Abstract
Understanding epigenetic differences that distinguish neurons and glia is of fundamental importance to the nascent field of neuroepigenetics. A recent study used genome-wide bisulfite sequencing to survey differences in DNA methylation between these two cell types, in both humans and mice. That study minimized the importance of cell type-specific differences in CpG methylation, claiming these are restricted to localized genomic regions, and instead emphasized that widespread and highly conserved differences in non-CpG methylation distinguish neurons and glia. We reanalyzed the data from that study and came to markedly different conclusions. In particular, we found widespread cell type-specific differences in CpG methylation, with a genome-wide tendency for neuronal CpG-hypermethylation punctuated by regions of glia-specific hypermethylation. Alarmingly, our analysis indicated that the majority of genes identified by the primary study as exhibiting cell type-specific CpG methylation differences were misclassified. To verify the accuracy of our analysis, we isolated neuronal and glial DNA from mouse cortex and performed quantitative bisulfite pyrosequencing at nine loci. The pyrosequencing results corroborated our analysis, without exception. Most interestingly, we found that gene-associated neuron vs. glia CpG methylation differences are highly conserved across human and mouse, and are very likely to be functional. In addition to underscoring the importance of independent verification to confirm the conclusions of genome-wide epigenetic analyses, our data indicate that CpG methylation plays a major role in neuroepigenetics, and that the mouse is likely an excellent model in which to study the role of DNA methylation in human neurodevelopment and disease.
Introduction
Methylation of cytosines in CpG dinucleotides is a highly stable epigenetic mechanism (1) with established roles in genomic imprinting, X-inactivation, and silencing of retrotransposons (2). Various data indicate that DNA methylation also plays crucial roles in brain development and transcriptional regulation, with implications for neural function and disease. Differentiation of neurons and glia (3–5), axon guidance (6), and synapse remodeling (6–8) are all associated with DNA methylation, and it has been suggested that DNA methylation mediates memory formation (8). Aberrant genomic imprinting (parent-of-origin specific expression) is the cause of major neurodevelopmental diseases including Prader–Willi syndrome (9) and Angelman syndrome (10). Further, age-related ‘epigenetic drift’ in DNA methylation is associated with the development of neurological diseases such as Alzheimer's (11) and Parkinson's (12).
Understanding the epigenetic etiology of neurological disease is complicated by the functional and epigenetic specialization of different regions of the brain, and by the epigenetic heterogeneity of different cell types. Cells in the central nervous system are dichotomized as either neurons or glia. Unlike neurons, glia do not generate action potentials but perform diverse functions including structural support, maintenance and homeostatic regulation (13). Two recent studies used the Illumina Infinium HumanMethylation450K (HM450) array to characterize neuron vs. glia differences in CpG methylation in the human brain. One reported that 14% of loci showed at least a 20% methylation difference between the two cell types (6). The other found that genomic regions of cell type-specific differential methylation are enriched at predicted enhancers (14). Using a different approach for reduced-representation methylation profiling, we recently reported major differences in CpG methylation between neuronal and non-neuronal cells in the mouse hypothalamus (15).
Improving upon previous studies using the HM450 array (6,14,16) or other reduced-representation platforms (15–17), Lister et al. recently performed genome-wide bisulfite sequencing (Bisulfite-seq) on neuronal and non-neuronal DNA isolated from several samples of mouse and human cortex (18). The breadth and depth of their dataset make it the presumptive gold standard for analysis of differential methylation between neurons and glia. Contrary to previous analyses, however, Lister et al. reported only minor cell type-specific CpG methylation (mCG) differences, and concluded that ‘differential mCG between neurons and glia was restricted to localized regions’. In trying to understand the reason for the discrepancy between their findings and those of previous studies, we noted that Lister et al. used a non-standard normalization procedure to analyze their Bisulfite-seq data. For each gene, they normalized intragenic methylation relative to the median methylation in a 50–100 kb flanking region. We postulated that this non-standard normalization approach may have led to a misinterpretation of their data.
We therefore reanalyzed the Bisulfite-seq data of Lister et al. (18) without normalization, and generated our own lists of genes with neuron vs. glia CpG methylation differences. To confirm the accuracy of our analysis, we used NeuN immunostaining and fluorescence-activated sorting to isolate neuronal and non-neuronal nuclei from mouse cortex, and performed extensive validation by quantitative bisulfite pyrosequencing. Contrary to the conclusions of Lister et al. (18), our re-analysis of their data and our independent verification indicate widespread and dramatic gene-associated CpG methylation differences between neurons and glia, in both mouse and human cortex. Most interestingly, our analysis additionally yielded the novel insight that these cell type-specific CpG methylation differences are remarkably well conserved across these two species.
Results
Widespread neuron vs. glia differences in CpG methylation in mouse cortex
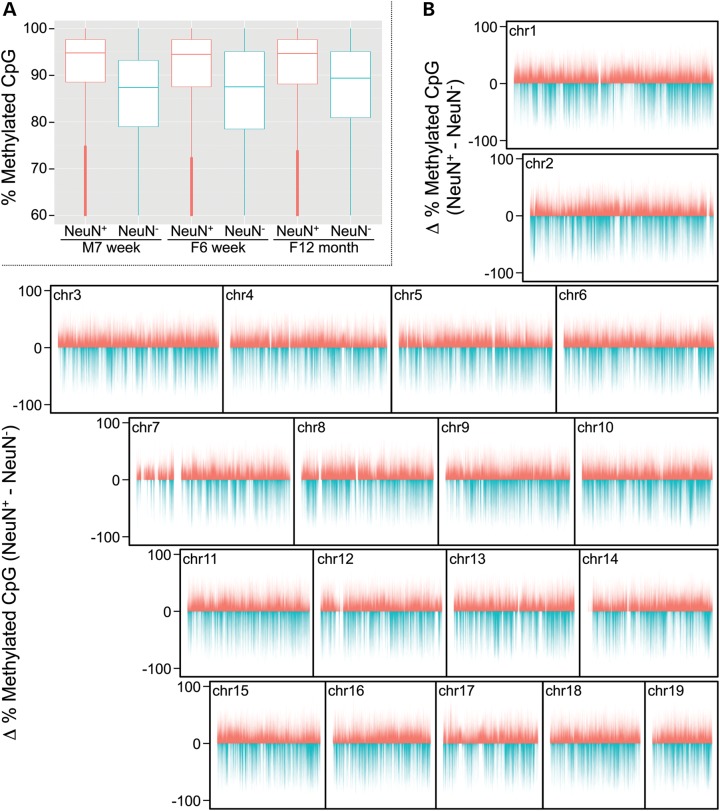
Lister et al. (18) isolated NeuN+ and NeuN− nuclei (neuronal and non-neuronal, respectively) from mouse and human cortex, and performed Bisulfite-seq. (Although the NeuN− nuclear fraction from brain includes a small fraction of other cell types, glia vastly predominate (14,15). For simplicity, we refer herein to the NeuN− fraction as representing glia.) We downloaded their Bisulfite-seq read count data and performed our own CpG methylation analysis. As previously described and validated (19), we performed our analysis at 200 bp resolution, focusing on all 200 bp genomic bins containing at least 2 CpG sites. We first evaluated an apparent discrepancy in Lister et al.'s data on mouse cortex. They showed that average CpG methylation genome-wide was substantially higher in NeuN+ relative to NeuN− cells in mouse cortex (their Fig. 2C) (18), consistent with our analysis (Fig. 1A). We were therefore surprised by their statement (18) that ‘differential mCG between neurons and glia was restricted to localized regions’. Our genome-wide analysis at 200 bp resolution (Fig. 1B) showed essentially the opposite, indicating widespread differences in CpG methylation between neurons and glia. The general tendency for neuronal hypermethylation relative to glia (N>G) across the genome is punctuated by regions of relative glial hypermethylation (G>N). Interestingly, although G>N bins were fewer in number, these methylation differences were generally of greater magnitude than N>G bins (P < 10−99).
