Abstract
Background
Coronary heart disease (CHD) is a cardiovascular disease characterized by high morbidity and mortality. Vascular endothelial growth factor (VEGF) and its receptor, named kinase insert domain-containing receptor (KDR, or VEGFR2), which are involved with angiogenesis and vascular repair, could partly contribute to the development of CHD. The aim of this study, therefore, was to investigate the potential correlations between genetic polymorphisms on VEGF and KDR and susceptibility to CHD, and the integrative role of SNPs combined on susceptibility to CHD were also studied.
Material/Methods
Venous blood samples gathered from 533 DCM patients and 533 healthy controls were used to genotype tag-SNPs of VEGF (rs699947, rs2010963, and rs3025010) and KDR (rs2071559, rs2305948, and rs1870377) by polymerase chain reaction (PCR) and SNaPshot assay. Investigations of potential haplotypes were conducted on the basis of SHEsis software. The odds ratio (ORs) and relevant 95% confidence intervals (95% CI) were used to estimate associations of SNPs/haplotypes with risk of CHD. Multivariate logistic regression was also performed, taking certain clinical characteristics (e.g., BMI, smoking, alcohol consumption, diabetes, and hypertension) into consideration. All statistical analyses were done with STATA Version 12.0 software.
Results
Our results suggest that rs699947 (T>C) on KDR are associated with susceptibility to CHD under the dominant model before (OR=1.35, 95% CI: 1.05–1.73, P=0.019) and after (OR=1.33, 95% CI: 1.01–1.76, P=0.044), allowing for clinical characteristics (e.g., BMI, smoking, alcohol consumption, diabetes, and hypertension). rs2305948 (G>A) and rs1870377 (A>T) on VEGF were also found to be associated with risk of CHD under the recessive model after adjustment with multivariate regression analyses (OR=1.21, 95% CI: 1.02–1.43, P=0.029; OR=2.54, 95% CI: 1.13–5.75, P=0.025); OR=2.83, 95% CI: 1.47–5.46, P=0.002, respectively). Additionally, haplotype analyses revealed that integration of 5 SNPs would either raise (e.g. C-C-T-G-T and T-G-T-G-T) or reduce (e.g. C-C-C-G-T, T-C-T-G-A, T-C-T-G-T, and T-G-T-G-A) risk of CHD.
Conclusions
Genetic polymorphisms on VEGF (rs699947) and KDR (rs2305948and rs1870377), as well as relevant haplotypes, may serve as genetic markers that might be useful in future investigations on the pathogenesis of CHD.
MeSH Keywords: Coronary Disease; Disease Susceptibility; Polymorphism, Genetic; Vascular Endothelial Growth Factor Receptor-2; Vascular Endothelial Growth Factor, Endocrine-Gland-Derived
Background
Coronary heart disease (CHD), also known as coronary artery disease (CAD), is a form of atherosclerosis-related cardiovascular disease with a global mortality rate of over 7 million deaths [1]. The disease progresses as plaques build up along the inner walls of heart arteries, narrowing the lumen of arteries and reducing blood flow to the heart. CHD usually occurs in middle-aged and elderly individuals and males are more vulnerable than females [2]. In light of the unfavorable consequences of CHD, the etiology of CHD has been extensively investigated. It was demonstrated that the cumulative effects of environmental and genetic factors could boost the incidence of the disease [3]. Furthermore, several case-control studies have suggested certain candidate genes that could be linked with CHD, and the heritability of CHD is estimated to be 50–60%. The genes have been demonstrated to be involved in inflammation, lipid metabolism, coagulation, and regulation of vascular tone, many of which have been deemed to be associated with various atherosclerosis-related phenotypes [4]. Previous studies have confirmed the association of raised level of serum and plasma VEGF with atherosclerosis [5] and CHD, respectively [6]. Polymorphisms of KDR have also been suggested to be related with risk of CHD via the role of KDR in the formation of blood vessel networks and atherosclerosis [7–9].
VEGF, typically expressed as a 46-kDa homodimer located on chromosome 6, can encode a mitogen that promotes vascular endothelial cell proliferation and angiogenesis [10], and the abnormal angiogenesis within blood vessels has been reported to be implicated in atherosclerosis and arterial diseases [11, 12]. Furthermore, VEGF are considered to be correlated with various signaling receptor complexes – VEGF binding receptors VEGFR-1(flt-1), VEGFR-2 (KDR/flk-1), and VEGFR-3 (also known as FLT-4) – while there also exist VEGF165 isoform-specific receptors, neurophilin-1 and neurophilin-2 [13–15]. As a receptor of VEGF, KDR is expressed in manifold cells, such as endothelial progenitor cells (EPCs), endothelial cells, and hematopoietic cells [9]. Additionally, KDR is regarded as a crucial receptor mediating angiogenesis and it seems to be essential to survival and integrity of endothelial cells [16,17].
Considering the potentially critical roles of VEGF and KDR in the pathogenesis of CHD, it is hypothesized that single-nucleotide polymorphisms (SNPs) located in the functional regions of VEGF and KDR might also facilitate CHD by influencing the expression of VEGF and KDR. Thus, the present case-control study was performed to explore the association of tagging SNPs (tag-SNPs) in VEGF (rs699947, rs2010963, and rs3025010) and KDR (rs2071559, rs2305948, and rs1870377) with risk of CHD in a Chinese Han population. We also investigated the synergic effects of SNPs combined in evolution of CHD
Material and Methods
Ethics statement
The current study was conducted in strict accordance with the protocol approved by the Ethics Committee of the Second Hospital of Dalian Medical University (Dalian, China). All participants were recruited by the Second Hospital of Dalian Medical University and they all (or their guardians) signed informed consents. Moreover, this research followed the tenets of the Declaration of Helsinki.
