Abstract
Zinc finger motifs are distributed amongst many eukaryotic protein families, directing nucleic acid–protein and protein–protein interactions. Zinc finger protein 106 (ZFP106) has previously been associated with roles in immune response, muscle differentiation, testes development and DNA damage, although little is known about its specific function. To further investigate the function of ZFP106, we performed an in-depth characterization of Zfp106 deficient mice (Zfp106−/−), and we report a novel role for ZFP106 in motor and sensory neuronal maintenance and survival. Zfp106−/− mice develop severe motor abnormalities, major deficits in muscle strength and histopathological changes in muscle. Intriguingly, despite being highly expressed throughout the central nervous system, Zfp106−/− mice undergo selective motor and sensory neuronal and axonal degeneration specific to the spinal cord and peripheral nervous system. Neurodegeneration does not occur during development of Zfp106−/− mice, suggesting that ZFP106 is likely required for the maintenance of mature peripheral motor and sensory neurons. Analysis of embryonic Zfp106−/− motor neurons revealed deficits in mitochondrial function, with an inhibition of Complex I within the mitochondrial electron transport chain. Our results highlight a vital role for ZFP106 in sensory and motor neuron maintenance and reveal a novel player in mitochondrial dysfunction and neurodegeneration.
Introduction
ZFP106 is a zinc finger protein with proposed roles in transcriptional control, RNA metabolism, the immune response, muscle development and differentiation and testes development (1–3). Phosphorylation of ZFP106 increases following DNA damage, which might suggest a possible role in the DNA damage response pathway (4). Recently, ZFP106 has been identified as a novel factor regulating transcription initiation by targeting RNA-polymerase I to the promoter of ribosomal RNA genes (5), linking for the first time ZFP106 function and RNA metabolism. Interestingly, the human gene encoding ZFP106 (ZFN106) is located within a familial amyotrophic lateral sclerosis (ALS) locus (ALS5) on human chromosome 15, while human ZFP106 is proposed to localize to the nucleolus (1).
Zfp106 encodes a 1888-amino acid protein with two N-terminal C2H2 zinc finger motifs, two C-terminal CWCH2 zinc finger motifs and seven WD40 repeats (2,3,6). Zinc finger motifs are important for protein–protein interactions and nucleic-acid binding (7), whilst WD40 repeats are involved in protein–protein interaction and facilitate the formation of large multi-protein complexes (8). Putative orthologues of Zfp106 are only found in mammals and exhibit a high degree of conservation between species.
Zfp106 mRNA expression is proposed to be driven from two promoters, with the resulting transcripts producing a number of isoforms (2). Full-length ZFP106 (ZNF106) mRNA is expressed ubiquitously and expression is particularly high in the heart, skeletal muscle and testis (2). Nuclear respiratory factor-1 (NRF-1) can activate Zfp106 mRNA expression, with expression patterns of full-length Zfp106 and Nrf1 mRNA coinciding in mouse embryos (2). A short isoform is specifically expressed in skeletal muscle, regulated by myogenin, and is strongly upregulated during myogenic differentiation (2).
While previous research on ZFP106 indicates different functions, in this study we have created the first ZFP106 animal model to further understand its in vivo functions. We have taken advantage of the resource provided by the international Knock Out Mouse Project (KOMP) (9) and the Sanger Mouse Genetics Project to further elucidate Zfp106 function. Mice homozygous for the knockout first promoter-less allele (tm1a(KOMP)Wtsi) in Zfp106 develop severe gait and motor abnormalities that deteriorate with age. The motor abnormalities exhibited by Zfp106−/− mice are likely due to a severe progressive adult-onset degenerative sensory-motor axonopathy.
Results
Zfp106 deficiency causes progressive motor abnormalities
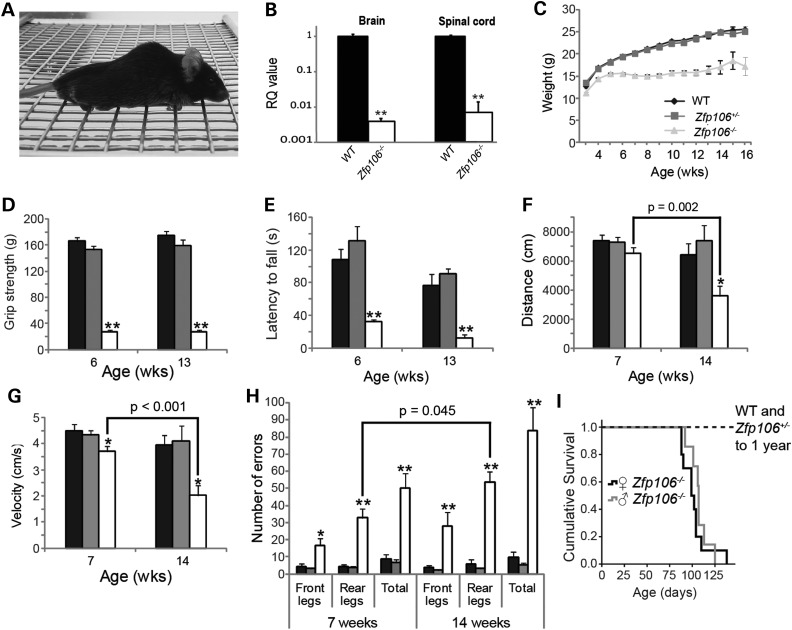
A knockout first promoter-less allele (tm1a(KOMP)Wtsi) within Zfp106 was created by The Sanger Mouse Genetics Project (http://www.sanger.ac.uk/mouseportal/) through KOMP (9). The construct targets Zfp106 intron 1 on chromosome 2 and includes a β-galactosidase (LacZ) reporter allele (see ‘Materials and Methods’). Mice homozygous for the tm1a(KOMP)Wtsi allele in Zfp106 (Zfp106−/−) are born in Mendelian ratios (data not shown) and both Zfp106−/−male and female mice develop an abnormal gait from ∼3 weeks of age, which deteriorates with age, causing animals to inaccurately place their limbs, especially their hindlimbs, when walking across a wire grate (Supplementary Material, Movie S1). Zfp106−/− animals also become severely kyphotic and develop limb grasping defects at 15-weeks of age whereby they pull all of their limbs into their body (Fig. 1A and Supplementary Material, Movie S1).
Figure 1.
Behavioural analyses of Zfp106−/− mice reveal severe motor abnormalities. Black bars: Zfp106−/−, grey bars: Zfp106+/−, white bars: Zfp106−/−. (A) Photograph of mutant Zfp106−/− mouse at 15-weeks of age displaying pronounced kyphosis and unable to coordinate hind limbs on wire grate. (B) qPCR analysis of Zfp106 expression levels in brain and spinal cord of 6-week-old male WT and Zfp106−/− littermates (n = 3 per genotype); expression in Zfp106−/− mice is normalized to Zfp106 expression in respective WT tissues, taken as a value of 1 (Log scale). (C) Female weights recorded weekly from ages 3 to 15 weeks; at least five mice were assessed per genotype per time point. Weight is diminished in Zfp106−/− female mice from 3-weeks of age (P = 0.04) and continues to significantly decrease, compared with WT and Zfp106+/− animals, to 15-weeks of age (P < 0.001). (D) Female grip strength and (E) accelerated Rotarod performance are reduced in Zfp106−/− mice at 6- and 13-weeks of age compared with WT and Zfp106+/− littermates (n≥ 6 per genotype). (F and G) Open field assessment of (F) distance moved, and (G) velocity for female mice at 7- and 14-weeks of age (see ‘Materials and Methods’). A reduction in distance moved (F), and velocity (G), was seen in 14-week-old female Zfp106−/− mice when compared with 7-week female Zfp106−/− mice; P values are indicated (n ≥ 5 per genotype and time point) and 14-week old WT littermates. (H) Female mice assessed for defects in coordination of front and rear legs at 7- and 14-weeks of age using the Locotronic® system (see ‘Materials and Methods’). Zfp106−/− mice made significantly more front and rear leg placement errors compared with WT littermates. Rear leg errors also significantly increased for 14-week-old Zfp106−/− mice compared with 7-week-old Zfp106−/− mice (n ≥ 5 per time point and genotype; P value indicated). Numbers and percentages shown represent the mean ± SEM. *P < 0.05; **P < 0.001. (I) Survival of male and female Zfp106−/− mice as determined by their humane endpoint (see ‘Materials and Methods’).
To assess Zfp106 expression we performed whole-genome transcriptomic analysis (RNA-seq) on spinal cord and qPCR on spinal cord and brain from adult wild-type (WT) mice and Zfp106−/− littermates. The unbiased RNA-seq expression data show that whilst some Zfp106 expression remains in spinal cords of Zfp106−/− mice, the expression levels are <20% of those identified in WT littermates (Table 1). qPCR of Zfp106 also revealed reduced expression in Zfp106−/− mouse tissues when compared with WT littermates; however, analysis via this technique indicated lower Zfp106 expression in Zfp106−/− mice when compared with RNA-seq analysis (0.4 and 0.7% of WT levels for brain and spinal cord, respectively) (Fig. 1B). This reduction in Zfp106 expression was also confirmed via northern-blot analysis from both brain and spinal cord (Supplementary Material, Fig. S1). The muscle-specific transcript was also downregulated in Zfp106−/− muscle via qPCR (Supplementary Material, Fig. S1). We also confirmed the expression pattern of Zfp106 using the LacZ reporter contained within the Zfp106 knockout cassette, with expression during embryogenesis and in many tissues in adulthood, including the central nervous system (CNS) and hind limb muscle (Supplementary Material, Fig. S1).
Table 1.
Top 10 statistically significant (adjusted P value <1E − 14) positively and negatively regulated genes from Zfp106−/− spinal cords via RNA-seq
| Gene | Fold difference |
|---|---|
| Calcoco2 | 0.15 |
| Zfp106 | 0.19 |
| Chodl | 0.38 |
| Calca | 0.55 |
| Pla2g3 | 0.60 |
| Slc10a4 | 0.61 |
| Mmp9 | 0.62 |
| Aif1l | 0.67 |
| Dhcr24 | 0.69 |
| Nefh | 0.76 |
| Mmp12 | 44.58 |
| Cst7 | 19.97 |
| Atp6v0d2 | 18.68 |
| Cxcl10 | 13.25 |
| Cxcl9 | 12.88 |
| Itgax | 12.23 |
| Tgm1 | 11.89 |
| Gpnmb | 11.63 |
| Lgals3 | 10.13 |
| Igfbpl1 | 13.25 |
We evaluated the behavioural phenotypes of male (Supplementary Material, Fig. S2) and female Zfp106−/− cohorts (Fig. 1) and found no sex differences. Body weight of female Zfp106−/− mice are significantly reduced from 3 weeks of age compared with WT and Zfp106+/− littermates (Fig. 1C; P = 0.04). When comparing grip strength of WT, Zfp106+/− and Zfp106−/− females we found a dramatic reduction in Zfp106−/− mice at 6 and 13 weeks of age (Fig. 1D). Rotarod testing revealed a similarly reduced motor performance in Zfp106−/− females compared with WT littermates (Fig. 1E). We also examined the behaviour of 6-week-old WT, Zfp106+/− and Zfp106−/− littermates using a modified SHIRPA analysis, which assesses a battery of simple phenotypic traits, with an emphasis on neurological function (10,11). These tests included: the presence/absence of tremors, limb grasping of front or rear legs, gait, the ability to hang from a wire (wire manoeuvre), and the ability to walk down a vertical wire grate (negative geotaxis, see ‘Materials and Methods’). Table 2 summarizes SHIRPA analysis from 6-week-old WT, Zfp106−/+ and Zfp106−/− littermates. ∼66% Zfp106−/− mice display mild to moderate tremors, >50% display limb grasping defects and all Zfp106−/− mice are severely impaired in wire hanging and are unable to maintain grip when made to walk down a vertical wire grate (negative geotaxis).
Table 2.
