Abstract
Dilated cardiomyopathy (DCM) is a leading cause of heart failure. In families with autosomal-dominant DCM, heterozygous missense mutations were identified in RNA-binding motif protein 20 (RBM20), a spliceosome protein induced during early cardiogenesis. Dermal fibroblasts from two unrelated patients harboring an RBM20 R636S missense mutation were reprogrammed to human induced pluripotent stem cells (hiPSCs) and differentiated to beating cardiomyocytes (CMs). Stage-specific transcriptome profiling identified differentially expressed genes ranging from angiogenesis regulator to embryonic heart transcription factor as initial molecular aberrations. Furthermore, gene expression analysis for RBM20-dependent splice variants affected sarcomeric (TTN and LDB3) and calcium (Ca2+) handling (CAMK2D and CACNA1C) genes. Indeed, RBM20 hiPSC-CMs exhibited increased sarcomeric length (RBM20: 1.747 ± 0.238 µm versus control: 1.404 ± 0.194 µm; P < 0.0001) and decreased sarcomeric width (RBM20: 0.791 ± 0.609 µm versus control: 0.943 ± 0.166 µm; P < 0.0001). Additionally, CMs showed defective Ca2+ handling machinery with prolonged Ca2+ levels in the cytoplasm as measured by greater area under the curve (RBM20: 814.718 ± 94.343 AU versus control: 206.941 ± 22.417 AU; P < 0.05) and higher Ca2+ spike amplitude (RBM20: 35.281 ± 4.060 AU versus control:18.484 ± 1.518 AU; P < 0.05). β-adrenergic stress induced with 10 µm norepinephrine demonstrated increased susceptibility to sarcomeric disorganization (RBM20: 86 ± 10.5% versus control: 40 ± 7%; P < 0.001). This study features the first hiPSC model of RBM20 familial DCM. By monitoring human cardiac disease according to stage-specific cardiogenesis, this study demonstrates RBM20 familial DCM is a developmental disorder initiated by molecular defects that pattern maladaptive cellular mechanisms of pathological cardiac remodeling. Indeed, hiPSC-CMs recapitulate RBM20 familial DCM phenotype in a dish and establish a tool to dissect disease-relevant defects in RBM20 splicing as a global regulator of heart function.
Introduction
Dilated cardiomyopathy (DCM) is the most common diagnosis leading to heart transplantation in the USA (1,2). While its underlying etiology is often idiopathic, genetic mechanisms are now recognized as a major contributor to DCM pathogenesis (3,4). Nearly one-third of individuals with DCM have familial disease (5–7) and pathogenic mutations have been identified in over 40 genes (8,9). In 2009, genetic linkage analysis in families with autosomal-dominant DCM led to the discovery of heterozygous missense mutations in an arginine/serine-rich domain of RNA-binding motif protein 20 (RBM20) (10). Mutations in RBM20 represent at least 3% of idiopathic DCM (11), are linked to younger age at diagnosis and are strongly associated with morbidity and mortality (10,12–15).
Compared with the majority of known DCM-causing genes encoding proteins responsible for structural integrity and contraction of cardiomyocytes, RBM20 is a functionally distinct gene, encoding an RNA-binding SR protein that regulates spliceosome formation and alternative splicing (11,12). Indeed, RBM20 DCM is a novel example of heart failure owing to a global defect in post-transcriptional regulation. RBM20 is predominantly expressed in striated muscle, with the highest expression in the heart (11). In vitro and in vivo animal studies have significantly contributed to characterizing the structural and functional pathogenesis of Rbm20 deficiency. A naturally occurring loss-of-function Rbm20 deletion in rats demonstrated that Rbm20 plays a major role in alternative splicing in cardiac adaptive responses mediated by titin (Ttn) and calcium/calmodulin-dependent protein kinase II (Camk2δ) (11,16). Deficient Rbm20 resulted in impaired sarcomere organization and ion transport in the sarcoplasmic reticulum and premature death. Additionally, knockdown studies in murine cells revealed Rbm20-deficient cardiogenesis attributable to early disruption of RNA processing and sarcomere remodeling, establishing its pathogenesis as a developmental disorder (16). However, the initial molecular derangements in RBM20-mediated DCM remain unknown in human cardiomyocytes.
Induced pluripotent stem cell (iPSC) technology offers a transformative approach to bioengineer pluripotent stem cells from patient-specific somatic cells (17–19). Bioengineered RBM20 patient-specific iPSC lines provide the novel opportunity to model heart disease, correlating clinical course with disease progression at a molecular level. Here we utilized the hiPSC model of RBM20 DCM for a patient-specific approach to map the primary transcriptome dysfunction and evaluate mechanistic corruption in disease pathogenesis. We hypothesized that RBM20 is an essential component of the RNA-processing machinery that is required to pattern normal structural and physiological integrity of nascent CMs. Insights gained from using patient-specific stem cells enable the anticipation of disease outcomes and targeting molecular therapy at the root cause of DCM.
Results
Generation of RBM20 familial DCM patient-specific hiPSCs
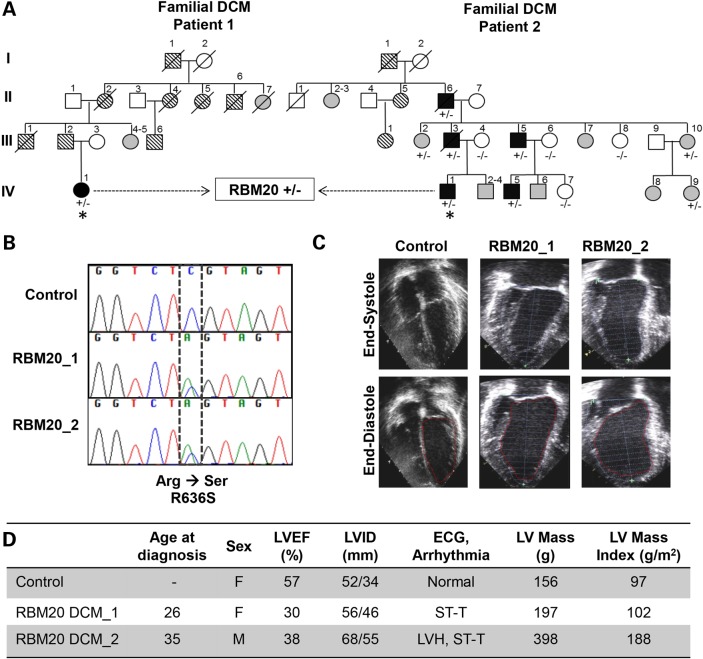
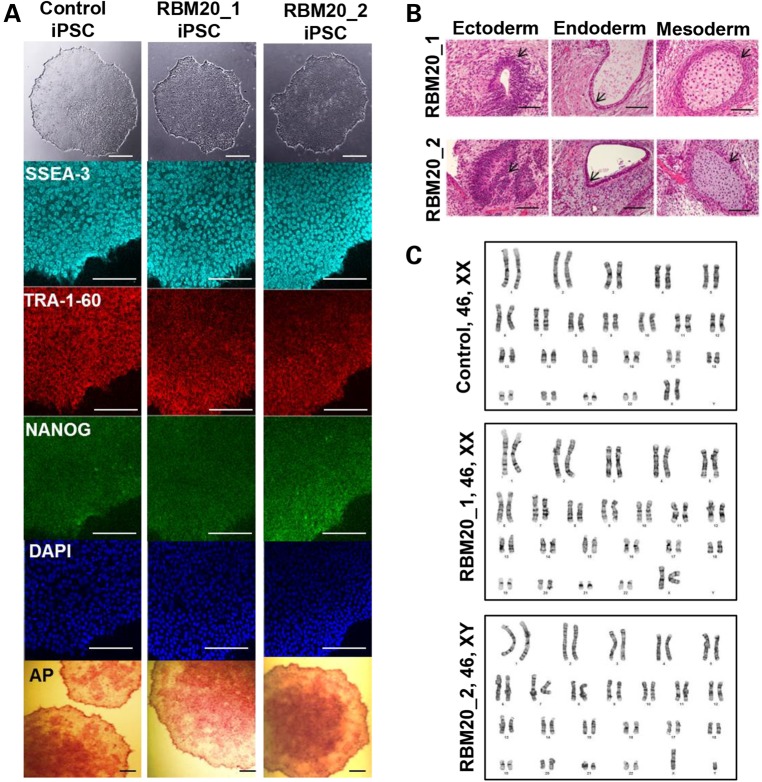
Dermal fibroblasts were obtained from two unrelated patients, a 26-year-old woman and a 35-year-old man, with familial DCM (Fig. 1A) (10). Both patients had a clinically aggressive form of DCM owing to an identical missense mutation in exon 9 of the RNA-binding motif 20 (RBM20) gene (c.1906C>A, R636S; Fig. 1B). Echocardiographic examination revealed left ventricular dilatation with reduced left ventricular ejection fraction (LVEF) and increased LV mass in both patients (Fig. 1C, D). Findings on electrocardiography were nonspecific. Patient-specific iPSCs were generated from primary fibroblasts of RBM20 patients through Sendi viral infection with the reprogramming factors OCT4, SOX-2, KLF-4, and c-MYC (17,20). Similar hiPSCs previously established from fibroblasts of a healthy individual using the identical reprogramming method served as controls. The selected RBM20-hiPSC colonies displayed characteristic human embryonic stem cell (ESC) morphology (Fig. 2A, top), exhibited positive immunostaining for ESC markers SSEA-3, TRA-1-60 and NANOG (Fig. 2A) and displayed alkaline phosphatase activity (Fig. 2A, bottom). Pluripotency of RBM20-hiPSCs was verified by in vitro teratoma formation assays, yielding cellular derivatives of all three germ layers including pigmented epithelium (ectoderm), columnar-lining epithelium (endoderm) and hyaline cartilage (mesoderm) (Fig. 2B) upon injection into the kidney capsules of immunodeficient mice. Finally, RBM20 and control-hiPSCs maintained normal karyotype (Fig. 2C), indicating appropriate nuclear reprogramming.