Study population
A total of 1066 specimens were obtained from the Second Hospital of Dalian Medical University from June 2011 to May 2014. In all, 533 unrelated CHD patients (250 males and 283 females) and 533 unrelated healthy volunteers (255 males and 278 females) were included in the present study, as outlined in Table 1. All enrolled subjects were of Chinese Han ethnicity.
Table 1.
Comparison of coronary heart disease patients and controls by selective characteristics.
| Clinical characteristics | CHD patients (n=533) | Healthy controls (n=533) | P-value | Adjusted P-value |
|---|---|---|---|---|
| Age (mean ±SD) | 62.3±10.2 | 61.5±9.3 | 0.181b | 0.212b |
| Male/female | 250/283 | 255/278 | 0.759a | 0.804a |
| BMI (kg/m2) | 24.03±3.33 | 22.41±2.77 | <0.001b | <0.001b |
| SBP (mmHg) | 131.12±22.51 | 129.71±21.12 | 0.187b | 0.259b |
| DBP (mmHg) | 78.98±14.31 | 80.11±13.21 | 0.182b | 0.180b |
| Pulse pressure (mmHg) | 51.33±16.41 | 50.12±14.88 | 0.208b | 0.207b |
| Smoking | ||||
| Yes | 256 | 233 | 0.157a | 0.141a |
| No | 277 | 300 | ||
| Alcohol consumption | ||||
| Yes | 242 | 144 | <0.001a | <0.001a |
| No | 291 | 389 | ||
| Hypertension | ||||
| Yes | 387 | 223 | <0.001a | <0.001a |
| No | 146 | 310 | ||
| Diabetes | ||||
| Yes | 149 | 69 | <0.001a | <0.001a |
| No | 384 | 464 | ||
CHD – coronary heart disease;
p value of student’s t test;
chi-square test; Adjusted P-value: significance after adjustment by multivariate-based logistic regression analysis.
Inclusion criteria were: (1) all the participants in the case group were diagnosed with CHD by coronary arteriography (CAG); (2) the coronary angiography reveals more than 50% narrowing of the lumen of at least 1 of the major coronary arteries; and (3) all subjects were at least 18 years old and they were fully informed about the research. Exclusion criteria were: (1) the participants were subject to chest pain due to other disorders, such as other cardiac disorders, severe neurosis, menopausal syndrome, cervical spondylosis, hyperthyroidism, and colopathy; (2) the subjects had severe dysfunctions of the liver, kidney, or lung, and hematopiesis; (3) their major coronary artery had no more than 20% stenosis; (4) participants were either under 18 years old or at the stage of pregnancy/lactation; (5) subjects had life-threatening diseases such as tumors and acquired immune deficiency syndrome (AIDS); and (6) the information of eligible subjects did not match the inclusion criterion or they were incomplete. Individuals who had a history of myocardial infarction (MI) or other vascular diseases and those who had undergone heart surgery were also excluded.
Risk factors of CHD
A complete series of clinical characteristics of CHD patients and healthy controls are shown in Table 1, including mean age and sex ratio of the participants. Among the mentioned features, certain vascular risk factors should be noted, such as body mass index (BMI), smoking status, alcohol consumption, hypertension, and diabetes status. The smoking status was defined as the consumption of at least 5 cigarettes per day or having a history of smoking in the last year. Diabetes was diagnosed when the subject had a fasting glucose above 7.8 mmol/l, or more than 11.1 mmol/l at 2 h after oral glucose challenge. Arterial hypertension was defined as the mean of 3 independent blood pressures as systolic pressure no less than 140 mmHg or diastolic pressure no less than 90 mmHg or the use of antihypertensive drugs. The alcohol consumption was defined as no less than 2 ounces of liquor per day or 4 ounce of beer per day. Systolic blood pressure (SBP) and diastolic blood pressure (DBP) were obtained using a sphygmomanometer of the same type. The difference of SBP and DBP equals the pulse pressure.
SNP selection
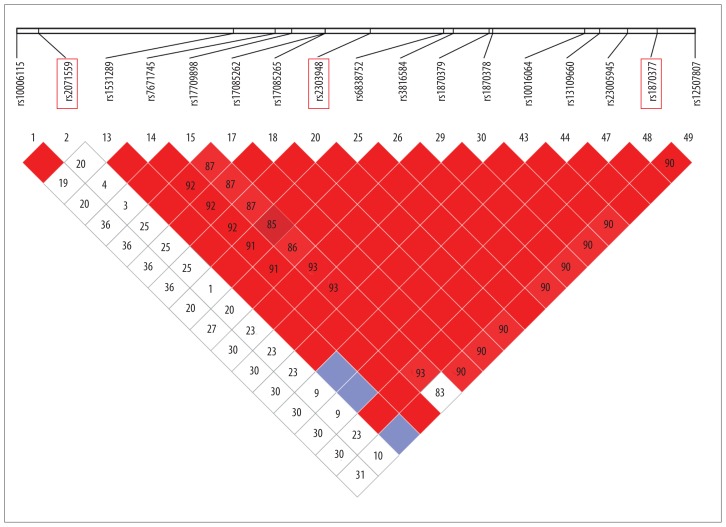
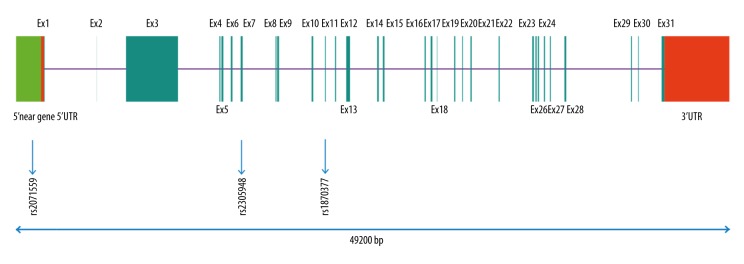
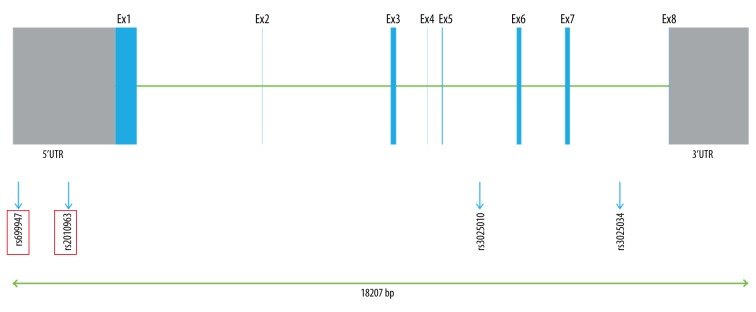
In the present study, the SNPs were obtained from an unrelated Chinese population in Shanghai with the public database (HapMap). Tag-SNPs were identified with use of the pair-wise option of Haploview 4.2 software and an r2 of 0.8 was used as a threshold for the analysis (Figure 1) [18]. Finally, 3 SNPs in VEGF (rs699947, rs2010963, and rs3025010) and 3 SNPs in KDR (rs2071559, rs2305948, and rs1870377) were selected. The relative SNP positions in KDR and VEGF are shown in Figures 2 and 3, respectively.
