymes, while there are differences between humans and rodent models in the expression and specificities of drug transporters [18, 19]. The regulation of xenobiotic-metabolizing enzymes and even drug/xenobiotic transporters can also differ between mice and humans [2, 20].
1.2. Use of genetically-modified mice
Genetically modified mouse models (including gene knockout mice and humanized models) have been used to study xenobiotic metabolism and xenobiotic-induced toxicity [21–23]. Through the knockout of specific genes encoding xenobiotic-metabolizing enzymes or xenobiotic receptors involved in the regulation genes encoding xenobiotic-metabolizing enzymes, the contribution of specific enzymes and receptors towards the toxicity of these chemicals can be determined. The creation of humanized mice through the insertion of the human genes into the mouse genome can, to a degree, circumvent the potential species difference for risk assessment and can be faithfully extrapolated to humans [21].
The use of genetically-modified mice models have been reviewed [21–25]. Genes encoding CYPs involved in the metabolism of drugs and other xenobiotics have been individually disrupted and mice lacking these gene have no deleterious phenotypes thus indicating that they are not involved in critical physiological functions or their functions are redundant. For example, mice with a knockout of the hepatic CYP reductase gene, required for the catalytic activity of CYPs, are viable [26, 27], thus indicating that CYPs involved in xenobiotic metabolism are not critical for reproduction and physiological homeostasis with minor alterations in liver function. More recently, in a monumental accomplishment, mice lacking almost all of the known xenobiotic-metabolizing enzyme members of the Cyp2c, Cyp2d, and Cyp3a gene clusters, a total of 30 genes, were created and these mice also were viable with a minor hepatic phenotype of unknown mechanism [28]. This latter study confirms and definitively established that the hepatic, non-mitochondrial CYPs are primarily involved in metabolism of foreign compounds and have no critical physiological role.
The successful application of Cyp gene knockout mice in the study of xenobiotic toxicity has been frequently reported. For example, CYP2E1 mediates the oxidative metabolism of a wide range of xenobiotics, that cause toxicity. CYP2E1 is the major rate-limiting enzyme that initiates the cascade of events leading to acetaminophen hepatotoxicity [2]. Cyp2e1-null mice were employed to demonstrate the role of CYPs in acetaminophen toxicity. These mice were resistant to the hepatoxic effects of the drug as revealed by LD50 analysis and various biochemical endpoints [29]. In another study, mice lacking expression of CYP1B1 were resistant to bone marrow toxicity when administered the carcinogen 7,12-dimethylbenz[a]anthracene [30]. Cyp1a1-null mice were employed to study the toxicity of orally-administered benzo[a]pyrene [31].
1.3. Metabolomics
In the last few years, metabolomics has been used to study xenobiotic metabolism and xenobiotic-mediated toxicity [32, 33]. Metabolomics, an area of investigation that measures changes in small molecules downstream of the genome, transcriptome, and proteome, captures the terminal alteration of endogenous chemicals. Metabolomics can also be used to identify and map metabolites derived from xenobiotics [32–34]. Unbiased determination of small molecules in biofluids, or extracts from cells, tissues, or organisms can give a clear description of the alteration of various biological processes. Metabolomics has been successfully introduced into the toxicological studies, as reviewed elsewhere [35–37]. A large number of studies have been performed employing metabolmics to study drug metabolism and toxicity in rats [38–40]. These studies have yielded much information on metabolism and markers for toxicity, including organ-specific toxicity [41]. However, there are not many genetically modified rat models and thus, many recept studies have focused on the study of xenobiotic-metabolism and toxicity using metabolomics and genetically modified mice. In addition, while there is a large literature on the use on 1H NMR in the study of drug metabolism and toxicity, as reviewed earlier [42], most studies in mice and genetically-modified mice have used liquid chromatography-mass spectrometry. The studies described below largely employ the ultra-performance liquid chromatography-electrospray ionization-quadrupole time-of-flight mass spectrometry (UPLC-ESI-QTOFMS) to examine the metabolism of xenobiotics and to find biomarkers for xenobiotic-induced hepatic toxicity [32].
2. Acetaminophen-induced hepatotoxicity
One of the most important CYPs for the metabolism of low MW xenobiotics, including industrial solvents and toxicants, is CYP2E1 [2]. It is highly expressed in liver, and to a muuch lower extent in extrahepatic tissues. CYP2E1 is conserved between humans, rats and mice, with no noted differences in catalytic activity between species. There are no known common polymorphisms that alter the expression or catalytic activity of CYP2E1. It is induced by some of its own substrates through a post-translational stabilization mechanism [43]. Despite, its high level of conservation between species and no polymoprhisms in humans, mice lacking expression of CYP2E1 are overtly normal with no notable developmental of physiological defects. It does not appear to influence glucose homeostasis in spite of its role in the metabolism of ketone bodies [44]. This mouse model is thus suitable for the study of the role of CYP2E1 in xenobiotic toxicity.
2.1 Metabolite mapping of acetaminophen
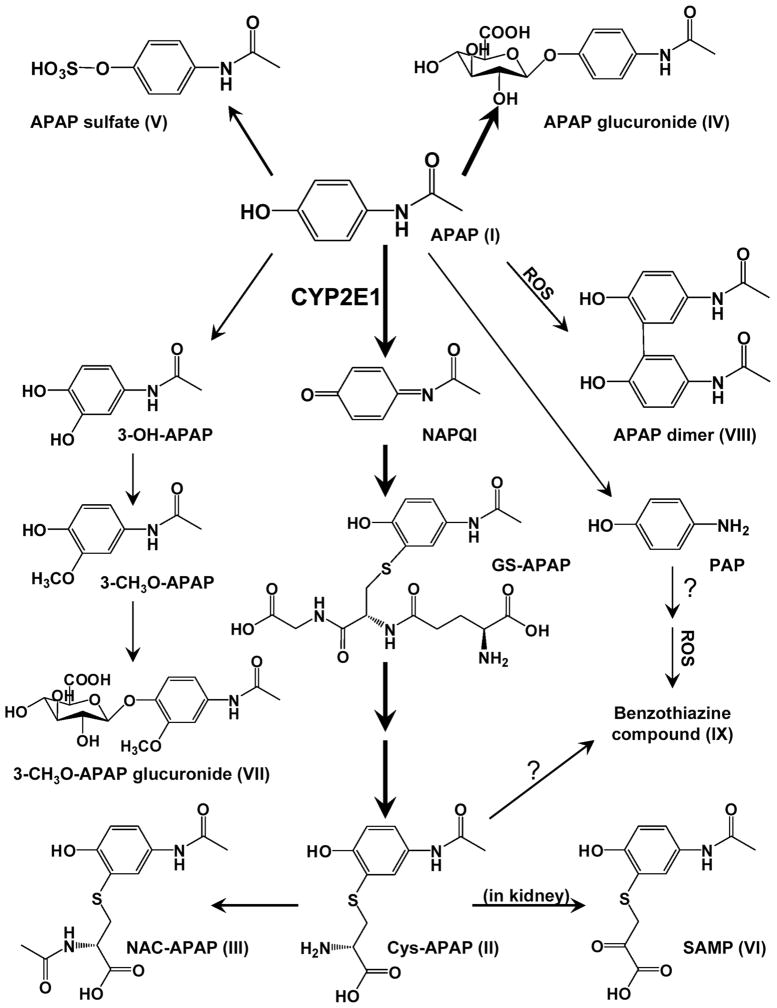
The value of gene knockout mice and metabolomics in the study of xenobiotic toxicity can be illustrated by a series of studies using the Cyp2e1-null mice and acetaminophen. Mice lacking CYP2E1 were resistance to the hepatotoxic effects of acetaminophen; mice without expression of both CYP2E1 and CYP1A2 were even more resistant to the drug as revealed by LD50 studies [29]. Metabolomics was used with wild-type and Cyp2e1-null to determine the role of CYP2E1-mediated metabolism and down-stream events in mechanism of acetaminophen-induced hepatotoxicity. Metabolite mapping, using the power of metabolomics, was performed and several derivatives of acetaminophen were found only in wild-type mice, with some metabolites diminished in Cyp2e1-null mice, indicating that they might be involved in acetaminophen toxicity [45]. Metabolomics revealed the complete metabolic map of nine metabolites of acetaminophen, including three novel metabolites, an acetaminophen dimer formed nonenzymatically in liver, 3-CH3-O-acetaminophen glucuronide and S-(5-acetylamino-2-hydroxyphenyl) mercaptopyruvic acid (SAMP) that is likely formed in the kidney through transamination of cysteine-acetaminophen (Figure 1). Metabolites derived from non-acetaminophen related endogenous compounds can also be discovered using metabolomics and these may serve as potential liver toxicity biomarkers [46]. These endogenous compounds might be of value as biomarkers for early toxicity and can offer clues to the mechanism of toxicity as noted below.
