Abstract
Hydrogen sulfide (H2S) functions in many physiological processes, including relaxation of vascular smooth muscles, mediation of neurotransmission, inhibition of insulin signaling, and regulation of inflammation. On the other hand, sulfane sulfur, which is a sulfur atom with six valence electrons but no charge, has the unique ability to bind reversibly to other sulfur atoms to form hydropersulfides (R-S-SH) and polysulfides (-S-Sn-S-). H2S and sulfane sulfur always coexist, and recent work suggests that sulfane sulfur species may be the actual signaling molecules in at least some biological phenomena. For example, one of the mechanisms of activity regulation of proteins by H2S is the S-sulfhydration of cysteine residues (protein Cys-SSH). In this review, we summarize recent progress on chemical tools for the study of H2S and sulfane sulfur, covering fluorescence probes utilizing various design strategies, H2S caged compounds, inhibitors of physiological H2S-producing enzymes (cystathionine γ-lyase, cystathionine β-synthase and 3-mercaptopyruvate sulfurtransferase), and labeling reagents. Fluorescence probes offer particular advantages as chemical tools to study physiological functions of biomolecules, including ease of use and real-time, nondestructive visualization of biological processes in live cells and tissues.
Keywords: hydrogen sulfide, sulfane sulfur, fluorescence probe, caged compound, enzyme inhibitor
Introduction
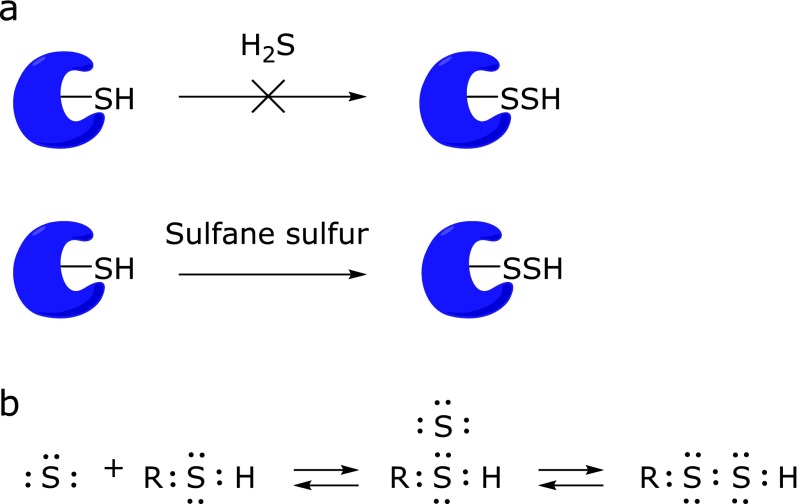
Hydrogen sulfide (H2S), a toxic gas smelling of rotten eggs, plays key roles in many physiological processes, including relaxation of vascular smooth muscles,(1,2) mediation of neurotransmission,(3,4) inhibition of insulin signaling,(5) and regulation of inflammation.(6,7) The most commonly used detection methods for H2S are the methylene blue method,(8) the electrode method(9) and the monobromobimane method.(10,11) However, these methods require destructive sampling, e.g., homogenization of biological samples. On the other hand, fluorescence detection has been widely used in biological studies to study the physiological roles of H2S, because this technology is easy to use, and enables real-time, nondestructive detection in living cells and tissues. Many selective fluorescence probes for H2S have been reported.(12–14) On the other hand, sulfane sulfur species consist of sulfur atom(s) with six valence electrons but no charge bound to other sulfur atom(s), as in hydropersulfides (R-S-SH) and polysulfides (-S-Sn-S-). They are attracting increasing interest, because it is reported that one of the mechanisms of activity regulation of proteins by H2S is S-sulfhydration of cysteine residues (SH→SSH).(2,15) Since H2S and polysulfide are redox partners, they would coexist in biological systems, and polysulfide seems likely to be much more effective than H2S in S-sulfhydration from a reactivity point of view.(16) Therefore, sulfane sulfur is considered to mediate at least some of the biological activities of H2S.
In this review, we summarize recent work on chemical tools (especially fluorescence probes) for the study of H2S and sulfane sulfur, and we briefly review their applications to biological studies.
Chemical Characteristics of H2S
H2S is highly water-soluble, with pKa1 of 6.8 and pKa2 of approximately 14;(17) consequently, 80% of H2S exists as strongly nucleophilic HS− at pH 7.4. In addition, H2S itself shows reducing ability, changing the oxidation state of sulfur from –2 (H2S) to 0 (S); for example, it can reduce azide and nitro groups.(18,19) These properties have been utilized to develop off/on-type fluorescence probes for H2S.