Figure 1.
Linkage disequilibrium (LD) plots of tag-SNPs in VEGF gene.
Figure 2.
Genetic location of the 3 tag-SNPs in KDR gene.
Figure 3.
Genetic location of the 3 tag-SNPs in VEGF gene.
Genetic analysis
A 5-ml venous blood sample was collected from each participant into a sterile tube containing heparin sodium. The mixtures of blood and heparin sodium were subsequently centrifuged at 3000 rpm for 10 min at room temperature, after which the separated plasma samples were stored at −20°C. Genomic DNA was extracted from frozen peripheral blood samples via a QIAmp Blood Mini Kit (Qiagen Inc., Valencia, California, United States) using the manufacturer’s protocols [19]. Tag SNPs were then amplified with the 9700 PCR System (Applied Biosystems) on the basis of the primers designed by Primer3 software (Table 2), after which the PCR products were purified by Shrimp Alkaline Phosphatase (SAP) method [18]. Finally, the SNaPshot assay (Applied Biosystems) was performed to confirm genotypes of 6 DNA samples. All genotyping procedures were carried out in a double-blind manner and the whole assays were proved to be reliable.
Table 2.
Primers of VEGF and KDR genetic polymorphisms for PCR amplification.
| SNP | Gen Pos | Alias name | Primers for PCR amplification | HWE | Global MAF |
|---|---|---|---|---|---|
| VEGF | |||||
| rs699947 | 5′UTR | 2055A>C(−2578) | F: 5′-GCTGTAGGCCAGACCCTG-3′ R: 3′-ACCCACCTATTAGTCTGAC-5′ |
0.280 | 0.325 |
| rs201096 | 5′UTR | 94C>G(−634) | F: 5′-TGATATTCATTGATCCGGGTTT -3′ R: 3′-CTGTCCCCGTTTCACTCA-5′ |
0.583 | 0.008 |
| rs3025010 | Intron 5 | 14625T>C | F: 5′-CCTCCTTTCTTCCCTGTG -3′ R: 3′-AACTTCCCCTGTCCGATG-5′ |
0.009 | 0.352 |
| KDR | |||||
| rs2071559 | 5′ near gene | −906T>C(−604T>C) | F: 5′-TTGGGAAATAGCGGGAAT-3′ R: 3′-GACCCGTTCACGCAAAAG-5′ |
0.705 | 0.499 |
| rs2305948 | Exon 7 | 889G>A(1192) | F: 5′-ACCCAAGTTCCTGACTACAA-3′ R: 3′-TTAGACCCGAAACCATTT-5′ |
0.088 | 0.153 |
| rs1870377 | Exon 11 | 1416A>T(1719) | F: 5′-CCATTCTTCACAAGGGTA -3′ R: 3′-CCTTTGAATCTTCGGTGA-5′ |
0.051 | 0.212 |
Statistical analysis
Data are presented as mean ± standard deviation (SD) or number (percentage). The chi-square test and Student’s t-test were used to compare case and control groups, as appropriate. The goodness-of-fit chi-square test was employed to assess Hardy-Weinberg equilibrium (HWE) for each tag-SNP. Four genetic models – the allelic (1/2 vs. 1/1), dominant (1/2+2/2 vs. 1/1), homozygous (2/2 vs. 1/1), and recessive (2/2 vs. 1/1+1/2) models – were used to assess the association of VEGF and KDR genetic polymorphisms with CHD susceptibility with odds ratios (ORs) and their 95% confidence intervals (CIs). Multivariate logistic regression was also used to estimate the association of the 6 tag-SNPs with risk of CHD after adjustment of age, sex, BMI, smoking status, hypertension, diabetes, and drinking status. All statistical analyses were conducted with STATA Version 12.0 software (Stata Corp, College Station, TX) and a 2-sided P value less than 0.05 was considered to be statistically significant.