Figure 1.
Clinical characteristics of RBM20 familial DCM. (A) Family pedigrees of two RBM20 DCM patients with an identical point mutation. Square = male; circle = female; black = affected; white = unaffected; gray = clinical status unknown; parallel diagonal lines = suspected DCM based on family history; slash through the symbol = deceased; + = mutant allele; − = wild-type allele; asterisk = proband whose skin fibroblasts were reprogrammed to hiPSCs. (B) Sequence chromatograms from RBM20 patients demonstrating the c.1906C>A, R636S missense mutation. (C) Representative four-chamber echocardiographic images of control (unaffected family member, IV-9) and patients with RBM20 DCM at end systole and diastole showing moderate LV dilatation. (D) Summary of clinical findings. LV, left ventricle; LVEF, left ventricular ejection fraction; LVID, left ventricular internal diameter; ECG, electrocardiogram; mm, millimeters; g, grams; g/m2, grams per millimeter squared; F, female; M, male; ST-T, nonspecific ST-T wave changes; LVH, left ventricular hypertrophy.
Figure 2.
Derivation and characterization of RBM20 familial DCM-hiPSCs. (A) Phase contrast (top) and immunostaining of undifferentiated control and RBM20 familial DCM-hiPSC colonies for the pluripotent markers SSEA-3, TRA-1-60 and NANOG with DAPI nuclear staining. RBM20 DCM-iPSC colonies stained positive for alkaline phosphatase (bottom) (44). (B) Teratoma formation in immunocompromised mice after injection of RBM20 DCM-hiPSCs. Note pigmented epithelium (ectoderm), columnar-lining epithelium (endoderm) and hyaline cartilage (mesoderm). Scale bars for A and B, 50 µm. (C) Representative karyotype analysis of control and RBM20 DCM-hiPSC clones from Patients 1 and 2.
Isoform changes in structural sarcomeric genes, TTN and LDB3
Using the monolayer differentiation strategy, healthy control and RBM20 hiPSC lines from both patients were differentiated into the cardiac lineage (Supplementary Material, Fig. S1a). Control and RBM20 hiPSC-CMs were defined as spontaneously contractile cells with a transcriptional profile that varied from their originating pluripotent stem cell state. Indeed, the atlas of discrete cardiogenic pathways showed localized interactions with the RBM20 hub gene by network analysis (Supplementary Material, Fig. S1b). To understand the time course of transcriptional changes occurring in these cells, we performed RT–PCR analysis of hiPSC-derived CMs as they differentiated over the course of 25 days into beating clusters (Supplementary Material, Fig. S1c). Expression of early-stage cardiac transcriptional factors (Nkx2.5, MEF2C) appeared between Days 5 and 10, and cardiac-specific markers (Myl2/Mlc2, TNNI3) appeared between Days 15 and 25. Furthermore, at Day 20 of differentiation, flow cytometry data identified 52.91% and 64.28% MF20+ cardiac cells derived from control and RBM20 hiPSC lines, respectively (Supplementary Material, Fig. S2). Control hiPSC-CMs displayed an average beating rate of 60 beats per minute, and RBM20 hiPSC-CMs had an average beating rate of 63 beats per minute (data not shown).
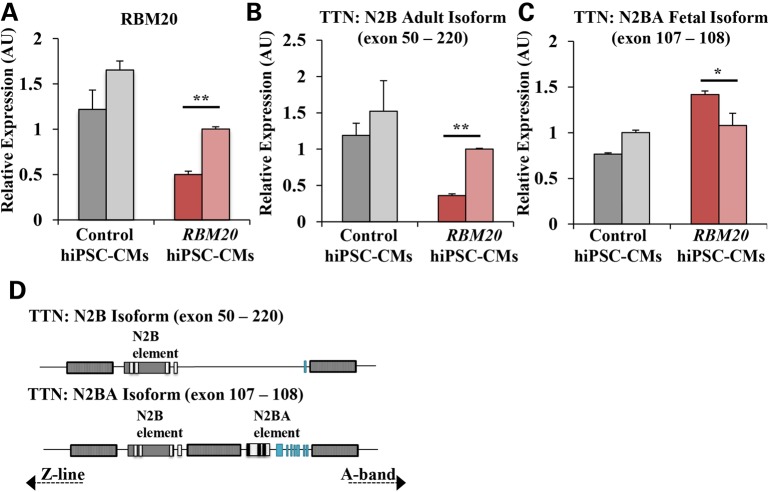
Endogenous RBM20 expression in RBM20 familial DCM hiPSC-CMs was significantly downregulated (RBM20: 0.752 ± 0.030 AU versus control: 1.436 ± 0.155 AU, averaged; P < 0.05, Fig. 3A). In accordance with previously reported Rbm20-dependent isoform changes, TTN N2B adult isoform (exon 50–220) was downregulated (RBM20: 0.681 ± 0.017 AU versus control: 1.356 ± 0.294 AU, averaged; P < 0.01, Fig. 3B) whereas TTN N2BA fetal isoform (exon 107–108) was upregulated (RBM20: 1.249 ± 0.086 AU versus control: 0.884 ± 0.019 AU, averaged; P < 0.05, Fig. 3C) in RBM20 hiPSC-CMs. This mis-regulation of normal titin splicing lead to a shift from the N2B adult to N2BA fetal isoform (Fig. 3D), resulting in an increased N2BA/N2B ratio (RBM20: 2.511 AU versus control: 0.650 AU and previously published cardiac tissue biopsy: 0.560 AU, averaged; P < 0.001) (21,22). It can be noted that an unspliced region of titin (exon 49–50) did not show statistically significant differences between control and RBM20 familial DCM hiPSC-CMs (Supplementary Material, Fig. S3a).
Figure 3.