Development of Fluorescence Probes for H2S
Fluorescence probes based on reduction of azide or nitro group to amino group
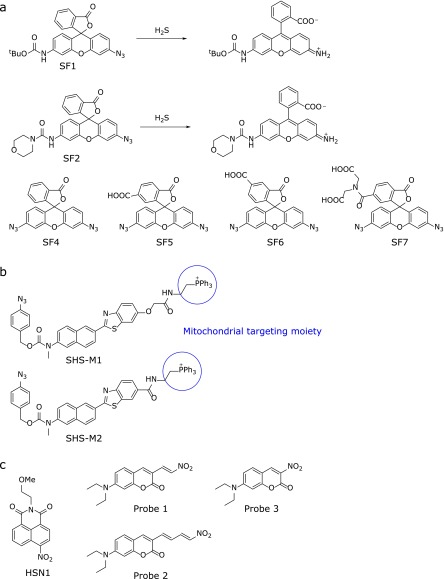
The first fluorescence probes utilizing azide reduction were SF1 and SF2, reported by Lippert et al.(20) (Fig. 1a). Reduction of the azide group of the xanthene moiety to amine leads to opening of the intramolecular spirocycle of the rhodamine scaffold; this restores the conjugated system of the xanthene moiety, leading to strong fluorescence (SF1 and SF2, ΦFl = 0.50 and 0.60, respectively). SF1 and SF2 showed 7- and 9-fold fluorescence increases, respectively, within 1 h after addition of 100 µM NaHS. The same authors subsequently developed improved probes (SF4-7) with enhanced sensitivity and cellular retention, using the same strategy (Fig. 1a).(21) Moreover, H2S fluorescence probes targeting specific organelles have also been developed. For example, Bae et al.(22) reported two fluorescence probes, SHS-M1 and SHS-M2, which incoporate a triphenylphosphonium group as a mitochondrial targeting moiety (Fig. 1b). Like the azide group, the nitro group can be reduced by H2S; Montoya et al.(23) reported a fluorescence probe based on this design concept (HSN1; Fig. 1c left), while Wu et al.(24) reported colorimetric and ratiometric probes (Fig. 1c middle and right). Thus, several types of H2S fluorescence probes utilizing azide or nitro group reduction have been reported.(12–14)
Fig. 1.
Fluorescence probes for H2S: (a) Probes utilizing reduction of an azide group. (b) Mitochondrially targeted probes utilizing reduction of azide. (c) Probes based on reduction of a nitro group.
Fluorescence probes based on the nucleophilicity of HS-
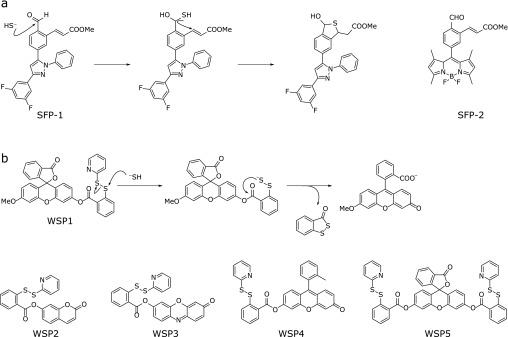
H2S (HS−) has strong nucleophilicity, which was utilized by Qian et al.(25) to design fluorescence probes SFP-1 and SFP-2 (Fig. 2a). In these probes, fluorescence off/on switching occurs via HS− addition to aldehyde, followed by Michael addition of the resulting intermediate to unsaturated methyl acrylate to form a thiohemiacetal under physiological conditions. The resulting stable tetrahydrothiophene shows strong fluorescence. Liu et al.(26) and Peng et al.(27) reported a fluorescein-based probe, WSP1-5 (Fig. 2b), in which the disulfide bond is cleaved by H2S followed by intramolecular nucleophilic attack of the persulfide group on the ester moiety; this releases the fluorophore, resulting in a large fluorescence increase.
Fig. 2.
Fluorescence probes for H2S utilizing nucleophilic reaction of HS−.
Fluorescence probes based on the quenching effect of copper ion (Cu2+)
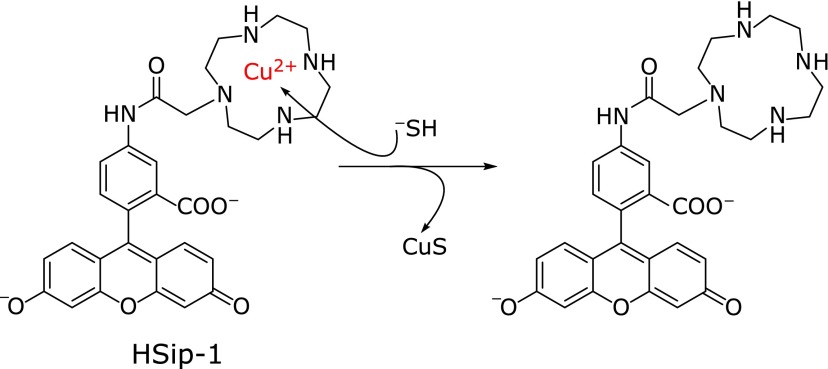
It is well known that heavy metal ions such as iron (III) ion (Fe3+) and Cu2+ quench the fluorescence of a nearby fluorophore,(28) and precipitation of CuS has been adapted for H2S detection by incorporating a Cu2+ complex moiety in the fluorophore. Choi et al.(29) reported that a dipicolylamine (DPA)–fluorescein complex with Cu2+ showed a turn-on fluorescence response to H2S, but without selectivity over other biothiols. Our group then designed and synthesized a H2S fluorescence probe, HSip-1, in which Cu2+ is complexed with an azamacrocyclic ring, which forms stable complex with Cu2+ (Fig. 3).(30) We expected that Cu2+ would be released from the azamacrocyclic ring when H2S binds to the Cu2+ center, resulting in a large fluorescence enhancement with selectivity against other biothiols. Indeed, HSip-1 showed a large and fast fluorescence increase after addition of 100 µM Na2S. It also has extremely high water-solubility, so that DMSO (dimethylsulfoxide) or detergent is not needed as a cosolvent. Further, although probes that release CuS may show cytotoxicity, HSip-1 was not toxic to living cells.(30) These properties make HSip-1 particularly suitable for biological studies both in vitro and in vivo.