Movement and neurological associated defects in Zfp106−/− mice, assessed using modified SHIRPA analysis
| Genotype | Sample sizea | Tremors (%)b | Limb grasp (%)b | Wire Manoeuvre deficits (%)b | Negative geotaxis deficits (%)b |
|---|---|---|---|---|---|
| WT | 6 | 0 | 0 | 0 | 0 |
| Zfp106+/− | 7 | 0 | 0 | 0 | 0 |
| Zfp106−/− | 9 | 66.7 | 55.6 | 100 | 100 |
aFemale animals assessed at 6 weeks of age.
b% of animals that displayed defects in each of the semi-quantitative measures, assessed during the modified SHIRPA test (see ‘Materials and Methods’).
WT, Zfp106+/− and Zfp106−/− littermates were assessed in an open field paradigm, whereby mouse movement is tracked within an arena for 30 min. We observed a progressive deterioration in the distance moved (Fig. 1F), the mean velocity of movement (Fig. 1G), and the percentage of time spent moving (data not shown) between 7- and 14-week-old Zfp106−/− mice. To characterize these progressive motor abnormalities further we analysed the number of placement errors made by either their front or rear legs using the Locotronic® system (Fig. 1H). At 7 weeks of age Zfp106−/− mice produce significantly more front and rear leg errors compared with WT littermates (front leg errors, WT 4.4 ± 1.4 versus Zfp106−/− 16.6 ± 4.3, P < 0.05; rear leg errors, WT 4.2 ± 1.1 versus Zfp106−/− 32.8 ± 5.2; P < 0.001). Importantly, the number of rear leg errors significantly increases in Zfp106−/− mice at 14 weeks of age to 53.5 ± 6.4 (P = 0.045) compared with 7-week-old Zfp106−/−, showing an age-dependent deterioration in motor coordination.
Zfp106+/− mice did not display any differences from WT littermates in any of the behavioural tests outlined above. Five one-year old Zfp106+/− mice were also analysed by modified SHIRPA with no obvious defects detected (data not shown).
Zfp106−/− mice have a reduced lifespan
Lifespan of Zfp106−/− mice was determined as the time to reach humane endpoint, defined as the time taken to lose 20% of peak body weight or an inability to right themselves within 20 s when placed on their sides. Out of 17 Zfp−/− mice examined, the majority were culled due to a 20% loss of peak body weight (n = 12), whilst a smaller proportion were culled due to an inability to right themselves impinging on their ability to feed (n = 5). All Zfp106−/− mice exhibited severe locomotor deficits at humane endpoint. The average lifespan of female Zfp106−/− mice was 102 ± 5 days (n = 10) and for males was 107 ± 4 days (n = 7) (Fig. 1I); there was no significant difference in survival between Zfp106−/− males and females (data not shown). WT and Zfp106+/− littermates survived with no abnormalities up to 365 days.
Hind limb muscle physiology of early and late stage Zfp106−/− mice
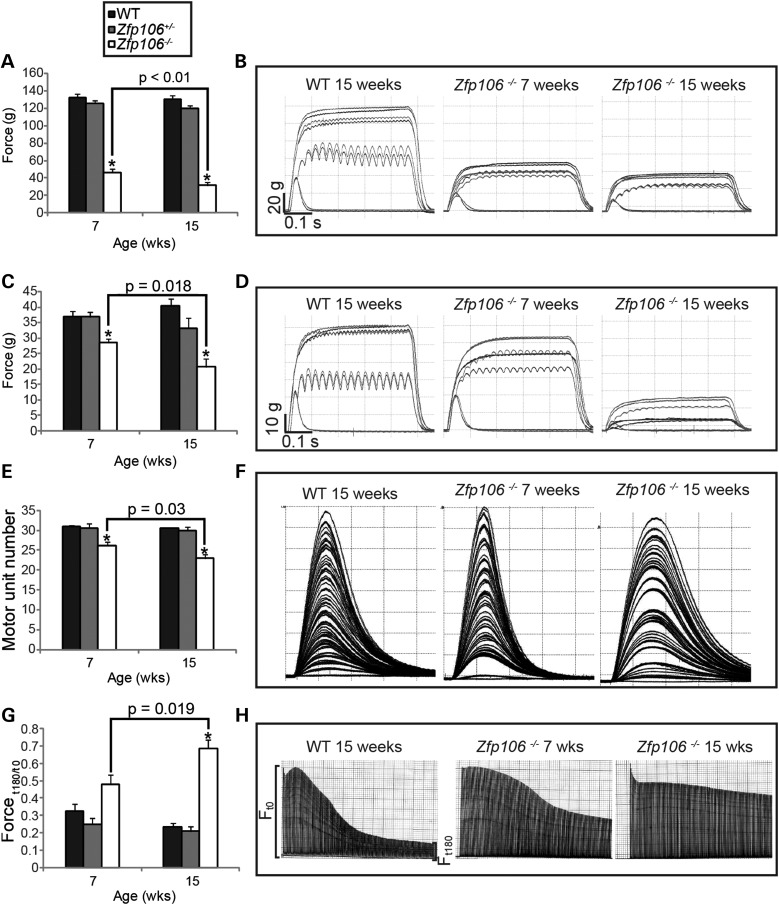
To further characterize the motor abnormalities in Zfp106−/− mice, we analysed in vivo the physiological characteristics of the fast twitch hind limb muscles tibialis anterior (TA) and extensor digitorum longus (EDL). The maximum isometric force (tetanic tension) was measured in the TA and EDL muscles from both legs of 7- and 15-week-old WT, Zfp106+/− and Zfp106−/− littermates (Fig. 2A–D). The maximum force generated from the TA of 7-week-old Zfp106−/− mice was ∼65% less than WT littermates and further reduced to ∼75% of WT littermates at 15-weeks of age (Fig. 2A and B). Similarly, the tetanic tension of EDL muscles from 7-week-old Zfp106−/− mice was ∼20% less than WT littermates and reduced by ∼50% in 15-week-old Zfp106−/− mice compared with WT littermates (Fig. 2C and D). Thus, TA muscles are affected earlier and more severely than the EDL muscles, as has been previously reported in mouse models of other neuromuscular disorders (12). Interestingly, both TA and EDL muscles displayed a clear deterioration in the amount of force generated as Zfp106−/− mice aged.
Figure 2.
Assessment of hind limb muscle force, motor unit survival and muscle fatigue on female mice. (A and B) Summary of the maximum tetanic muscle force generated from the TA muscle. (A) TA tetanic muscle force generated by Zfp106−/− mice at 7-weeks of age (47 g ± 4 g) is reduced compared with WT littermates (133 g ± 4 g). TA tetanic tension is further reduced in 15-week-old Zfp106−/− mice (32 g ± 4 g) compared with 7-week-old Zfp106−/− mice. (B) Representative traces of TA tetanic tension from WT and Zfp106−/− mice at 7- and 15-weeks of age. (C and D) Tetanic muscle force generated from the EDL at 7- and 15-weeks of age (see ‘Materials and Methods’). (C) EDL tetanic muscle force generated by Zfp106−/− mice at 7-weeks (29 g ± 1 g) is reduced compared with WT littermates (37 g ± 2 g) and becomes further reduced in 15-week-old Zfp106−/− mice (21 g ± 3 g). (D) Representative traces of EDL tetanic tension from WT, 7- and 15-week old Zfp106−/− mice. (E and F) The number of surviving motor units in the EDL muscle of female mice at 7- and 15-weeks of age; motor units are reduced in 7-week-old Zfp106−/− mice (26 ± 0.9) compared with WT littermates (31 ± 0.4), and are further reduced in 15-week-old Zfp106−/− mice (23 ± 1). (F) Representative motor unit traces from EDL muscles of WT, 7- and 15-week old Zfp106−/− mice. Each twitch trace recording is a single motor unit. (G and H) FI (see ‘Materials and Methods’ section) of EDL muscle for female mice at 7 and 15-weeks of age. FI of 7-week old Zfp106−/− mice (0.48 ± 0.05) is increased compared with WT littermates (0.32 ± 0.04) but not significantly (P = 0.07). FI increases further in 15-week-old Zfp106−/− mice (0.69 ± 0.05) compared with 7-week old Zfp106−/− mice. (H) Representative fatigue traces from WT, 7- and 15-week Zfp106−/− mice, produced by repetitive stimulation of the EDL muscle for 180 s. Each line in the trace represents a single tetanic tension. Results are presented as the mean ± SEM. At least 10 legs were assessed per genotype per time point. *P ≤ 0.001, or indicated within each chart when comparing Zfp106−/− mice from 7- and 15-weeks of age.
We next determined the number of functional motor units innervating EDL muscles in 7- and 15-week-old WT, Zfp106+/− and Zfp106−/− mice (Fig. 2E and F). In 7-week-old Zfp106−/− mice, there was a small but significant reduction in motor unit survival, such that 26 ± 0.9 motor units (n = 12) innervated the EDL in Zfp106−/− mice compared with 31 ± 0.4 motor units (n = 10) in WT littermates (P < 0.001). By 15 weeks, fewer motor units survived in Zfp106−/− mice, with 23 ± 1 motor units (n = 12) surviving compared with 31 ± 0.5 motor units (n = 10) in WT littermates (P < 0.001), suggesting a progressive loss of motor neurons.
Fast twitch muscles such as EDL normally fatigue rapidly and cannot maintain force when repeatedly stimulated. We therefore examined the fatigue characteristics of EDL muscles in 7- and 15-week-old Zfp106−/− mice, by undertaking a fatigue test in which the muscles were repeatedly stimulated (Fig. 2G and H). Typical fatigue traces from EDL muscles of WT and Zfp106−/− littermates are depicted in Figure 2H showing that in Zfp106−/− mice there was a clear shift in the fatigue pattern of EDL, becoming fatigue resistant as the mice aged. From traces such as these, a fatigue index (FI) was calculated for each muscle, where an FI of 1.0 indicates that a muscle is completely fatigue resistant, as previously described (13). Our findings revealed that 7-week-old WT mice had a FI of 0.32 ± 0.04 (n = 10) (Fig. 2G and H) compared with 0.48 ± 0.06 in 7-week-old Zfp106−/− (n = 12), an increase of 33.3%, although this difference was not statistically significant (P = 0.07). However, by 15 weeks of age, the FI of EDL in Zfp106−/− mice was 0.69 ± 0.05 (n = 12), compared with 0.23 ± 0.02 (n = 10) in WT littermates, a statistically significant increase of 66.7% (P < 0.001), and a statistically significant increase of 30.4% over 7-week-old Zfp106−/− mice (P = 0.019). These results show that in Zfp106−/− mice, EDL muscles are significantly more fatigue resistant than those of WT littermates, a characteristic usually observed in slow twitch muscles.