Isoform switching of structural sarcomeric gene, TTN, owing to alternative splicing defect in RBM20 familial DCM hiPSC-CMs. (A) qRT-PCR showing endogenous expression of RBM20 in control and RBM20 hiPSC-CMs at Day 20. (B) qRT-PCR expression of TTN N2B isoform (exon 50–220). (C) qRT-PCR expression of TTN N2BA isoform (exon 107–108). Two distinct clones were examined for control and RBM20_1 hiPSC lines, represented as distinct bars. Values were normalized to the housekeeping gene GAPDH and expressed as mean ± SEM. Expression values are relative to control hiPSC-CMs (n = 3 biological repetitions with three technical replicates for each biological repetition). (D) Schematic representation of TTN N2B adult and N2BA fetal isoform, showing the region of alternative splicing (*P < 0.05; **P < 0.01; ***P < 0.001, paired Student's t test between control and RBM20).
Additionally, we found that LDB3 (LIM domain binding 3; also known as cypher), another gene that maintains sarcomere structure (23), was significantly downregulated in exon 2–6 region in RBM20 familial DCM hiPSC-CMs (RBM20: 1.050 ± 0.108 AU versus control: 3.918 ± 0.571 AU, averaged, P < 0.05; Supplementary Material, Fig. S3b), in agreement with previously reported changes in exon 4 of LDB3 (11). To evaluate potential changes owing to hiPSC-CM maturity level, standardized cardiomyocyte marker, MYL7, was examined and no marked differences were found between control and RBM20 familial DCM hiPSC-CMs (Supplementary Material, Fig. S3c).
Structural assessment of RBM20 hiPSCs-CMs
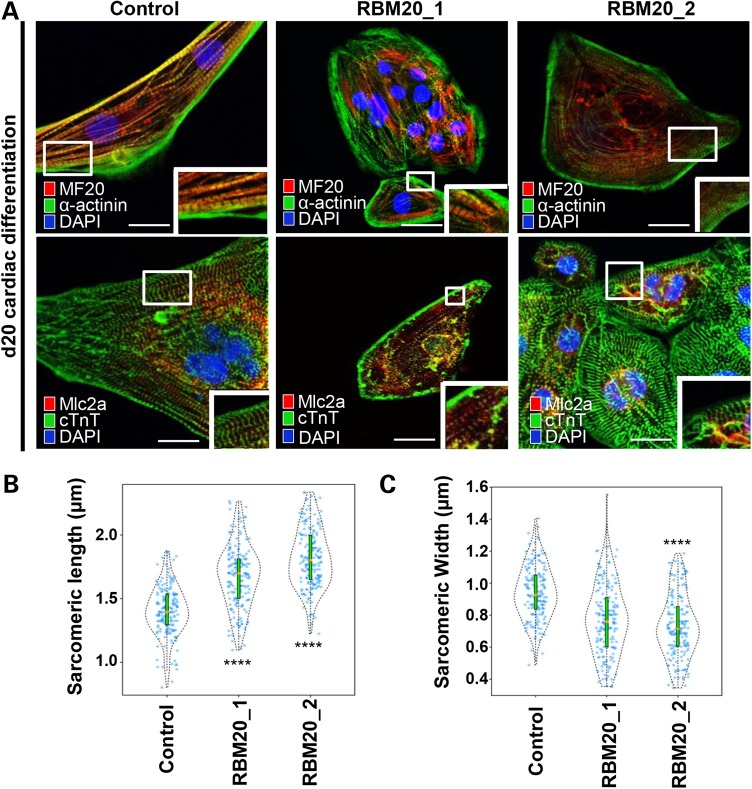
We next analyzed the sarcomeric structure of RBM20 hiPSC-CMs by immunostaining and transmission electron microscopy (TEM). At Day 20 of differentiation, both control and RBM20-deficient CMs showed expression of structural markers, α-actinin, MF20 (anti-myosin heavy chain), myosin light chain 2a and 2v (Mlc2a/2v), and cardiac troponin T (cTnT) (Fig. 4A), confirming the cardiomyocyte phenotype of these cell lines. In accordance with previous mouse Rbm20-deficient studies (16), we observed disorganized actinin fibers in addition to elongated and narrow sarcomeres. To quantify the degree of sarcomere abnormalities in human cells, we performed blinded quantitative morphometric assessment of sarcomeric dimensions in both RBM20 familial DCM and control CMs at Day 20. This assay revealed a significant increase in sarcomere length [RBM20: 1.747 ± 0.238 µm (n = 350; average RBM20 Patient 1 and RBM20 Patient 2) versus control: 1.404 ± 0.194 µm (n = 175); P < 0.0001] (Fig. 4B) and a decrease in sarcomere width [RBM20: 0.791 ± 0.609 µm (n = 350; average RBM20 Patient 1 and RBM20 Patient 2) versus control: 0.943 ± 0.166 µm (n = 175); P < 0.0001] (Fig. 4C) in RBM20 hiPSC-CMs compared with healthy control CMs. In agreement, this abnormal pattern in sarcomere morphology was also observed in previously reported RBM20 patient-derived samples from cardiac biopsy (16). TEM experiments verified the atypical sarcomere geometry in RBM20 hiPSC-CMs compared with healthy control cardiomyocytes with length [RBM20: 1.653 ± 0.171 µm (n = 3) versus control: 0.863 ± 0.203 µm (n = 3)] and width [RBM20: 0.227 ± 0.032 µm (n = 3) versus control: 0.461 ± 0.019 µm (n = 3)] (Supplementary Material, Fig. S4).
Figure 4.
RBM20 familial DCM hiPSC-CMs display abnormal sarcomere geometry. (A) Immunostainings of RBM20 hiPSCs-derived single-cell CMs for MF20, α-actinin, MLC2A and cTnT at Day 20. Note variable expression levels of cardiac-specific markers in control versus RBM20 DCM-hiPSCs-CMs (MF20 = Myosin II heavy chain; MLC2A = Myosin light chain 2A; cTnT = cardiac troponin T). Abnormal sarcomere geometry present in RBM20-deficient hiPSCs-CMs. Scale bars: 20 µm. (B) and (C) Violin plots show difference in sarcomere geometry of control versus RBM20 DCM hiPSCs-CMs. Orange dot = median of sarcomeric length and width (****P < 0.0001, n = 175). Scale bars: 0.5 µm.
Isoform changes in calcium-handling genes, CACNA1C and CAMK2D
Differential splicing of CACNA1C, which encodes the α1C subunit of the L-type calcium (Ca2+) channel, and CAMK2D, which regulates Ca2+ homeostasis in the heart, are associated with RBM20 mutation (11). Moreover, it was reported that the persistence of the unspliced form of Camk2d is independently linked to the DCM phenotype (24). Concurrently, we found downregulated expression of both CACNA1C in exon 8–10 (RBM20: 1.019 ± 0.059 AU versus control: 1.796 ± 0.155 AU, averaged, P < 0.01; Supplementary Material, Fig. S5a) and CAMK2D in exon 12–16 (RBM20: 1.005 ± 0.027 AU versus control: 2.459 ± 0.091 AU, averaged, P < 0.05; Supplementary Material, Fig. S5b) in RBM20 familial DCM hiPSC-CMs compared with healthy control hiPSC-CMs.
To further validate the loss-of-function associated with RBM20 deficiency, we assessed the splicing activity of established target, CAMK2D at Day 20 with GAPDH gene serving as a loading control (Supplementary Material, Fig. S5c). Agarose gel electrophoresis revealed retention of unspliced exons 15 and 16 in the CAMK2D transcript resulting in abnormal persistence of isoform B (199 bp; ∼83% RBM20) owing to the RBM20 DCM-specific mutation (Supplementary Material, Fig. S5c). Hence, mis-splicing of Ca2+-handling genes, CACNA1C and CAMK2D, in RBM20 familial DCM hiPSC-CMs could contribute to disruptions in intracellular Ca2+ homeostasis and abnormal excitation–contraction coupling at the sarcomere level.