Fig. 3.
Fluorescence probes based on the fluorescence quenching effect of Cu2+.
Development of Photocontrollable Hydrogen Sulfide Donors
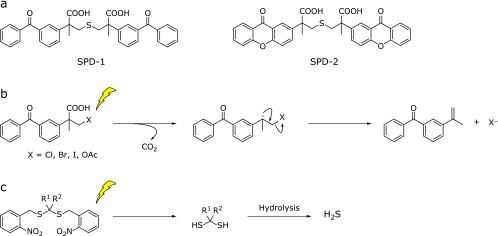
In biological research on H2S, inorganic sulfide salts have been widely used as H2S donors. However, H2S generation from these salts is rapid and does not mimic endogenous H2S release. Instead, controllable H2S donors are required for detailed investigation of the physiological functions of H2S. Fukushima et al.(31) reported a photolysis-induced H2S donor, SPD-1, which enables precise control of the location, timing and dosage of H2S release by means of light irradiation (Fig. 4a). SPD-1 is a caged compound, in which H2S is directly protected by ketoprofenate, and generates H2S proportionally to the irradiation time and light intensity, with simultaneous release of a photoproduct, 2-propenylbenzophenone (Fig. 4b).(32) They subsequently developed an improved H2S donor, SPD-2, in which xanthone is used as a photolabile protecting group (Fig. 4a).(33) SPD-2 has longer absorbance wavelength than SPD-1, and the generation of H2S can be precisely controlled by irradiation in the UVA range (325–385 nm). SPD-2 shows more efficient H2S production than SPD-1, presumably because of its absorption wavelength range. Devarie-Baez et al.(34) also reported a caged H2S donor based on the structure of geminal-dithiols (Fig. 4c), which are unstable in aqueous solutions, releasing H2S. They protected the free SH of gem-dithiol with 2-nitrobenzyl, a widely used type of caging chromophore, to obtain a stable gem-dithiol-based H2S donor. The free gem-dithiol intermediate is produced upon light irradiaton and hydrolysis of this intermediate affords H2S.
Fig. 4.
(a) Chemical structures of SPD-1 and SPD-2. (b) Photoreaction mechanism of ketoprofenate-based caged compounds. (c) Reaction mechanism of caged gem-dithiol.
Physiological Functions of Sulfane Sulfur
H2S has been suggested to be an endogenous signaling molecule, and one of its regulatory mechanisms is thought to be S-sulfhydration of protein cysteine residues (SH→SSH).(2,15) However, H2S is a fully reduced sulfur species and is basically a reductant, whereas S-sulfhydration of cysteine by H2S is an oxidation reaction. Further, thiols and H2S are formally at the same oxidation state (2–), and would not react with each other.(17) Therefore, it is thought that S-sulfhydration is not mediated by H2S directly (Fig. 5a). On the other hand, sulfane sulfur is a form of sulfur with six valence electrons and no charge (S0), which has the unique ability to reversibly bind to other sulfur atoms, as seen in elemental sulfur (S8), persulfides (R-S-SH) and polysulfides (-S-Sn-S-). Thus, it is considered that proposed S-sulfhydration reactions involving H2S may actually be mediated by sulfane sulfur as shown in Fig. 5a and b. It has long been known that some sulfane sulfur species exist endogenously in biological systems,(35–37) but in general, their occurrence and function in cells and tissues remain unclear. Recently, Ida et al.(38) demonstrated that the enzymes cystathionine γ-lyase (CSE) and cystathionine β-synthase (CBS) are capable of directly generating cysteine persulfide, Cys-SSH from cystine. Moreover some researchers explain that polysulfides with various numbers of sulfur atoms may be generated via oxidized H2S(39,40) and it has also been shown that polysulfides induce Ca2+ influx by activating transient receptor potential (TRP)A1 channels in rat astrocytes much more efficiently than H2S.(41) Thus, sulfane sulfur is a potentially important signaling/effector species, and much of the reported biological activity associated with H2S may actually be due to sulfane sulfur. It has also been proposed that H2S can be generated by degradation of persulfide, i.e., sulfane sulfur may be a precursor to biological H2S in the presence of thiols.(38,42)
Fig. 5.
S-Sulfhydration reaction: (a) Sulfane sulfur is much more effective for the illustrated reaction than H2S. (b) Proposedmechanism of S-sulfhydration reaction mediated by sulfane sulfur and tautomerization of hydropersulfide.
Development of Fluorescence Probes for Sulfane Sulfur
The increasing recognition of the importance of sulfane sulfur in biological systems has led to the development of fluorescence probes for sulfane sulfur. Hydropersulfide (R-SSH) has significantly different chemical properties from structurally related thiols (R-SH). The pKa values of hydropersulfides are lower than that of H2S, so hydropersulfides should be stronger and more reactive nucleophiles than thiols. In addition, sulfane sulfur is electrophilic, and can react with nucleophiles. Taking advantage of these properties, several off/on type fluorescence probes for sulfane sulfur have been designed and developed.