Figure 1.
Differential CpG methylation between murine neurons and glia. (A) Global levels of methyl-CpG are substantially higher in neuronal (NeuN+) compared with non-neuronal (NeuN−) cells in mouse cortex. (Shown are the six samples analyzed by Lister et al. and presented in their Fig. 2C.) (B) Chromosome plots of cell type-specific methylation differences (neuronal minus non-neuronal) reveal a general pattern of neuron-specific hypermethylation punctuated by regions of dramatic glia-specific hypermethylation.
Of the 1.8 M 200 bp bins with sufficient coverage, 554 691 showed N>G (absolute average difference of at least 10% methylation) and 226 369 showed G>N. Altogether, therefore, we found nearly half (43.6%) of informative bins to be differentially methylated. To gain a clearer view of the CpG methylation landscape distinguishing neurons and glia, we visually examined our representation of Lister et al.'s data on the UCSC genome browser. (Users can browse the genome-wide data via a link included in Supplementary Material). Surveying mouse chromosomes 1, 13 and 17, for example, led to the identification of many large genomic regions with nearly exclusive N>G or G>N (Supplementary Material, Fig. S1). Regions of N>G (Supplementary Material, Fig. S1A, C, G, I and J) tended to be large (often spanning several Mb) and in some cases encompassed clusters of functionally-related small genes (Supplementary Material, Fig. S1I and J). G>N regions, on the other hand, were generally smaller than N>G regions (<500 kb) and tended to overlap precisely with genes, such as Satb2, Pbx1, Phactr1, Atxn1, Mef2c, Fbxl17 and Nrxn1 (Supplementary Material, Fig. S1B, D, E, F, H, K and L, respectively).
Gene-centered analysis highlights specific discrepancies
A centerpiece of the Lister et al. analysis (their Fig. 4A) was a heat-map representation of 25 260 genes clustered on the basis of CpG and non-CpG methylation (18). In their heat map, intragenic methylation was normalized relative to the median methylation in a 50–100 kb region flanking each gene. Although the rationale for this normalization approach was not stated, one potential reason would be to correct for sample-specific differences in bisulfite conversion efficiency. Since CpC dinucleotides are rarely methylated in mammalian cells, observed CpC methylation can be used as an indicator of conversion efficiency (20). Our analysis of CpC methylation in the Lister et al. data, however, indicated genome-wide bisulfite conversion efficiency of greater than 99.5% in all the murine neuron and glia samples (Supplementary Material, Table S9).
To compare our CpG methylation results directly with those of Lister et al., we too performed a gene-centered analysis. We contacted the authors to obtain lists of genes corresponding to the clusters delineated by the horizontal lines in panel B of their Figure 4, which we designated A through O (top to bottom). Based on the text describing the boxes in their Figure 4A, genes from that figure were categorized as G>N if they belonged to clusters A or B, and N>G if they belonged to cluster H. In our analysis, we classified a gene as differentially methylated if the average absolute neuron vs. glia methylation difference across all bins informative within that gene was greater than 10% (see Methods and Supplementary Material, Table S1).