Functional assessment of RBM20 hiPSCs-CMs
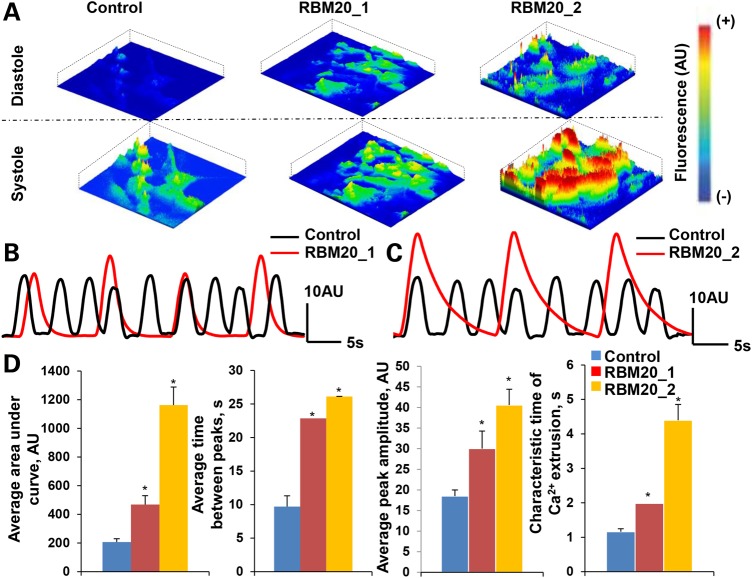
We next assayed the Ca2+ handling properties at Day 20 of in vitro cardiogenesis by characterizing spontaneously occurring intracellular Ca2+ transients. Fluorescent activity of Fluo-4AM-labeled control and RBM20 hiPSC-CMs were recorded to analyze spontaneous Ca2+ transients. Intracellular Ca2+ activity during diastole and systole revealed amplified Ca2+ transients in RBM20 hiPSC-CMs compared with control CMs (Fig. 5A). Indeed, the spontaneous Ca2+ tracing denotes defective Ca2+ machinery in both Patient 1 (Fig. 5B) and Patient 2 (Fig. 5C) with RBM20 familial DCM compared with healthy control.
Figure 5.
Defective calcium handling of RBM20 familial DCM hiPSC-CMs. (A) Representative images of spontaneous Ca2+ transients recorded from Fluo4-AM-labeled hiPSC-CMs during diastole and systole at Day 20. Scale: red = high fluorescence; blue = low fluorescence. (B) and (C) Measurements of spontaneous Ca2+ transients in control and RBM20 DCM hiPSCs-CMs from RBM20 patient 1 (b) and RBM20 patient 2 (c). (D) RBM20 DCM hiPSCs-CMs from both patients reveal abnormal Ca2+ handling as demonstrated by enhanced area under the curve and peak amplitude of calcium transients, as well as prolonged time between calcium spikes (*P < 0.05, n = 3). AU: arbitrary units; s: seconds.
Specifically, Ca2+ spike amplitude, area under the curve, time between peaks and characteristic time of Ca2+ extrusion varied between RBM20 hiPSC-CMs and control CMs (Fig. 5D). Notably, compared with healthy control CMs, RBM20 hiPSC-CMs exhibited greater area under the curve [RBM20: 814.718 ± 94.343 AU (n = 6; average RBM20 Patient 1 and RBM20 Patient 2) versus control: 206.941 ± 22.417 AU (n = 3); P < 0.05] and higher Ca2+ spike amplitude [RBM20: 35.281 ± 4.060 AU(n = 6; average RBM20 Patient 1 and RBM20 Patient 2) versus control:18.484 ± 1.518 AU(n = 3); P < 0.05], correlating with a higher concentration of Ca2+ release into the cytoplasm. Furthermore, individual Ca2+ transients suggest that the time between peaks was longer [RBM20: 24.472 ± 0.029s (n = 6; averaged RBM20 Patient 1 and RBM20 Patient 2) versus control: 9.703 ± 1.603s (n = 3); P < 0.05] resulting in slower beating activity. In addition, the characteristic time of Ca2+ extrusion, corresponding with the duration to remove Ca2+ from the cytoplasm, was longer in cardiomyocytes from RBM20 familial DCM patients than control cells [RBM20: 3.188 ± 0.462s (n = 6; average RBM20 Patient 1 and RBM20 Patient 2) versus control: 1.160 ± 0.088s (n = 3); P < 0.05], indicating disruptions in the Ca2+-handling machinery.
Effect of chronotropic stress
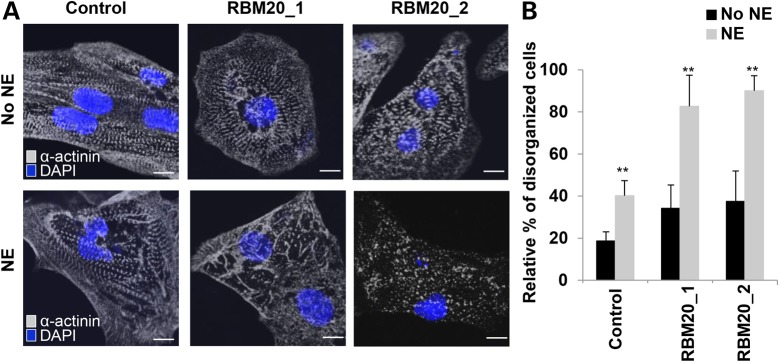
We next examined whether treatment with β-adrenergic agonist, a positive inotropic reagent, can induce the phenotypic stress response of RBM20 familial DCM hiPSC-CMs. Indeed, in vitro treatment with 10 µm norepinephrine (NE) markedly increased the number of CMs with punctate sarcomeric α-actinin distribution from RBM20 familial DCM-iPSC clones (Fig. 6A). Several RBM20 hiPSC-CMs showed complete myofilament degeneration following NE treatment, which was not present in control hiPSC-CMs (Fig. 6A). Disorganized sarcomeric pattern, defined as ≥25% punctate appearance, was more evident in RBM20 hiPSC-CMs [RBM20: 86 ± 10.5% (n = 63; averaged RBM20 Patient 1 and RBM20 Patient 2) versus control: 40 ± 7% (n = 60); P < 0.001] (Fig. 6B). Additionally, average cell area (mm2) decreased in RBM20 hiPSC-CMs following NE treatment (Supplementary Material, Fig. S6). These findings indicate that RBM20 hiPSC-CMs are susceptible to chronotropic stress induced by in vitro β-adrenergic stimulation, corresponding with previous findings in TNNT2 familial DCM hiPSC-CMs (18).
Figure 6.
RBM20 familial DCM hiPSC-CMs are susceptible to structural damage upon β-adrenergic stimulation with norepinephrine. (A) Representative images of sarcomeric α-actinin immunostaining of single control and RBM20 DCM hiPSCs-CMs after treatment with and without NE at Day 20. Compared with controls, NE treatment-induced sarcomeric disorganization in single RBM20 DCM hiPSCs-CMs. Scale bar: 20 μm. (B) Percentage of hiPSCs-CMs with disorganized sarcomeric α-actinin staining patterns (defined as ≥25% punctate appearance) with or without NE treatment (**P < 0.001). mm2, millimeters squared.
Microarray and gene ontology analysis
RBM20 is known to regulate cardiac gene expression owing to its role in RNA processing (10,25). To map the RBM20 transcriptome, hiPSC lines from RBM20 familial DCM patients and healthy controls were profiled across cardiogenesis including stages that preceded cellular phenotypes consistent with maladaptation. Biological triplicate samples were collected from pluripotent cells to beating cardiomyocytes at five different time points (Day 0, Day 10, Day 15, Day 20 and Day 25) (Supplementary Material, Fig. S7a) and subjected to microarray analysis. Unsupervised hierarchical clustering based on all the probe sets in the microarrays, representing relative distance among samples, defined developmental units grouping pluripotent samples (Day 0), early cardiac samples (Day 10 and Day 15) and late cardiac samples (Day 20 and Day 25) (Supplementary Material, Fig. S7b). Indeed, genome-wide expression profiles varied depending on the stage of differentiation and the health status of the patient as shown by Principal Component Analysis (Supplementary Material, Fig. S7c). Comparison of transcriptome profiles of control and RBM20 familial DCM patient-specific lines revealed 128 differential genes (Supplementary Material, Table S1) with 50 of them showing consistent differential expression patterns between the two disease cell lines upon in vitro differentiation (Supplementary Material, Fig. S7d). While RBM20 was not detected in the top 50 prioritized genes, its expression was downregulated in both patient lines compared with healthy control (Supplementary Material, Fig. S7e).