Fluorescence probes based on sulfane sulfur attachment to thiol
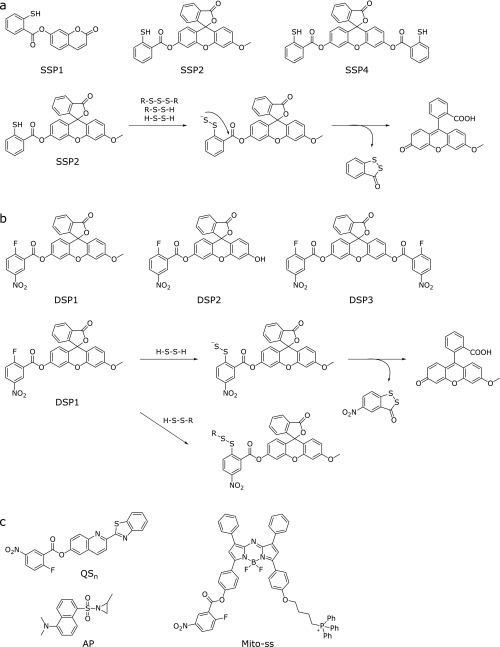
Chen et al.(43) reported the SSP series of fluorescence probes for sulfane sulfur (Fig. 6a). The design strategy for these probes is as follows. First, sulfane sulfur reacts with the sulfur atom of a thiol group of the probe, affording persulfide (R-SS−). Then, intramolecular nucleophilic attack of persulfide (R-SS−) on the ester moiety occurs, releasing the fluorophore (Fig. 6a). Thus, the probe shows a large fluorescence increase upon reaction with sulfane sulfur.
Fig. 6.
Chemical structures and reaction mechanisms of (a) SSPs and (b) DSPs with sulfane sulfur and hydrogen persulfide. (c) Two-photon excitation and NIR fluorescence probes for hydrogen persulfide utilizing 2-fluoro-5-nitrobenzoate as the H2S2/H2Sn recognition moiety, and a fluorescence probe for hydrogen persulfide utilizing an aziridine ring as a H2S2/H2Sn recognition moiety.
Fluorescence probes based on the nucleophilicity of H2S2/H2Sn
Liu et al.(44) reported a fluorescence probe for hydrogen polysulfides (H2Sn, n>1), which can be considered as oxidized forms of H2S (Fig. 6b). In this molecular design, 2-fluoro-5-nitrobenzoate (the H2S2/H2Sn recognition moiety) and fluorescein (the fluorophore) are linked together. As shown in Fig. 6b, nucleophilic aromatic substitution of H2S2/H2Sn with the 2-fluoro-5-nitrobenzoate moiety of the probe affords the persulfide intermediate, and then intramolecular nucleophilic attack of the persulfide (R-SS−) on the ester moiety releases the fluorophore. This probe can also react with biothiols to form thioether products, but these compounds do not undergo intramolecular cyclization, and therefore the probe shows high specificity for H2Sn or end-free persulfide (−S-Sn-S−, n≥0). This approach can also be applied to other fluorophores: for example, Zeng et al.(45) reported a two-photon excited fluorescence probe for H2S2/H2Sn, QSn, using 2-benzothiazol-2-yl-quinolin-6-ol as a two-photon fluorophore and 2-fluoro-5-nitrobenzoate as a H2S2/H2Sn recognition moiety (Fig. 6c). They successfully visualized both exogenous and endogenous H2S2/H2Sn in living cells and zebrafish embryo. Moreover, Gao et al.(46) developed a near-infrared (NIR) fluorescence probe, Mito-ss, which has aza-BODIPY as the NIR fluorophore, 2-fluoro-5-nitrobenzoate as the H2S2/H2Sn recognition moiety and the triphenylphosphonium group as a mitochondrial targeting moiety (Fig. 6c). Chen et al.(47) also reported a fluorescence probe, AP, based on an aziridine ring-opening reaction of H2Sn (Fig. 6c).
Other Chemical Tools Including Enzyme Inhibitors and Labeling Reagents
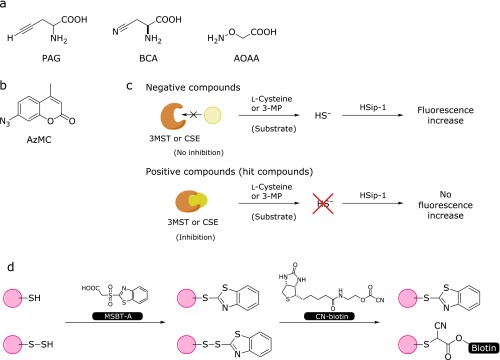
Physiological H2S is enzymatically produced by CSE, CBS and 3-mercaptopyruvate sulfurtransferase (3MST), and inhibitors of these enzymes have been widely used in biological studies. d,l-Propargylglycine (PAG) and β-cyano-l-alanine (BCA) are specific inhibitors of CSE, and aminooxyacetic acid (AOAA) inhibits both CSE and CBS (Fig. 7a).(48,49) However, these inhibitors do not have sufficiently high selectivity for the target enzyme, and no inhibitor for 3MST has been reported. So, selective and sensitive inhibitors for these enzymes are still required. Inhibitor screening assay based on fluorescence detection has great advantages in terms of speed and convenience, and can be applied to high-throughput screening (HTS) of large chemical libraries. Thorson et al.(50) performed CBS inhibitor screening of 1,900 chemical compounds by using 7-azide-4-methylcoumarin (AzMC) (Fig. 7b), and successfully identified several novel CBS inhibitors. Our group also performed HTS of a chemical library containing about 160,000 compounds for inhibitors of CSE and 3MST by using HSip-1.(51) In the assay, purified 3MST or CSE and the corresponding substrate [3-mercaptopyruvate (3-MP) or cysteine] were used, and HSip-1 reacted with H2S released by the enzymatic reaction. Compounds that suppressed the fluorescence increase of HSip-1 in response to H2S were selected as candidate inhibitors (Fig. 7c). After further examination of the selectivity of the hit compounds, we identified both 3MST- and CSE-selective inhibitors. We are currently investigating the possible utility of these inhibitors for biological studies.