Results
The samples included in the present study were obtained from 533 CHD subjects and 533 healthy controls with aid of the Second Hospital of Dalian Medical University (Dalian, China). Genotype frequencies in both groups conformed to Hardy-Weinberg equilibrium (HWE) (Table 2). The selected characteristics of cases and controls are shown in Table 1. Specifically, no statistically significant difference was observed for age (P=0.954), sex (P=0.759), BMI (P=0.709), SBP (P=0.912), DBP (P=0.872), pulse pressure (P=0.892), or smoking status (P=0.157). Nonetheless, several clinical characteristics, including hypertension, diabetes status, and alcohol consumption, displayed statistically remarkable distinctions between CHD patients and healthy controls (P<0.001).
Associations between VEGF gene polymorphisms and the risk of CHD
As shown in Table 3, there existed no significant distinctions in genotype and allele distribution of rs3025010 (T>C) between CHD patients and controls. Furthermore, rs2010963 (C>G) was simply linked with risk of CHD before influences of clinical characteristics (e.g., BMI, smoking, alcohol consumption, diabetes, and hypertension) were considered. Subjects carrying variant genotypes (CG/GG) had a decreased CHD risk when compared with wild genotype CC (OR=0.78, 95%CI=0.62–0.99, P=0.049). However, rs699947 (T>C) in the 5′UTR of VEGF was significantly associated with CHD risk when confounding factors investigated in this study were all allowed for. Compared with individuals with homozygote TT genotype, for instance, subjects carrying the variant genotypes (TC/CC) had a notably increased CHD risk (OR=1.33, 95%CI=1.01–1.76, P=0.044).
Table 3.
Associations of three common polymorphisms of VEGF and KDR with risk of coronary heart disease.
| SNP/genotype | CHD patients (n=533) | Healthy controls (n=533) | OR (95% CI) | χ2 | P value | Adjusted OR (95% CI) | Adjusted P value |
|---|---|---|---|---|---|---|---|
| rs699947 (T>C) | |||||||
| TT | 180 | 217 | Ref. | ||||
| TC | 250 | 237 | 1.28 (0.98–1.68) | 3.357 | 0.067 | 2.05 (1.49, 2.82) | <0.001 |
| CC | 103 | 79 | 1.68 (1.18–2.24) | 6.325 | 0.012 | 2.45 (1.62, 3.70) | <0.001 |
| Dominant (CC+TC vs. TT) | 1.35 (1.05–1.73) | 5.495 | 0.019 | 1.33 (1.01, 1.76) | 0.044 | ||
| Recessive (CC vs. TT+TC) | 1.38 (0.99–1.90 | 3.816 | 0.051 | 1.56 (1.09, 2.24) | 0.015 | ||
| Allele (C allele vs. T allele) | 1.27 (1.07–1.51) | 7.277 | 0.007 | –– | –– | ||
| rs2010963 (C>G) | |||||||
| CC | 250 | 223 | Ref. | ||||
| CG | 233 | 239 | 0.85 (0.66–1.10) | 1.508 | 0.219 | 0.78 (0.58, 1.05) | 0.100 |
| GG | 50 | 71 | 0.55 (0.37–0.84) | 7.910 | 0.005 | 0.76 (0.48, 1.19) | 0.229 |
| Dominant (CG+GG vs. CC) | 0.78 (0.62–0.99) | 3.884 | 0.049 | 0.780 (0.595, 1.023) | 0.073 | ||
| Recessive (GG vs. CC+CG) | 0.60 (0.40–0.89) | 6.539 | 0.011 | 0.83 (0.54, 1.27) | 0.378 | ||
| Allele (G allele vs. C allele) | 0.78 (0.65–0.94) | 7.134 | 0.008 | –– | –– | ||
| rs3025010 (T>C) | |||||||
| TT | 142 | 160 | Ref. | ||||
| TC | 287 | 290 | 1.12 (0.84–1.47) | 0.587 | 0.444 | –– | –– |
| CC | 104 | 94 | 1.25 (0.87–1.79) | 1.450 | 0.229 | –– | –– |
| Dominant (CC+TC vs. TT) | 1.15 (0.88–1.50) | 1.024 | 0.312 | –– | –– | ||
| Recessive (CC vs. TT+TC) | 1.16 (0.85–1.58) | 0.895 | 0.344 | –– | –– | ||
| Allele (C allele vs. T allele) | 1.11 (0.93–1.31) | 1.360 | 0.244 | –– | –– | ||
| rs2071559 (T>C) | |||||||
| TT | 122 | 143 | Ref. | ||||
| TC | 253 | 261 | 1.29 (0.95–1.74) | 0.700 | 0.100 | 1.25 (0.89, 1.76) | 0.192 |
| CC | 158 | 129 | 1.47 (1.04–2.06) | 4.804 | 0.028 | 1.36 (0.93, 1.99) | 0.117 |
| Dominant (CC+TC vs. TT) | 1.38 (1.04–1.83) | 4.954 | 0.026 | 1.24 (0.91, 1.70) | 0.169 | ||
| Recessive (CC vs. TT+TC) | 1.24 (0.94–1.63) | 2.288 | 0.130 | 1.16 (0.85, 1.57) | 0.347 | ||
| Allele (C allele vs. T allele) | 1.21 (1.02–1.43) | 4.777 | 0.029 | –– | –– | ||
| rs2305948 (G>A) | |||||||
| GG | 388 | 416 | Ref. | ||||
| GA | 122 | 105 | 1.25 (0.93–1.67) | 2.131 | 0.144 | 1.25 (0.90, 1.76) | 0.189 |
| AA | 23 | 12 | 2.06 (1.01–4.19) | 4.089 | 0.043 | 3.04 (1.29, 7.17) | 0.011 |
| Dominant (AA+GA vs. GG) | 1.33 (1.00–1.76) | 3.967 | 0.046 | 1.39 (1.02, 1.91) | 0.040 | ||
| Recessive (AA vs. GA+GG) | 1.96 (0.96–3.98) | 5.575 | 0.059 | 2.54 (1.13, 5.75) | 0.025 | ||
| Allele (G allele vs. A allele) | 1.36 (1.06–1.74) | 5.950 | 0.015 | –– | –– | ||
| rs1870377 (A>T) | |||||||
| AA | 311 | 325 | Ref. | ||||
| AT | 183 | 192 | 1.23 (0.96–1.59) | 0.063 | 0.802 | 0.99 (0.74, 1.32) | 0.938 |
| TT | 39 | 16 | 2.00 (1.05–3.84) | 4.525 | 0.033 | 2.66 (1.35, 5.24) | 0.005 |
| Dominant (TT+AT vs. AA) | 1.29 (1.01–1.65) | 4.196 | 0.041 | 1.15 (0.88, 1.52) | 0.311 | ||
| Recessive (TT vs. AA+AT) | 1.84 (0.97–3.51) | 3.569 | 0.059 | 2.83 (1.47, 5.46) | 0.002 | ||
| Allele (T allele vs. A allele) | 1.27 (1.04–1.56) | 5.374 | 0.020 | –– | –– | ||
CHD – coronary heart disease; OR – odds ratio; CI – confidence interval; Adjusted OR – odds ratio after adjustment by multivariate-based logistic regression analysis; Adjusted P-value – significance after adjustment by multivariate-based logistic regression analysis.