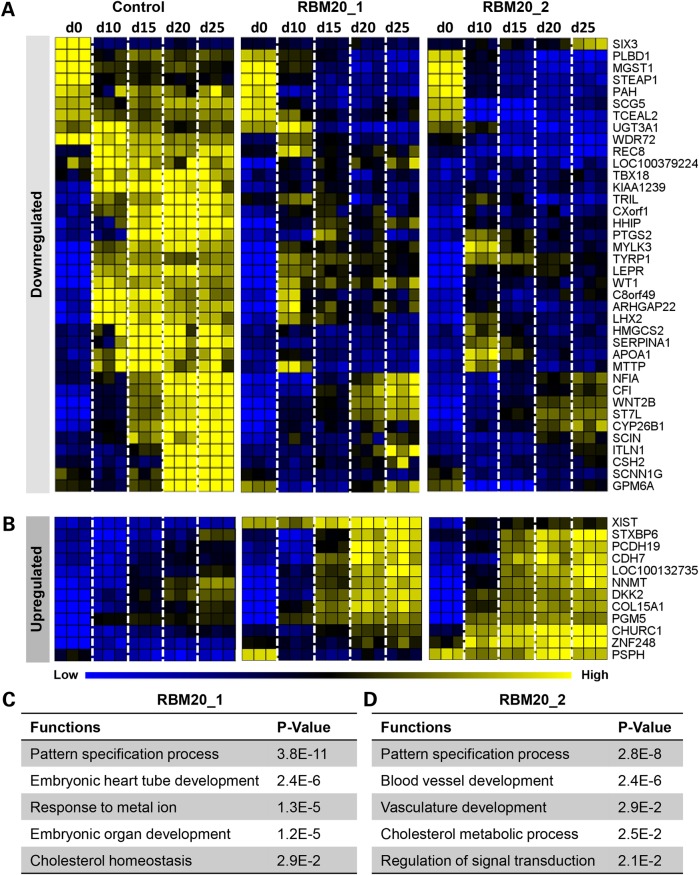
In the differentially expressed RBM20 profile, 38 of the 50 (76%) genes were downregulated (Fig. 7A) and 12 of the 50 (24%) genes were upregulated (Fig. 7B) according to the time-course microarray data set (five time points from hiPSC to adult cardiomyocyte). Gene ontology (GO) enrichment analysis of these 50 genes identified key functions including the pattern specification process (TBX18, CYP26B1, HHIP and LHX2), embryonic heart tube development, response to metal ion (PTGS2 and SERPINA1), embryonic organ development and cholesterol homeostasis (HMGCS2, APOA1 and MTTP) from RBM20 Patient 1 [P ≤ 2.9E−2 calculated based on hypergeometric distribution from Database for Annotation, Visualization and Integrated Discovery (DAVID, v6.7)] (Fig. 7C). Furthermore, RBM20 patient 2 demonstrated GO enrichment analysis with prioritized defects in blood vessel development (ARHGAP22, WT1, LEPR and COL15A1), vasculature development (ARHGAP22, WT1, LEPR and COL15A1), cholesterol metabolic process (HMGCS2, APOA1, LEPR and TYRP1) and regulation of signal transduction (SIX3 and DKK2) (P ≤ 2.1E−2 calculated based on hypergeometric distribution from DAVID, v6.7) (Fig. 7D). The most significantly enriched function conserved between both RBM20 patients centered on the pattern specification process (TBX18, CYP26B1, HHIP and LHX2) (P ≤ 2.8E−8 calculated based on hypergeometric distribution from DAVID, v6.7), which govern cell response and instruction to differentiate during early development (26). These results highlight the conserved developmental defects across two families with identical mutations linked to the molecular etiology of RBM20 familial DCM leading to dysfunctional cardiac gene expression during early stages of stem cell-derived cardiogenesis.
Figure 7.
Aberrant transcriptome profile contributes to defective RBM20 cardiogenesis. (A) and (B) Comparative transcriptome analysis of control (n = 3, biological repetitions with three technical replicates for each biological repetition, at each time point) and RBM20 familial DCM (n = 3, biological repetitions with three technical replicates for each biological repetition, at each time point) hiPSC lines during in vitro cardiogenesis identified the top 50 divergent gene expression profiles that were downregulated (A) or upregulated (B) in the RBM20 DCM group compared with control. (C) and (D) GO enrichment analysis revealed corresponding functions in RBM20 patient 1 (C) and RBM20 patient 2 (D), showing a common function of the pattern specification process in both RBM20 familial DCM patients.
Discussion
Familial DCM is a progressive heart disease associated with heart failure and risk of sudden death. In the current study, we evaluated the ability of hiPSC technology to model familial DCM caused by RBM20 mutation and provided mechanistic insights by transcriptome evaluation. To our knowledge, mutations in RBM20 are the first genetic abnormalities of post-transcriptional modification that results in non-syndromic cardiomyopathy. Remarkably, the originally reported RBM20 missense mutations were concentrated in a hotspot comprised of five amino acids within an arginine/serine-rich region (10). Here, patient-specific hiPSCs were created from two unrelated patients with familial DCM caused by the identical R636S missense mutation in the RBM20 gene.
While penetrance of familial DCM is age-dependent and variable, RBM20 mutations have been described to be highly penetrant and associated with earlier age at diagnosis of DCM (10,14,15). Brauch et al. found only 5 of 44 at-risk family members who carried an RBM20 mutation but did not fulfill DCM diagnostic criteria (10). Moreover, four of these five subjects had LV dilatation, a known precursor to DCM (5,7). Currently, the pathogenesis of RBM20 DCM at the molecular and cellular level is not sufficiently understood to account for variable, age-dependent penetrance. Whether hiPSC technology and the ability to derive patient-specific CMs can provide information that predicts DCM penetrance and outcome remains to be determined. Our sample size is too small to make definitive correlations between hiPSC-CM and in situ cardiac phenotypes. However, it is intriguing that Patient 2, with more severe LV remodeling (i.e. increased LV dilation and mass) than Patient 1, had exaggerated sarcomeric disarray and Ca2+ dysregulation. Both were treated with standard carvedilol pharmacotherapy. Additional studies of hiPSC clones from phenotypically characterized RBM20 mutation carriers, accounting for variables of age, sex and medical treatment, will be required to potentially identify modifying genes or epigenetic mechanisms.
By assessing CMs differentiating from RBM20 DCM-hiPSCs and comparing them with healthy control cells, we were able to (i) confirm sarcomeric abnormalities in both electron microscopy studies and α-actinin immunostaining analysis, (ii) identify calcium-handling properties with Fluo4-AM-labeled fluorescent video imaging, (iii) demonstrate susceptibility to chronotropic stress from β-adrenergic stimulation with norepinephrine, (iv) map dynamic gene expression profile of RBM20 DCM-hiPSC-CMs during in vitro cardiogenesis and (v) pinpoint divergent cardiac gene networks that disrupt the early patterning of RBM20 cardiogenesis.
Collectively, these findings indicate that hiPSC model system is sufficient to recapitulate molecular defects in patients with DCM owing to mutations in RBM20 and reveal mechanistic insights to reversible pathogenesis. These findings are consistent with recent studies that demonstrated pathological changes in Rbm20-deficient rats with impaired splicing of the sarcomeric protein, Titin and the calcium regulator, Camk2d, leading to structural and functional defects (11). In our study, the in vitro RBM20 familial DCM hiPSC-CM model from quality-controlled and selected clones of both patients reproduced the structural (sarcomeric alterations) and functional (abnormal calcium handling) properties, thought to be central to DCM pathogenesis. This was manifested in the ultrastructural analysis of the RBM20 DCM hiPSC-CMs by the elongation and narrowing present in sarcomere geometry. Previously, cardiac biopsy tissues derived directly from patients with RBM20 mutations were described to have an analogous sarcomeric morphological structure (16). Herein, we show for the first time that altered structure of sarcomere unit in patient-derived cardiac biopsy tissues with RBM20-dependent DCM can be recapitulated in vitro with patient-specific RBM20 DCM hiPSC disease model.