Fig. 7.
(a) Chemical structures of inhibitors of CSE and CBS. (b) Chemical structure of AzMC. (c) Screening schemes for 3MST and CSE inhibitors. (d) Tag-switching technique for detecting protein Cys S-sulfhydration.
In order to study the role of S-sulfhydration (SH→SSH) as an oxidative post-translational modification, Zhang et al.(52) reported a selective tag-switch method that can be used to label protein S-sulfhydrated residues. Since thiols and persulfides in proteins have similar reactivity, it is difficult to achieve selective detection of S-sulfhydrated residues among protein residues. They resolved this problem by using two reagents, one nucleophilic reagent and one reporter, which selectively label protein persulfides (protein Cys-SSH) in a two-step reaction as shown in Fig. 7d. The first step is to block both -SH and -SSH with methylsulfonyl benzothiazole (MSBT).(53) The corresponding adducts, -S-benzothiazole and -S-S-benzothiazole, have different reactivities towards nucleophiles; specifically, only the -S-S-benzothiazole moiety is highly reactive with carbon-based nucleophile, which was also developed by Zhang et al.(53) By using a carbon-based nucleophile conjugated with biotin, only protein persulfide residues can be labeled.
Biological Applications of Developed Chemical Tools for H2S and Sulfane Sulfur
Next, we introduce some recent reports on the biological applications of these chemical tools. Ida et al. (38) used a fluorescence probe for sulfane sulfur, SSP2 (Fig. 6a), to study CBS and CSE activities inside cells, as well as a MS-based quantification of sulfane sulfur. They first used SSP2 to evaluate increased total sulfane sulfur level inside living A549 cells in which CSE/CBS was overexpressed, and then they assessed individual sulfane sulfur species in these cells by means of LC-MS/MS. Overexpression of CSE or CBS led to a dramatic increase of Cys–SSH and Cys–SSSH, and CBS knockdown caused a decrease of Cys–SSH. They proposed that facile sulfur atom transfers from low-molecular-weight species such as Cys–SSH to protein thiols result in protein Cys-S-polythiolation. In order to further examine this hypothesis in cells, they also used a tag-switch assay, in which protein Cys-SSH is selectively detected with a biotin-CN-labeled probe (a carbon-based nucleophile). They found that protein polysulfide residues were greatly increased after overexpression of CBS and CSE, and proteomic analysis identified several Cys-polythiolated proteins.
The role of sulfane sulfur in detoxification pathways is also a topic of increasing interest. For instance, Abiko et al.(54) reported that the electrophile methylmercury (MeHg) was metabolized to bismethylmercury sulfide (MeHg)2S by endogenous persulfides such as GSH persulfides and protein cysteine persulfides in biological systems. They used liver and heart cytosolic fractions prepared from WT and CSE-knockout mice, and detected (MeHg)2 only in the fractions of WT mice. In another paper, they showed that the Keap1-Nrf2 pathway is activated by 1,2-dihydroxynaphthalene-4-sulfenic acid (1,2-NQH2-SOH), generated by reaction of 1,2-NQ (1,2-naphthoquinone) with sulfane sulfur under oxidative stress, through the modification of Cys171 of Keap1.(55) Activation of the transcriptional factor Nrf2 causes up-regulation of downstream genes, such as glutamate-cysteine ligase and multidrug resistance-associated proteins (MRPs), facilitating the excretion of polar metabolites into the extracellular space. By using fluorescence probes, Marutani et al.(56) established that H2S-donor compounds increased intracellular sulfane sulfur levels and had a cytoprotective effect. They designed unique hybrid molecules bearing a H2S-releasing moiety as well as a N-methyl-d-aspartate receptor antagonist moiety. These hybrid molecules exhibited protective effects against oxygen and glucose deprivation-induced death of primary cortical neurons. They were also cytoprotective against cell death induced by 1-methyl-4-phenylpyridinium [MMP+, a metabolite of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)]. MPTP induces neurodegeneration of dopaminergic neurons in mammalian midbrain, which leads to Parkinson’s disease-like symptoms.(57) To dissect the cytoprotective effect of the hybrid molecule, they separately assessed H2S and sulfane sulfur levels in SH-SY5Y cells and in the culture medium without cells upon addition of the hybrid molecule by utilizing their specific fluorescence probes, HSip-1 and SSP4 (Fig. 3 and 6a). Interestingly, the results suggested that the cytoprotective effect of the hybrid molecule against MPP+-induced toxicity may correlate with the ability to produce intracellular sulfane sulfur rather than H2S, albeit this molecule releases H2S but not sulfane sulfur. Although it remains unclear how such H2S-releasing hybrid molecules affect the intracellular level of sulfane sulfur, compounds that increase intracellular sulfane sulfur levels may be potentially useful as neuroprotective agents to treat neurodegenerative diseases.