Figure 1.
Metabolic map of acetaminophen. Thick lines represent major pathways, and thin lines denote minor pathways. Adapted from [45].
2.2 Biomarker for acetaminophen-induced hepatotoxicity
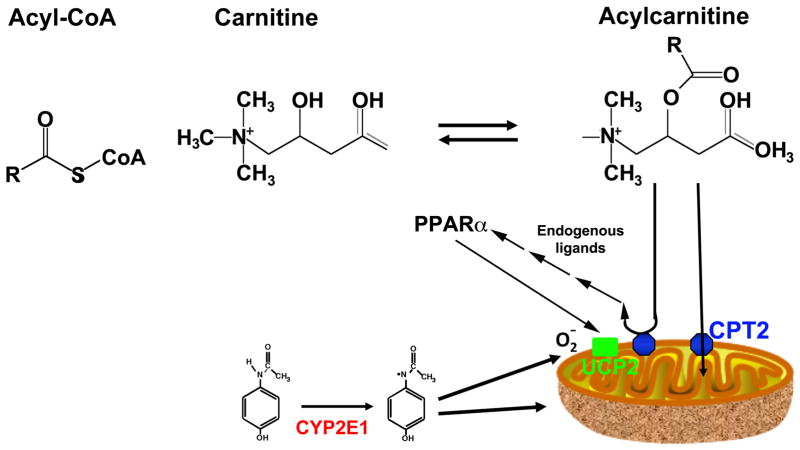
Metabolomics was then employed to discover biomarkers for acetaminophen-induced hepatotoxicity, importantly, early biomarkers for the onset of liver damage. Through comparsion of the metabolomics results of serum biosamples between wild-type and Cyp2e1-null mice after treatment with a toxic dose of acetaminophen, a differential, time-dependent alteration of serum acylcarnitines was found [47]. Metabolomics revealed mouse strain- and acetaminophen-dependent separation of groups early after exposure to toxic doses of acetaminophen. Notably, the separation was driven by a difference in acylcarnitines that were markedly increased at high acetaminophen doses in wild-type and and not Cyp2e1-null mice. While remaining high in wild-type mice, serum acylcarnitine levels gradually returned to normal in Cyp2e1-null mice at the end of the 24 h treatment. The early increase in serum acylcarnetines correlated with upregulation of the nuclear receptor peroxisome proliferator-activated receptor α (PPARα) signaling following acetaminophen treatment [47]. PPARα a nuclear receptor that is involved in the control of lipid homeostasis in the liver and kidney and to some extent, in other tissues [48]. It is highly induced by xenobiotic ligands and by endogenous fatty acid metabolites under constions of food restrictions. PPARα controls the expression of genes involved in lipid transport and fatty acid β-oxidation in the peroxisomes and mitochondria. This study suggest that mitochondrial damage was a very early event in the acetaminophen toxicity since acylcarnitine accumulation in the cytosol and leakage into the serum is due in large part to a lack of mitochondrial uptake of these metabolites by mitochondrial transporter carnitine acyltransferases (CPT) [49]. The resultant liver damage leads to acylcarnitine leakage to the blood where they were detected (Figure 2). In addition, the upregulation of PPARα target genes is likely due to an increase in endogenous metabolites, probably derived from fatty acids, produced as a result of the mitochondrial damage, that are agonist for PPARα. In humans, the extent of mitochondrial damage, as revealed by serum biomarkers, correlated with survival after acetaminophen overdose [3].
Figure 2.
Role of PPARα in acetaminophen-induced hepatotoxicity. Fatty acylcarnitines formed from acyl-CoA and carnitine are required for uptake of fatty acids to the mitochondria by carnitine palmitoyltransferase 2 (CPT2) for fatty acid β-oxidation to occur. Upon mitochondrial damage by an electrophilic metabolite of acetaminophen, NAPQI, the mitochondria is damaged leading to lack of acylcarnitines uptake and leakage to the cytoplasm and serum upon liver cell death. Endogenous ligands from this leakage can activate PPARα that in turn induced fatty acid β-oxidation by peroxisomes and induction of uncoupling protein 2 (UCP2) as a defense against oxidative stress. Data taken from [50].
2.3 Mechanism of acetaminophen hepatotoxicity
The transient increase in PPARα signaling may be a defensive mechanism by which fatty acid catabolism is increased along with elevated mitochondrial uncoupling protein 2 (UCP2), encoded buy the PPARα target gene Ucp2, as a defense against increased reactive oxygen species produced by acetaminophen overdose (Figure 2) [50]. UCP2 is a membrane protein that transports protons and anions, such as Cl-, Br-, and NO3- across the mitochondrial inner membrane [51]. Activation of PPARα, using the synthetic agonist Wy-14,643, protected mice from acetaminophen-induced hepatotoxicity as a result of indution of Ucp2. The mechanism of this protection is likely due to a decrease of acetaminophen-induced reactive oxygen species (ROS) by UCP2, and an explanation by which CYP2E1-mediated metabolic activation of acetaminophen through mitochondrial damage, inhibited mitochondrial fatty acid oxidation [50].
3. Alcohol-induced liver disease
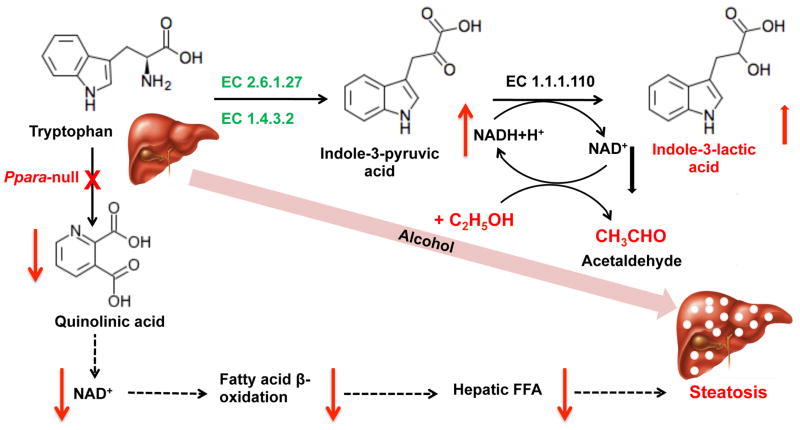
Alcohol-induced liver disease (ALD) is a leading cause of nonaccident-related deaths in the United States due in large part to liver injury, notably alcoholic liver disease and liver cirrhosis [52]. Mice lacking PPARα are higly susceptible to ALD, thus making them an excellent model to study the mechanisms of this disease [53]. Metabolomic comparison of alcohol-treated wild-type and Ppara-null mice showed that levels of indole-3-lactic acid were markedly elevated in urine, indicating a mechanism where disruption of the PPARα gene impairs conversion of tryptophan to NAD+ through quinolinic acid leading to decreased NAD+-dependent fatty acid β-oxidation [54]. NAD+ is absolutely required for proper cellular functions and thus lower levels of this metabolite would have deleterious consequences. Oxidation of alcohol to acetaldehyde and acetic acid further decreases the NAD+/NADH ratio leading to free fatty acid accumulation in the liver and hepatic steatosis (Figure 3). The buildup of NADH drives the reduction of indole-3-pyruvic acid to indole-3-lactic acid, resulting in an increase in urinary excretion of this compound [54, 55]. Additionally, this metabolite could be a prediction biomarker for alcohol-induced liver disease as revealed in the Ppara-null mouse model. The role of PPARα in protection against ALD suggests a potential overlap in the mechanism of toxicity between acetaminophen and ALD that needs to be explored by additonal experimentations.
Figure 3.
Mechanism of generation of biomarkers that are associated with alcohol and hepatic steatosis. EC 2.6.1.27, tryptophan transaminase; EC 1.4.3.2, L-amino-acid oxidase; EC 1.1.1.110, indolelactate dehydrogenase. Derived from [55].
4. Hepatotoxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin
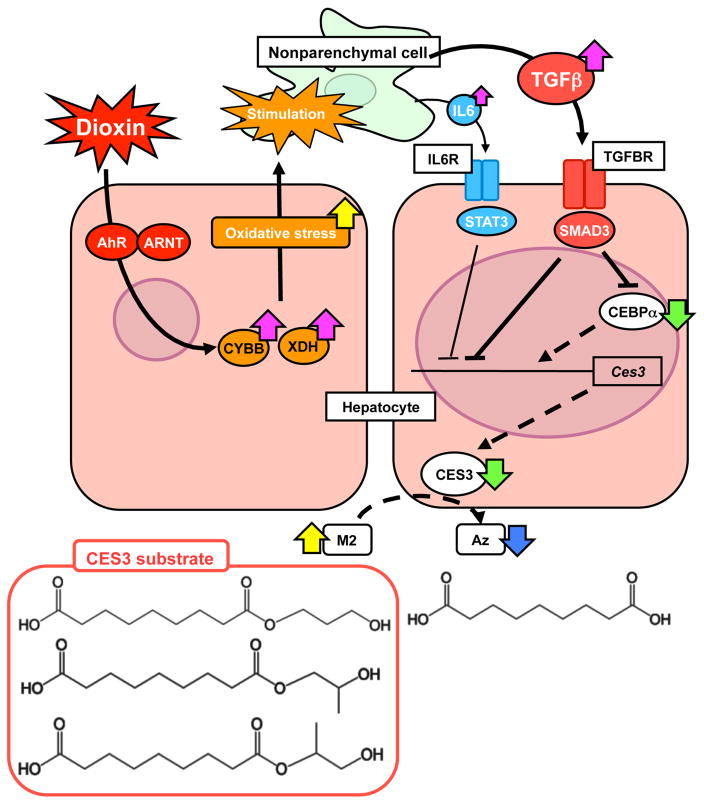
The aryl-hydrocarbon receptor (AhR) is a basic helix-loop-helix trasnsription factor that controls a large number of genes including those encoding CYP1A1, CYP1A2 and CYP1B1 [56]. It is the receptor that medicates the biological activities of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) which is among the most potent environmentally toxic compounds. TCDD causes immunotoxicity, chloracne in humans and liver toxicity, but the detailed toxicity mechanism by which it causes liver injury remains unclear. Comparison of serum metabolomics profiles between wild-type mice and aryl-hydrocarbon receptor (Ahr)-null mice identified azelaic acid monoesters as significantly increased through downregulation of hepatic carboxylesterase 3 (CES3), an enzyme that hydrolyzes these esters, after treatment with TCDD, in a AhR-dependent manner [57]. Methionine- and choline-deficient (MCD), an experimental model for non-alcoholic steatohepatitis (NASH), was also associated with increased azelaic acid monoesters that result from downregulation of CES3, thus indicating a link between xenobiotic toxicity and NASH. However, no change in CES3 levels and azelaic acid monoesters was found in simple hepatosteatosis. Increased inflammatory cytokines produced by both TCDD and MCD-induced NASH caused suppression of expression of the Ces3 gene via TGFβ-SMAD3 and IL6-STAT3 signaling pathways (Figure 4). This study established a role for the AhR signalling pathway in TCDD-induced steatohepatitis and revealed a similarity in mechanism of liver toxicity between TCDD-induced toxicity and NASH.
Figure 4.
Mechanism by which TCDD increases liver steatosis and azelaic acid esters. TCDD (dioxin) enters hepatocytes and elevates oxidative stress that increases production of proinflammatory cytokines such as IL6 and TGFβ in neighboring Kupffer cells. IL6 and TGFβ act on hepatocytes to suppress STAT3 and SMAD3 signaling resulting in suppression of the Ces3 gene encoding carboxylesterase 3 (CES3) which converts azelaic acid (Az) esters to Az. Lower CES3 expression also results on increase hepatic lipids due to lower hydrolysis of long-chain fatty acid esters and thioesters which may have a role in the elimination of hepatic lipid stores. Derived from [57].
5. Hepatoxicity of isoniazid and rifampicin
Cyp2e1-null mice and metabolomics were used to probe the mechanism of toxicity of isoniazid, a first-line medication used in the treatment of tuberculosis [58]. Metabolomic analysis showed that production of metabolites such as p-cresol glucuronide and p-cresol sulfate were elevated in wild-type mice but not in Cyp2e1-null mice. This suggests that the drug altered the gut microbiota population in a CYP2E1-dependent fashion, since these metabolites are derived from the gut flora [59]. The higher abundance of bile acids, bile acid metabolites, carnitine and carnitine derivatives in isoniazid-treated wild-type mice was also not detected in Cyp2e1-null mice. These results indicated that CYP2E1 might play a role in isoniazid-induced cholestasis through alteration of gut bacteria, enhancement of bile acid accumulation and mitochondria β-oxidation although a mechanism that remains to be determined.
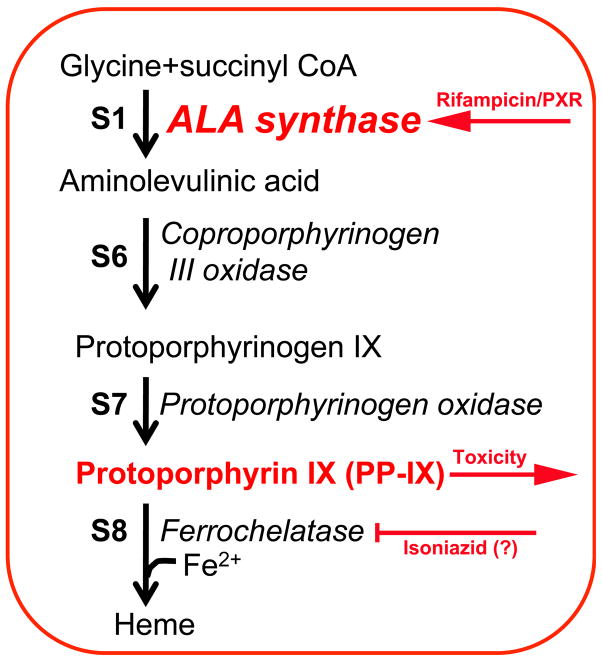
Rifampicin is usually combined with isoniazid for the treatment of tuberculosis. However, co-therapy with these two drugs results in mild to severe liver toxicity [60]. Since rifampicin is a human PXR specific ligand, PXR-humanized mice were used to study the mechanism of hepatotoxicity in mice co-treated with rifampicin and isoniazid. Toxicity, which was characterized by increased serum ALT and alkaline phosphatase, markers for hepatocyte and bile duct injury, respectively, was only observed in PXR-humanized mice co-treated with both drugs, and not found in wild-type mice and Pxr-null mice [61]. Mild liver toxicity was noted upon treatment with rifampicin alone but not with mono-treatment with isoniazid. In addition, occluded bile ducts were obvious in the livers of treated PXR-humanized mice. To investigate the nature of these occlusions, metabolomics was carried out in the bile ducts revealing the presence of high levels of protoporphyrin IX, an intermediate in porphyrin and heme synthesis, in PXR-humanized mice; increased protoporphyrin IX was also found in hepatocytes. Rifampicin was found to markedly induce the gene encoding aminolevulinic acid synthase in these mice indicating that the increased in protoporphyrin IX is due in part to its increased synthesis. Treatment with the product of aminolevulinic acid synthase, aminolevulinic acid, was found to potentiate the rifampicin and isoniazid hepatotoxicity. The detailed mechanism by which isoniazid contributes this toxic event remains unknown. However, one possibility is that isoniazid inhibits ferrochelatase activity resulting in the accumulation of protoporphyrin IX as a result of blocking the heme synthesis pathway (Figure 5). This study illustrates the value of PXR-humanized in unraveling the mechanism of sporadic hepatic toxicity of a widely used drug therapy. These data may help in the prediction of patients that my exhibit toxicity when treated with rifampicin and isoniazid.
Figure 5.
Proposed mechanism for rifampicin and isoniazid co-therapy-induced hepatotoxicity. Rifampicin induces ALA synthase resulting in increased protoporphyrin IX. Isoniazid, through a yet to be determined mechanism, might inhibit ferrochelatase resulting in blockage of further metabolism of this intermediate of heme synthesis. Data taken from [61].