Zfp106 deficiency causes progressive motor neuron degeneration with reactive gliosis and loss of motor and sensory axons
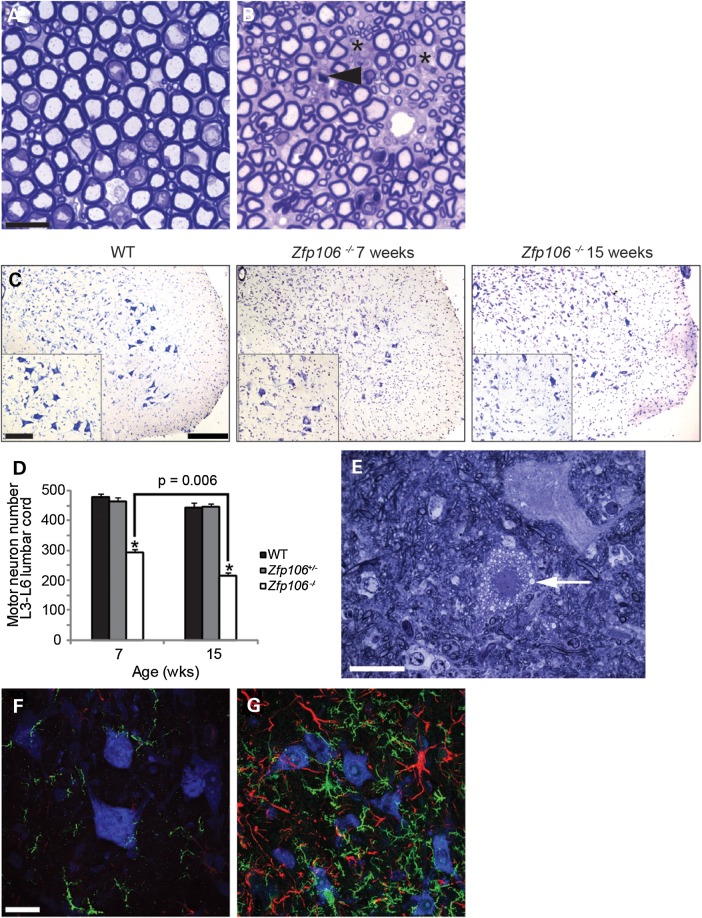
Behavioural, physiological and histopathological studies of Zfp106−/− mice all indicate possible neuronal defects. Therefore, we examined the morphology of motor neurons and their axons from WT and Zfp106−/− littermates. Toluidine blue stained semi-thin sections from the lumbar L4 and L5 ventral roots revealed widespread loss of fibres, active degeneration of motor axons with no clear signs of regeneration and apparently normal myelination (Fig. 3A and B). To assess motor neuron survival, we counted the number of motor neurons from the sciatic pool in the lumbar spinal cord (L3-L6) of 7- and 15-week-old WT, Zfp106+/− and Zfp106−/− littermates (Fig. 3C and D). The number of motor neurons of 7-week-old Zfp106−/− mice is reduced by ∼35% compared with WT littermates and decreased by a further ∼15% at 15 weeks of age, corresponding to a total motor neuron loss of 50% in 15-week-old Zfp106−/− mice (Fig. 3C and D). Interestingly, visualization of toluidine blue stained semi-thin sections of the lumbar region of Zfp106−/− spinal cord showed the occasional presence of vacuolated motor neurons, likely to be undergoing cell death (Fig. 3E). Immunohistochemical staining of the lumbar spinal cord of 15-week-old WT and Zfp106−/− littermates using antibodies to GFAP (astrocytosis) and IBA-1 (microgliosis) also revealed widespread reactive gliosis, coincidental with over 50% motor neuron loss, not seen in WT or Zfp106+/− littermates (Fig. 3F and G, and data not shown).
Figure 3.
Motor neuron and axonal degeneration and gliosis in Zfp106−/− mouse spinal cord. (A and B) Representative toluidine blue stained sections of lumbar L4 and L5 ventral roots from 15-week old WT (A) and Zfp106−/− (B) mice. Zfp106−/− mice have fewer motor axons than WT littermates (spaces are marked with asterisks), and have degenerating axons, indicated with arrow head. Scale bar is 20 μm. (C) Representative images of the ventral horn of lumbar spinal cord sections stained for Nissl from WT, 7- and 15-week-old Zfp106−/− mice. The number of motor neurons in the sciatic pool (inset) was counted for each genotype and age to calculate motor neuron survival (D). Scale bars: main images 200 μm inset 100 μm. (D) The survival of motor neurons in the sciatic pool in female mice at 7- and 15-weeks of age. Motor neuron survival in 7-week-old Zfp106−/− mice (294 ± 11) is reduced compared with WT littermates (481 ± 9) and further reduced in 15-week-old Zfp106−/− mice (217 ± 9). Numbers represent the mean ± SEM. At least five animals were assessed per genotype per time point (*P < 0.001). (E) Representative toluidine blue stained section of 15-week-old Zfp106−/− lumbar spinal cord showing an example of a vacuolated (arrow) motor neuron. (F and G) Lumbar spinal cord sections of 15-week-old WT (F) and Zfp106−/− (G) mice stained for IBA-1 (green), GFAP (red) and Nissl (blue). Immunoreactivity for micro- and astrogliosis is dramatically increased in the lumbar of 15-week-old Zfp106−/− mice compared with WT littermates. Scale bar is 20 μm.
We performed p62 staining of spinal cord sections from Zfp106−/− mice because p62 is commonly found localized into aggregates in neurodegenerative disease (14). However, we did not observe any altered localization or aggregation of p62 in the lumbar region of 15-week-old Zfp106−/− mice (data not shown). Terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) staining on ventral spinal cord of 15-week-old WT and Zfp106−/− mice showed cells undergoing active cell death in Zfp106−/− mice (Supplementary Material, Fig. S3A and B).
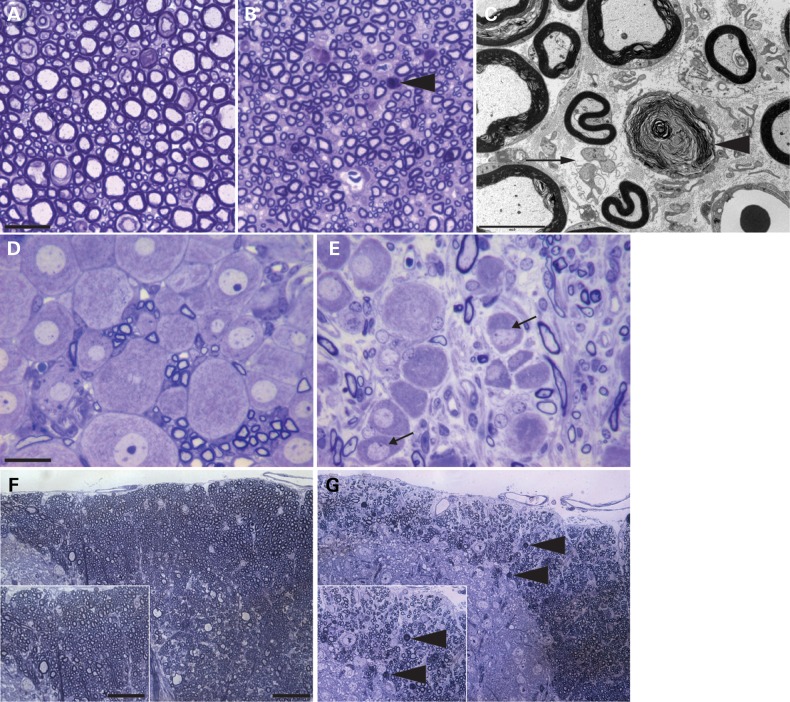
Given the striking loss of lower motor neurons, we next examined the sensory system. Assessment of L4 and L5 dorsal roots from 15-week-old female WT and Zfp106−/− littermates revealed a loss of both large and small sensory axons (Fig. 4A and B). We also observed degeneration of myelinated and unmyelinated axons and no evidence of axonal regeneration or demyelination (Fig. 4C). In addition, numerous L4 and L5 dorsal root ganglia (DRG) of 15-week-old female Zfp106−/− mice were smaller compared with WT littermates, some showing signs of chromatolysis (Fig. 4D and E). Examination of dorsal fasciculi in the spinal cord revealed degeneration of ascending sensory axons (Fig. 4F and G). However, we did not observe any morphological or pathological defects in the descending spinal cord tracts (data not shown). Thus, Zfp106−/− mice have deficits in both the motor and sensory neuronal systems of the spinal cord.
Figure 4.
Axonal degeneration of sensory neurons. (A, B) Representative toluidine blue stained sections of lumbar L4 and L5 dorsal roots from WT (A) and Zfp106−/− (B) mice. Zfp106−/− mice have fewer sensory axons, and degenerating axons, indicated with arrow head. Scale bar is 20 μm. (C) Electron micrograph of Zfp106−/− dorsal root showing degenerating myelinated axons (arrow head) and evidence of loss of unmyelinated axons with denervated Schwann cells (arrow). Scale bar is 5 μm. (D and E) Representative toluidine blue stained section of lumbar DRG from WT (D) and Zfp106−/− (E) mice. Numerous DRG neurons from Zfp106−/− mice (E) are shrunken and irregular and some show signs of chromatolysis, with flattened nuclei (indicated with arrows). Scale bar is 20 μm. (F and G) Representative toluidine blue stained sections of the dorsal fasciculi from the lumbar spinal cord from WT (F) and Zfp106−/− (G) mice showing fewer axons and axonal degeneration (arrow heads). Scale bar is 20 μm. All images are of 15-week-old WT or Zfp106−/− mice, images representative of three animals per genotype.
Zfp106−/− neuronal loss is not developmental
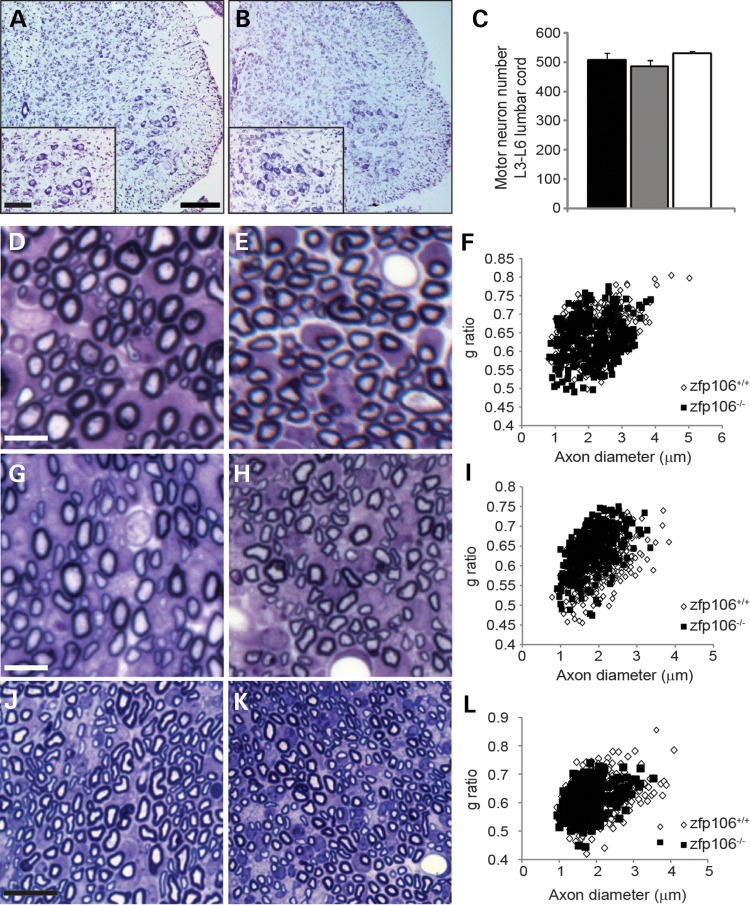
Following the identification of progressive neurodegeneration in both motor and sensory neurons of the spinal cord, we next sought to establish whether neuronal abnormalities are present during development. Motor neuron survival was unaffected in P9 Zfp106−/− mice compared with WT littermates (Fig. 5A–C) and DRG from Zfp106−/− mice were also indistinguishable from WT littermates at this age (data not shown). Axonal morphology was quantified on toluidine blue stained semi-thin sections from the lumbar L4 and L5 ventral and dorsal roots, and from sciatic nerve sections of P9 animals (Fig. 5D–L and Supplementary Material, Fig. S4). Axonal morphology from ventral roots was similar between WT controls and Zfp106−/− mice (Fig. 5D–F). However, there was a significant reduction in axonal diameter on the sciatic nerve of Zfp106−/− mice when compared to WT controls (Fig. 5J–L and Supplementary Material, Fig. S4). This is accompanied with a significant shift in the frequency distribution towards a thinner axon diameter and an increased g-ratio in Zfp106−/− dorsal roots when compared with WT (Fig. 5G–I). Thus, deficiency of Zfp106 leads to mild early changes in axonal morphology but does not cause neuronal loss at P9.
Figure 5.