Conclusions and Prospects
Here, we have reviewed fluorescence probes for H2S and sulfane sulfur based on a variety of design strategies, caged compounds of H2S, inhibitors of CSE, CBS and 3MST, and labeling reagents for protein S-sulfhydrated residues, and their biological applications. It is well established that H2S acts as a signaling and effector molecule, but its chemical mechanisms of action largely remain to be established. However, studies with recently developed fluorescence probes and detection methods for H2S and sulfane sulfur have indicated that sulfane sulfur may be the actual signaling molecule mediating at least some of the physiological functions of H2S. Further development and application of chemical tools for H2S and sulfane sulfur studies, especially to detect endogenous H2S or sulfane sulfur in vivo, are expected to throw light on the role of these mediators in the control mechanisms of various important physiological functions.
Acknowledgments
This work was supported in part by MEXT (Grant Nos. 24689003, 24659042 and 26104509 to K.H.). K.H. was also supported by Mochida Memorial Foundation for Medical and Pharmaceutical Research, The Naito Foundation, The Asahi Glass Foundation, and Takeda Science Foundation.
References
- 1.Yang G, Wu L, Jiang B, et al. H2S as a physiologic vasorelaxant: hypertension in mice with deletion of cystathionine γ-lyase. Science. 2008;322:587–590. doi: 10.1126/science.1162667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paul BD, Snyder SH. H2S signalling through protein sulfhydration and beyond. Nat Rev Mol Cell Biol. 2012;13:499–507. doi: 10.1038/nrm3391. [DOI] [PubMed] [Google Scholar]
- 3.Abe K, Kimura H. The possible role of hydrogen sulfide as an endogenous neuromodulator. J Neurosci. 1996;16:1066–1071. doi: 10.1523/JNEUROSCI.16-03-01066.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimura H. Physiological role of hydrogen sulfide and polysulfide in the central nervous system. Neurochem Int. 2013;63:492–497. doi: 10.1016/j.neuint.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Kaneko Y, Kimura Y, Kimura H, Niki I. l-Cysteine inhibits insulin release from the pancreatic β-cell: possible involvement of metabolic production of hydrogen sulfide, a novel gasotransmitter. Diabetes. 2006;55:1391–1397. doi: 10.2337/db05-1082. [DOI] [PubMed] [Google Scholar]
- 6.Li L, Bhatia M, Zhu YZ, et al. Hydrogen sulfide is a novel mediator of lipopolysaccharide-induced inflammation in the mouse. FASEB J. 2005;19:1196–1198. doi: 10.1096/fj.04-3583fje. [DOI] [PubMed] [Google Scholar]
- 7.Lo Faro ML, Fox B, Whatmore JL, Winyard PG, Whiteman M. Hydrogen sulfide and nitric oxide interactions in inflammation. Nitric Oxide. 2014;41:38–47. doi: 10.1016/j.niox.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Kubáň V, Dasgupta PK, Marx JN. Nitroprusside and methylene blue methods for silicone membrane differentiated flow injection determination of sulfide in water and wastewater. Anal Chem. 1992;64:36–43. doi: 10.1021/ac00025a008. [DOI] [PubMed] [Google Scholar]
- 9.Tsai DM, Kumar AS, Zen JM. A highly stable and sensitive chemically modified screen-printed electrode for sulfide analysis. Anal Chim Acta. 2006;556:145–150. doi: 10.1016/j.aca.2005.05.038. [DOI] [PubMed] [Google Scholar]
- 10.Klingerman CM, Trushin N, Prokopczyk B, Haouzi P. H2S concentrations in the arterial blood during H2S administration in relation to its toxicity and effects on breathing. Am J Physiol Regul Integr Comp Physiol. 2013;305:R630–R638. doi: 10.1152/ajpregu.00218.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishida M, Sawa T, Kitajima N, et al. Hydrogen sulfide anion regulates redox signaling via electrophile sulfhydration. Nat Chem Biol. 2012;8:714–724. doi: 10.1038/nchembio.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu F, Han X, Chen L. Fluorescent probes for hydrogen sulfide detection and bioimaging. Chem Commun (Camb) 2014;50:12234–12249. doi: 10.1039/c4cc03312d. [DOI] [PubMed] [Google Scholar]
- 13.Lippert AR. Designing reaction-based fluorescent probes for selective hydrogen sulfide detection. J Inorg Biochem. 2014;133:136–142. doi: 10.1016/j.jinorgbio.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 14.Shimamoto K, Hanaoka K. Fluorescent probes for hydrogen sulfide (H2S) and sulfane sulfur and their applications to biological studies. Nitric Oxide. 2015;46:72–79. doi: 10.1016/j.niox.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Mustafa AK, Gadalla MM, Sen N, et al. H2S signals through protein S-sulfhydration. Sci Signal. 2009;2:ra72. doi: 10.1126/scisignal.2000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toohey JI. Sulfur signaling: is the agent sulfide or sulfane? Anal Biochem. 2011;413:1–7. doi: 10.1016/j.ab.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 17.Ono K, Akaike T, Sawa T, et al. Redox chemistry and chemical biology of H2S, hydropersulfides, and derived species: implications of their possible biological activity and utility. Free Radic Biol Med. 2014;77:82–94. doi: 10.1016/j.freeradbiomed.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kazemi F, Kiasat AR, Sayyahi S. Chemoselective reduction of azides with sodium sulfide hydrate under solvent free conditions. Phosphorus Sulfur. 2004;179:1813–1817. [Google Scholar]