6. Bile acid-induced hepatotoxicity
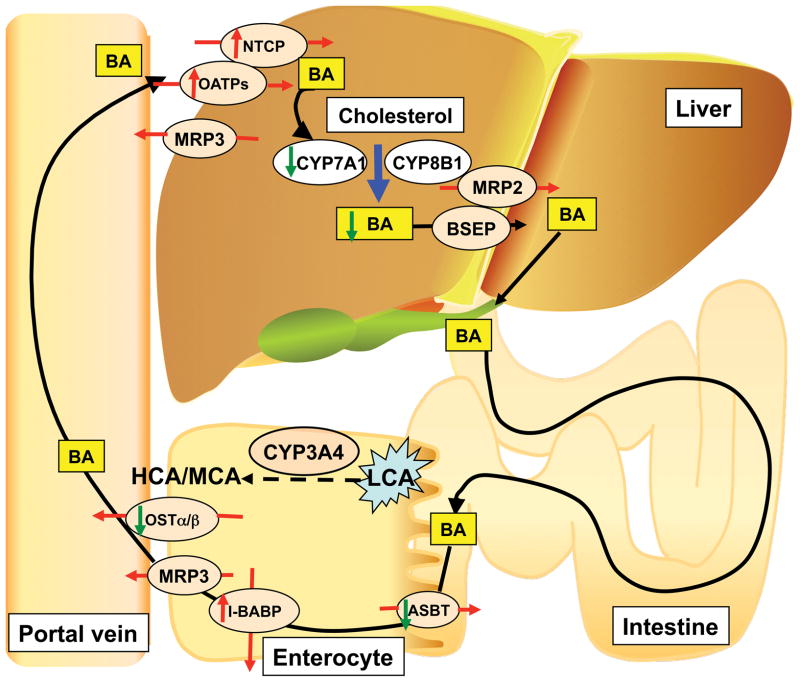
Bile acids are synthesized in the liver from cholesterol by CYP7A1 and CYP8B1. They are conjutated in mice with taruine (and by glycine in humans) and transported through the cannaliculus to the bile duct and into the intestine where they function to help digestion through the solubilization of dietary fats and vitamins [62, 63]. Bile acids are alsom metabolized by gut bacteria to secondary bile acids and >90% are reabsorbed to the blood stream and return to the liver through a process called enteroheptic circulation that is regulated by the farnesoid X receptor (FXR) [64, 65]. When bile acid synthesis and transport are disruped byt liver disease, marked hepatotoxocity can result in part through the hepatic accumulation of certain bile acid metabolites. Lithocholic acid (LCA) is a known hepatotoxin that accumulates in liver diseases such as cholestasis [66]. In another example of the value of gene knockout and humanized mice in exploring the mechanism of hepatotoxicity, mice lacking vitamin D receptor (VDR) expression in intestine and expressing human CYP3A4 in the inestine and liver (VdrΔIEpC/3A4) were used to investigate the role of intestine in modulation of bile acid homeostasis and toxicity in liver [67]. CYP3A4 can metabolize and detoxify LCA in liver [68] and earlier studies revealed that LCA can induced CYP3A4 expression in liver through activating VDR [69]. Compared with LCA-treated VdrΔIEpC mice which do not express CYP3A4, LCA-treated VdrΔIEpC/3A4 mice that express CYP3A4 showed decreased hepatotoxicity. Metabolomics analysis revealed that this protection was associated with increased LCA metabolism and detoxification, and suppression of bile acid transporter expression in the small intestine [67]. Thus, the small intestine, through VDR signaling, has a role in the protection from the hepatotoxicity of bile acids (Figure 6). This study, using genetically modified mice, shows how the intestine can, under certain circumstances, serve as a defense against bile acid-induced hepatotoxicity
Figure 6.
Proposed mechanism for the role of VDR and CYP3A4 in the protection against the hepatotoxicity of lithocholic acid. Abbreviations: ASBT, apical sodium-dependent bile acid transporter; BA, bile acid; BSEP, bile salt export protein; HCA, hydroxycholic acid; I-BABP, intestinal bile acid-binding protein; LCA, lithocholic acid; MCA, muricholic acid; MRP, multidrug-resistance protein; NTCP, Na+-taurocholate cotransporting polypeptide; OATP, organic ion transporting polypeptide; OST, organic solute and steroid transporter. Data taken from [64] and [67].
7. Ritonavir metabolism
Ritonavir (RTV) is an important component of protease inhibitor (PI) regimens for antiretroviral therapy. RTV inhibits CYP3A-mediated metabolism of PIs, thus increasing plasma concentrations of PIs [66] and making these RTV-boosted PI regimens are highly effective [70]. However, RTV-containing regimens increase the risk of liver injury and the detailed mechanisms of this RTV-induced liver injury remain unknown [71, 72]. Drug metabolism and bioactivation are associated with drug toxicity [73], and previous studies identified multiple metabolic pathways of RTV, including hydroxylation of the isopropyl side chain, N-demethylation, and cleavage of the terminal thiazole and isopropylthiazole group [74, 75]. However, it remains unclear whether RTV bioactivation pathways are associated with RTV-induced liver injury.
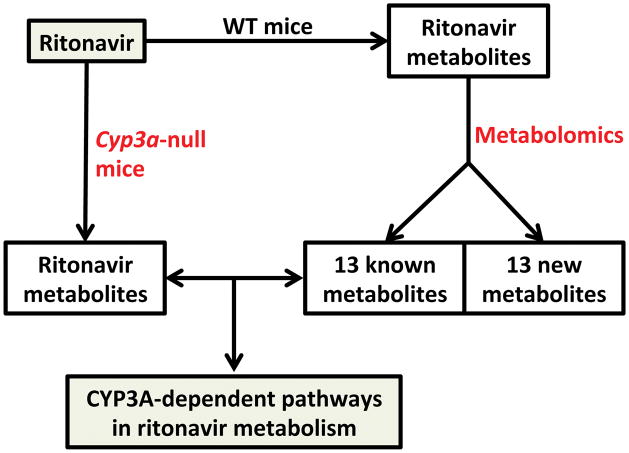
A recent study explored RTV metabolism using a metabolomic approach [76]. Twenty-six RTV metabolites were identified, including 13 previously reported metabolites and 13 novel metabolites. Among these new RTV metabolites, five were associated with RTV bioactivation [76]. RTV is a potent CYP3A inhibitor, but it is also a CYP3A substrate [74]. Thus, the role of CYP3A in RTV metabolism cannot be fully determined by in vitro experiments because RTV inhibits CYP3A. Therefore, the Cyp3a-null mouse model was employed, providing an ideal tool to determine the contribution of CYP3A to RTV metabolism in vivo. Compared to wild-type mice, several metabolic pathways of RTV, including the bioactivation, were decreased significantly in Cyp3a-null mice, indicating that these metabolic pathways of RTV are CYP3A-dependent [76]. CYP3A is highly expressed in the liver and small intestine [14], and the expression pattern of CYP3A may explain the gastrointestinal adverse effects of RTV, such as diarrhea, nausea, vomiting, and abdominal pain [71, 77]. CYP3A expression is highly inducible, and pre- or co-treatment with a CYP3A inducer may increase RTV bioactivation and potentiate RTV toxicity. Indeed, the hepatotoxicity of RTV-boosted PI regimens was observed in subjects who were pre-treated with rifampicin or efavirenz, a ligand and agonist of PXR that induces CYP3A expression [78–81]. This metabolomic analysis extended the metabolic map of RTV, which identified 13 known and 13 novel RTV metabolites, including 5 bioactivation pathways. Furthermore, CYP3A-dependent metabolic pathways of RTV were determined by using Cyp3a-null mice [76]. These data can be used to predict and prevent the potential drug-drug interactions and adverse drug reactions associated with RTV. This study also highlights the power of genetically engineered mouse models and the metabolomic approach in studying of drug metabolism (Figure 7).
Figure 7.
Determine the role of CYP3A in ritonavir metabolism using a metabolomic approach and Cyp3a-null mice. The metabolomic analysis extended the metabolic map of ritonavir. The CYP3A-dependent pathways in ritonavir metabolism were confirmed by using Cyp3a-null mice. Data taken from [76].