Neuronal survival and axonal morphology at P9. (A and B) Representative images of the ventral horn of lumbar spinal cord sections stained for Nissl from P9 WT and Zfp106−/− mice. The number of motor neurons in the sciatic pool (inset) was counted for each genotype. Scale bars: main images 200 μm, inset 100 μm. (C) Survival of motor neurons in the sciatic pool in female mice at P9. Motor neuron survival is unaffected in P9 Zfp106−/− mice (530 ± 15) compared with WT littermates (508 ± 23). Numbers represent the mean ± SEM. At least five animals were assessed per genotype per time point. (D and E) Representative toluidine blue stained sections of lumbar L4 and L5 ventral roots from P9 WT (D) and Zfp106−/− (E) mice. (F) Distribution graph relating g-ratio to axon diameter of all myelinated axons analysed from ventral roots show no differences between WT and Zfp106−/−. Scale bar is 20 μm. (G and H) Representative toluidine blue stained sections of lumbar L4 and L5 dorsal roots from P9 WT (G) and Zfp106−/− (H) mice. Scale bar is 20 μm. (I) Distribution graph relating g-ratio to axon diameter from dorsal roots. Zfp106−/− mice have a significant shift in the frequency distribution towards a thinner axon diameter and an increased g-ratio compared with WT (P < 0.001). (J and K) Representative toluidine blue stained sections of the sciatic nerve from P9 WT (J) and Zfp106−/− (K) mice. Scale bar is 10 μm. (L) Distribution graph relating g-ratio to axon diameter for the sciatic nerve. Zfp106−/− mice show a significant shift in the frequency distribution towards a smaller axon diameter compared with WT animals (P < 0.001) but no change in g-ratio.
Zfp106 deficiency does not cause major abnormalities in the brain
We performed an immunohistological examination on the brains of female 15-week-old WT and Zfp106−/− littermates using the following stains and protein markers: H&E, GFAP, IBA-1, p62, ubiquitin, myelin basic protein and neurofilament. There were no major abnormalities in the anatomy of the brain or differences in reactive gliosis, p62, MBP or neurofilament staining between WT and Zfp106−/− mice (data not shown). Therefore, deficiency in Zfp106 causes specific defects in the spinal cord and PNS but not the brain, despite being expressed at similar levels in the brain (Fig. 1B).
Muscle histopathology in Zfp106−/− mice
The changes in the physiological characteristics of the TA and EDL muscles of Zfp106−/− mice led us to perform a more in-depth muscle histopathological analysis. Examination of gross morphology of Zfp106−/− mice clearly showed a reduced mass and diminished size of muscles (Supplementary Material, Fig. S5A). Analysis of 6- and 14-week-old WT and Zfp106−/− littermates revealed reduced TA, gastrocnemius and soleus muscle weight in 14-week but not 6-week-old Zfp106−/− mice when compared with WT littermates (P < 0.01). However, no differences were found in EDL muscle weights at either 6 or 14 weeks of age (Fig. 6A and data not shown) or in tibia length from 14-week old WT and Zfp106−/− littermates (Supplementary Material, Fig. S5B). Histopathological analysis of muscles (gastrocnemius, soleus and EDL) from 6-week-old Zfp106−/− littermates was comparable to that seen in WT littermates (data not shown). However, Haematoxylin and eosin (H&E) staining of the gastrocnemius from 14-week-old Zfp106−/− mice, which had atrophied by ∼80% at that stage, revealed extensive muscle fibre atrophy, with areas of increased nuclearity, and evidence of reorganization and fibre regeneration (Fig. 6B and C). Extensive areas of the gastrocnemius were also fibrotic (Fig. 6D and E). We also assessed the distribution of fibre types within the gastrocnemius and soleus using NADH–tetrazolium reductase (NADH-TR) and ATPase stainings and found increased type I/highly oxidative fibres in the gastrocnemius and soleus of 14-week-old Zfp106−/− mice compared with WT littermates (Fig. 6F –I). Moreover, in all sections of soleus muscle from Zfp106−/− mice examined there was a greater spread of fibre size, ranging from very small to very large muscle fibres, compared with WT soleus muscle (Supplementary Material, Fig. S5C). In addition, the soleus of 14-week-old Zfp106−/− mice had fewer fibres when compared with WT littermates (Fig. 3J). Thus, Zfp106 deficiency causes progressive muscle pathology.
Figure 6.
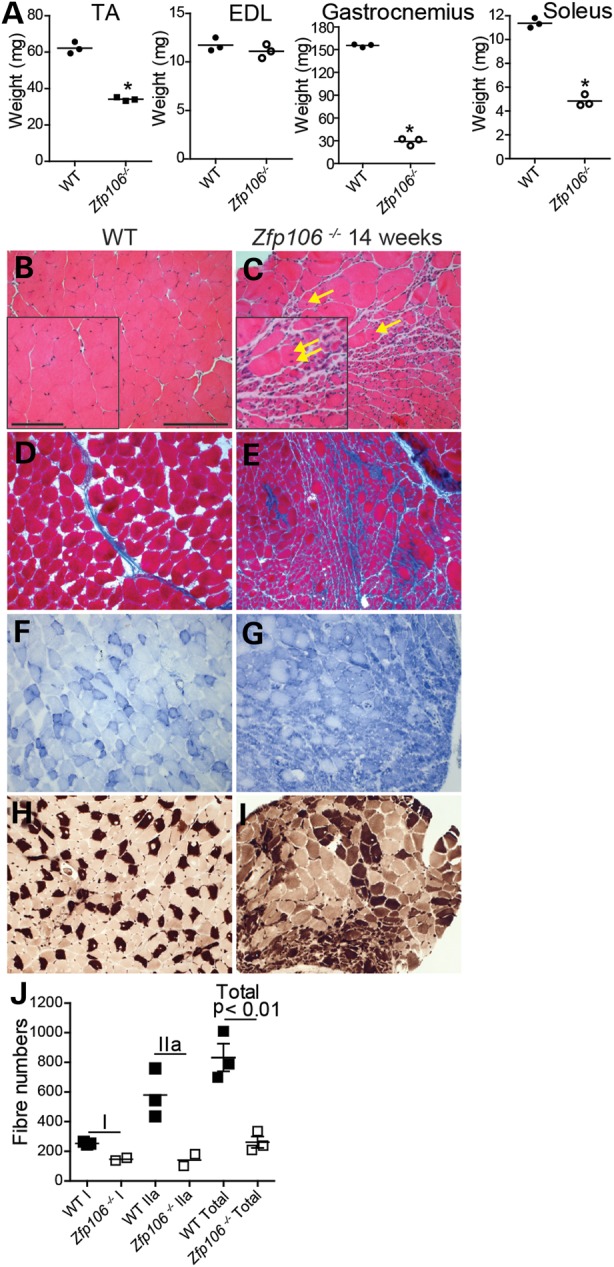
Muscle histology of WT and Zfp106−/− male mice. (A) Muscle weights for the TA (WT, 62 mg ± 1 mg; Zfp106−/−, 34 mg ± 1 mg), EDL (WT, 11 mg ± 0.4 mg; Zfp106−/−, 11 mg ± 0.4 mg), gastrocnemius (WT, 155 mg ± 1 mg; Zfp106−/−, 29 mg ± 23 mg) and soleus (WT, 11 mg ± 0.2 mg; Zfp106−/−, 5 mg ± 0.3 mg) from male 14-week old WT and Zfp106−/− littermates (n = 3). TA, gastrocnemius and soleus weights are significantly different from WTs (*P < 0.01). (B–I) Representative images of muscle histopathology from 14-week-old WT and Zfp106−/− mice: (B and C) H&E staining of the gastrocnemius shows significant atrophy, reduced fibre size and centralized nuclei (yellow arrows) in Zfp106−/− muscle compared with WT littermates; (D and E) Masson's trichome staining of gastrocnemius muscle reveals fibrosis in Zfp106−/− muscle (E), evident with increased blue staining; (F and G) NADH-TR staining of the gastrocnemius shows greatly increased dark blue staining in Zfp106−/− muscle (G) compared with WT littermates, indicative of a higher proportion of slower twitch, type I muscle fibres; (H and I) ATPase staining of Zfp106−/− soleus muscle (I) showing an increase in darkly stained fibres, indicating an increase in the proportion of type I fibres overall. Scale bars: main images 20 μm, inset 10 μm. (J) Type I (WT, 253 ± 6 fibres; Zfp106−/−, 147 ± 10 fibres), type IIa (WT, 579 ± 95 fibres; Zfp106−/−, 141 ± 40 fibres) and total fibre number (WT, 833 ± 92 fibres; Zfp106−/−, 261 ± 38 fibres) were counted for the soleus muscle from 14-week-old WT and Zfp106−/− littermates using ATPase staining. The soleus from Zfp106−/− mice has significantly fewer muscle fibres than that of WT mice (P < 0.01).
Zfp106 deficiency affects genome-wide transcription
A role in transcriptional control has been recently proposed for ZFP106 (5). To assess how Zfp106 deficiency might affect transcriptional regulation, we analysed genome-wide transcriptional changes from the spinal cords of 6-week-old Zfp106−/− mice using RNA-seq. At this stage, Zfp106−/− mice already display motor and sensory neuron pathology (Figs 3 and 4). Analysis of gene expression revealed a large number of differentially expressed genes, the majority of which were upregulated in Zfp106−/− when compared with WT littermate spinal cords (Table 1 and Supplementary Material, Fig. S6A). Pathway analysis of differentially expressed genes showed a variety of affected cellular processes, prominently including the inflammatory response, metabolic disease, cell-to-cell signalling and interaction, hematological system function, immune cell trafficking and cell motility.
Zfp106 deficiency causes mitochondrial abnormalities
To investigate whether mitochondrial dysfunction may be a feature of Zfp106−/− neurons, we examined the mitochondrial properties of embryonic motor neurons derived from WT, Zfp106+/− and Zfp106−/− littermates.
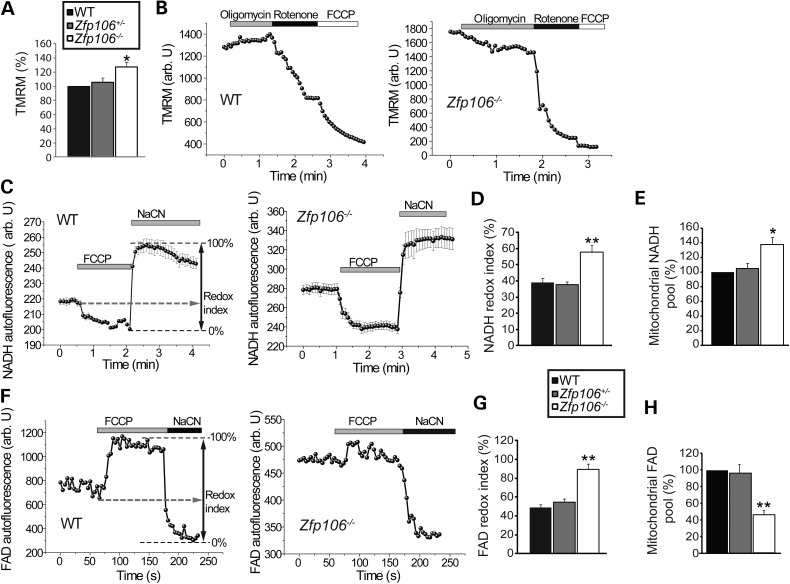
We first assessed mitochondrial membrane potential (Δψm), an indicator of the mitochondrial energetic state, measured using tetramethylrhodamine methylester (TMRM), a cell-permeable, potentiometric, fluorescent dye that is readily sequestered by active mitochondria (15,16). TMRM is a positively charged molecule which accumulates within mitochondria in inverse proportion to Δψm; hyperpolarized mitochondria will accumulate more dye, and hypopolarized mitochondria will accumulate less dye. Fluorescent dye accumulation in mitochondria is then optically detected by confocal microscopy. We found that mitochondria in motor neurons derived from Zfp106−/− mice were associated with a significant increase (27% ± 7%, P < 0.001) in the TMRM signal compared with mitochondria from WT motor neurons (n = 34 cells) (Fig. 7A). In Zfp106+/− motor neurons, values of TMRM fluorescence were not significantly different from WT (Fig. 7A). These data suggest that Zfp106 deficiency is associated with increased basal Δψm in embryonic motor neurons, at least in vitro.
Figure 7.