- 19.Shiao MJ, Lai LL, Ku WS, Lin PY, Hwu JR. Chlorotrimethylsilane in combination with sodium sulfide as the equivalent of sodium trimethylsilanethiolate in organic reactions. J Org Chem. 1993;58:4742–4744. [Google Scholar]
- 20.Lippert AR, New EJ, Chang CJ. Reaction-based fluorescent probes for selective imaging of hydrogen sulfide in living cells. J Am Chem Soc. 2011;133:10078–10080. doi: 10.1021/ja203661j. [DOI] [PubMed] [Google Scholar]
- 21.Lin VS, Lippert AR, Chang CJ. Cell-trappable fluorescent probes for endogenous hydrogen sulfide signaling and imaging H2O2-dependent H2S production. Proc Natl Acad Sci USA. 2013;110:7131–7135. doi: 10.1073/pnas.1302193110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bae SK, Heo CH, Choi DJ, et al. A ratiometric two-photon fluorescent probe reveals reduction in mitochondrial H2S production in Parkinson’s disease gene knockout astrocytes. J Am Chem Soc. 2013;135:9915–9923. doi: 10.1021/ja404004v. [DOI] [PubMed] [Google Scholar]
- 23.Montoya LA, Pluth MD. Selective turn-on fluorescent probes for imaging hydrogen sulfide in living cells. Chem Commun (Camb) 2012;48:4767–4769. doi: 10.1039/c2cc30730h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu MY, Li K, Hou JT, Huang Z, Yu XQ. A selective colorimetric and ratiometric fluorescent probe for hydrogen sulfide. Org Biomol Chem. 2012;10:8342–8347. doi: 10.1039/c2ob26235e. [DOI] [PubMed] [Google Scholar]
- 25.Qian Y, Karpus J, Kabil O, et al. Selective fluorescent probes for live-cell monitoring of sulphide. Nat Commun. 2011;2:495. doi: 10.1038/ncomms1506. [DOI] [PubMed] [Google Scholar]
- 26.Liu C, Pan J, Li S, et al. Capture and visualization of hydrogen sulfide by a fluorescent probe. Angew Chem Int Ed Engl. 2011;50:10327–10329. doi: 10.1002/anie.201104305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng B, Chen W, Liu C, et al. Fluorescent probes based on nucleophilic substitution-cyclization for hydrogen sulfide detection and bioimaging. Chem Euro J. 2014;20:1010–1016. doi: 10.1002/chem.201303757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Silva AP, Gunaratne HQN, Gunnlaugsson T, et al. Signaling recognition events with fluorescent sensors and switches. Chem Rev. 1997;97:1515–1566. doi: 10.1021/cr960386p. [DOI] [PubMed] [Google Scholar]
- 29.Choi MG, Cha S, Lee H, Jeon HL, Chang SK. Sulfide-selective chemosignaling by a Cu2+ complex of dipicolylamine appended fluorescein. Chem Commun (Camb) 2009:7390–7392. doi: 10.1039/b916476f. [DOI] [PubMed] [Google Scholar]
- 30.Sasakura K, Hanaoka K, Shibuya N, et al. Development of a highly selective fluorescence probe for hydrogen sulfide. J Am Chem Soc. 2011;133:18003–18005. doi: 10.1021/ja207851s. [DOI] [PubMed] [Google Scholar]
- 31.Fukushima N, Ieda N, Sasakura K, et al. Synthesis of a photocontrollable hydrogen sulfide donor using ketoprofenate photocages. Chem Commun (Camb) 2014;50:587–589. doi: 10.1039/c3cc47421f. [DOI] [PubMed] [Google Scholar]
- 32.Lukeman M, Scaiano JC. Carbanion-mediated photocages: rapid and efficient photorelease with aqueous compatibility. J Am Chem Soc. 2005;127:7698–7699. doi: 10.1021/ja0517062. [DOI] [PubMed] [Google Scholar]
- 33.Fukushima N, Ieda N, Kawaguchi M, et al. Development of photo-controllable hydrogen sulfide donor applicable in live cells. Bioorg Med Chem Lett. 2015;25:175–178. doi: 10.1016/j.bmcl.2014.11.084. [DOI] [PubMed] [Google Scholar]
- 34.Devarie-Baez NO, Bagdon PE, Peng B, Zhao Y, Park CM, Xian M. Light-induced hydrogen sulfide release from “caged” gem-dithiols. Org Lett. 2013;15:2786–2789. doi: 10.1021/ol401118k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamanishi T, Tuboi S. The mechanism of the l-cystine cleavage reaction catalyzed by rat liver γ-cystathionase. J Biochem. 1981;89:1913–1921. doi: 10.1093/oxfordjournals.jbchem.a133393. [DOI] [PubMed] [Google Scholar]
- 36.Iciek M, Włodek L. Biosynthesis and biological properties of compounds containing highly reactive, reduced sulfane sulfur. Pol J Pharmacol. 2001;53:215–225. [PubMed] [Google Scholar]