Furthermore, this structural abnormality of the sarcomere units could be in part attributed to RBM20-dependent mis-splicing of LDB3 and TTN, which is required for proper maintenance of striated muscle independent of physiological stress (22,23). Indeed, we observed TTN isoform switching from the N2B adult to N2BA fetal isoform. Notably, increased N2BA/N2B ratio has independently been identified in the setting of heart failure (22,27). Distinguishing splice variants at the protein level deserves further investigation as these structural changes may contribute to a reduction in cardiac contractility of DCM patients.
In addition to the sarcomeric structural aberration, RBM20 DCM hiPSC-CMs revealed dysregulation of Ca2+ homeostasis compared with control hiPSC-CMs. Previously, genome-wide transcriptome analyses have identified a cohort of RBM20-dependent genes, including CACNA1C and CAMK2D (11,28). CAMK2D and CACNA1C are involved in the highly coordinated process of translating electrical signals into mechanical conduction in the heart (28). Additionally, the persistence of the unspliced form of CAMK2D was independently reported to be associated with the DCM phenotype (24). Our collective splicing analysis suggests that RBM20 has a crucial role in multiple aspects of cardiac function through effects on genes involved in biomechanics (TTN and LDB3), ion homeostasis and electrical activity (CAMK2D and CACNA1C) and signal transduction (CAMK2D). Additionally, changes in isoform expression of LDB3 and CACNA1C have been independently associated with DCM and arrhythmia, respectively (29,30).
In accordance with this link between Ca2+ machinery alterations and cardiomyopathy, Fluo4-AM recordings of the RBM20 DCM hiPSC-CMs showed greater area under the curve and peak amplitude of Ca2+ transients, as well as prolonged time between Ca2+ spikes resembling those previously reported in murine models (16). While imbalances in Ca2+ homeostasis have been reported as a key characteristic in several cardiomyopathies (31,32), it remains to be elucidated whether these abnormalities are a symptom of DCM or the causal factor. This in vitro data suggest Ca2+ is likely causative as this phenotype is demonstrated early and in the absence of physiological stress.
RBM20 DCM hiPSC-CMs, in addition to being less capable of maintaining their sarcomeric structural and Ca2+ handling functional integrity, are more susceptible to positive chronotropic (heart rate) stress. Previous studies have shown that DCM phenotype in mouse transgenic models of DCM can be induced by positive inotropic (contractile) stress (33,34). Indeed, increased contraction force can exacerbate symptoms in clinical DCM patients (35). Higher sensitivity to chronotropic stress was noted in RBM20 DCM hiPSC-CMs leading to increased percentage of disorganized cells. This finding is in agreement with the work of Lan et al., who observed a similar response to norepinephrine treatment in hiPSC lines from familial hypertrophic cardiomyopathy (36). Furthermore, inotropic and chronotropic agents such as dobutamine are currently used in stress echocardiography to establish prognosis of patients with idiopathic DCM (37). For patients with RBM20 DCM, monitoring in vitro response to inotropes yields a robust and consistent readout, indicating a previously unknown mechanism of stress tolerance. Future studies could utilize this in vitro stress test model to provide dose-dependent information and aid as an independent predictor of DCM onset and prognosis for at-risk individuals identified by genetic screening.
Recent studies recognized familial DCM as a developmental disorder that is patterned during early cardiogenesis and propagated with cellular mechanisms of pathological cardiac remodeling (16). Here, we established a comprehensive microarray time course of human RBM20 DCM and healthy, control cardiac development based on natural differentiation stages, including pluripotent, early cardiac and late cardiac phases. By overlapping unbiased RBM20 and control transcriptome profiles from hiPSC lines, we derived a set of differentially expressed genes with a majority of downregulated substrates. Indeed, RBM20 is involved in regulating spliceosome formation and alternative splicing of cardiac-specific genes (12). Previous studies using RNA deep-sequencing identified >30 Rbm20-dependent genes as alternatively mis-spliced (11). Here, we show that dysregulation of a wide range of cardiac-related genes and pathways can be effectively detected using the patient-specific hiPSC disease model system. Specifically, we identified novel functions including the pattern specification process, which regulates cellular differentiation during development and early patterning of mesoderm (38). We believe these results are the first report to demonstrate early molecular perturbations in human RBM20 cardiogenesis.
Despite the advantages of the hiPSC approach to model cardiogenesis of inherited cardiac disorders, this strategy possesses inherent limitations including the inability to evaluate pathological changes at the tissue and systemic levels such as interstitial fibrosis, scarring and myocyte disarray (36). This could be important, for example, in studying the potential physiological role of chamber dilatation and systolic function in DCM (10). Advancements in tissue engineering, however, could provide a solution to study three-dimensional properties and external mechanical forces of patient-specific heart tissue (39). Another limitation is that hiPSC-CMs have been described as developmentally immature with similar gene expression profiles as fetal cardiomyocytes (36,40). Ongoing efforts in the field include the development of strategies to induce hiPSC-CM maturation (41). Variances in maturity as well as sex could account for differences in GO analysis between the RBM20 patients. Finally, several unique missense mutations including R634Q, R636S, R636H, S637G and P638L (10) have been identified to cause RBM20 DCM, and our findings will need to be confirmed as common pathways before molecular therapies could be designed.
The findings in this study are limited to quality-controlled and selected clones based on efficient reprogramming, cardiac differentiation potential and beating activity. Results may be further focused with a more diverse cohort to decipher disease-causing pathways. Utilizing multiple clones from isogenic control iPSC lines generated by correcting RBM20 mutations using genome editing tools would be ideal, particularly for future studies profiling gene expression studies between control and disease iPSC-derived cardiac cells at different developmental stages. Additionally, quantifying stage-specific percentage of CMs, smooth muscle cells (SMCs) and endothelial cells (ECs) in the total differentiated population could allow for better interpretation of the microarray results.
In conclusion, our data demonstrate the unique ability of the hiPSC technology to model RBM20 DCM. Using this approach, we demonstrated that some of the key features of RBM20 DCM including sarcomere-related pathology, abnormal Ca2+ handling properties and chronotropic susceptibility of RBM20 DCM-hiPSC-CMs are discernable in vitro. This study also provides important dysfunctional molecular networks in early RBM20 cardiogenesis, offering a distinctive advantage for genotype-phenotype analysis to decipher initial disease-causing mechanisms using patient-specific stem cells. Future studies can utilize disease-specific hiPSC-CM models of RBM20 DCM for pharmacological screening to identify molecules that can halt or reverse dysfunctional cardiogenesis.
Materials and Methods
An expanded description of the methods used for teratoma formation, immunofluorescence, gene expression, TEM, norepinephrine (NE) challenge, microarray and statistical data analysis is provided in Supplementary Material.
Patient-specific hiPSC derivation
The study protocol was approved by the Mayo Clinic Institutional Review Board (Mayo Clinic IRB 12-007154), and subjects were enrolled following informed written consent. Dermal fibroblasts were isolated from a punch skin biopsy obtained from two unrelated patients in whom familial pathogenic RBM20 mutations had previously been identified (RBM20 Patient 1, 26-year-old female, and RBM20 Patient 2, 35-year-old male) (10). Fibroblasts were similarly isolated from a 21-year-old healthy female control subject without RBM20 mutation. Disease and control patient-specific hiPSC lines were generated by Sendai virus reprogramming with OCT4, c-MYC, SOX2 and KLF4 as previously described (17,20). hiPSC clones were maintained in mTeSR1 media (Stemcell Technologies, Vancouver, Canada) on geltrex-coated culture dishes (Life Technologies, Grand Island, NY).
hiPSC cardiac differentiation
CM differentiation of RBM20 (passage ≤ 35 to 40) and control (passage ≤ 30) hiPSCs was initiated using the chemically defined medium, three components (CDM3) protocol as previously described (42). Briefly, undifferentiated hiPSCs were dispersed into single-cell suspension using TrypLE Express (Life Technologies) and plated as a monolayer on geltrex-coated culture dishes. At 80% to 90% confluence, media was changed to CDM3, consisting of RPMI 1640 medium (Life Technologies), 75 mg/ml O. sativa-derived recombinant human albumin (Sigma–Aldrich, St. Louis, MO) and 64 mg/ml L-ascorbic acid 2-phosphate (Sigma–Aldrich) (42). On Day 0, medium was supplemented with 12 µm CHIR99021 (LC Laboratories, Woburn, MA) and 10 ng/ml activin A (R&D Systems, Minneapolis, MN) for 24 h. On Day 3, medium was supplemented with 5 mm IWP2 (Sigma–Aldrich) and 20 ng/ml BMP4 (R&D Systems) for 48 h. Spontaneously contracting areas were noted from Day 10. All experiments utilized Day 20 hiPSC-CMs for analysis.