Mitochondrial abnormalities in Zfp106−/− embryonic motor neurons. (A) Resting Δψm from embryonic WT, Zfp106+/− and Zfp106−/− motor neurons, estimated by live cell imaging using TMRM. Data are normalized to resting Δψm for WT (100%) and represent the mean ± SEM. (B) Representative traces of Δψm (TMRM, arbitrary units) in response to oligomycin (2 μg/ml) (inhibitor of the F1FO-ATP synthase), rotenone (5 μm) (inhibitor of complex I) and the minimum TMRM fluorescence achieved after addition of FCCP 1 μm (uncoupling agent) for WT and Zfp106−/− embryonic motor neurons. Following oligomycin addition WT motor neuron Δψm hyperpolarises, whilst Zfp106−/− depolarises. (C) NADH autofluorescence monitored in WT and Zfp106−/− motor neurons, the addition of FCCP maximizes mitochondrial respiration, thus minimizes mitochondrial NADH. NaCN was added to block mitochondrial respiration and therefore maximize mitochondrial NADH. Redox index (the initial redox level expressed as a percentage of the range) and mitochondrial NADH level (the difference in absolute values between the minimum and maximum NADH autofluorescence) are described graphically. (D) NADH redox index for WT, Zfp106+/− and Zfp106−/− motor neurons calculated from (C). NADH redox index is increased in Zfp106−/− embryonic motor neurons (58% ± 4%) compared with WT littermates (39% ± 3%). (E) Mitochondrial pool of NADH represented as % of WT. NADH levels are increased in Zfp106−/− (138% ± 9.1%) motor neurons when compared with WT. (F) FAD autofluorescence monitored in WT and Zfp106−/− motor neurons, the addition of FCCP maximizes respiration, increasing FAD autofluorescence to maximal levels, whilst NaCN inhibits respiration, reducing FAD autofluorescence to minimal levels. (G) FAD redox index for WT, Zfp106+/− and Zfp106−/− motor neurons, calculated by normalizing the FCCP response to 100% and the NaCN response to 0%. FADH redox index is increased in Zfp106−/− embryonic motor neurons (89% ± 6%) compared with WT littermates (49% ± 3%). (H) Mitochondrial pool of FAD represented as % of WT. FAD levels are decreased in Zfp106−/− motor neurons (45.6% ± 5.2%) when compared with WT. Data from (A–H) are represented as mean ± SEM; n ≥ 34 motor neurons per genotype, per condition. *P < 0.05; **P < 0.01.
We next investigated how Zfp106 deficiency results in an increase in Δψm in embryonic motor neurons from Zfp106−/− mice when compared with Zfp106+/− and WT littermates. In cells with normal oxidative phosphorylation, Δψm is maintained by the proton pump activity of the respiratory chain. However, if oxidative phosphorylation is impaired, then hydrolysis of ATP by the F1FO-ATP synthase (Complex V) can pump protons across the inner membrane and so maintain Δψm (17). Application of the inhibitor of the F1FO-ATP synthase, oligomycin, to WT motor neurons increased Δψm by 3 ± 0.1% (n = 23) (Fig. 7B), and in Zfp106+/− motor neurons by 5 ± 0.3% (n = 27). However, exposure of Zfp106−/− motor neurons to oligomycin caused a profound decrease in Δψm (TMRM signal fell by 17% ± 3%, P < 0.001, n = 32) (Fig. 7B). Thus, in Zfp106−/− motor neurons, the impaired activity of the respiratory chain switches the F1FO complex to an ATP consumption mode which serves to maintain Δψm.
The redox state of NADH or FAD is a function of respiratory chain activity and the rate of substrate supply. We measured the resting level of NADH and FAD++ autofluorescence and generated the ‘redox index’; a ratio of the maximally oxidized (response to FCCP) and maximally reduced (response to sodium cyanide (NaCN)) signals. The redox index of NADH in Zfp106−/− motor neurons was significantly more oxidized than in WT littermates (WT: 39% ± 3%, n = 35 versus Zfp106−/−: 58% ± 4%, n = 46; P < 0.001) (Fig. 7C and D). We also estimated the levels of the mitochondrial pool of NADH, the substrate for complex I. NADH levels were significantly increased in Zfp106−/− motor neurons (Fig. 7E) when compared with WT controls. Thus, the combination of accumulating substrate and a diminished redox index in Zfp106−/− motor neurons suggest that Zfp106 deficiency leads to functional inhibition of the NADH-dependent (Complex I) mitochondrial respiration.
In addition, FAD++ levels were increased in Zfp106−/− motor neurons compared with WT littermates (WT: 49% ± 3% versus Zfp106−/− 89% ± 6%; n = 47; P < 0.001) (Fig. 7F and G). The levels of the mitochondrial pool of FAD were also estimated; FAD levels were significantly lower in Zfp106−/− motor neurons when compared with WT controls (Fig. 7H). These findings suggest that Zfp106 deficiency results in activation of Complex II respiration. The FAD redox state in WT motor neurons (n = 37) was indistinguishable from Zfp106+/− motor neurons (n = 44) (Fig. 7F and G). Thus, the striking abnormalities in motor neuron mitochondrial properties may underlie the neurodegenerative effects caused by Zfp106 deficiency.
Discussion
Zfp106 is expressed during development and ubiquitously in adulthood, however, its functions remain unknown. Here, we report a new Zfp106-deficient mouse strain, which presents with reduced body weight from 3 weeks of age, a reduction in grip strength, the appearance of progressive gait abnormalities and the development of a severe kyphosis compared with WT and Zfp106+/− littermates (Fig. 1 and Supplementary Material, Movie S1). These phenotypes were associated with neuromuscular deficits. Physiological and histopathological analyses revealed that Zfp106−/− mice undergo a progressive loss of motor neurons, degeneration of motor and sensory axons together with progressive, severe muscle atrophy (Figs 2, 3, 4 and 6). Importantly, we established that neurodegeneration is progressive and does not occur during development, as motor and sensory neurons were intact at P9. However, mild changes in axonal morphology were present at this early stage (Fig. 5), suggesting that the neurodegeneration process may be already starting.
Interestingly, despite Zfp106 being expressed throughout the CNS (2), abnormalities in Zfp106−/− mice are confined to lower motor neurons, sensory neurons and their peripheral axons, and in the dorsal spinal cord ascending tracts (Figs 3 and 4). No major morphological or pathological abnormalities were observed in the descending spinal cord tracts or in other brain regions such as cerebellum, basal ganglia, thalamus, hippocampus or cortex. Thus, preservation of Zfp106 expression levels is essential for the maintenance and survival of spinal cord residing neurons and their axons, suggesting that spinal cord residing neurons, with their exceptionally long axons, are particularly susceptible to changes in Zfp106 expression. It may be possible that neurons from other brain areas may also be compromised, but the rapid degeneration of spinal cord neurons may preclude pathology potentially appearing in other areas. Future studies conditionally deleting Zfp106 specifically in different brain areas will be required to address this issue.
Histopathological assessment of young (6-week-old) Zfp106−/− muscle did not reveal overt pathological abnormalities (data not shown), whilst 14-week-old Zfp106−/− mice display varying degrees of muscle involvement ranging from no significant atrophy in the EDL to an ∼80% weight loss of the gastrocnemius (Fig. 6). This suggests that the observed muscle pathology may be secondary to the neuronal degeneration. However, it is possible that deficiency of Zfp106 in muscle also contributes to the neurodegeneration, and future studies that conditionally remove Zfp106 from neurons or muscle will clarify this point.
In humans, ZFP106 is localized to the nucleus, and with the presence of N- and C-terminal zinc finger motifs, is likely to regulate gene expression (1). Although several commercial antibodies are available against human ZFP106, none of them recognize endogenous mouse ZFP106, at least from brain and spinal cord (data not shown). Thus, we used RNA-seq analysis to gain insight into the functional consequences of Zfp106 deficiency, showing a number of genes misregulated due to Zfp106 deficiency. Pathway analysis of the differentially regulated genes supports a role for Zfp106 in neuronal maintenance and survival. Previous research has shown that NRF-1, a transcription factor linked to the regulation of mitochondrial function (18) and neurite outgrowth (19), is important in the transcription of full length ZFP106 (2). Nrf1 null mice are embryonic lethal, however, mice in which Nrf1 is specifically deleted from the CNS (CNS conditional Nrf1−/− mice) develop a juvenile aggressive neurodegenerative phenotype (20,21). Neurodegeneration is more widespread in CNS conditional Nrf1−/− mice compared with Zfp106−/− mice, involving alterations in the hippocampus and cerebellum but, importantly, CNS conditional Nrf1−/− mice display pathology in the ventral horn of the spinal cord, including increased oxidative stress. Whether Zfp106 plays a role in the development of the Nrf1−/− conditional knock out phenotype remains uncertain, however, the effect of Nrf1−/− on spinal cord neuronal morphology has similarities to the abnormalities observed here in Zfp106−/− mice. The contribution of ZFP106 to neuronal survival within the spinal cord and PNS may suggest a specific role as a downstream regulator of NRF-1.
Mitochondrial dysfunction has been proposed to be an instigating factor in many neurodegenerative conditions (22). We show here that a deficiency in Zfp106 causes profound mitochondrial abnormalities in embryonic motor neurons (Fig. 7). Deficiency of Zfp106 induces inhibition of Complex I within the electron transport chain of mitochondria and activation of Complex II. It has been previously shown that Complex II can become activated in order to compensate for Complex I inhibition, leading to the hydrolysis of ATP by the F1FO-ATP synthase (Complex V) and the pumping of protons across the inner mitochondrial membrane (23,24). Overall, these abnormalities result in an increase in Δψm, as observed in Zfp106−/− embryonic motor neurons. Interestingly, elevated Δψm can lead to an increase in the production of reactive oxygen species (ROS) and can be a cause of neuronal cell death (25,26). Furthermore, inhibition of Complex I and the resulting activation of Complex II have been shown to reverse the flow of electrons within the electron transport chain and produce free radicals (27), which can in turn lead to mitochondrial and cellular damage. Although we provide strong evidence of mitochondrial dysfunction due to Zfp106 deficiency, whether this dysfunction is directly or indirectly mediated by ZFP106 remains an open question.
Mitochondrial abnormalities have been reported in a variety of neurodegenerative disorders affecting motor and sensory neurons in the spinal cord and their axons, including motor neuron diseases such ALS (28–30), spinal muscular atrophy (SMA) (31) and prominently in peripheral neuropathies, including Charcot-Marie-Tooth (CMT) (32). A number of causative CMT genes are associated with mitochondrial function, including: MFN2, GDAP1 or MT-ATP6 (33–35) and the Δψm of CMT patients carrying mutations in MFN2 or GDAP1 is disrupted (36,37). Furthermore, peripheral neuropathy is observed in patients with mitochondrial diseases, such as Leigh's disease, mitochondrial neurogastrointestinal encephalopathy (MNGIE), neuropathy, ataxia and retinitis pigmentosa (NARP) (38) and spastic paraplegia with optic atrophy and neuropathy (SPG55) (39).
Any possible role for ZFP106 in human disease remains to be established, as disease-causing mutations in ZFN106 have not yet been identified. The neuro-anatomical phenotypes observed in Zfp106−/− mice have striking similarities with human axonal sensory-motor neuropathies, such as CMT type 2 (CMT2). CMT2 prevalently affect the sensory and motor nerves, but also show loss of MNs, spinal cord gliosis and degeneration of the fasciculus gracilis, as documented here for Zfp106−/− mice. Recessive forms of CMT2 usually have an earlier onset, occurring in the first decade of life and frequently before the age of two (40,41). These characteristics are similar to the deficits observed in Zfp106−/− mice, which start developing symptoms at weaning (Fig. 1 and Supplementary Material, Movie S1). However, since the function of human ZFP106 is not yet clear, it will be important to screen for ZFN106 mutations in early-onset CMT2 cases (42), but also milder forms of CMT, as well as other neurodegenerative disease in which there is degeneration of either motor or sensory neurons, such as ALS and progressive muscular atrophy (PMA).