8. Endogenous biomarkers for xenobiotic toxicity
Xenobiotic-induced toxicity responses can be monitored by the altered levels of endogenous compounds that are produced after toxic insult [41]. For example acyl carnitines are found in the serum after acetaminophen overdose that resulted from disruption of mitochondrial function [47]. Metabolomics analysis can be used to both determine the extent of xenobiotic metabolism and the production of altered endogenous metabolites. However, exposure to high doses of xenobiotics resulted in metabolites derived from the xenobiotics that can interfere with the separation of the endogenous non-xenobiotic metabolites in the control vehicle-treated group and the xenobiotic-treated group. Therefore, discrimination of the contribution of xenobiotic metabolites from xenobiotics-induced endogenous compounds needs to be established in order to find toxicity biomarkers and reveal the detailed toxicity mechanisms. This can be accomplished by filtering out the xenobiotic and its known metabolites during the data analysis step. Stable isotope-based metabolomics is another possible method to achieve this task where it is feasible to synthesize the xenobiotic with a stable isotope such as deuterium or 13C [46]. Through comparing urinary ions from ethanol (C2H6O) and deuterated ethanol (C2D6O) treatment, a new metabolite of ethanol N-acetyltaurine was discovered that can function as a biomarker of hyperlacetatemia [82]. Stable isotope metabolomics can be used to discriminate xenobiotic-derived metabolites from xenobiotic-induced endogenous metabolites. For example, a metabolic map of ten tempol metabolites was uncovered along with other metabolites that resulted from the action of tempol on the intestinal microbiota [83].
9. Expert opinion
Genetically-modified mice have distinct advantages in the study of xenobiotic metabolism. Notably, they offer a means to precisely define the roles of specific enzymes in the metabolism of xenobiotics in vivo. The CYPs and phase 2 enzymes are ideally amenable to this technology since they are largely dispensible for normal development and physiological homeostasis. One or multiple enzymes can be disrupted and the mice are otherwise normal and can be used to study xenobiotic metabolism [28]. Similarly, the receptors such as AHR [84, 85], PXR [86, 87] and constitutive androstane receptor (CAR) [88] can be knocked out and the mice are viable, although abnormal immune, hepatic and reproductive phenotypes are present in the Ahr-null mice [84, 85]. These mouse models are now widely used and distributed by Jackson Laboratories and other commercial sources. Mice with disrupted phase 2 enzymes such as glutathione S-transferases [89], [90] and UDP-glucuronosyltransferases [91], have also been produced but discussion of these models is beyond the scope of this review.
This review largely focussed on the hepatic toxicity of xenobiotics that is largely driven by metabolism, and the use of metabolomics and genetically-modified mice to detemine the metabolic maps of xenobiotics, to uncover the mechanisms of xenobiotic-induced toxicities, define the genes involved in mediating toxicities and find biomarkers for toxicities. However, xenobiotics can also cause toxicity at other organ sites, such as the central nervous system, intestine, muscle (heart), bladder, kindey, lung and even skin. The mechanism of these toxicities in various organs can be diverse and may not be due to metabolism. While studies using 1H-NMR metabonomics have been used to find organ-specific endogenous metabolites that can be biomarkers for xenobiotic induced toxicities [41], the use of a more sensitive UPLC-ESI-QTOFMS metabolomics platform has not been exploited to find, using known organ-speficie toxicants, the metabolite biomarkers that reflect xenobiotic toxicities in specific organs.
To produce mouse lines that can be used to predict human xenobiotic metabolism in vivo, human CYP and nuclear receptor genes have been introduced into their respective null mouse counterparts. While mouse models humanized for specific CYPs and nuclear receptors have been generated [21], mice in which the complete complement of hepatic regulatory genes and xenobiotic-metabolizing genes have been replaced with their human counterparts have not been developed, although there are ongoing efforts in this area [28, 92, 93]. Ideally, however, it would be of great utility to have a rodent model in which the whole human liver is introduced. It should be noted that the genetically-modified mouse models, in particular the mice expressing human CYPs and xenobiotic receptors, offer a better predictive model for humans than do wild-type mice, they still have many differences from human liver. Thus, studies with human hepatocytes and liver extracts are vital. One possible solution, that has been studies for a number of years, are the chimeric liver replacement mouse models that use hepatocyte transplantation into immunocompromised mice that undergo liver failure [94–96]. These mice are highly predictive of human liver xenobiotic metabolism. However, these mice are expensive since each mouse needs to be custome made and cannot be propagated. They are not viable enough for longer term toxicity studies and they are still dependent on the liver used for the mouse liver replacement. There are other issues from this model, including differences in liver architecture between mice and humans, and influence of the immune system, which shows marked species differences [97], in xenobiotic-mediated toxicity [98].
As noted above, each human liver has a different levels of the various xenobiotic-metabolizing enzymes. One possibel solution to the variability of human livers is to use inducible pluripotent (iPS) cells to make a reliable and reneable source for hepatoctyes to use directly for xenobiotic metabolism studies and for reconstituting mice using the chimeric mouse model. Liver iPS cells is an area of active investigation which will likely lead to cells that are of value for xenobiotic metabolism and toxicity studies [6, 94, 95, 97–99]
As a key component of systems biology, metabolomics plays an increasingly important role in mechanistic elucidation of xenobiotic toxicity. Genetically-modified (including drug-metabolizing enzymes and nuclear receptors) mice can clearly elucidate the role of specific genes in xenobiotics-induced toxicity. The combination of these two key technologies can further promote the understanding of mechanisms of xenobitoic toxicity through analyzing the influence of specific genes towards the xenobiotics-induced alteration of endogenous compounds through the use of metabolic network modeling using a number of commercial and public data bases and programs. However, limitations exist for the application of metabolomics for genetically-modified mouse-based xenobiotic toxicity studies. Therefore, the following steps are recommended for use of metabolomics and genetically-modified mouse models into the study of xenobiotic metabolism and xenobiotic-induced toxicity: 1) Thorough comparison of metabolomics profiles between wild-type and genetically-modified mice, searching for genotype-dependent endogenous metabolites; 2) Based on the structures of a xenobiotic, searching and elucidation of metabolites derived from xenobiotics; 3) Discovery of specific alteration of endogenous compounds induced by xenobiotics-induced toxicity.
Article highlights box.
Genetically modified mouse models have served as ideal tools to determine the role of a specific gene in xenobiotic metabolism and xenobiotic-induced toxicity.
Metabolomics has been used to investigate xenobiotic metabolism and endobiotic metabolism and their associations with xenobiotic-mediated toxicity.
Transgenic mice and metabolomics provide deep insights for the mechanism of acetaminophen-induced hepatotoxicity.
Transgenic mice and metabolomics provide novel insights into the toxicities of alcohol; 2,3,7,8-tetrachlorodibenzo-p-dioxin; isoniazid and rifampicin; bile acid; and ritonavir.
Transgenic mice and metabolomics provide the state-of-art appraoches for research in Pharmacology and Toxicology.
Footnotes
Financial and competing interests disclosure
The authors were supported in part by the National Cancer Institute Intramural Research Program, National Institute of Diabetes and Digestive and Kidney Diseases [DK090305] and the National Institute of Allergy and Infectious Diseases [AI095425]. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Papers of special note have been highlighted as either of importance (*) or of considerable importance (**) to the research field.