Associations between KDR gene polymorphisms and the risk of CHD
It was obvious that all the 3 tag-SNPs were associated with CHD risk (Table 3) before multivariate-based logistic regression was applied. Regarding rs2071559 (T>C) in the 5′ near gene of KDR, the subjects carrying the variant genotypes (TC/CC) had a signally increased CHD risk compared to carriers of TT (OR=1.38, 95%CI=1.04–1.83, P=0.026) before multivariate-based regression analysis. Nonetheless, 2 additional tag-SNPs – rs2305948 and rs1870377, located in the Exon 7 and Exon 11 of KDR respectively – were both associated with CHD risk even after exclusion of interferences, because the subjects carrying the variant genotypes AA/GA and TT separately had an higher CHD risk than the corresponding carriers of GG (OR=3.04, 95%CI=1.29–7.17, P=0.011) and AA/AT (OR=2.83, 95%CI=1.47–5.46, P=0.002).
Association between haplotypes of 5 SNPs involved and susceptibility to CHD
Among the 14 haplotypes ultimately studied after removing ones with frequency lower than 0.03, we observed that carriers of C-C-C-G-T, T-C-T-G-T, T-G-T-G-A, and T-G-T-G-T were less prone to CHD than carriers with other haplotypes (OR=0.42, 95% CI: 0.22–0.78; OR=0.65, 95% CI: 0.50–0.85; OR=0.52, 95% CI: 0.32–0.87; OR=0.59, 95% CI: 0.41–0.85, respectively). In contrast, haplotypes C-C-T-G-T and T-G-T-G-T were closely associated with incremental risk of CHD in comparison with other haplotypes (OR=7.02, 95% CI: 3.19–15.47; OR=2.99, 95% CI: 1.54–5.84, respectively).
Discussion
In the current case-control study, we found a significant association of 5 polymorphisms on VEGF and KDR with susceptibility to CHD in a Chinese Han population. In particular, mutants of the 3 SNPs (rs699947, rs2305948 and rs1870377) were still risk factors for CHD development after the effects of certain clinical characteristics (e.g., BMI, smoking, alcohol consumption, diabetes, and hypertension) were eliminated. Simultaneously, haplotype analyses revealed that integration of 5 SNPs would either raise or reduce risk of CHD.
VEGF, the renowned series of glycoprotein, is largely composed of secretory materials of the vascular endothelial cells, as well as additional cellular types [20,21]. The appropriate binding of VEGF and its matching KDR, uniquely expressed and present in the endothelial cells, could primarily contribute to mediating neovascularization of atherosclerosis plaques by supplementing nutrition for the plaques and thus enlarging them [22,23]. The VEGF/KDR axis, furthermore, could also account for the occurrence of inflammatory responses in relation to vessel walls through inducing the generation of relevant cytokines (e.g., IL-6, IL-8, and GRO-α), roles of which have been demonstrated to be of significance in 3 interconnected pathophysiological durations involved with evolution of CHD: formation of atherosclerosis, rupture of plaques, and coronary spasm [24–27]. It still remains debated, nonetheless, whether VEGF serves as a pro-atherosclerosis factor because VEGF has been conceived as a cause of stepped-up re-endothelialization, thereby resisting intimal thickening and thrombus development [28,29]. The controversy could be construed as distinct phenomena resulting from relatively higher or lower ratio of various isoforms of VEGF. Double isoforms of VEGF165, VEGF 165a, and VEGF 165b possess pro-angiogenic and anti-angiogenic properties, and higher ratios of a/b or b/a would either promote or inhibit the evolution of atherosclerosis, with regulation controlled by insulin-like growth factor (IGF) [30]. The roles of polymorphisms mentioned in the current study obviously tended to facilitate the progression of atherosclerosis with consequently high a-to-b ratios of VEGF isoforms. Because type II diabetes (T2D) is thought to advance the development of atherosclerosis and the accompanying cardiac complications [31,32], the genetic mutants making predisposition to T2D and myocardial infarctions possible could also be potentially associated with the occurrence of CHD, such as polymorphisms of rs699947, rs201096, rs3025010, rs2071559, rs2305948, and rs1870377 [PMID: 17264508; PMID: 25128838].