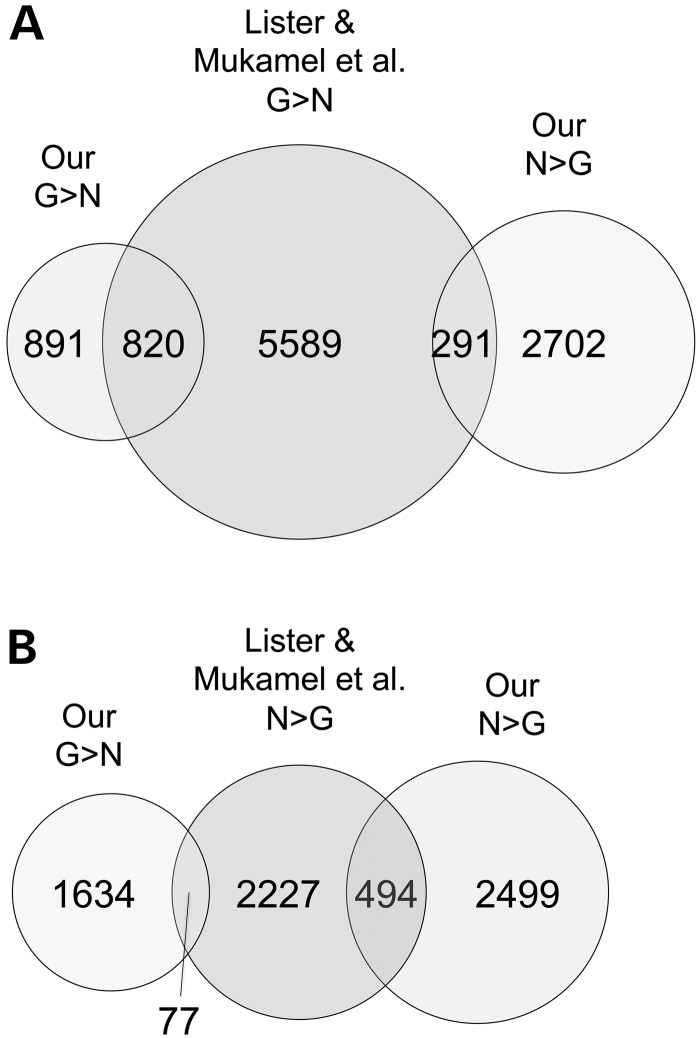
Of the 1711 genes we classified as CpG-hypermethylated in glia relative to neurons (G>N), only about half were similarly categorized by Lister et al. (Fig. 2A). Of the 2993 genes we classified as CpG-hypermethylated in neurons relative to glia (N>G), the agreement with Lister et al. was even poorer; only 17% overlapped those they identified as such (Fig. 2B). A compelling example is provided by the 3 Mb region of mouse chromosome 17 with general N>G (Supplementary Material, Fig. S1I). Of a cluster of 21 Vmn2r genes (Vmnr2r90-110) within this region, none was identified as N>G by Lister et al., whereas our analysis (Supplementary Material, Table S1) identified 20 as N>G. We noted that genes with low CpG density were more likely to be classified differently in the two analyses. Most of the genes classified only by Lister et al. as G>N were among the lowest three quartiles of CpG density, whereas most of the genes classified only by us as N>G were among the lowest quartile of CpG density (Supplementary Material, Fig. S2A and B, respectively). Alarmingly, many of the genes we identified as showing neuron vs. glia CpG methylation differences were categorized by Lister et al. as showing the opposite cell type-specific difference (291 and 77) (Fig. 2A and B, respectively).
Figure 2.
Genes classified as hypermethylated in neurons or in glia by our analysis overlap poorly with those similarly classified by Lister et al. (A) Of the 1711 glia-hypermethylated genes from our analysis, less than half were classified as such by Lister et al. Additionally, of 6700 genes from their analysis with that classification, 291 were classified oppositely by our analysis. (B) About 17% of the 2993 genes we classified as hypermethylated in neurons were similarly classified by Lister et al.; of the 2798 neuron-hypermethylated genes from their analysis, 77 were classified oppositely by our analysis.
Quantitative bisulfite pyrosequencing validates our analysis
Since both our analysis and that of Lister et al. were based on the same Bisulfite-seq data, such marked discrepancies indicate that at least one of them is unreliable. We therefore performed independent validation by quantitative bisulfite pyrosequencing (15), focusing on genes categorized discordantly by the two analyses. We isolated nuclei from mouse cortex, immunostained them for the neuron-specific marker NeuN, and performed fluorescence-activated sorting to separate neuronal and non-neuronal DNA (Supplementary Material, Fig. S3).
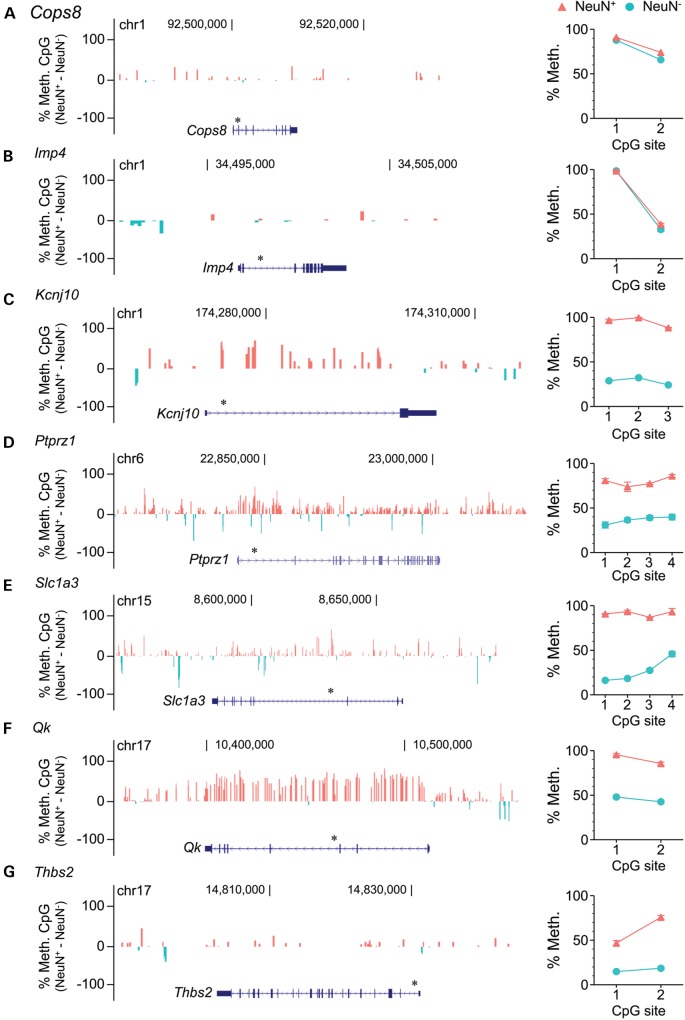
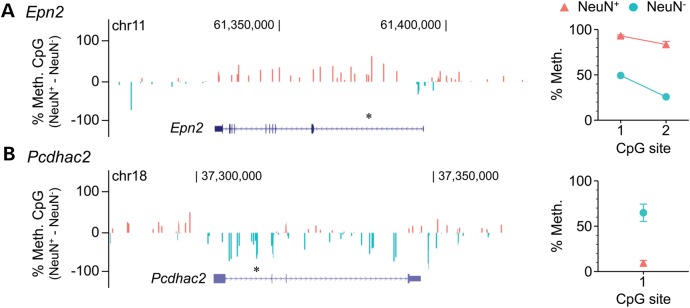
Two genes (Cops8 and Imp4) included by Lister et al. in a cluster of genes with ‘intragenic mCG enrichment in glia and depletion in neurons’ (their cluster B) showed no cell type-specific methylation differences in our analysis. The pyrosequencing data (Fig. 3A–B) agreed with our assessment. Five genes (Kcnj10, Ptprz1, Slc1a3, Qk and Thbs2) classified by Lister et al. as showing no cell type methylation differences (i.e. included in their clusters G and M) (18) were classified by our analysis as hypermethylated in neurons relative to glia. Again, in all five cases, the pyrosequencing data (Fig. 3C–G) corroborated our classification. We also examined two genes characterized by Lister et al. as having ‘intragenic mCG and mCH hypomethylation in neurons but not glia’ (Epn2, their cluster A), or ‘neuronal mCG and mCH hypermethylation, and glial mCG and mCH hypomethylation’ (Pcdhac2, their cluster H). These were classified by our analysis as showing the exact opposite cell type-specific differences. In both cases, the pyrosequencing results (Fig. 4A–B) supported our classification. Hence, in 9 out of 9 genes tested, independent quantitative measurements in mouse cortex verified the reliability of our non-normalized cell type-specific methylation analysis.