- 37.Toohey JI, Cooper AJL. Thiosulfoxide (sulfane) sulfur: new chemistry and new regulatory roles in biology. Molecules. 2014;19:12789–12813. doi: 10.3390/molecules190812789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ida T, Sawa T, Ihara H, et al. Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc Natl Acad Sci U S A. 2014;111:7606–7611. doi: 10.1073/pnas.1321232111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greiner R, Pálinkás Z, Bäsell K, et al. Polysulfide links H2S to protein thiol oxidation. Antioxid Redox sign. 2013;19:1749–1765. doi: 10.1089/ars.2012.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimura H. Hydrogen sulfide and polysulfides as biological mediators. Molecules. 2014;19:16146–16157. doi: 10.3390/molecules191016146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kimura Y, Mikami Y, Osumi K, Tsugane M, Oka J, Kimura H. Polysulfides are possible H2S-derived signaling molecules in rat brain. FASEB J. 2013;27:2451–2457. doi: 10.1096/fj.12-226415. [DOI] [PubMed] [Google Scholar]
- 42.Kawamura S, Otsuji Y, Nakabayashi T, Kitao T, Tsurugi J. Aralkyl hydrodisulfides. IV. The reaction of benzyl hydrodisulfide with several nucleophiles. J Org Chem. 1965;30:2711–2714. [Google Scholar]
- 43.Chen W, Liu C, Peng B, Zhao Y, Pacheco A, Xian M. New fluorescent probes for sulfane sulfurs and the application in bioimaging. Chem Sci. 2013;4:2892–2896. doi: 10.1039/C3SC50754H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu C, Chen W, Shi W, et al. Rational design and bioimaging applications of highly selective fluorescence probes for hydrogen polysulfides. J Am Chem Soc. 2014;136:7257–7260. doi: 10.1021/ja502968x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng L, Chen S, Xia T, Hu W, Li C, Liu Z. Two-photon fluorescent probe for detection of exogenous and endogenous hydrogen persulfide and polysulfide in living organisms. Anal Chem. 2015;87:3004–3010. doi: 10.1021/acs.analchem.5b00172. [DOI] [PubMed] [Google Scholar]
- 46.Gao M, Yu F, Chen H, Chen L. Near-infrared fluorescent probe for imaging mitochondrial hydrogen polysulfides in living cells and in vivo. Anal Chem. 2015;87:3631–3638. doi: 10.1021/ac5044237. [DOI] [PubMed] [Google Scholar]
- 47.Chen W, Rosser EW, Zhang D, et al. A specific nucleophilic ring-opening reaction of aziridines as a unique platform for the construction of hydrogen polysulfides sensors. Org Lett. 2015;17:2776–2779. doi: 10.1021/acs.orglett.5b01194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whiteman M, Le Trionnaire S, Chopra M, Fox B, Whatmore J. Emerging role of hydrogen sulfide in health and disease: critical appraisal of biomarkers and pharmacological tools. Clin Sci (Lond) 2011;121:459–488. doi: 10.1042/CS20110267. [DOI] [PubMed] [Google Scholar]
- 49.Asimakopoulou A, Panopoulos P, Chasapis CT, et al. Selectivity of commonly used pharmacological inhibitors for cystathionine β synthase (CBS) and cystathionine γ lyase (CSE) Br J Pharmacol. 2013;169:922–932. doi: 10.1111/bph.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thorson MK, Majtan T, Kraus JP, Barrios AM. Identification of cystathionine β-synthase inhibitors using a hydrogen sulfide selective probe. Angew Chem Int Ed. 2013;52:4641–4644. doi: 10.1002/anie.201300841. [DOI] [PubMed] [Google Scholar]
- 51.Shimamoto K, Hanaoka K, Sasakura K, et al. Inhibitor screening for 3-mercaptopyruvate sulfurtrasferase (3MST) and cystathionine γ-lyase (CSE) Nitric Oxide. 2014;39:S49. [Google Scholar]
- 52.Zhang D, Macinkovic I, Devarie-Baez NO, et al. Detection of protein S-sulfhydration by a tag-switch technique. Angew Chem Int Ed Engl. 2014;53:575–581. doi: 10.1002/anie.201305876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang D, Devarie-Baez NO, Li Q, Lancaster JR, Jr, Xian M. Methylsulfonyl benzothiazole (MSBT): a selective protein thiol blocking reagent. Org Lett. 2012;14:3396–3399. doi: 10.1021/ol301370s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abiko Y, Yoshida E, Ishii I, Fukuto JM, Akaike T, Kumagai Y. Involvement of reactive persulfides in biological bismethylmercury sulfide formation. Chem Res Toxicol. 2015;28:1301–1306. doi: 10.1021/acs.chemrestox.5b00101. [DOI] [PubMed] [Google Scholar]
- 55.Shinkai Y, Abiko Y, Ida T, et al. Reactive sulfur species-mediated activation of the Keap1-Nrf2 pathway by 1,2-naphthoquinone through sulfenic acids formation under oxidative stress. Chem Res Toxicol. 2015;28:838–847. doi: 10.1021/tx500416y. [DOI] [PubMed] [Google Scholar]
- 56.Marutani E, Kosugi S, Tokuda K, et al. A novel hydrogen sulfide-releasing N-methyl-d-aspartate receptor antagonist prevents ischemic neuronal death. J Biol Chem. 2012;287:32124–32135. doi: 10.1074/jbc.M112.374124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marutani E, Sakaguchi M, Chen W, et al. Cytoprotective effects of hydrogen sulfide-releasing N-methyl-d-aspartate receptor antagonists mediated by intracellular sulfane sulfur. Medchemcommun. 2014;5:1577–1583. doi: 10.1039/C4MD00180J. [DOI] [PMC free article] [PubMed] [Google Scholar]