Calcium imaging
Intracellular calcium (Ca2+) dynamics in control and RBM20 DCM hiPSC-CMs at Day 20 were assessed as previously described (16,43). Briefly, cells were dissociated into small clumps using collagenase II (480 U/ml for 15 min; Worthington, Lakewood, NJ) and plated on geltrex-coated 35-mm glass-bottom dishes (Thermo Scientific, Waltham, MA) overnight. Following attachment, cells were loaded with the Ca2+-fluorescent probe Fluo-4 AM (Invitrogen, Carlsbad, CA) for 2 h in Tyrode's solution containing 140 mm NaCl, 5.4 mm KCl, 1 mm MgCl2, 10 mm glucose, 1.5 mm CaCl2 and 10 mm HEPES (pH 7.4) and imaged with a Zeiss LSM Live 5 laser confocal microscope (Zeiss, Oberkochen, Germany). Spontaneous Ca2+ transients were recorded at 37°C. A custom Matlab program (Mathworks, Natick, MA) was implemented to calculate the mean amplitude of Ca2+ spikes corresponding to the mean area under the curve of spontaneous Ca2+ transients and the mean time between spikes from 8 to 12 consecutive Ca2+ cycles (16). To determine the characteristic time of Ca2+ extrusion (τ), the relaxation phase of each transient (from peak amplitude to baseline) was fitted to the exponential function:
where y is fluorescence in AU, t is time and τ is the time to reach 63% of exponential relaxation. Ca2+ analysis was performed in replicates at Day 20.
Supplementary Material
Funding
This study was supported by the National Institutes of Health (MSTP/T32/65841, NCATS/TL1/TR000137 and OD007015-01), Todd and Karen Wanek Family Foundation for Hypoplastic Left Heart Syndrome, and Minnesota Regenerative Medicine Grant (Graduate Scholar MRM/2015/GSCH/003).
Supplementary Material
Acknowledgements
We thank Lois Rowe for assistance with histological analysis and also Jonathan Nesbitt. We are grateful for the help from Jeanne Theis, Muhammad Qureshi, Boyd Rasmussen, Matthew Hoplin, Jennifer Miller and Traci Paulson. We thank Maria Caruso-Keller, Scott Gamb, Kristin Mantz and James Tarara for help with imaging. Fibroblasts and nuclear reprogramming services were provided by ReGen Theranostics in Rochester, MN.
Conflict of Interest statement: Mayo Clinic has financial interests and ownership in ReGen Theranostics.
References
- 1.Lund L.H., Edwards L.B., Kucheryavaya A.Y., Dipchand A.I., Benden C., Christie J.D., Dobbels F., Kirk R., Rahmel A.O., Yusen R.D. et al. (2013) The Registry of the International Society for Heart and Lung Transplantation: thirtieth official adult heart transplant report--2013; focus theme: age. J. Heart Lung Transplant., 32, 951–964. [DOI] [PubMed] [Google Scholar]
- 2.Dipchand A.I., Kirk R., Edwards L.B., Kucheryavaya A.Y., Benden C., Christie J.D., Dobbels F., Lund L.H., Rahmel A.O., Yusen R.D. et al. (2013) The Registry of the International Society for Heart and Lung Transplantation: sixteenth official pediatric heart transplantation report--2013; focus theme: age. J. Heart Lung Transplant., 32, 979–988. [DOI] [PubMed] [Google Scholar]
- 3.Fatkin D., Otway R., Richmond Z. (2010) Genetics of dilated cardiomyopathy. Heart Fail. Clin., 6, 129–140. [DOI] [PubMed] [Google Scholar]
- 4.Hershberger R.E., Lindenfeld J., Mestroni L., Seidman C.E., Taylor M.R., Towbin J.A. (2009) Genetic evaluation of cardiomyopathy--a Heart Failure Society of America practice guideline. J. Card. Fail., 15, 83–97. [DOI] [PubMed] [Google Scholar]
- 5.Michels V.V., Moll P.P., Miller F.A., Tajik A.J., Chu J.S., Driscoll D.J., Burnett J.C., Rodeheffer R.J., Chesebro J.H., Tazelaar H.D. (1992) The frequency of familial dilated cardiomyopathy in a series of patients with idiopathic dilated cardiomyopathy. N. Engl. J. Med., 326, 77–82. [DOI] [PubMed] [Google Scholar]
- 6.Mestroni L., Krajinovic M., Severini G.M., Pinamonti B., Di Lenarda A., Giacca M., Falaschi A., Camerini F. (1994) Familial dilated cardiomyopathy. Br. Heart J., 72, S35–S41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baig M.K., Goldman J.H., Caforio A.L., Coonar A.S., Keeling P.J., McKenna W.J. (1998) Familial dilated cardiomyopathy: cardiac abnormalities are common in asymptomatic relatives and may represent early disease. J. Am. Coll. Cardiol., 31, 195–201. [DOI] [PubMed] [Google Scholar]
- 8.Hershberger R.E., Siegfried J.D. (2011) Update 2011: clinical and genetic issues in familial dilated cardiomyopathy. J. Am. Coll. Cardiol., 57, 1641–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olson TM, C.D. (2013) In Allen H.D., D.D., Shaddy R.E., Feltes T.F. (eds), Moss and Adams’ Heart Disease in Infants, Children, and Adolescents: Including the Fetus and Young Adult. Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia, Vol. II, pp. 1235–1246. [Google Scholar]
- 10.Brauch K.M., Karst M.L., Herron K.J., de Andrade M., Pellikka P.A., Rodeheffer R.J., Michels V.V., Olson T.M. (2009) Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J. Am. Coll. Cardiol., 54, 930–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo W., Schafer S., Greaser M.L., Radke M.H., Liss M., Govindarajan T., Maatz H., Schulz H., Li S., Parrish A.M. et al. (2012) RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat. Med., 18, 766–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S., Guo W., Dewey C.N., Greaser M.L. (2013) Rbm20 regulates titin alternative splicing as a splicing repressor. Nucl. Acids Res., 41, 2659–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Millat G., Bouvagnet P., Chevalier P., Sebbag L., Dulac A., Dauphin C., Jouk P.S., Delrue M.A., Thambo J.B., Le Metayer P. et al. (2011) Clinical and mutational spectrum in a cohort of 105 unrelated patients with dilated cardiomyopathy. Eur. J. Med. Genet., 54, e570–e575. [DOI] [PubMed] [Google Scholar]
- 14.Li D., Morales A., Gonzalez-Quintana J., Norton N., Siegfried J.D., Hofmeyer M., Hershberger R.E. (2010) Identification of novel mutations in RBM20 in patients with dilated cardiomyopathy. Clin. Transl. Sci., 3, 90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wells Q.S., Becker J.R., Su Y.R., Mosley J.D., Weeke P., D'Aoust L., Ausborn N.L., Ramirez A.H., Pfotenhauer J.P., Naftilan A.J. et al. (2013) Whole exome sequencing identifies a causal RBM20 mutation in a large pedigree with familial dilated cardiomyopathy. Circ. Cardiovasc. Genet., 6, 317–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beraldi R., Li X., Martinez Fernandez A., Reyes S., Secreto F., Terzic A., Olson T.M., Nelson T.J. (2014) Rbm20-deficient cardiogenesis reveals early disruption of RNA processing and sarcomere remodeling establishing a developmental etiology for dilated cardiomyopathy. Hum. Mol. Genet., 23, 3779–3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell, 131, 861–872. [DOI] [PubMed] [Google Scholar]