Figure 3.
Quantitative bisulfite pyrosequencing validates our analysis of Lister et al.'s Bisulfite-seq data. (A, B) Cops8 and Imp4, classified by Lister et al. as showing ‘intragenic mCG enrichment in glia and depletion in neurons' showed no evidence of cell type-specific methylation differences, either in our analysis of their Bisulfite-seq data (left) or in our pyrosequencing assays (right). (C–G) Five genes (Kcnj10, Ptprz1, Slc1a3, Qk and Thbs2) classified by Lister et al. as showing no cell type-specific methylation differences all showed neuron-specific hypermethylation, both in our analysis of their data (left) and in our pyrosequencing assays (right). Asterisks above gene diagrams indicate genomic regions covered by pyrosequencing. Pyrosequencing data represent the mean ± SEM of 6–7 samples per cell type.
Figure 4.
At genes with discrepant cell type-specific methylation calls between the two analyses, quantitative bisulfite pyrosequencing validates our analysis. (A) Epn2, classified by Lister et al. as showing ‘intragenic mCG and mCH hypomethylation in neurons but not glia’ is actually hypermethylated in neurons relative to glia, according to both their raw Bisulfite-seq data (left) and our bisulfite pyrosequencing (right). (B) Pcdhac, classified by Lister et al. as showing ‘neuronal mCG and mCH hypermethylation, and glial mCG and mCH hypomethylation’ is actually hypermethylated in glia relative to neurons, according to both their raw Bisulfite-seq data (left) and our bisulfite pyrosequencing (right). Asterisks above gene diagrams indicate genomic regions covered by pyrosequencing. Pyrosequencing data represent the mean ± SEM of 6–7 samples per cell type.
Comparisons with other analytical approaches, and an independent data set, also corroborate our analysis
In addition to our pyrosequencing studies, we sought complementary means by which to test the validity of our analysis. We compared our lists of differentially methylated genes with those identified by MOABS (21), a statistically-based and easily reproducible method for identifying differentially methylated regions (DMRs) from Bisulfite-seq data. (MOABS identifies ‘credible differences’ in methylation based on confidence intervals around the mean methylation in two samples.) Although we employed settings intended to enable the identification of large DMRs (e.g. by allowing up to 5 kb between ‘adjacent’ differentially methylated CpGs), MOABS identified 182 138 DMRs, most of which were under 20 kb in size (Supplementary Material, Fig. S4). Consistent with our analysis (Fig. 1B), MOABS identified more N>G than G>N DMRs, and G>N DMRs showed greater credible differences (Supplementary Material, Fig. S5). Only 856 MOABS DMRs were larger than 20 kb, and the largest was 307 kb. Many of the multiple-Mb N>G tracts we identified by eye (Supplementary Material, Fig. S1) were subdivided into discrete DMRs by MOABS; for example, within the 6 Mb N>G region on chromosome 17 (Supplementary Material, Fig. S1J), MOABS identified 203 DMRs together encompassing only 869 kb. Nonetheless, to compare genic methylation differences identified by MOABS with those we identified, we compiled all the genes located within MOABS DMRs. By this approach, MOABS identified 288 genes as N>G, and 132 as G>N. Over 1/3 of the genes MOABS identified as either N>G or G>N were likewise classified by our analysis (Supplementary Material, Fig. S6). Most importantly, unlike in the comparison of our results with those of Lister et al. (Fig. 2), no genes identified by MOABS as either N>G or G>N were classified oppositely by our analysis (Supplementary Material, Fig. S6).
Guintivano et al. (6) recently reported DNA methylation profiling (Illumina HM450) in neuronal and glial DNA from human cortex from 58 individuals, providing an additional opportunity to test the validity of our approach for calling differentially methylated genes. We downloaded Lister et al.'s Bisulfite-seq data on neurons and glia from human cortex and used our bin-specific approach to identify differentially methylated genes. We contacted Zachary Kaminsky, the senior author of the Guintivano et al. study, who kindly provided us with an annotated list of all the HM450 probes with statistically significant (FDR corrected) methylation differences between neurons and glia in their data set (6). Of the 902 genes identified as G>N from the Guintivano et al. data, approximately 1/3 overlapped with G>N genes in our analysis of the Lister et al. data (Supplementary Material, Fig. S7A). Likewise, over 1/4 of the 835 genes identified as N>G from the Guintivano et al. data overlapped with N>G genes in our analysis of the Lister et al. data (Supplementary Material, Fig. S7B). Again, unlike in the comparison of our analysis with that of Lister et al. (Fig. 2), opposite classification of genes as G>N or N>G between Guintivano et al. and our analysis of the Lister et al. human data was negligible (Supplementary Material, Fig. S7). Together, the comparisons with the MOABS results and with the Guintivano et al. data set reinforce the validity of our analysis.
Our analysis uncovers remarkable conservation of cell type-specific CpG methylation differences from mouse to human
Lister et al. used their Bisulfite-seq data on neurons and glia from mouse and human cortex to identify CG-DMRs (CpG–differentially methylated regions) in both species, and provided a gene-functional analysis of those they found in mouse. They did not, however, report a detailed analysis of the conservation of neuron vs. glia CpG methylation differences across species. We therefore set out to determine the degree to which cell type-specific differences in CpG methylation are conserved across mouse and human cortex. We analyzed the Bisulfite-seq data of Lister et al. to identify genes exhibiting neuron vs. glia CpG methylation differences in cortex in both species. A nominal absolute methylation threshold was set for cell-type differences (≥10%); to focus on robust differences, only genes with at least five informative bins were included (Supplementary Material, Table S2).