- 1.Hornberg JJ, Mow T. How can we discover safer drugs? Future Med Chem. 2014;6:481–3. doi: 10.4155/fmc.14.15. [DOI] [PubMed] [Google Scholar]
- 2.Gonzalez FJ. The 2006 Bernard B. Brodie Award Lecture. Cyp2e1. Drug metabolism and disposition: the biological fate of chemicals. 2007;35:1–8. doi: 10.1124/dmd.106.012492. [DOI] [PubMed] [Google Scholar]
- 3.McGill MR, Staggs VS, Sharpe MR, Lee WM, Jaeschke H. Serum mitochondrial biomarkers and damage-associated molecular patterns are higher in acetaminophen overdose patients with poor outcome. Hepatology. 2014 doi: 10.1002/hep.27265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh SS. Preclinical pharmacokinetics: an approach towards safer and efficacious drugs. Current drug metabolism. 2006;7:165–82. doi: 10.2174/138920006775541552. [DOI] [PubMed] [Google Scholar]
- 5.Greer ML, Barber J, Eakins J, Kenna JG. Cell based approaches for evaluation of drug-induced liver injury. Toxicology. 2010;268:125–31. doi: 10.1016/j.tox.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz RE, Fleming HE, Khetani SR, Bhatia SN. Pluripotent stem cell-derived hepatocyte-like cells. Biotechnology advances. 2014;32:504–13. doi: 10.1016/j.biotechadv.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khetani SR, Berger DR, Ballinger KR, Davidson MD, Lin C, Ware BR. Microengineered Liver Tissues for Drug Testing. Journal of laboratory automation. 2015 doi: 10.1177/2211068214566939. [DOI] [PubMed] [Google Scholar]
- 8.Monks TJ, Lau SS. Reactive intermediates and their toxicological significance. Toxicology. 1988;52:1–53. doi: 10.1016/0300-483x(88)90195-3. [DOI] [PubMed] [Google Scholar]
- 9.Leblanc A, Shiao TC, Roy R, Sleno L. Improved detection of reactive metabolites with a bromine-containing glutathione analog using mass defect and isotope pattern matching. Rapid Commun Mass Spectrom. 2010;24:1241–50. doi: 10.1002/rcm.4507. [DOI] [PubMed] [Google Scholar]
- 10.Li F, Lu J, Ma X. Profiling the reactive metabolites of xenobiotics using metabolomic technologies. Chemical research in toxicology. 2011;24:744–51. doi: 10.1021/tx200033v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li AP. In vitro human hepatocyte-based experimental systems for the evaluation of human drug metabolism, drug-drug interactions, and drug toxicity in drug development. Current topics in medicinal chemistry. 2014;14:1325–38. doi: 10.2174/1568026614666140506114411. [DOI] [PubMed] [Google Scholar]
- 12.Wilkinson GR. Cytochrome P4503A (CYP3A) metabolism: prediction of in vivo activity in humans. Journal of pharmacokinetics and biopharmaceutics. 1996;24:475–90. doi: 10.1007/BF02353475. [DOI] [PubMed] [Google Scholar]
- 13.Guengerich FP, Gillam EM, Shimada T. New applications of bacterial systems to problems in toxicology. Critical reviews in toxicology. 1996;26:551–83. doi: 10.3109/10408449609037477. [DOI] [PubMed] [Google Scholar]
- 14.Guengerich FP. Cytochrome P-450 3A4: regulation and role in drug metabolism. Annual review of pharmacology and toxicology. 1999;39:1–17. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- 15.McKinnon RA, Burgess WM, Hall PM, Roberts-Thomson SJ, Gonzalez FJ, McManus ME. Characterisation of CYP3A gene subfamily expression in human gastrointestinal tissues. Gut. 1995;36:259–67. doi: 10.1136/gut.36.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Granvil CP, Yu AM, Elizondo G, Akiyama TE, Cheung C, Feigenbaum L, et al. Expression of the human CYP3A4 gene in the small intestine of transgenic mice: in vitro metabolism and pharmacokinetics of midazolam. Drug metabolism and disposition: the biological fate of chemicals. 2003;31:548–58. doi: 10.1124/dmd.31.5.548. [DOI] [PubMed] [Google Scholar]
- 17.Cheng J, Ma X, Gonzalez FJ. Pregnane X receptor- and CYP3A4-humanized mouse models and their applications. British journal of pharmacology. 2011;163:461–8. doi: 10.1111/j.1476-5381.2010.01129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kroetz DL, Yee SW, Giacomini KM. The pharmacogenomics of membrane transporters project: research at the interface of genomics and transporter pharmacology. Clinical pharmacology and therapeutics. 2010;87:109–16. doi: 10.1038/clpt.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan Q. Membrane transporters and drug development: relevance to pharmacogenomics, nutrigenomics, epigenetics, and systems biology. Methods in molecular biology. 2010;637:1–21. doi: 10.1007/978-1-60761-700-6_1. [DOI] [PubMed] [Google Scholar]
- 20.Jones SA, Moore LB, Shenk JL, Wisely GB, Hamilton GA, McKee DD, et al. The pregnane X receptor: a promiscuous xenobiotic receptor that has diverged during evolution. Molecular endocrinology. 2000;14:27–39. doi: 10.1210/mend.14.1.0409. [DOI] [PubMed] [Google Scholar]
- 21.Cheung C, Gonzalez FJ. Humanized mouse lines and their application for prediction of human drug metabolism and toxicological risk assessment. The Journal of pharmacology and experimental therapeutics. 2008;327:288–99. doi: 10.1124/jpet.108.141242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheer N, Snaith M, Wolf CR, Seibler J. Generation and utility of genetically humanized mouse models. Drug Discov Today. 2013;18:1200–11. doi: 10.1016/j.drudis.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Scheer N, Wolf CR. Genetically humanized mouse models of drug metabolizing enzymes and transporters and their applications. Xenobiotica; the fate of foreign compounds in biological systems. 2014;44:96–108. doi: 10.3109/00498254.2013.815831. [DOI] [PubMed] [Google Scholar]
- 24.Shen HW, Jiang XL, Gonzalez FJ, Yu AM. Humanized transgenic mouse models for drug metabolism and pharmacokinetic research. Current drug metabolism. 2011;12:997–1006. doi: 10.2174/138920011798062265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boverhof DR, Chamberlain MP, Elcombe CR, Gonzalez FJ, Heflich RH, Hernandez LG, et al. Transgenic animal models in toxicology: historical perspectives and future outlook. Toxicological sciences: an official journal of the Society of Toxicology. 2011;121:207–33. doi: 10.1093/toxsci/kfr075. [DOI] [PubMed] [Google Scholar]
- 26.Henderson CJ, Otto DM, Carrie D, Magnuson MA, McLaren AW, Rosewell I, et al. Inactivation of the hepatic cytochrome P450 system by conditional deletion of hepatic cytochrome P450 reductase. The Journal of biological chemistry. 2003;278:13480–6. doi: 10.1074/jbc.M212087200. [DOI] [PubMed] [Google Scholar]
- 27.Wu L, Gu J, Weng Y, Kluetzman K, Swiatek P, Behr M, et al. Conditional knockout of the mouse NADPH-cytochrome p450 reductase gene. Genesis. 2003;36:177–81. doi: 10.1002/gene.10214. [DOI] [PubMed] [Google Scholar]
- 28.Scheer N, McLaughlin LA, Rode A, Macleod AK, Henderson CJ, Wolf CR. Deletion of 30 murine cytochrome P450 genes results in viable mice with compromised drug metabolism. Drug metabolism and disposition: the biological fate of chemicals. 2014;42:1022–30. doi: 10.1124/dmd.114.057885. [DOI] [PubMed] [Google Scholar]
- 29.Zaher H, Buters JT, Ward JM, Bruno MK, Lucas AM, Stern ST, et al. Protection against acetaminophen toxicity in CYP1A2 and CYP2E1 double-null mice. Toxicology and applied pharmacology. 1998;152:193–9. doi: 10.1006/taap.1998.8501. [DOI] [PubMed] [Google Scholar]
- 30.Heidel SM, MacWilliams PS, Baird WM, Dashwood WM, Buters JT, Gonzalez FJ, et al. Cytochrome P4501B1 mediates induction of bone marrow cytotoxicity and preleukemia cells in mice treated with 7,12-dimethylbenz[a]anthracene. Cancer research. 2000;60:3454–60. [PubMed] [Google Scholar]
- 31.Nebert DW, Shi Z, Galvez-Peralta M, Uno S, Dragin N. Oral benzo[a]pyrene: understanding pharmacokinetics, detoxication, and consequences--Cyp1 knockout mouse lines as a paradigm. Molecular pharmacology. 2013;84:304–13. doi: 10.1124/mol.113.086637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson CH, Patterson AD, Idle JR, Gonzalez FJ. Xenobiotic metabolomics: major impact on the metabolome. Annual review of pharmacology and toxicology. 2012;52:37–56. doi: 10.1146/annurev-pharmtox-010611-134748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang ZZ, Gonzalez FJ. LC-MS-based metabolomics: an update. Arch Toxicol. 2014;88:1491–502. doi: 10.1007/s00204-014-1234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen C, Ma X, Malfatti MA, Krausz KW, Kimura S, Felton JS, et al. A comprehensive investigation of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) metabolism in the mouse using a multivariate data analysis approach. Chemical research in toxicology. 2007;20:531–42. doi: 10.1021/tx600320w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beger RD, Sun J, Schnackenberg LK. Metabolomics approaches for discovering biomarkers of drug-induced hepatotoxicity and nephrotoxicity. Toxicology and applied pharmacology. 2010;243:154–66. doi: 10.1016/j.taap.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 36.Geenen S, Taylor PN, Snoep JL, Wilson ID, Kenna JG, Westerhoff HV. Systems biology tools for toxicology. Arch Toxicol. 2012;86:1251–71. doi: 10.1007/s00204-012-0857-8. [DOI] [PubMed] [Google Scholar]