- 18.Sun N., Yazawa M., Liu J., Han L., Sanchez-Freire V., Abilez O.J., Navarrete E.G., Hu S., Wang L., Lee A. et al. (2012) Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci. Transl. Med., 4, 130ra147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson T.J., Martinez-Fernandez A., Terzic A. (2010) Induced pluripotent stem cells: developmental biology to regenerative medicine. Nat. Rev. Cardiol., 7, 700–710. [DOI] [PubMed] [Google Scholar]
- 20.Folmes C.D., Martinez-Fernandez A., Perales-Clemente E., Li X., McDonald A., Oglesbee D., Hrstka S.C., Perez-Terzic C., Terzic A., Nelson T.J. (2013) Disease-causing mitochondrial heteroplasmy segregated within induced pluripotent stem cell clones derived from a patient with MELAS. Stem Cells, 31, 1298–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neagoe C., Kulke M., del Monte F., Gwathmey J.K., de Tombe P.P., Hajjar R.J., Linke W.A. (2002) Titin isoform switch in ischemic human heart disease. Circulation, 106, 1333–1341. [DOI] [PubMed] [Google Scholar]
- 22.Nagueh S.F., Shah G., Wu Y., Torre-Amione G., King N.M., Lahmers S., Witt C.C., Becker K., Labeit S., Granzier H.L. (2004) Altered titin expression, myocardial stiffness, and left ventricular function in patients with dilated cardiomyopathy. Circulation, 110, 155–162. [DOI] [PubMed] [Google Scholar]
- 23.Arimura T., Inagaki N., Hayashi T., Shichi D., Sato A., Hinohara K., Vatta M., Towbin J.A., Chikamori T., Yamashina A. et al. (2009) Impaired binding of ZASP/Cypher with phosphoglucomutase 1 is associated with dilated cardiomyopathy. Cardiovasc. Res., 83, 80–88. [DOI] [PubMed] [Google Scholar]
- 24.Xu X., Yang D., Ding J.H., Wang W., Chu P.H., Dalton N.D., Wang H.Y., Bermingham J.R. Jr., Ye Z., Liu F. et al. (2005) ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell, 120, 59–72. [DOI] [PubMed] [Google Scholar]
- 25.Guo W., Pleitner J.M., Saupe K.W., Greaser M.L. (2013) Pathophysiological defects and transcriptional profiling in the RBM20−/− rat model. PLoS One, 8, e84281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine M., Davidson E.H. (2005) Gene regulatory networks for development. Proc. Natl Acad. Sci. USA, 102, 4936–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morano I., Hadicke K., Grom S., Koch A., Schwinger R.H., Bohm M., Bartel S., Erdmann E., Krause E.G. (1994) Titin, myosin light chains and C-protein in the developing and failing human heart. J. Mol. Cell Cardiol., 26, 361–368. [DOI] [PubMed] [Google Scholar]
- 28.Maatz H., Jens M., Liss M., Schafer S., Heinig M., Kirchner M., Adami E., Rintisch C., Dauksaite V., Radke M.H. et al. (2014) RNA-binding protein RBM20 represses splicing to orchestrate cardiac pre-mRNA processing. J. Clin. Invest., 124, 3419–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng H., Zheng M., Peter A.K., Kimura K., Li X., Ouyang K., Shen T., Cui L., Frank D., Dalton N.D. et al. (2011) Selective deletion of long but not short Cypher isoforms leads to late-onset dilated cardiomyopathy. Hum. Mol. Genet., 20, 1751–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang Z.Z., Sharma S., Zheng S., Chawla G., Nikolic J., Black D.L. (2011) Regulation of the mutually exclusive exons 8a and 8 in the CaV1.2 calcium channel transcript by polypyrimidine tract-binding protein. J. Biol. Chem., 286, 10007–10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fatkin D., McConnell B.K., Mudd J.O., Semsarian C., Moskowitz I.G., Schoen F.J., Giewat M., Seidman C.E., Seidman J.G. (2000) An abnormal Ca(2+) response in mutant sarcomere protein-mediated familial hypertrophic cardiomyopathy. J. Clin. Invest., 106, 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Semsarian C., Ahmad I., Giewat M., Georgakopoulos D., Schmitt J.P., McConnell B.K., Reiken S., Mende U., Marks A.R., Kass D.A. et al. (2002) The L-type calcium channel inhibitor diltiazem prevents cardiomyopathy in a mouse model. J. Clin. Invest., 109, 1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gramlich M., Michely B., Krohne C., Heuser A., Erdmann B., Klaassen S., Hudson B., Magarin M., Kirchner F., Todiras M. et al. (2009) Stress-induced dilated cardiomyopathy in a knock-in mouse model mimicking human titin-based disease. J. Mol. Cell Cardiol., 47, 352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandar S., Yeo L.S., Leimena C., Tan J.C., Xiao X.H., Nikolova-Krstevski V., Yasuoka Y., Gardiner-Garden M., Wu J., Kesteven S. et al. (2010) Effects of mechanical stress and carvedilol in lamin A/C-deficient dilated cardiomyopathy. Circ. Res., 106, 573–582. [DOI] [PubMed] [Google Scholar]
- 35.Borow K.M., Lang R.M., Neumann A., Carroll J.D., Rajfer S.I. (1988) Physiologic mechanisms governing hemodynamic responses to positive inotropic therapy in patients with dilated cardiomyopathy. Circulation, 77, 625–637. [DOI] [PubMed] [Google Scholar]
- 36.Lan F., Lee A.S., Liang P., Sanchez-Freire V., Nguyen P.K., Wang L., Han L., Yen M., Wang Y., Sun N. et al. (2013) Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell, 12, 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pratali L., Picano E., Otasevic P., Vigna C., Palinkas A., Cortigiani L., Dodi C., Bojic D., Varga A., Csanady M. et al. (2001) Prognostic significance of the dobutamine echocardiography test in idiopathic dilated cardiomyopathy. Am. J. Cardiol., 88, 1374–1378. [DOI] [PubMed] [Google Scholar]
- 38.Lekven A.C., Thorpe C.J., Waxman J.S., Moon R.T. (2001) Zebrafish wnt8 encodes two wnt8 proteins on a bicistronic transcript and is required for mesoderm and neurectoderm patterning. Dev. Cell., 1, 103–114. [DOI] [PubMed] [Google Scholar]
- 39.Masumoto H., Ikuno T., Takeda M., Fukushima H., Marui A., Katayama S., Shimizu T., Ikeda T., Okano T., Sakata R. et al. (2014) Human iPS cell-engineered cardiac tissue sheets with cardiomyocytes and vascular cells for cardiac regeneration. Sci. Rep., 4, 6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burridge P.W., Keller G., Gold J.D., Wu J.C. (2012) Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell, 10, 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caspi O., Huber I., Gepstein A., Arbel G., Maizels L., Boulos M., Gepstein L. (2013) Modeling of arrhythmogenic right ventricular cardiomyopathy with human induced pluripotent stem cells. Circ. Cardiovasc. Genet., 6, 557–568. [DOI] [PubMed] [Google Scholar]
- 42.Burridge P.W., Matsa E., Shukla P., Lin Z.C., Churko J.M., Ebert A.D., Lan F., Diecke S., Huber B., Mordwinkin N.M. et al. (2014) Chemically defined generation of human cardiomyocytes. Nat. Methods, 11, 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edmondson D.G., Lyons G.E., Martin J.F., Olson E.N. (1994) Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development, 120, 1251–1263. [DOI] [PubMed] [Google Scholar]
- 44.Wang K., McCarter R., Wright J., Beverly J., Ramirez-Mitchell R. (1991) Regulation of skeletal muscle stiffness and elasticity by titin isoforms: a test of the segmental extension model of resting tension. Proc. Natl Acad. Sci. USA, 88, 7101–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.