Of the 3 eligible SNPs in VEGF, rs3025010 is situated in the intron area, is cleaved after being transcribed into mRNAs and hence does not usually strongly affect protein functions. In contrast, rs699947 and rs2010963 are located in the 5′UTR region, affecting the transcription and expression of VEGF. The dissimilarity can, to a certain extent, account for the differentiations in associations between SNPs and susceptibility to CHD. More specifically, rs2010963 (94C>G) is probably a functional polymorphism, because serum VEGF levels in subjects with CC genotype have been demonstrated to be remarkably higher than in other genotypes [33–35]. Additionally, the 3 SNPs of KDR, located in the 5′ near gene and exon regions, could affect differential expression of KDR and thus affect risk of CHD through interfering with the efficiency of VEGF binding. According to Shahbazi et al., the wild-type allele VEGF 2055CC is correlated with elevated VEGF concentration, seemingly conferring protection against CHD [36,37]. Howell et al. also verified that VEGF 2055AA genotype could be considered as a risk factor for atherosclerosis, which is consistent with the regulation of VEGF in the endothelial integrity of coronary artery walls [38]. All the above results are in line with the fact that CHD is closely related with the expression level of VEGF and the binding efficiency of KDR, indicating the crucial roles of SNPs on KDR and VEGF in CHD pathogenesis (Table 3).
However, as CHD development was primarily attributed to the complicated interplay of genetic and environmental factors, there might sometimes appear illusions that certain polymorphisms elevate risk of CHD, but in fact it was concomitant disorders or unhealthy lifestyles of subjects that were really correlated with risk of CHD. Specifically, among the 5 SNPs (rs699947, rs2010963, rs2071559, rs2305948, and rs1870377) previously regarded as the underlying etiology of CHD development, mutants of VEGF (rs699947) and KDR (rs2305948 and rs1870377) could predispose to CHD after excluding the effects of other factors (BMI, SBP, pulse, DBP, smoking, alcohol consumption, hypertension, and diabetes) via analysis of multivariate logistic regression (Table 3).
As for the synergic reactions of all 5 SNPs involved, it is interesting to discover that carriers of 4 haplotypes (C-C-C-G-T, T-C-T-G-A, T-C-T-G-T, and T-G-T-G-A) seem to be less susceptible to CHD, while carriers of other haplotypes (C-C-T-G-T and T-G-T-G-T) tended to suffer from CHD more readily. In fact, the protective role of T allele of rs699947 and G allele of rs2305948 were highlighted in terms of haplotype T-C-T-G-A and T-C-T-G-T when compared with T/A alleles of rs1870377 (Table 4). Moreover, the significance of rs1870377 was pronounced when considering haplotypes T-G-T-G-A and T-G-T-G-T, since the mutant T allele of rs1870377 enables carriers of T-G-T-G-T to be less vulnerable to CHD, while the A allele appears to play the opposite role. It could be supposed from the 4 haplotypes mentioned above that the function of rs1870377 (A>T) would overwhelm that of mixed effects of rs699947 (T>C) and rs2305948 (G>A) in the presence of T-G-T-G composed of another 4 SNPs, yet the relationship was reversed with haplotype T-C-T-G. Finally, the positive effect of rs2305948 was relatively outstanding in haplotype C-C-C-G-T for CHD development, while combined actions of rs699947 and rs1870377 in view of C-C-T-G-T. Admittedly, the conclusion mentioned above might not be applicable to other groups, as investigations would differ according to ethnic background of subjects and sample size. A possibility of selection bias also could not be ruled out, in that our case-control study was hospital-based.
Table 4.
Haplotype analysis for three polymorphisms of VEGF (rs699947 and rs201096) and KDR (rs2071559, rs2305948 and rs1870377).
| Haplotpe | Case | Control | χ2 | P (Fisher) | P (Pearson) | OR (95% CI) |
|---|---|---|---|---|---|---|
| C-C-C-G-A | 94 (0.088) | 80 (0.075) | 1.283 | 0.257 | 0.257 | 1.20 (0.88, 1.64) |
| C-C-C-G-T | 14 (0.013) | 33 (0.031) | 7.828 | 0.005 | 0.005 | 0.42 (0.22, 0.78) |
| C-C-T-G-A | 118 (0.111) | 101 (0.095) | 1.727 | 0.189 | 0.189 | 1.21 (0.91, 1.61) |
| C-C-T-G-T | 48 (0.045) | 7 (0.007) | 31.239 | <0.001 | <0.001 | 7.02 (3.19, 15.47) |
| C-G-C-G-A | 43 (0.041) | 44 (0.042) | 0.002 | 0.968 | 0.968 | 0.99 (0.65, 1.52) |
| C-G-T-G-A | 41 (0.039) | 45 (0.042) | 0.145 | 0.703 | 0.703 | 0.92 (0.60, 1.42) |
| T-C-C-A-A | 33 (0.032) | 24 (0.023) | 1.799 | 0.180 | 0.180 | 1.44 (0.84, 2.45) |
| T-C-C-G-A | 150 (0.141) | 141 (0.132) | 0.497 | 0.481 | 0.481 | 1.10 (0.85, 1.41) |
| T-C-C-G-T | 56 (0.053) | 38 (0.036) | 3.853 | 0.050 | 0.050 | 1.52 (0.99, 2.32) |
| T-C-T-G-A | 107 (0.100) | 157 (0.147) | 10.378 | 0.001 | 0.001 | 0.65 (0.50, 0.85) |
| T-C-T-G-T | 24 (0.022) | 45 (0.042) | 6.473 | 0.011 | 0.011 | 0.52 (0.32, 0.87) |
| T-G-C-G-A | 76 (0.071) | 89 (0.084) | 1.020 | 0.313 | 0.313 | 0.85 (0.62, 1.17) |
| T-G-T-G-A | 49 (0.046) | 81 (0.076) | 8.057 | 0.005 | 0.005 | 0.59 (0.41, 0.85) |
| T-G-T-G-T | 34 (0.032) | 12 (0.011) | 11.356 | 0.001 | 0.001 | 2.99 (1.54, 5.84) |
OR – odds ratio; CI – confidence interval.