- 37.Bouhifd M, Hartung T, Hogberg HT, Kleensang A, Zhao L. Review: toxicometabolomics. Journal of applied toxicology: JAT. 2013;33:1365–83. doi: 10.1002/jat.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindon JC, Keun HC, Ebbels TM, Pearce JM, Holmes E, Nicholson JK. The Consortium for Metabonomic Toxicology (COMET): aims, activities and achievements. Pharmacogenomics. 2005;6:691–9. doi: 10.2217/14622416.6.7.691. [DOI] [PubMed] [Google Scholar]
- 39.Boudonck KJ, Rose DJ, Karoly ED, Lee DP, Lawton KA, Lapinskas PJ. Metabolomics for early detection of drug-induced kidney injury: review of the current status. Bioanalysis. 2009;1:1645–63. doi: 10.4155/bio.09.142. [DOI] [PubMed] [Google Scholar]
- 40.Kamp H, Fabian E, Groeters S, Herold M, Krennrich G, Looser R, et al. Application of in vivo metabolomics to preclinical/toxicological studies: case study on phenytoin-induced systemic toxicity. Bioanalysis. 2012;4:2291–301. doi: 10.4155/bio.12.214. [DOI] [PubMed] [Google Scholar]
- 41.Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nature reviews Drug discovery. 2002;1:153–61. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- 42.Coen M, Holmes E, Lindon JC, Nicholson JK. NMR-based metabolic profiling and metabonomic approaches to problems in molecular toxicology. Chemical research in toxicology. 2008;21:9–27. doi: 10.1021/tx700335d. [DOI] [PubMed] [Google Scholar]
- 43.Song BJ, Veech RL, Park SS, Gelboin HV, Gonzalez FJ. Induction of rat hepatic N-nitrosodimethylamine demethylase by acetone is due to protein stabilization. The Journal of biological chemistry. 1989;264:3568–72. [PubMed] [Google Scholar]
- 44.Koop DR, Casazza JP. Identification of ethanol-inducible P-450 isozyme 3a as the acetone and acetol monooxygenase of rabbit microsomes. The Journal of biological chemistry. 1985;260:13607–12. [PubMed] [Google Scholar]
- 45.Chen C, Krausz KW, Idle JR, Gonzalez FJ. Identification of novel toxicity-associated metabolites by metabolomics and mass isotopomer analysis of acetaminophen metabolism in wild-type and Cyp2e1-null mice. The Journal of biological chemistry. 2008;283:4543–59. doi: 10.1074/jbc.M706299200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen C, Gonzalez FJ, Idle JR. LC-MS-based metabolomics in drug metabolism. Drug Metab Rev. 2007;39:581–97. doi: 10.1080/03602530701497804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen C, Krausz KW, Shah YM, Idle JR, Gonzalez FJ. Serum metabolomics reveals irreversible inhibition of fatty acid beta-oxidation through the suppression of PPARalpha activation as a contributing mechanism of acetaminophen-induced hepatotoxicity. Chemical research in toxicology. 2009;22:699–707. doi: 10.1021/tx800464q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonzalez FJ. Animal models for human risk assessment: the peroxisome proliferator-activated receptor alpha-humanized mouse. Nutrition reviews. 2007;65:S2–6. doi: 10.1111/j.1753-4887.2007.tb00323.x. [DOI] [PubMed] [Google Scholar]
- 49.Jogl G, Hsiao YS, Tong L. Structure and function of carnitine acyltransferases. Annals of the New York Academy of Sciences. 2004;1033:17–29. doi: 10.1196/annals.1320.002. [DOI] [PubMed] [Google Scholar]
- 50.Patterson AD, Shah YM, Matsubara T, Krausz KW, Gonzalez FJ. Peroxisome proliferator-activated receptor alpha induction of uncoupling protein 2 protects against acetaminophen-induced liver toxicity. Hepatology. 2012;56:281–90. doi: 10.1002/hep.25645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Donadelli M, Dando I, Fiorini C, Palmieri M. UCP2, a mitochondrial protein regulated at multiple levels. Cellular and molecular life sciences: CMLS. 2014;71:1171–90. doi: 10.1007/s00018-013-1407-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neuman MG, French SW, French BA, Seitz HK, Cohen LE, Mueller S, et al. Alcoholic and non-alcoholic steatohepatitis. Experimental and molecular pathology. 2014 doi: 10.1016/j.yexmp.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakajima T, Kamijo Y, Tanaka N, Sugiyama E, Tanaka E, Kiyosawa K, et al. Peroxisome proliferator-activated receptor alpha protects against alcohol-induced liver damage. Hepatology. 2004;40:972–80. doi: 10.1002/hep.20399. [DOI] [PubMed] [Google Scholar]
- 54.Manna SK, Patterson AD, Yang Q, Krausz KW, Li H, Idle JR, et al. Identification of noninvasive biomarkers for alcohol-induced liver disease using urinary metabolomics and the Ppara-null mouse. J Proteome Res. 2010;9:4176–88. doi: 10.1021/pr100452b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manna SK, Patterson AD, Yang Q, Krausz KW, Idle JR, Fornace AJ, et al. UPLC-MS-based urine metabolomics reveals indole-3-lactic acid and phenyllactic acid as conserved biomarkers for alcohol-induced liver disease in the Ppara-null mouse model. J Proteome Res. 2011;10:4120–33. doi: 10.1021/pr200310s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murray IA, Patterson AD, Perdew GH. Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nature reviews Cancer. 2014;14:801–14. doi: 10.1038/nrc3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsubara T, Tanaka N, Krausz KW, Manna SK, Kang DW, Anderson ER, et al. Metabolomics identifies an inflammatory cascade involved in dioxin- and diet-induced steatohepatitis. Cell Metab. 2012;16:634–44. doi: 10.1016/j.cmet.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stehr M, Elamin AA, Singh M. Filling the pipeline - new drugs for an old disease. Current topics in medicinal chemistry. 2014;14:110–29. doi: 10.2174/1568026613666131113152908. [DOI] [PubMed] [Google Scholar]
- 59.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A. 2009;106:14728–33. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saukkonen JJ, Cohn DL, Jasmer RM, Schenker S, Jereb JA, Nolan CM, et al. An official ATS statement: Hepatotoxicity of antituberculosis therapy. Amer J Resp Crit Care Med. 2006;174:935–52. doi: 10.1164/rccm.200510-1666ST. [DOI] [PubMed] [Google Scholar]
- 61.Li F, Lu J, Cheng J, Wang L, Matsubara T, Csanaky IL, et al. Human PXR modulates hepatotoxicity associated with rifampicin and isoniazid co-therapy. Nat Med. 2013;19:418–20. doi: 10.1038/nm.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hofmann AF. Bile acids: trying to understand their chemistry and biology with the hope of helping patients. Hepatology. 2009;49:1403–18. doi: 10.1002/hep.22789. [DOI] [PubMed] [Google Scholar]
- 63.Chiang JY. Bile acid metabolism and signaling. Comprehensive Physiology. 2013;3:1191–212. doi: 10.1002/cphy.c120023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gonzalez FJ. Nuclear receptor control of enterohepatic circulation. Comprehensive Physiology. 2012;2:2811–28. doi: 10.1002/cphy.c120007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matsubara T, Li F, Gonzalez FJ. FXR signaling in the enterohepatic system. Mol Cell Endocrinol. 2013;368:17–29. doi: 10.1016/j.mce.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bierman WF, van Agtmael MA, Nijhuis M, Danner SA, Boucher CA. HIV monotherapy with ritonavir-boosted protease inhibitors: a systematic review. Aids. 2009;23:279–91. doi: 10.1097/QAD.0b013e32831c54e5. [DOI] [PubMed] [Google Scholar]
- 67.Cheng J, Fang ZZ, Kim JH, Krausz KW, Tanaka N, Chiang JY, et al. Intestinal CYP3A4 protects against lithocholic acid-induced hepatotoxicity in intestine-specific VDR-deficient mice. J Lipid Res. 2014;55:455–65. doi: 10.1194/jlr.M044420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deo AK, Bandiera SM. 3-ketocholanoic acid is the major in vitro human hepatic microsomal metabolite of lithocholic acid. Drug metabolism and disposition: the biological fate of chemicals. 2009;37:1938–47. doi: 10.1124/dmd.109.027763. [DOI] [PubMed] [Google Scholar]