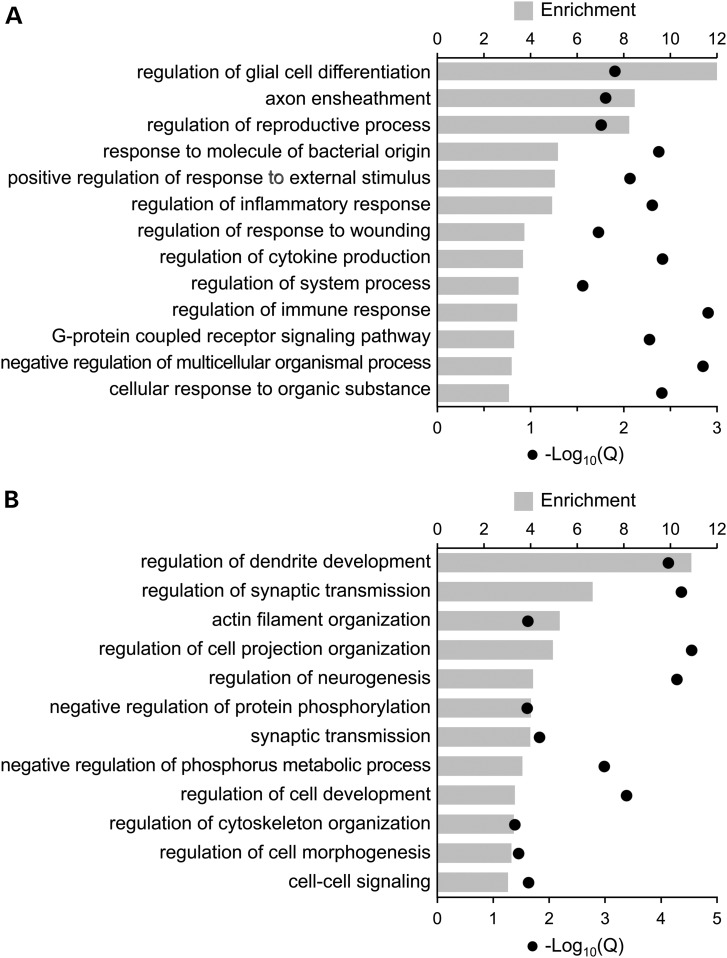
We were surprised by the high degree of conservation across species. Of genes classified in either mouse or human as N>G, 86% were classified as such in both species (Supplementary Material, Table S3). Likewise, of genes classified in either mouse or human as G>N, 87% were classified as such in both species (Supplementary Material, Table S6). Interestingly, conservation of cell type-specific methylation differences was higher in gene bodies than in promoter regions; 93% of genes with gene body neuron vs. glia methylation differences in both species were hypermethylated in the same cell type, compared with 75% for promoters (χ2, P = 5.6 × 10−13). Not surprisingly, conservation of cell type-specific methylation differences was positively associated with conservation at the gene sequence level (Supplementary Material, Fig. S8). We performed a gene ontology (GO) analysis to evaluate the functional relevance of these highly conserved methylation differences, and found strong and highly significant enrichments specific to processes and components involved in neurogenesis and brain development. Notably, genes with conserved hypermethylation in neurons relative to glia were enriched for glia-related processes including axon ensheathment, and regulation of glial cell differentiation and immune response (Fig. 5A and Supplementary Material, Tables S4 and S5). Conversely, genes with conserved hypermethylation in glia relative to neurons were enriched for neuron-related processes including regulation of neurogenesis and dendrite development, and neuron components such as synapse part, neuron projection, and dendrite (Fig. 5B and Supplementary Material, Tables S7 and S8).
Figure 5.
Gene ontology analysis of genes with conserved neuron vs. glia methylation differences between mouse and human reveals strong and highly significant enrichments for major neural processes. (A) Genes with conserved neuronal hypermethylation are significantly enriched for glia-specific processes such as glial differentiation, axon ensheathment and immune system functions. (B) Genes with conserved glial hypermethylation are significantly enriched for neuron-related processes such as dendrite development, synaptic transmission and actin filament organization. ‘Enrichment’ is the fold enrichment of the GO term relative to the background set. ‘Q’ represents the P-value of the enrichment corrected for multiple testing by GOrilla.
Discussion
We have independently reanalyzed the neuron- and glia-specific genome-wide CpG methylation data of Lister et al. (18) and come to conclusions substantially different from theirs. Our interpretation is that the non-standard approach they used to normalize their Bisulfite-seq data (in both their Figs. 4 and 5) introduced nonexistent cell type-specific methylation differences and obscured or even reversed real differences. Contrary to their assertion that ‘differential mCG between neurons and glia [is] restricted to localized regions’, our analysis demonstrates that widespread differences in CpG methylation distinguish neurons and glia. Further, thousands of genes we characterized as showing intragenic methylation differences between neurons and glia were classified otherwise by Lister et al.
Unlike Lister et al., we validated our analysis by independently sorting NeuN+ and NeuN− cortical nuclei and performing quantitative bisulfite pyrosequencing. Because Bisulfite-seq experiments directly yield quantitative estimates of percent methylation, it is not uncommon for studies employing Bisulfite-seq to forgo independent verification of their results (18,22,23). The potential for systematic errors (such as those caused by PCR biases following bisulfite modification (24)), however, combined with the complex and non-standardized analytical methods used in these types of studies, necessitate independent verification of Bisulfite-seq analyses to confirm their reliability (20). Clearly, normalization of intragenic methylation based on flanking region methylation should be avoided. Moreover, normalization of Bisulfite-seq data is likely inappropriate in general; these data provide methylated and unmethylated read counts at each methylation site, so are effectively normalized at the CpG (or CpH) level.
Few previous studies have explored neuroepigenetic conservation between human and mouse on a genomic scale. One analysis, using the Methyl-MAPS procedure (25), found that CpG methylation is weakly correlated across human and mouse brain, both in the promoters of nearly 9000 orthologous genes and in regions with high sequence conservation. An analysis of over 900 orthologous genes in visual cortex in human and mouse found that most genes showed similar expression between the two species (26). In a study comparing human and mouse gene expression microarray data in three brain regions, over 80% of the top 500 genes with expression differences between caudate nucleus and cerebellum in humans showed similar expression differences in mice (27). Importantly, none of these previous studies performed separate analyses in neuronal and glial fractions.