Conclusions
In conclusion, genetic polymorphisms on VEGF (rs699947) and KDR (rs2305948and rs1870377), as well as relevant haplotypes, may serve as genetic markers that might be useful in future investigations on the pathogenesis of CHD.
Further investigations are, therefore, required to validate the functional relationship between SNPs of VEGF and KDR and susceptibility to CHD in terms of ethnic differences, which might contribute to the development of novel methods for diagnosis and treatment of CHD in the future.
Footnotes
Conflict of interest
Author’s disclosures of potential conflicts of interest: none for all authors.
Source of support: Departmental sources
References
- 1.Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finegold JA, Asaria P, Francis DP. Mortality from ischaemic heart disease by country, region, and age: statistics from World Health Organisation and United Nations. Int J Cardiol. 2013;168:934–45. doi: 10.1016/j.ijcard.2012.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng C, Wang Q, You W, et al. MiRNAs as biomarkers of myocardial infarction: A meta-analysis. PLoS One. 2014;9:e88566. doi: 10.1371/journal.pone.0088566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howell WM, Ali S, Rose-Zerilli MJ, Ye S. VEGF polymorphisms and severity of atherosclerosis. J Med Genet. 2005;42:485–90. doi: 10.1136/jmg.2004.025734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim HS, Blann AD, Chong AY, et al. Plasma vascular endothelial growth factor, angiopoietin-1, and angiopoietin-2 in diabetes: implications for cardiovascular risk and effects of multifactorial intervention. Diabetes Care. 2004;27:2918–24. doi: 10.2337/diacare.27.12.2918. [DOI] [PubMed] [Google Scholar]
- 6.Petrovic D. The role of vascular endothelial growth factor gene as the genetic marker of atherothrombotic disorders and in the gene therapy of coronary artery disease. Cardiovasc Hematol Agents Med Chem. 2010;8:47–54. doi: 10.2174/187152510790796183. [DOI] [PubMed] [Google Scholar]
- 7.Fong GH, Rossant J, Gertsenstein M, Breitman ML. Role of the Flt-1 receptor tyrosine kinase in regulating the assembly of vascular endothelium. Nature. 1995;376:66–70. doi: 10.1038/376066a0. [DOI] [PubMed] [Google Scholar]
- 8.Shalaby F, Rossant J, Yamaguchi TP, et al. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature. 1995;376:62–66. doi: 10.1038/376062a0. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Zheng Y, Zhang W, et al. Polymorphisms of KDR gene are associated with coronary heart disease. J Am Coll Cardiol. 2007;50:760–67. doi: 10.1016/j.jacc.2007.04.074. [DOI] [PubMed] [Google Scholar]
- 10.Zhang W, Sun K, Zhen Y, et al. VEGF receptor-2 variants are associated with susceptibility to stroke and recurrence. Stroke. 2009;40:2720–26. doi: 10.1161/STROKEAHA.109.554394. [DOI] [PubMed] [Google Scholar]
- 11.Yla-Herttuala S, Rissanen TT, Vajanto I, Hartikainen J. Vascular endothelial growth factors: biology and current status of clinical applications in cardiovascular medicine. J Am Coll Cardiol. 2007;49:1015–26. doi: 10.1016/j.jacc.2006.09.053. [DOI] [PubMed] [Google Scholar]
- 12.Khurana R, Simons M, Martin JF, Zachary IC. Role of angiogenesis in cardiovascular disease: a critical appraisal. Circulation. 2005;112:1813–24. doi: 10.1161/CIRCULATIONAHA.105.535294. [DOI] [PubMed] [Google Scholar]
- 13.Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9:669–76. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 14.Ruhrberg C. Growing and shaping the vascular tree: multiple roles for VEGF. Bioessays. 2003;25:1052–60. doi: 10.1002/bies.10351. [DOI] [PubMed] [Google Scholar]
- 15.Tammela T, Zarkada G, Wallgard E, et al. Blocking VEGFR-3 suppresses angiogenic sprouting and vascular network formation. Nature. 2008;454:656–60. doi: 10.1038/nature07083. [DOI] [PubMed] [Google Scholar]
- 16.Waltenberger J, Claesson-Welsh L, Siegbahn A, et al. Different signal transduction properties of KDR and Flt1, two receptors for vascular endothelial growth factor. J Biol Chem. 1994;269:26988–95. [PubMed] [Google Scholar]
- 17.Gerber HP, McMurtrey A, Kowalski J, et al. Vascular endothelial growth factor regulates endothelial cell survival through the phosphatidylinositol 3′-kinase/Akt signal transduction pathway. Requirement for Flk-1/KDR activation. J Biol Chem. 1998;273:30336–43. doi: 10.1074/jbc.273.46.30336. [DOI] [PubMed] [Google Scholar]
- 18.Cao Q, Wang J, Zhang M, et al. Genetic variants in RKIP are associated with clear cell renal cell carcinoma risk in a Chinese population. PLoS One. 2014;9:e109285. doi: 10.1371/journal.pone.0109285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui QT, Li Y, Duan CH, et al. Further evidence for the contribution of the vascular endothelial growth factor gene in coronary artery disease susceptibility. Gene. 2013;521:217–21. doi: 10.1016/j.gene.2013.03.091. [DOI] [PubMed] [Google Scholar]