- 69.Matsubara T, Yoshinari K, Aoyama K, Sugawara M, Sekiya Y, Nagata K, et al. Role of vitamin D receptor in the lithocholic acid-mediated CYP3A induction in vitro and in vivo. Drug metabolism and disposition: the biological fate of chemicals. 2008;36:2058–63. doi: 10.1124/dmd.108.021501. [DOI] [PubMed] [Google Scholar]
- 70.Merry C, Barry MG, Mulcahy F, Ryan M, Heavey J, Tjia JF, et al. Saquinavir pharmacokinetics alone and in combination with ritonavir in HIV-infected patients. Aids. 1997;11:F29–33. doi: 10.1097/00002030-199704000-00001. [DOI] [PubMed] [Google Scholar]
- 71.Sulkowski MS. Hepatotoxicity associated with antiretroviral therapy containing HIV-1 protease inhibitors. Seminars in liver disease. 2003;23:183–94. doi: 10.1055/s-2003-39949. [DOI] [PubMed] [Google Scholar]
- 72.Bruno R, Sacchi P, Maiocchi L, Patruno S, Filice G. Hepatotoxicity and antiretroviral therapy with protease inhibitors: A review. Digestive and liver disease: official journal of the Italian Society of Gastroenterology and the Italian Association for the Study of the Liver. 2006;38:363–73. doi: 10.1016/j.dld.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 73.Guengerich FP, MacDonald JS. Applying mechanisms of chemical toxicity to predict drug safety. Chemical research in toxicology. 2007;20:344–69. doi: 10.1021/tx600260a. [DOI] [PubMed] [Google Scholar]
- 74.Kumar GN, Rodrigues AD, Buko AM, Denissen JF. Cytochrome P450-mediated metabolism of the HIV-1 protease inhibitor ritonavir (ABT-538) in human liver microsomes. The Journal of pharmacology and experimental therapeutics. 1996;277:423–31. [PubMed] [Google Scholar]
- 75.Denissen JF, Grabowski BA, Johnson MK, Buko AM, Kempf DJ, Thomas SB, et al. Metabolism and disposition of the HIV-1 protease inhibitor ritonavir (ABT-538) in rats, dogs, and humans. Drug metabolism and disposition: the biological fate of chemicals. 1997;25:489–501. [PubMed] [Google Scholar]
- 76.Li F, Lu J, Ma X. Metabolomic screening and identification of the bioactivation pathways of ritonavir. Chemical research in toxicology. 2011;24:2109–14. doi: 10.1021/tx2004147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Struble KA, Pratt RD, Gitterman SR. Toxicity of antiretroviral agents. The American journal of medicine. 1997;102:65–7. doi: 10.1016/s0002-9343(97)00065-x. discussion 68–9. [DOI] [PubMed] [Google Scholar]
- 78.Hariparsad N, Nallani SC, Sane RS, Buckley DJ, Buckley AR, Desai PB. Induction of CYP3A4 by efavirenz in primary human hepatocytes: comparison with rifampin and phenobarbital. Journal of clinical pharmacology. 2004;44:1273–81. doi: 10.1177/0091270004269142. [DOI] [PubMed] [Google Scholar]
- 79.Haas DW, Koletar SL, Laughlin L, Kendall MA, Suckow C, Gerber JG, et al. Hepatotoxicity and gastrointestinal intolerance when healthy volunteers taking rifampin add twice-daily atazanavir and ritonavir. Journal of acquired immune deficiency syndromes. 2009;50:290–3. doi: 10.1097/QAI.0b013e318189a7df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jamois C, Riek M, Schmitt C. Potential hepatotoxicity of efavirenz and saquinavir/ritonavir coadministration in healthy volunteers. Archives of drug information. 2009;2:1–7. doi: 10.1111/j.1753-5174.2009.00016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nijland HM, L’Homme RF, Rongen GA, van Uden P, van Crevel R, Boeree MJ, et al. High incidence of adverse events in healthy volunteers receiving rifampicin and adjusted doses of lopinavir/ritonavir tablets. Aids. 2008;22:931–5. doi: 10.1097/QAD.0b013e3282faa71e. [DOI] [PubMed] [Google Scholar]
- 82.Shi X, Yao D, Chen C. Identification of N-acetyltaurine as a novel metabolite of ethanol through metabolomics-guided biochemical analysis. The Journal of biological chemistry. 2012;287:6336–49. doi: 10.1074/jbc.M111.312199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li F, Pang X, Krausz KW, Jiang C, Chen C, Cook JA, et al. Stable isotope- and mass spectrometry-based metabolomics as tools in drug metabolism: a study expanding tempol pharmacology. J Proteome Res. 2013;12:1369–76. doi: 10.1021/pr301023x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fernandez-Salguero P, Pineau T, Hilbert DM, McPhail T, Lee SS, Kimura S, et al. Immune system impairment and hepatic fibrosis in mice lacking the dioxin-binding Ah receptor. Science. 1995;268:722–6. doi: 10.1126/science.7732381. [DOI] [PubMed] [Google Scholar]
- 85.Schmidt JV, Su GH, Reddy JK, Simon MC, Bradfield CA. Characterization of a murine Ahr null allele: involvement of the Ah receptor in hepatic growth and development. Proc Natl Acad Sci U S A. 1996;93:6731–6. doi: 10.1073/pnas.93.13.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A. 2001;98:3369–74. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xie W, Barwick JL, Downes M, Blumberg B, Simon CM, Nelson MC, et al. Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature. 2000;406:435–9. doi: 10.1038/35019116. [DOI] [PubMed] [Google Scholar]
- 88.Wei P, Zhang J, Egan-Hafley M, Liang S, Moore DD. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature. 2000;407:920–3. doi: 10.1038/35038112. [DOI] [PubMed] [Google Scholar]
- 89.Henderson CJ, Smith AG, Ure J, Brown K, Bacon EJ, Wolf CR. Increased skin tumorigenesis in mice lacking pi class glutathione S-transferases. Proc Natl Acad Sci U S A. 1998;95:5275–80. doi: 10.1073/pnas.95.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Henderson CJ, Wolf CR. Knockout and transgenic mice in glutathione transferase research. Drug Metab Rev. 2011;43:152–64. doi: 10.3109/03602532.2011.562900. [DOI] [PubMed] [Google Scholar]
- 91.Nguyen N, Bonzo JA, Chen S, Chouinard S, Kelner MJ, Hardiman G, et al. Disruption of the ugt1 locus in mice resembles human Crigler-Najjar type I disease. The Journal of biological chemistry. 2008;283:7901–11. doi: 10.1074/jbc.M709244200. [DOI] [PubMed] [Google Scholar]
- 92.Scheer N, Ross J, Rode A, Zevnik B, Niehaves S, Faust N, et al. A novel panel of mouse models to evaluate the role of human pregnane X receptor and constitutive androstane receptor in drug response. J Clin Invest. 2008;118:3228–39. doi: 10.1172/JCI35483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Luisier R, Lempiainen H, Scherbichler N, Braeuning A, Geissler M, Dubost V, et al. Phenobarbital induces cell cycle transcriptional responses in mouse liver humanized for constitutive androstane and pregnane x receptors. Toxicological sciences: an official journal of the Society of Toxicology. 2014;139:501–11. doi: 10.1093/toxsci/kfu038. [DOI] [PubMed] [Google Scholar]
- 94.Tateno C, Yoshizane Y, Saito N, Kataoka M, Utoh R, Yamasaki C, et al. Near completely humanized liver in mice shows human-type metabolic responses to drugs. Am J Pathol. 2004;165:901–12. doi: 10.1016/S0002-9440(10)63352-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kakuni M, Morita M, Matsuo K, Katoh Y, Nakajima M, Tateno C, et al. Chimeric mice with a humanized liver as an animal model of troglitazone-induced liver injury. Toxicology letters. 2012;214:9–18. doi: 10.1016/j.toxlet.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 96.Kitamura S, Sugihara K. Current status of prediction of drug disposition and toxicity in humans using chimeric mice with humanized liver. Xenobiotica; the fate of foreign compounds in biological systems. 2014;44:123–34. doi: 10.3109/00498254.2013.868062. [DOI] [PubMed] [Google Scholar]
- 97.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–8. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 98.Takai S, Higuchi S, Yano A, Tsuneyama K, Fukami T, Nakajima M, et al. Involvement of immune- and inflammatory-related factors in flucloxacillin-induced liver injury in mice. Journal of applied toxicology: JAT. 2014 doi: 10.1002/jat.3002. [DOI] [PubMed] [Google Scholar]
- 99.Asgari S, Moslem M, Bagheri-Lankarani K, Pournasr B, Miryounesi M, Baharvand H. Differentiation and transplantation of human induced pluripotent stem cell-derived hepatocyte-like cells. Stem cell reviews. 2013;9:493–504. doi: 10.1007/s12015-011-9330-y. [DOI] [PubMed] [Google Scholar]