Conservation of tissue- and cell type-specific methylation differences from mouse to human has previously been reported for several genes (28–31), and one recent study reported the conservation of neuron-specific CpG methylation at Cacna1c (32). Our reanalysis of the Lister et al. data enabled the discovery that gene-associated neuron vs. glia CpG methylation differences are well conserved from mouse to human. This is the first report of conservation of these differences on a genome-wide scale, and the degree of this conservation—over 85%—is remarkable. Although many genes were excluded due to naming ambiguity or lack of coverage, it is clear that these cell type-specific epigenetic differences are generally consistent between mouse and human.
Moreover, GO analysis of these conserved CpG methylation differences provided powerful support for the hypothesis that CpG methylation plays a central role in neural differentiation. Conserved hypermethylation in glia relative to neurons was associated with neuronal functions such as regulation of dendrite development and neurogenesis, as well as actin filament organization, which is required for development of long-term memory (33). GO analysis of genes with conserved hypermethylation in neurons relative to glia found associations with axon ensheathment and glial development, as might be expected, but also revealed a strong enrichment for immune response-related genes. This finding is consistent with the role of glial cells as mediators of immunity in the CNS (34–36). Our analysis therefore suggests that CpG methylation plays an important role in regulating the CNS immune response. In particular, given that dysregulation of the glial immune response is a contributing factor to neurodegenerative diseases (37,38), the high degree of conservation of neuron vs. glia CpG methylation differences within immune response genes supports the use of mouse models to gain insights into epigenetic mechanisms underlying neurodegeneration.
Overall, our findings suggest that CpG methylation plays an important role in cell type-specific epigenetic regulation in the brain. Moreover, the extremely high conservation of neuron vs. glia epigenetic differences from mouse to human indicates that the mouse is likely to be an excellent animal model of human neuroepigenetic development and disease.
Materials and Methods
Reanalysis of Bisulfite-seq data
We downloaded the mapped read count data from Lister et al.'s Bisulfite-seq experiments for NeuN+ and NeuN− fractions from three murine cortex samples (male, 7 week; female, 6 week; female, 12 month) (GSM1173786-91 on GEO) and two human cortex samples (female, 53 years; male, 55 years) (GSM1173773-4, GSM 1173776-7). Coordinates from human experiments were lifted over to hg19. For robust quantitation we considered only CpG sites covered by at least 10 reads. As previously described (19) we performed our analysis at a 200 bp resolution, focusing on all 200 bp bins genome-wide containing ≥2 CpG sites, using the same criteria for bin coverage. The quantitative accuracy of our Bisulfite-seq analytical pipeline has been validated by bisulfite pyrosequencing (19). For each cell type, average percent methylation was calculated across all samples at the 200 bp bin level, from which average cell type-specific methylation differences (neuron minus glia) were determined. (The samples from the 6 week-old female mouse were excluded from these and all subsequent calculations due to low coverage relative to the other two mice). Only bins covered across all samples were considered. Intragenic methylation differences were defined as the average methylation difference across all covered bins within each gene.
Estimation of bisulfite conversion efficiency
The methylation percentage of each non-CpG (i.e. CpH) read from Lister et al.'s data on GEO was determined, as well as the identity of the non-guanine base in the CpH dinucleotide. Overall percentages for CpC, CpA and CpT methylation were calculated for each of the four murine samples (Supplementary Material, Table S9).
Gene-centric comparison with the findings of Lister et al.
We contacted Ryan Lister and Eran Mukamel, who provided us with lists of genes indicated in the 15 clusters of Figure 4A of their paper (18). (To avoid confusion with the numbered boxes in their Fig. 4A, clusters were designated as A-O, from top to bottom, delineated by the horizontal lines in their Fig. 4B). Using a 10% intragenic methylation difference threshold, we categorized all genes with at least one informative bin as neuron-hypermethylated, glia-hypermethylated, or not differentially methylated (Supplementary Material, Table S1). We then compared our lists of genes showing cell type-specific differences with those identified by Lister et al. (18). For the analysis presented in Supplementary Material, Figure S2, CpG density for each RefSeq gene was calculated by dividing the number of CpG sites within the gene body by the number of dinucleotides in the gene body. Density scores were averaged for genes with multiple transcripts. Genes were split into quartiles by CpG density score, and the numbers of genes in each quartile from the N>G and G>N gene lists were plotted.
Independent verification of methylation differences
We used immunostaining for the neuron-specific marker NeuN, followed by fluorescence-activated sorting to isolate neuronal and non-neuronal nuclei (15) from C57BL/6J mouse cortex at postnatal age 140 days (P140) (Supplementary Material, Fig. S3). Quantitative measurement of CpG site-specific DNA methylation was performed by bisulfite pyrosequencing (15) (N = 7 NeuN− and 6 NeuN+ samples). Prior to use, each pyrosequencing assay's linearity and quantitative accuracy was verified using methylation standards (39) (Supplementary Material, Fig. S9).
Unbiased statistical search for differentially methylated regions
We used MOABS (21) to find differentially methylated regions genome-wide. Since we were interested in neuron vs. glia differences, and MOABS can only be used to make pair-wise comparisons, the two neuron and the two glia samples were combined in a single MOABS run. The following parameters were used: minimum credible difference: 0.1; minimum nominal difference: 0.1; maximum distance between consecutive DMCs: 5000 bp; DMR-finding method 2. To verify that combining both samples into a single MOABS run did not introduce technical errors, MOABS was run with the same settings, except the combined male neuronal and female glia samples were run against the combined male glial and female neuronal samples. MOABS found a relatively small number of DMRs using the mixed inputs (N = 2870 vs. 182 138 for the neuron vs. glia run), implying that the combination of the male and female reads for the neuron vs. glia analysis had a minimal effect on the DMR finding. Overlaps of DMRs with the regions visible in the genome browser (Supplementary Material, Fig. S1) and with genes identified as N>G or G>N in our analysis of the Lister et al. data (Supplementary Material, Table S1), as well as other various BED file processing steps, were performed using bedtools (40).