ZFN106 is located on human chromosome 15 within the critical region of a previously mapped familial juvenile ALS linked region (ALS5) (43). Although a recent report suggests that mutations in Spatacsin (SPG11) are causative of ALS5, it is not clear if SPG11 mutations explain all available ALS5-linked kindreds (43–45). Interestingly, a recent report identified ZFP106 as a novel factor regulating the initiation of transcription by targeting RNA-polymerase I to the promoter region of ribosomal RNA genes (5). This, together with ZFP106 proposed localization to the nucleolus links ZFP106 function with nucleolus function and RNA metabolism abnormalities, which together with mitochondrial dysfunction, are three proposed common pathomechanisms leading to ALS (46,47).
In conclusion, our findings indicate that Zfp106 deficiency impairs mitochondrial function and is required for the maintenance and survival of specific motor and sensory neurons in vivo. Although no ZFN106 mutations have yet been found in human disease, the phenotype of Zfp106−/− mice, comprising spinal cord specific neuronal deficits and mitochondrial dysfunction, raises the intriguing and novel possibility that ZFP106 is involved in neurodegenerative disease.
Materials and Methods
Mice
Zfp106−/− mice (Zfp106tm1a(KOMP)Wtsi) were generated through the Knockout Mouse Project (KOMP), which aims to produce embryonic stem cells with knockouts in a number of mouse genes all of which will be publically available (http://www.komp.org/geneinfo.php?geneid=87357). The Zfp106−/− mice were generated by targeting C57BL/6N ES cells using a knockout first, promoter-less allele (tm1a(KOMP)Wtsi) (http://www.komp.org/alleles.php#conditional-promoterless-csd). The construct targeted Zfp106 intron 1, inserting a Neomycin resistance and a LacZ reporter cassette flanked by flippase recombinase (Flp) target sites (FRT). LoxP sites were also engineered flanking Zfp106 exon 2, allowing for the conversion to a conditional allele via Flp recombination, followed by crossing to appropriate Cre lines. We confirmed the location of the tm1a(KOMP)Wtsi allele in the Zfp106 gene of Zfp106−/− mice using Southern blotting (data not shown).
WT, Zfp106+/− and Zfp106−/− mice were on a hybrid C57BL/6NTac/Den;C57BL/6J-Tyrc-Brd;C57BL/6N background and assessed as littermates. Experiments were performed blind to genotype and the humane endpoint was defined as a loss of 20% body weight or an inability to right after 20 s. Animals were assessed daily and weighed weekly.
Behavioural analysis
WT, Zfp106+/− and Zfp106−/− littermate female only mice were assessed for grip strength (BioSeb, Chaville, France) of all four limbs, as per the manufacturer's instructions. Grip strength readings were taken twice each at 6- and 13-weeks of age and the mean of both values for each time point per animal was used. Cohort sizes per time point were as follows: 6-weeks: 16 WT, 16 Zfp106+/− and 14 Zfp106−/− animals; 13-weeks: 13 WT, 9 Zfp106+/− and 7 Zfp106−/− animals.
WT, Zfp106+/− and Zfp106−/− littermate female mice were placed on an accelerating rotarod (Ugo Basile) twice a day, three times a week, at 6- and 12-week time points, as described previously (48). Cohort sizes per time point were as follows: 7-weeks: 6 WT, 10 Zfp106+/− and 14 Zfp106−/− animals; 12-weeks: 7 WT, 8 Zfp106+/− and 8 Zfp106−/− animals.
Modified SHIRPA methods were performed as described previously (10,11). Tremors were qualitatively assessed by observation in a viewing jar and recorded as having tremors (1) or not (0). Limb grasping was assessed as either clasping one or more limbs (1) or not (0). The wire manoeuvre was assessed by hanging animals from a wire by their forelimbs and recording the degree to which they were able to hold their body weight. Animals were only scored as defective if they could not hang onto the wire for more than 1 s. Negative geotaxis involves animals walking down a vertical grate. Animals that slipped down at least half of the wire grate were scored as defective. Mice were subject to modified SHIRPA analysis at 6-weeks of age and included: 15 WT, 15 Zfp106+/− and 17 Zfp106−/− animals.
WT, Zfp106+/− and Zfp106−/− littermate female mice were monitored in an open field paradigm (Noldus Ethovision 3.1). Animals were placed in an arena and monitored using Ethovision 3.1 tracking software for 30 min at 7- and 14-week time points. The parameters distance moved (cm) and velocity (cm/s) were averaged per genotype and time point. Cohort sizes per time point were as follows: 7 weeks: 15 WT, 13 Zfp106+/− and 14 Zfp106−/− animals; and 14 weeks: 5 WT, 9 Zfp106+/− and 6 Zfp106−/− animals.
WT, Zfp106+/− and Zfp106−/− littermate female mice accuracy of paw placement was analysed using Locotronic® (Intelli-Bio, France). Briefly, animals begin in a starting area, illuminated with white light and move down a corridor consisting of a flat ladder (bars: 3 mm diameter, spaced by 7 mm) with three infrared sensors above and below each bar space, which registered movement errors 100 times a second. Animals were assessed three times each at 7- and 14-weeks of age; averages were taken per animal and at each time point. Cohort sizes per time point were as follows: 7 weeks: 10 WT, 12 Zfp106+/− and 14 Zfp106−/− animals; and 14 weeks: 6 WT, 7 Zfp106+/− and 5 Zfp106−/− animals.
Physiological assessment of hindlimb muscle force, motor units and FI
Muscle force, motor unit number and FI for female WT, Zfp106+/− and Zfp106−/− littermates were examined at 7- and 15-weeks of age, as described previously (49). Cohort sizes per time point were as follows: 7 weeks: 5 WT, 6 Zfp106+/− and 6 Zfp106−/− animals; and 15 weeks: 5 WT, 5 Zfp106+/− and 6 Zfp106−/− animals. Briefly, mice were anesthetized (4.5% chlorohydrate, 1 ml/100 g of body weight), the distal tendons of the TA and EDL muscles in both hindlimbs attached to isometric force transducers (Dynamometer UFI Devices, Welwyn Garden City, UK), and the sciatic nerve exposed and sectioned. Isometric contractions were elicited by stimulating the nerve to the TA or EDL using squarewave pulses of 0.02 ms duration at supra-maximal intensity, via silver wire electrodes. Contractions were elicited by trains of stimuli at frequencies of 40, 80 and 100 Hz and measured using Scope software. The number of motor units innervating the EDL was determined by stimulating the motor nerve with stimuli increasing intensity, resulting in incremental increases in twitch tension due to successive recruitment of motor axons. The number of increments was counted giving an estimate of the number of functional motor units present in EDL muscle. The FI for the EDL muscle was examined by stimulating the EDL at 40 Hz for 250 ms every second for 180 s. Contractions were recorded with a pen recorder (Lectromed Multitrace 2) and the FI calculated as a measure of muscle fatigability and expressed as a ratio: FI = Ft180/Ft0.
Morphological assessment of muscle and neuronal tissue
Ten micrometer cross-sections of frozen gastrocnemius, soleus and EDL were examined with H&E, Masson's trichrome and NADH-TR activity to determine the oxidative capacity of the muscle fibres, as described previously (50). Then, 10 μm cross-sections of the soleus were subjected to ATPase staining (acid preincubation at pH 4.47) (51) and the total number of fibres and proportion of fibre types within the soleus were assessed (52). Muscle morphology was analysed in WT and Zfp106−/− littermates at 6- and 14-weeks of age (male, n = 3 per genotype).
For assessment of motor neuron survival, the animals were terminally anaesthetized with 4.5% chlorohydrate (1 ml/100 g of body weight), transcardially perfused with 4% PFA and the lumbar region of the spinal cord removed, post-fixed in 4% PFA and cryopreserved in 30% sucrose. Serial 20 μm transverse sections were cut on a cryostat, stained with gallocyanin, a Nissl stain (53) and the number of positively stained motor neurons in the sciatic motor pool of every third section between the L3 and L6 levels of the spinal cord was counted, as previously described (12,13). Only large, polygonal neurons with a clearly identifiable nucleus and nucleolus were included in counts. This protocol avoids the possibility of counting the same cells twice in consecutive sections. A total of 40 sections were examined per spinal cord and at least five female mice were analysed from each experimental group.
For analysis of ventral and dorsal roots, DRG, and corticospinal tracts animals were perfused transcardially with saline followed with 3% glutaraldehyde in 0.05 M sodium cacodylate. Right L4 and L5 DRG with the attached dorsal and ventral roots, and spinal cords were dissected, post-fixed in 1% osmium tetroxide (Agar Scientific) and processed into araldite CY212 epoxy resin (Agar Scientific) through graded alcohols and propylene oxide using a standard protocol. Semi-thin sections (0.8 μm) were cut on an Ultracut E ultra-microtome (Leica), stained with 1% toluidine blue containing 1% borax (BDH), and examined with a Dialux light microscope. Neuronal morphology was analysed from 15-week-old WT and Zfp106−/− littermates (female, n = 3 per genotype).
Axon morphology
Images of three semi-thin sections at P9 from lumbar L4 and L5 dorsal and ventral roots and sciatic nerve sections were selected randomly per animal. Three animals per genotype were used. To calculate axon diameter and g-ratios of myelinated axons, three random regions from each picture were analysed. All myelinated axons within each region were measured to calculate the g-ratio and axon diameter. In total, approximately 40 axons within each section and more than 120 axons per animal were analysed using AxioVision LE Rel. 4.2 software. The examiner was blind to genotype.
Electron microscopy
WT and Zfp106−/− littermates were perfused as above (3% glutaraldehyde in 0.05 M sodium cacodylate) and dorsal roots dissected. Ultrathin sections 80 nm thick were cut from selected areas of the tissue, stained with uranyl acetate and lead citrate, and examined at 80 kV in a Philips CM10 electron microscope. Images were taken using a Megaview 3 digital camera and Olympus iTEM software. Morphology was analysed from 15-week-old WT and Zfp106−/− littermates (female, n = 3 per genotype).
Immunocytochemistry
For the analysis of brain histopathology 15-week-old WT and Zfp106−/− mice were perfused transcardially with saline followed with 4% PFA, brain transferred to formalin and embedded in paraffin wax. Staining with H&E and anti- GFAP, IBA-1, p62, MBP and neurofilament antibodies was undertaken using a Ventana automated immunohistochemical staining machine (Ventana Medical Systems, Tuscon, AZ, USA). Paraffin-embedded sections once incubated with appropriate secondary antibodies were developed using 3,30-diaminobenzidine and counterstained with haematoxylin.
Twenty micrometer transverse sections from the lumbar region of fixed spinal cords collected onto glass slides were immuno-fluorescently stained with anti-GFAP (Cy-conjugated mouse monoclonal, Sigma; used at 1:1000) and anti-IBA1 (rabbit polyclonal, Abcam; used at 1:500), visualized using a secondary AlexaFluor488-conjugated antibody (Invitrogen) and counterstained with NeuroTrace® 435⁄455 fluorescent Nissl stain (Invitrogen). A C-terminal p62 antibody (Progen, Germany; 1:300) was also used to stain lumbar spinal cord sections. Confocal images were taken using a Zeiss 710 microscope. Active cell death was analysed using TUNEL staining (Promega) following manufacturer recommendations.
Immunoblot
Fresh frozen spinal cords from female mice at 6 and 16 weeks of age were homogenized in RIPA buffer with protease inhibitors (complete mini EDTA-free, Roche). After centrifuging the homogenate at 12 000×g at 4°, the supernatant was used for western-blot analysis using pre-cast SDS gels (Invitrogen). The primary antibodies assessed included rabbit anti-actin (Sigma), LC3II (Nanotools) and ZFP106 (Novus, Santa Cruz, Bethyl). The Odyssey infrared imaging system (Li-Cor) was used for band detection and quantification.