- 20.Berse B, Brown LF, Van de Water L, et al. Vascular permeability factor (vascular endothelial growth factor) gene is expressed differentially in normal tissues, macrophages, and tumors. Mol Biol Cell. 1992;3:211–20. doi: 10.1091/mbc.3.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Namiki A, Brogi E, Kearney M, et al. Hypoxia induces vascular endothelial growth factor in cultured human endothelial cells. J Biol Chem. 1995;270:31189–95. doi: 10.1074/jbc.270.52.31189. [DOI] [PubMed] [Google Scholar]
- 22.Moulton KS, Vakili K, Zurakowski D, et al. Inhibition of plaque neovascularization reduces macrophage accumulation and progression of advanced atherosclerosis. Proc Natl Acad Sci USA. 2003;100:4736–41. doi: 10.1073/pnas.0730843100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srivatsa SS, Edwards WD, Boos CM, et al. Histologic correlates of angiographic chronic total coronary artery occlusions: influence of occlusion duration on neovascular channel patterns and intimal plaque composition. J Am Coll Cardiol. 1997;29:955–63. doi: 10.1016/s0735-1097(97)00035-1. [DOI] [PubMed] [Google Scholar]
- 24.Hao Q, Wang L, Tang H. Vascular endothelial growth factor induces protein kinase D-dependent production of proinflammatory cytokines in endothelial cells. Am J Physiol Cell Physiol. 2009;296:C821–27. doi: 10.1152/ajpcell.00504.2008. [DOI] [PubMed] [Google Scholar]
- 25.Katayama N, Nakao K, Horiuchi K, et al. [Disease activities and serum C-reactive protein levels in patients with vasospastic angina pectoris]. J Cardiol. 2005;46:63–70. [in Japanese] [PubMed] [Google Scholar]
- 26.Li JJ, Nie SP, Xu B, et al. Inflammation in variant angina: is there any evidence? Med Hypotheses. 2007;68:635–40. doi: 10.1016/j.mehy.2006.05.068. [DOI] [PubMed] [Google Scholar]
- 27.Troseid M, Seljeflot I, Hjerkinn EM, Arnesen H. Interleukin-18 is a strong predictor of cardiovascular events in elderly men with the metabolic syndrome: synergistic effect of inflammation and hyperglycemia. Diabetes Care. 2009;32:486–92. doi: 10.2337/dc08-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grosskreutz CL, Anand-Apte B, Duplaa C, et al. Vascular endothelial growth factor-induced migration of vascular smooth muscle cells in vitro. Microvasc Res. 1999;58:128–36. doi: 10.1006/mvre.1999.2171. [DOI] [PubMed] [Google Scholar]
- 29.Keavney B, McKenzie C, Parish S, et al. Large-scale test of hypothesised associations between the angiotensin-converting-enzyme insertion/deletion polymorphism and myocardial infarction in about 5000 cases and 6000 controls. International Studies of Infarct Survival (ISIS) Collaborators. Lancet. 2000;355:434–42. doi: 10.1016/s0140-6736(00)82009-7. [DOI] [PubMed] [Google Scholar]
- 30.Perrin RM, Konopatskaya O, Qiu Y, et al. Diabetic retinopathy is associated with a switch in splicing from anti- to pro-angiogenic isoforms of vascular endothelial growth factor. Diabetologia. 2005;48:2422–27. doi: 10.1007/s00125-005-1951-8. [DOI] [PubMed] [Google Scholar]
- 31.Grundy SM, Benjamin IJ, Burke GL, et al. Diabetes and cardiovascular disease: a statement for healthcare professionals from the American Heart Association. Circulation. 1999;100:1134–46. doi: 10.1161/01.cir.100.10.1134. [DOI] [PubMed] [Google Scholar]
- 32.Moreno PR, Fuster V. New aspects in the pathogenesis of diabetic atherothrombosis. J Am Coll Cardiol. 2004;44:2293–300. doi: 10.1016/j.jacc.2004.07.060. [DOI] [PubMed] [Google Scholar]
- 33.Petrovic D, Verhovec R, Globocnik Petrovic M, et al. Association of vascular endothelial growth factor gene polymorphism with myocardial infarction in patients with type 2 diabetes. Cardiology. 2007;107:291–95. doi: 10.1159/000099064. [DOI] [PubMed] [Google Scholar]
- 34.Awata T, Inoue K, Kurihara S, et al. A common polymorphism in the 5′-untranslated region of the VEGF gene is associated with diabetic retinopathy in type 2 diabetes. Diabetes. 2002;51:1635–39. doi: 10.2337/diabetes.51.5.1635. [DOI] [PubMed] [Google Scholar]
- 35.Inoue M, Itoh H, Ueda M, et al. Vascular endothelial growth factor (VEGF) expression in human coronary atherosclerotic lesions: possible pathophysiological significance of VEGF in progression of atherosclerosis. Circulation. 1998;98:2108–16. doi: 10.1161/01.cir.98.20.2108. [DOI] [PubMed] [Google Scholar]
- 36.Renner W, Kotschan S, Hoffmann C, et al. A common 936 C/T mutation in the gene for vascular endothelial growth factor is associated with vascular endothelial growth factor plasma levels. J Vasc Res. 2000;37:443–48. doi: 10.1159/000054076. [DOI] [PubMed] [Google Scholar]
- 37.Shahbazi M, Fryer AA, Pravica V, et al. Vascular endothelial growth factor gene polymorphisms are associated with acute renal allograft rejection. J Am Soc Nephrol. 2002;13:260–64. doi: 10.1681/ASN.V131260. [DOI] [PubMed] [Google Scholar]
- 38.De Winter RJ, Klomp M. Understanding the role of endothelial progenitor cells in cardiovascular disease, coronary artery lesion progression, and in-stent restenosis. JACC Cardiovasc Interv. 2010;3:87–89. doi: 10.1016/j.jcin.2009.11.006. [DOI] [PubMed] [Google Scholar]