Comparison of gene-based analysis with published human data
We calculated genic methylation scores from Guintivano et al.'s Illumina HM450 data (6) by averaging the neuron minus glia methylation difference of all probes which were labeled as ‘body’ in the gene annotations field and had an FDR P-value <0.05. Genes were considered to be hypermethylated in one cell type if they contained at least 5 FDR-significant probes and had at least a 10% methylation difference between neurons and glia (N = 835 for N>G and N = 902 for G>N). The lists of differentially methylated genes were compared to our lists of human genes containing at least five informative bins and having a 10% methylation difference (N = 1326 for N>G and N = 1017 for G>N) (Supplementary Material, Fig. S7).
Comparison of cell type-specific methylation differences across human and mouse
The list of human and mouse gene name conversions was downloaded from HGNC Comparison of Orthology Predictions website (41). To remove ambiguity, only genes having a one-to-one correspondence in gene name between the two species were considered in this analysis. From these unambiguous genes (N = 10 460), only those with solid informativity (i.e. containing ≥5 intragenic bins with coverage in both species) were kept, leaving 7998 genes. Genes for which both species had at least a 10% intragenic methylation difference were classified as showing cell type-specific hypermethylation (N = 383; Supplementary Material, Table S2). Of these, genes were classified as ‘congruent’ if they were hypermethylated in the same cell type in both species. Gene ontology analysis was performed with GOrilla (42) using the congruent genes (N = 167 for neuron-hypermethylated genes, N = 189 for glia-hypermethylated genes, respectively) as the target set and the 7998 informative genes as the background set (Supplementary Material, Tables S3 and S6). This analysis was repeated using promoter methylation (promoter defined as the 3 kb window centered on the TSS), except the number of required informative bins was relaxed to two to account for the generally smaller size of promoters compared to genes. This yielded 5912 informative promoters; of these, 704 had cell type differences of at least 10% in both species, and 529 genes (N = 249 for neuron-hypermethylated genes and N = 280 for glia-hypermethylated genes) had congruent differences.
Comparing methylation conservation with sequence conservation
To investigate the relationship between conserved methylation and sequence-level conservation, exonic sequence conservation data were downloaded in bulk from NCBI Homologene (43) and extracted using a custom script. Of the 7998 informative genes from above, 7720 had a sequence conservation score in Homologene; these genes were then split into quartiles by Homologene score. The Homologene quartile distribution of the 338 genes which had consistent cross-species methylation differences (i.e. the subset of the 383 genes from above which had a Homologene score) was then determined (Supplementary Material, Fig. S8). Since the Homologene score is derived from intragenic sequence comparisons, this analysis was not performed on the promoter methylation conservation data.
Statistical analyses
To determine whether there was a difference in the distribution of methylation differences between neuron- and glia-hypermethylated bins, a two-tailed unequal variances t-test was performed (N = 554 691 neuron-hypermethylated bins and 226 369 glia-hypermethylated bins). The significance of the difference in inter-species congruence between promoter and gene body methylation was determined by chi-squared test.
Supplementary Material
Funding
This work was supported by grants from the National Institutes of Health Roadmap Epigenomics Program (grant number U01DA025956) and the U.S. Department of Agriculture (USDA) (grant numbers CRIS 6250-51000-055 and CRIS 3092-5-001-059).
Supplementary Material
Acknowledgements
We thank Adam Gillum (Baylor College of Medicine, USDA/ARS Children's Nutrition Research Center) for assistance with the figures.
Conflict of Interest statement. None declared.
References
- 1.Cedar H., Bergman Y. (2009) Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet., 10, 295–304. [DOI] [PubMed] [Google Scholar]
- 2.Bestor T.H., Edwards J.R., Boulard M. (2014) Notes on the role of dynamic DNA methylation in mammalian development. Proc. Natl. Acad. Sci. USA, 112, 6796–6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hatada I., Namihira M., Morita S., Kimura M., Horii T., Nakashima K. (2008) Astrocyte-Specific genes are generally demethylated in neural precursor cells prior to astrocytic differentiation. PLoS ONE, 3, e3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma D.K., Marchetto M.C., Guo J.U., Ming G.L., Gage F.H., Song H. (2010) Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat. Neurosci., 13, 1338–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takizawa T., Nakashima K., Namihira M., Ochiai W., Uemura A., Yanagisawa M., Fujita N., Nakao M., Taga T. (2001) DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev. Cell, 1, 749–758. [DOI] [PubMed] [Google Scholar]
- 6.Guintivano J., Aryee M.J., Kaminsky Z.A. (2013) A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics, 8, 290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng J., Zhou Y., Campbell S.L., Le T., Li E., Sweatt J.D., Silva A.J., Fan G. (2010) Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci., 13, 423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller C.A., Sweatt J.D. (2007) Covalent modification of DNA regulates memory formation. Neuron, 53, 857–869. [DOI] [PubMed] [Google Scholar]
- 9.Cassidy S.B., Schwartz S., Miller J.L., Driscoll D.J. (2012) Prader-Willi syndrome. Genet. Med., 14, 10–26. [DOI] [PubMed] [Google Scholar]
- 10.Meng L., Ward A.J., Chun S., Bennett C.F., Beaudet A.L., Rigo F. (2015) Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature, 518, 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Jager P.L., Srivastava G., Lunnon K., Burgess J., Schalkwyk L.C., Yu L., Eaton M.L., Keenan B.T., Ernst J., McCabe C. et al. (2014) Alzheimer's disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat. Neurosci., 17, 1156–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masliah E., Dumaop W., Galasko D., Desplats P. (2013) Distinctive patterns of DNA methylation associated with Parkinson disease. Epigenetics, 8, 1030–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallo V., Deneen B. (2014) Glial development: the crossroads of regeneration and repair in the CNS. Neuron, 83, 283–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kozlenkov A., Roussos P., Timashpolsky A., Barbu M., Rudchenko S., Bibikova M., Klotzle B., Byne W., Lyddon R., Di Narzo A.F. et al. (2014) Differences in DNA methylation between human neuronal and glial cells are concentrated