RNA-seq
Whole spinal cords from male, 6-week old Zfp106tm1a(KOMP)Wtsi mice (WT, n = 6; Zfp106−/−, n = 5) were homogenized and total RNA extracted using an RNeasy fibrous tissue extraction kit (Qiagen). RNA integrity was confirmed and concentration measured (∼0.2 mg/ml) using a Bioanalyzer (Agilent). cDNA libraries were created from 5 μg total RNA as follows. Using a TruSeq RNA Sample Prep v2 kit (Illumina), poly-A tailed RNA (mRNA) was purified from total RNA using an oligo dT magnetic bead pull-down. The resulting mRNA was fragmented using metal ion-catalysed hydrolysis and random priming used to synthesize double-stranded cDNA. End repair was performed with a combination of fill-in reactions and exonuclease activity to produce blunt ends. A single ‘A’ base was added to blunt ends followed by ligation to Illumina Paired-End Sequencing adapters containing unique index sequences, allowing samples to be pooled. The resulting libraries were amplified through 10 cycles of PCR using KAPA Hifi Polymerase, products were pooled based on a post-PCR Agilent Bioanalyzer, then the pool was size-selected using the LabChip XT Caliper (200–300 bp range). The multiplexed library was sequenced on the Illumina HiSeq 2000 (75 bp paired-end read length) aiming for >3 Gigabases of data per sample. Sequencing reads were aligned against the NCBI build 37.2 of the Mus musculus reference genome using the software TopHat (54). The TopHat alignment incorporated the reference genes annotations (GTF format) provided as part of the Illumina iGenomes project. Duplicate reads were filtered using the PICARD tools. Read counts per gene and per exon were computed using the python scripts provided with the DESeq/DEXSeq packages. Differential expression at the gene level was computed using the R package DESeq (55) and at the exon level using the R package DEXSeq (56).
qPCR
Gene expression of Zfp106 from 6-week-old WT and Zfp106−/− mice (female, n = 3 per genotype): ∼20–25 mg of brain or spinal cord was homogenized and RNA extracted using a Qiagen RNeasy fibrous tissue extraction kit. 200 ng of total RNA was used in a 10 μl reaction using the TaqMan RNA-to-CT One Step kit (Applied Biosystems). A TaqMan probe flanking the Zfp106 splice acceptor region was used with GAPDH as an endogenous control (Applied Biosystems) in a multiplex primer-limited reaction. Reactions were performed in triplicate on an ABI Viia7 qPCR machine and analysed with Viia7 1.1 software. Gene expression data for Zfp106−/− tissues were analysed using the ΔΔCT method and normalized using GAPDH as endogenous reference gene (which expression is not changed by Zfp106 deficiency via RNA-seq, data not shown) relative to WT tissues (57).
Northern blot
Total mRNA was isolated from brains and spinal cords using the Qiagen RNeasy lipid kit. RNA concentration was estimated using Nanodrop (Thermo scientific). 3′UTR probes were cloned from WT brain or spinal cord genomic DNA into a TOPO-TA (Invitrogen) vector by PCR. Digoxigenin-labeled antisense RNA 3′UTR probes were generated through in vitro transcription (Roche). RNA and an RNA ladder (Invitrogen) were run on formaldehyde gels (Ambion) for 2–3 h, then transferred upright for 4 h to N+ nylon membrane. RNA was crosslinked with a stratalinker, then the membrane was pre-hybridized with Ambion hybridization solution at 68°C. Probe was denatured and added to hybridization solution, and hybridization was carried out overnight. After membrane washings, CDP-Star development solution was used to detect bound probe. Membranes were exposed to X-ray film and developed.
Embryonic motor neuron culture
Motor neuron cultures were prepared from WT, Zfp106+/− and Zfp106−/− littermate embryos as previously described (58). Briefly, embryonic spinal cords (E13.5) were removed, ventral horns isolated, motor neurons plated at 40 000 cells/cm2 onto cover-slips; cover-slips were coated with 1.5 mg/ml polyornithine (Sigma Aldrich) in sterile water for a minimum of 2 h followed with 1 mg/ml laminin (Sigma Aldrich) in L-15 medium (Sigma Aldrich) for 2 h. Motor neurons were maintained in complete neurobasal medium (CNB); 200 ml of which contains: 191 ml neurobasal medium, 4 ml B27 supplement (1 unit/ml; Gibco), 4 ml horse serum (PAA Laboratories), 500 µl 0.5 mm l-glutamine (Gibco), 100 µl 0.05% 2-mercaptoethanol (Gibco), 20 µl ciliarly neurotrophic factor (CNTF; 500 pg/ml; Alomone Labs), 2 µl glial-derived neurotrophic factor (GDNF; 100 pg/ml; Alomone Labs), 2 µl brain-derived neurotrophic factor (BDNF; 100 pg/ml), 2 ml P/S, in a 37°C, 5% CO2 humidified incubator for a minimum of 7 days.
Mitochondrial function
Embryonic motor neurons (identified as having at least three processes) were loaded for 40 min with 20 nm TMRM (Molecular Probes) in a HEPES-buffered salt solution (HBSS) composed (mm): 156 NaCl, 3 KCl, 2MgSO4, 1.25 KH2PO4, 2 CaCl2, 10 glucose and 10 HEPES, pH adjusted to 7.3 with NaOH. The dye was present at the same concentration in all solutions throughout the experiment.
TMRM measurements were made using a Zeiss 710 CLSM equipped with a META detection system and a 40× oil immersion objective. Illumination intensity was kept to a minimum (at 0.1–0.2% of laser output) to avoid phototoxicity and the pinhole set to give an optical slice of ∼2 μm. TMRM was excited using the 565 nm laser line and fluorescence measured above 580. FAD++ autofluorescence was excited at 458 and measured at 510–530 nm.
The autofluorescence of NADH (and NAD(P)H) in motor neuron cultures was imaged using an epifluorescence inverted microscope equipped with a 40× fluorite objective. Excitation light was provided by a Xenon arc lamp, and the beam passed through a monochromator (Cairn Research, UK). Emitted fluorescence light was reflected through a long-pass filter to a cooled CCD camera (Retiga, QImaging, Canada) and digitized to 12 bit resolution. All imaging data were collected and analysed using software from Andor (UK). The blue autofluorescence emitted by the pyridine nucleotides NADH and NAD(P)H in their reduced form was excited with a 360 nm and emission was measured at 435–485 nm.
For assessment of the redox state, the dynamic range of NADH autofluorescence was measured. The dynamic range of the signals was defined by obtaining the maximally oxidized signal following the response to 1 μm trifluorocarbonylcyanide phenylhydrazone (FCCP, which stimulates maximal respiration and fully oxidizes the mitochondrial NADH pool). In these conditions, mitochondrial NADH is taken as 0%. The maximally reduced signal was then defined as the response to 1 mm sodium cyanide (NaCN) which fully inhibits respiration), preventing NADH oxidation, and so promoting maximal NADH reduction. In these conditions, mitochondrial NADH is taken as 100%.
All data presented were obtained from at least five cover-slips and two to three different cell preparations.
Statistical analysis
An ANOVA test was used to compare between WT, Zfp106+/− and Zfp106−/− genotypes per time point followed by Bonferroni's multiple comparisons testing correction for weight, grip strength, rotarod, open field parameters distance moved and velocity, soleus muscle fibre types, and mitochondrial data. The Mann–Whitney test was used to compare WT with Zfp106−/− littermates per time point or Zfp106−/− cohorts between time points for TA and EDL maximum muscle force, surviving motor units innervating the EDL, FI and motor neuron survival. Cumulative frequencies for axon distribution were compared using the Kolmogorov–Smirnov test. Two-tailed tests were used in all instances and significance level was set at P < 0.05.
Ethical approval
All experiments were performed under licence from the UK Home Office regulations. All applicable international, national and/or institutional guidelines for the care and use of animals were followed. All procedures performed in mice were in accordance with the ethical standards of the institutions or practice at which the studies were conducted.
Supplementary Material
Funding
This work was funded by the UK Medical Research Council (MRC) to A.A.-A. and a Motor Neurone Disease Association (MNDA) project grant to A.A.-A. and EMCF. D.L.H.B. is a Wellcome Trust Senior Clinical Scientist Fellow and P.F. is a MRC/MNDA Lady Edith Wolfson Clinician Scientist Fellow. Funding to pay the Open Access publication charges for this article was provided by the MRC grant number: MC_UP_A390_1106.
Supplementary Material
Acknowledgements
We are indebted to Jim Humphries, Jenny Corrigan, Liz Darley, Elizabeth Joynson, Natalie Walters, Sara Wells and the whole necropsy, histology, genotyping and MLC ward 6 teams at MRC Harwell for excellent technical assistance. We thank the staff of the WTSI Illumina Bespoke Team for the RNA-seq data, the Sanger Mouse Genetics Project for the initial mouse characterization and Dr David Adams for critical reading of the manuscript. We also thank KOMP for the mouse embryonic stem cells carrying the knockout first promoter-less allele (tm1a(KOMP)Wtsi) within Zfp016.
Conflict of Interest statement. None declared.