in enhancers and non-CpG sites. Nucleic Acids Res., 42, 109–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li G., Zhang W., Baker M.S., Laritsky E., Mattan-Hung N., Yu D., Kunde-Ramamoorthy G., Simerly R.B., Chen R., Shen L. et al. (2014) Major epigenetic development distinguishing neuronal and non-neuronal cells occurs postnatally in the murine hypothalamus. Hum. Mol. Genet., 23, 1579–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montano C.M., Irizarry R.A., Kaufmann W.E., Talbot K., Gur R.E., Feinberg A.P., Taub M.A. (2013) Measuring cell-type specific differential methylation in human brain tissue. Genome Biol., 14, R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwamoto K., Bundo M., Ueda J., Oldham M.C., Ukai W., Hashimoto E., Saito T., Geschwind D.H., Kato T. (2011) Neurons show distinctive DNA methylation profile and higher interindividual variations compared with non-neurons. Genome Res., 21, 688–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lister R., Mukamel E.A., Nery J.R., Urich M., Puddifoot C.A., Johnson N.D., Lucero J., Huang Y., Dwork A.J., Schultz M.D. et al. (2013) Global epigenomic reconfiguration during mammalian brain development. Science, 341, 1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunde-Ramamoorthy G., Coarfa C., Laritsky E., Kessler N.J., Harris R.A., Xu M., Chen R., Shen L., Milosavljevic A., Waterland R.A. (2014) Comparison and quantitative verification of mapping algorithms for whole-genome bisulfite sequencing. Nucleic Acids Res., 42, e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bock C. (2012) Analysing and interpreting DNA methylation data. Nat. Rev. Genet., 13, 705–719. [DOI] [PubMed] [Google Scholar]
- 21.Sun D., Xi Y., Rodriguez B., Park H., Tong P., Meong M., Goodell M., Li W. (2014) MOABS: model based analysis of bisulfite sequencing data. Genome Biol., 15, R38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen P.Y., Feng S., Joo J.W., Jacobsen S.E., Pellegrini M. (2011) A comparative analysis of DNA methylation across human embryonic stem cell lines. Genome Biol., 12, R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meissner A., Mikkelsen T.S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B.E., Nusbaum C., Jaffe D.B. et al. (2008) Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature, 454, 766–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen L., Guo Y., Chen X., Ahmed S., Issa J.P.J. (2007) Optimizing annealing temperature overcomes bias in bisulfite PCR methylation analysis. Biotechniques, 42, 48–58. [DOI] [PubMed] [Google Scholar]
- 25.Xin Y., O'Donnell A.H., Ge Y., Chanrion B., Milekic M., Rosoklija G., Stankov A., Arango V., Dwork A.J., Gingrich J.A. et al. (2011) Role of CpG context and content in evolutionary signatures of brain DNA methylation. Epigenetics, 6, 1308–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng H., Shen E.H., Hohmann J.G., Oh S.W., Bernard A., Royall J.J., Glattfelder K.J., Sunkin S.M., Morris J.A., Guillozet-Bongaarts A.L. et al. (2012) Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell, 149, 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strand A.D., Aragaki A.K., Baquet Z.C., Hodges A., Cunningham P., Holmans P., Jones K.R., Jones L., Kooperberg C., Olson J.M. (2007) Conservation of regional gene expression in mouse and human brain. PLoS Genet., 3, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ching T.T., Maunakea A.K., Jun P., Hong C., Zardo G., Pinkel D., Albertson D.G., Fridlyand J., Mao J.H., Shchors K. et al. (2005) Epigenome analyses using BAC microarrays identify evolutionary conservation of tissue-specific methylation of SHANK3. Nat. Genet., 37, 645–651. [DOI] [PubMed] [Google Scholar]
- 29.Lindsey J.C., Kawauchi D., Schwalbe E.C., Solecki D.J., Selby M.P., McKinnon P.J., Olson J.M., Hayden J.T., Grundy R.G., Ellison D.W. et al. (2014) Cross-species epigenetics identifies a critical role for VAV1 in SHH subgroup medulloblastoma maintenance. Oncogene, 34, 4746–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y., Wang M., Jiang S., Lu Y., Tao D., Yang Y., Ma Y., Zhang S. (2014) Demethylation of CpG islands in the 5′ upstream regions mediates the expression of the human testis-specific gene MAGEB16 and its mouse homolog Mageb16. BMB Rep., 47, 86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merbs S.L., Khan M.A., Hackler L. Jr., Oliver V.F., Wan J., Qian J., Zack D.J. (2012) Cell-specific DNA methylation patterns of retina-specific genes. PLoS One, 7, e32602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishioka M., Shimada T., Bundo M., Ukai W., Hashimoto E., Saito T., Kano Y., Sasaki T., Kasai K., Kato T. et al. (2013) Neuronal cell-type specific DNA methylation patterns of the Cacna1c gene. Int. J. Dev. Neurosci., 31, 89–95. [DOI] [PubMed] [Google Scholar]
- 33.Huang W., Zhu P.J., Zhang S., Zhou H., Stoica L., Galiano M., Krnjevic K., Roman G., Costa-Mattioli M. (2013) mTORC2 controls actin polymerization required for consolidation of long-term memory. Nat. Neurosci., 16, 441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen C., Massie A., De Keyser J. (2013) Immune players in the CNS: the astrocyte. J. Neuroimmune Pharmacol., 8, 824–839. [DOI] [PubMed] [Google Scholar]
- 35.Kettenmann H., Hanisch U.-K., Noda M., Verkhratsky A. (2011) Physiology of microglia. Physiol. Rev., 91, 461–553. [DOI] [PubMed] [Google Scholar]
- 36.Kim S.U., de Vellis J. (2005) Microglia in health and disease. J. Neurosci. Res., 81, 302–313. [DOI] [PubMed] [Google Scholar]
- 37.Cox D.J., Field R.H., Williams D.G., Baran M., Bowie A.G., Cunningham C., Dunne A. (2015) DNA sensors are expressed in astrocytes and microglia in vitro and are upregulated during gliosis in neurodegenerative disease. Glia, 63, 812–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norden D.M., Muccigrosso M.M., Godbout J.P.. Microglial priming and enhanced reactivity to secondary insult in aging, and traumatic CNS injury, and neurodegenerative disease. Neuropharmacology, 96, 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris R.A., Wang T., Coarfa C., Nagarajan R.P., Hong C., Downey S.L., Johnson B.E., Fouse S.D., Delaney A., Zhao Y. et al. (2010) Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol., 28, 1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quinlan A.R., Hall I.M. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eyre T.A., Wright M.W., Lush M.J., Bruford E.A. (2007) HCOP: a searchable database of human orthology predictions. Brief. Bioinform., 8, 2–5. [DOI] [PubMed] [Google Scholar]
- 42.Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z. (2009) GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics, 10, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.NCBI Resource Coordinators. (2015) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 43, D6–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.