References
- 1.Grasberger H., Bell G.I. (2005) Subcellular recruitment by TSG118 and TSPYL implicates a role for zinc finger protein 106 in a novel developmental pathway. Int. J. Biochem. Cell. Biol., 37, 1421–1437. [DOI] [PubMed] [Google Scholar]
- 2.Grasberger H., Ye H., Mashima H., Bell G.I. (2005) Dual promoter structure of ZFP106: regulation by myogenin and nuclear respiratory factor-1. Gene, 344, 143–159. [DOI] [PubMed] [Google Scholar]
- 3.Zuberi A.R., Christianson G.J., Mendoza L.M., Shastri N., Roopenian D.C. (1998) Positional cloning and molecular characterization of an immunodominant cytotoxic determinant of the mouse H3 minor histocompatibility complex. Immunity, 9, 687–698. [DOI] [PubMed] [Google Scholar]
- 4.Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R. 3rd, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y. et al. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science, 316, 1160–1166. [DOI] [PubMed] [Google Scholar]
- 5.Ide S., Dejardin J. (2015) End-targeting proteomics of isolated chromatin segments of a mammalian ribosomal RNA gene promoter. Nat. Commun., 6, 6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatayama M., Aruga J. (2010) Characterization of the tandem CWCH2 sequence motif: a hallmark of inter-zinc finger interactions. BMC Evol. Biol., 10, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mackay J.P., Crossley M. (1998) Zinc fingers are sticking together. Trends Biochem. Sci., 23, 1–4. [DOI] [PubMed] [Google Scholar]
- 8.Yaffe M.B., Elia A.E. (2001) Phosphoserine/threonine-binding domains. Curr. Opin. Cell Biol., 13, 131–138. [DOI] [PubMed] [Google Scholar]
- 9.Lloyd K.C. (2011) A knockout mouse resource for the biomedical research community. Ann. N Y Acad. Sci., 1245, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rogers D.C., Fisher E.M., Brown S.D., Peters J., Hunter A.J., Martin J.E. (1997) Behavioral and functional analysis of mouse phenotype: SHIRPA, a proposed protocol for comprehensive phenotype assessment. Mamm. Genome, 8, 711–713. [DOI] [PubMed] [Google Scholar]
- 11.Rogers D.C., Peters J., Martin J.E., Ball S., Nicholson S.J., Witherden A.S., Hafezparast M., Latcham J., Robinson T.L., Quilter C.A. et al. (2001) SHIRPA, a protocol for behavioral assessment: validation for longitudinal study of neurological dysfunction in mice. Neurosci. Lett., 306, 89–92. [DOI] [PubMed] [Google Scholar]
- 12.Kalmar B., Novoselov S., Gray A., Cheetham M.E., Margulis B., Greensmith L. (2008) Late stage treatment with arimoclomol delays disease progression and prevents protein aggregation in the SOD1 mouse model of ALS. J. Neurochem., 107, 339–350. [DOI] [PubMed] [Google Scholar]
- 13.Kieran D., Greensmith L. (2004) Inhibition of calpains, by treatment with leupeptin, improves motoneuron survival and muscle function in models of motoneuron degeneration. Neuroscience, 125, 427–439. [DOI] [PubMed] [Google Scholar]
- 14.Bartlett B.J., Isakson P., Lewerenz J., Sanchez H., Kotzebue R.W., Cumming R.C., Harris G.L., Nezis I.P., Schubert D.R., Simonsen A. et al. (2011) p62, Ref(2)P and ubiquitinated proteins are conserved markers of neuronal aging, aggregate formation and progressive autophagic defects. Autophagy, 7, 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abramov A.Y., Duchen M.R. (2010) Impaired mitochondrial bioenergetics determines glutamate-induced delayed calcium deregulation in neurons. Biochim. Biophys. Acta, 1800, 297–304. [DOI] [PubMed] [Google Scholar]
- 16.Duchen M.R., Surin A., Jacobson J. (2003) Imaging mitochondrial function in intact cells. Methods Enzymol., 361, 353–389. [DOI] [PubMed] [Google Scholar]
- 17.Campanella M., Casswell E., Chong S., Farah Z., Wieckowski M.R., Abramov A.Y., Tinker A., Duchen M.R. (2008) Regulation of mitochondrial structure and function by the F1Fo-ATPase inhibitor protein, IF1. Cell Metab., 8, 13–25. [DOI] [PubMed] [Google Scholar]
- 18.Biswas M., Chan J.Y. (2010) Role of Nrf1 in antioxidant response element-mediated gene expression and beyond. Toxicol. Appl. Pharmacol., 244, 16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang W.T., Chen H.I., Chiou R.J., Chen C.Y., Huang A.M. (2005) A novel function of transcription factor alpha-Pal/NRF-1: increasing neurite outgrowth. Biochem. Biophys. Res. Commun., 334, 199–206. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi A., Tsukide T., Miyasaka T., Morita T., Mizoroki T., Saito Y., Ihara Y., Takashima A., Noguchi N., Fukamizu A. et al. (2011) Central nervous system-specific deletion of transcription factor Nrf1 causes progressive motor neuronal dysfunction. Genes Cells, 16, 692–703. [DOI] [PubMed] [Google Scholar]
- 21.Lee C.S., Lee C., Hu T., Nguyen J.M., Zhang J., Martin M.V., Vawter M.P., Huang E.J., Chan J.Y. (2011) Loss of nuclear factor E2-related factor 1 in the brain leads to dysregulation of proteasome gene expression and neurodegeneration. Proc. Natl. Acad. Sci. USA, 108, 8408–8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lezi E., Swerdlow R.H. (2012) Mitochondria in neurodegeneration. Adv. Exp. Med. Biol., 942, 269–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee I., Bender E., Arnold S., Kadenbach B. (2001) New control of mitochondrial membrane potential and ROS formation—a hypothesis. Biol. Chem., 382, 1629–1636. [DOI] [PubMed] [Google Scholar]
- 24.Yao Z., Gandhi S., Burchell V.S., Plun-Favreau H., Wood N.W., Abramov A.Y. (2011) Cell metabolism affects selective vulnerability in PINK1-associated Parkinson's disease. J. Cell Sci., 124, 4194–4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abramov A.Y., Smulders-Srinivasan T.K., Kirby D.M., Acin-Perez R., Enriquez J.A., Lightowlers R.N., Duchen M.R., Turnbull D.M. (2010) Mechanism of neurodegeneration of neurons with mitochondrial DNA mutations. Brain, 133, 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kussmaul L., Hirst J. (2006) The mechanism of superoxide production by NADH:ubiquinone oxidoreductase (complex I) from bovine heart mitochondria. Proc. Natl. Acad. Sci. USA, 103, 7607–7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valko M., Leibfritz D., Moncol J., Cronin M.T., Mazur M., Telser J. (2007) Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol., 39, 44–84. [DOI] [PubMed] [Google Scholar]
- 28.Bilsland L.G., Nirmalananthan N., Yip J., Greensmith L., Duchen M.R. (2008) Expression of mutant SOD1 in astrocytes induces functional deficits in motoneuron mitochondria. J. Neurochem., 107, 1271–1283. [DOI] [PubMed] [Google Scholar]
- 29.Faes L., Callewaert G. (2011) Mitochondrial dysfunction in familial amyotrophic lateral sclerosis. J. Bioenerg. Biomembr., 43, 587–592. [DOI] [PubMed] [Google Scholar]
- 30.Joyce P.I., Fratta P., Fisher E.M., Acevedo-Arozena A. (2011) SOD1 and TDP-43 animal models of amyotrophic lateral sclerosis: recent advances in understanding disease toward the development of clinical treatments. Mamm. Genome, 22, 420–448. [DOI] [PubMed] [Google Scholar]
- 31.Acsadi G., Lee I., Li X., Khaidakov M., Pecinova A., Parker G.C., Huttemann M. (2009) Mitochondrial dysfunction in a neural cell model of spinal muscular atrophy. J. Neurosci. Res., 87, 2748–2756. [DOI] [PubMed] [Google Scholar]
- 32.Burchell V.S., Gandhi S., Deas E., Wood N.W., Abramov A.Y., Plun-Favreau H. (2010) Targeting mitochondrial dysfunction in neurodegenerative disease: Part II. Expert Opin. Ther. Targets, 14, 497–511. [DOI] [PubMed] [Google Scholar]
- 33.Cuesta A., Pedrola L., Sevilla T., Garcia-Planells J., Chumillas M.J., Mayordomo F., LeGuern E., Marin I., Vilchez J.J., Palau F. (2002) The gene encoding ganglioside-induced differentiation-associated protein 1 is mutated in axonal Charcot-Marie-Tooth type 4A disease. Nat. Genet., 30, 22–25. [DOI] [PubMed] [Google Scholar]
- 34.Zuchner S., Mersiyanova I.V., Muglia M., Bissar-Tadmouri N., Rochelle J., Dadali E.L., Zappia M., Nelis E., Patitucci A., Senderek J. et al. (2004) Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat. Genet., 36, 449–451. [DOI] [PubMed] [Google Scholar]
- 35.Pitceathly R.D., Murphy S.M., Cottenie E., Chalasani A., Sweeney M.G., Woodward C., Mudanohwo E.E., Hargreaves I., Heales S., Land J. et al. (2012) Genetic dysfunction of MT-ATP6 causes axonal Charcot-Marie-Tooth disease. Neurology, 79, 1145–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baxter R.V., Ben Othmane K., Rochelle J.M., Stajich J.E., Hulette C., Dew-Knight S., Hentati F., Ben Hamida M., Bel S., Stenger J.E. et al. (2002) Ganglioside-induced differentiation-associated protein-1 is mutant in Charcot-Marie-Tooth disease type 4A/8q21. Nat. Genet., 30, 21–22. [DOI] [PubMed] [Google Scholar]
- 37.Loiseau D., Chevrollier A., Verny C., Guillet V., Gueguen N., Pou de Crescenzo M.A., Ferre M., Malinge M.C., Guichet A., Nicolas G. et al. (2007) Mitochondrial coupling defect in Charcot-Marie-Tooth type 2A disease. Ann. Neurol., 61, 315–323. [DOI] [PubMed] [Google Scholar]
- 38.Ouvrier R., Grew S. (2010) Mechanisms of disease and clinical features of mutations of the gene for mitofusin 2: an important cause of hereditary peripheral neuropathy with striking clinical variability in children and adults. Dev. Med. Child Neurol., 52, 328–330. [DOI] [PubMed] [Google Scholar]
- 39.Shimazaki H., Takiyama Y., Ishiura H., Sakai C., Matsushima Y., Hatakeyama H., Honda J., Sakoe K., Naoi T., Namekawa M. et al. (2012) A homozygous mutation of C12orf65 causes spastic paraplegia with optic atrophy and neuropathy (SPG55). J. Med. Genet., 49, 777–784. [DOI] [PubMed] [Google Scholar]
- 40.Gabreels-Festen A.A., Joosten E.M., Gabreels F.J., Jennekens F.G., Gooskens R.H., Stegeman D.F. (1991) Hereditary motor and sensory neuropathy of neuronal type with onset in early childhood. Brain, 114 (Pt 4), 1855–1870. [DOI] [PubMed] [Google Scholar]
- 41.Ouvrier R.A., McLeod J.G., Morgan G.J., Wise G.A., Conchin T.E. (1981) Hereditary motor and sensory neuropathy of neuronal type with onset in early childhood. J. Neurol. Sci., 51, 181–197. [DOI] [PubMed] [Google Scholar]
- 42.Baets J., Deconinck T., De Vriendt E., Zimon M., Yperzeele L., Van Hoorenbeeck K., Peeters K., Spiegel R., Parman Y., Ceulemans B. et al. (2011) Genetic spectrum of hereditary neuropathies with onset in the first year of life. Brain, 134, 2664–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hentati A., Ouahchi K., Pericak-Vance M.A., Nijhawan D., Ahmad A., Yang Y., Rimmler J., Hung W., Schlotter B., Ahmed A. et al. (1998) Linkage of a commoner form of recessive amyotrophic lateral sclerosis to chromosome 15q15-q22 markers. Neurogenetics, 2, 55–60. [DOI] [PubMed] [Google Scholar]
- 44.Daoud H., Zhou S., Noreau A., Sabbagh M., Belzil V., Dionne-Laporte A., Tranchant C., Dion P., Rouleau G.A. (2012) Exome sequencing reveals SPG11 mutations causing juvenile ALS. Neurobiol Aging, 33, 839, e835–839. [DOI] [PubMed] [Google Scholar]
- 45.Orlacchio A., Babalini C., Borreca A., Patrono C., Massa R., Basaran S., Munhoz R.P., Rogaeva E.A., St George-Hyslop P.H., Bernardi G. et al. (2010) SPATACSIN mutations cause autosomal recessive juvenile amyotrophic lateral sclerosis. Brain, 133, 591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ling S.C., Polymenidou M., Cleveland D.W. (2013) Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron, 79, 416–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haeusler A.R., Donnelly C.J., Periz G., Simko E.A., Shaw P.G., Kim M.S., Maragakis N.J., Troncoso J.C., Pandey A., Sattler R. et al. (2014) C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature, 507, 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Acevedo-Arozena A., Kalmar B., Essa S., Ricketts T., Joyce P., Kent R., Rowe C., Parker A., Gray A., Hafezparast M. et al. (2011) A comprehensive assessment of the SOD1G93A low-copy transgenic mouse, which models human amyotrophic lateral sclerosis. Dis. Model. Mech., 4, 686–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ricketts T., McGoldrick P., Fratta P., de Oliveira H.M., Kent R., Phatak V., Brandner S., Blanco G., Greensmith L., Acevedo-Arozena A. et al. (2014) A nonsense mutation in mouse Tardbp affects TDP43 alternative splicing activity and causes limb-clasping and body tone defects. PLoS One, 9, e85962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corrochano S., Mannikko R., Joyce P.I., McGoldrick P., Wettstein J., Lassi G., Raja Rayan D.L., Blanco G., Quinn C., Liavas A. et al. (2014) Novel mutations in human and mouse SCN4A implicate AMPK in myotonia and periodic paralysis. Brain, 137, 3171–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brooke M.H., Kaiser K.K. (1970) Muscle fiber types: how many and what kind? Arch. Neurol., 23, 369–379. [DOI] [PubMed] [Google Scholar]
- 52.Wang L.C., Kernell D. (2001) Quantification of fibre type regionalisation: an analysis of lower hindlimb muscles in the rat. J. Anat., 198, 295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kellett B.S. (1963) Gallocyanin-chrome alum: a routine stain for nissl substance in paraffin Sections. J. Med. Lab. Technol., 20, 196–198. [PubMed] [Google Scholar]
- 54.Trapnell C., Pachter L., Salzberg S.L. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics, 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anders S., Huber W. (2010) Differential expression analysis for sequence count data. Genome Biol., 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anders S., Reyes A., Huber W. (2012) Detecting differential usage of exons from RNA-seq data. Genome Res., 22, 2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Livak K.J., Schmittgen T.D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- 58.Joyce P.I., McGoldrick P., Saccon R.A., Weber W., Fratta P., West S.J., Zhu N., Carter S., Phatak V., Stewart M. et al. (2015) A novel SOD1-ALS mutation separates central and peripheral effects of mutant SOD1 toxicity. Human Mol. Genet., 24, 1883–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.