Abstract
The lipid phosphatase FIG4 is a subunit of the protein complex that regulates biosynthesis of the signaling lipid PI(3,5)P2. Mutations of FIG4 result in juvenile lethality and spongiform neurodegeneration in the mouse, and are responsible for the human disorders Charcot–Marie–Tooth disease, Yunis–Varon syndrome and polymicrogyria with seizures. We previously demonstrated that conditional expression of a wild-type FIG4 transgene in neurons is sufficient to rescue most of the abnormalities of Fig4 null mice, including juvenile lethality and extensive neurodegeneration. To evaluate the contribution of the phosphatase activity to the in vivo function of Fig4, we introduced the mutation p.Cys486Ser into the Sac phosphatase active-site motif CX5RT. Transfection of the Fig4Cys486Ser cDNA into cultured Fig4−/− fibroblasts was effective in preventing vacuolization. The neuronal expression of an NSE-Fig4Cys486Ser transgene in vivo prevented the neonatal neurodegeneration and juvenile lethality seen in Fig4 null mice. These observations demonstrate that the catalytically inactive FIG4 protein provides significant function, possibly by stabilization of the PI(3,5)P2 biosynthetic complex and/or localization of the complex to endolysosomal vesicles. Despite this partial rescue, later in life the NSE-Fig4Cys486Ser transgenic mice display significant abnormalities that include hydrocephalus, defective myelination and reduced lifespan. The late onset phenotype of the NSE-Fig4Cys486Ser transgenic mice demonstrates that the phosphatase activity of FIG4 has an essential role in vivo.
Introduction
FIG4 encodes a lipid phosphatase that regulates biogenesis of the signaling lipid PI(3,5)P2 (1–4). The important role of FIG4 in the mammalian nervous system was first indicated by the spongiform degeneration of the brain and peripheral nervous system (PNS) caused by a loss of function mutation of Fig4 in the plt mouse (5). A similar phenotype results from mutation of the associated protein VAC14 (6). FIG4 is localized on the cytoplasmic surface of endolysosomal vesicles in a complex that also includes the scaffold protein VAC14 and the PI(3)P kinase PIKFYVE (6–8). The close association between the kinase that generates PI(3,5)P2 from PI3P and the phosphatase that removes the 5-phosphate results in transient, spatially localized production of PI(3,5)P2 that is thought to regulate vesicle fusion and trafficking events.
In addition to its phosphatase activity toward the substrate PI(3,5)P2 (9), there is evidence that the FIG4 protein can stabilize the PI(3,5)P2 biosynthetic complex and contribute to localization at the surface of endolysosomal vesicles. Deletion of yeast FIG4 impairs the localization of VAC14 and PIKFYVE to the vacuole surface (6,9). The phosphatase-inactive yeast mutant fig4-1 (9) stabilizes the association of VAC14 and PIKFYVE (7). Mammalian FIG4 protein is destabilized in mutant mice lacking VAC14 (10), although VAC14 and PIKFYVE proteins are stable in mice lacking FIG4 (10,11). The human patient mutation p.Ile41Thr impairs interaction of FIG4 with VAC14 resulting in destabilization of the FIG4 protein (10). Thus the lethal phenotype of Fig4 null mice, which lack FIG4 protein, could result either from loss of FIG4 enzymatic activity or from loss of its stabilization functions, which indirectly impair the biosynthesis of PI(3,5)P2. To distinguish between these functions, we generated a mutant FIG4 with an active-site mutation that produces a stable FIG4 protein lacking catalytic activity. We evaluated the ability of this mutant to rescue cellular vacuolization and survival of the Fig4 null mouse.
Fig4 is a member of the Sac phosphatase family that contains the active-site motif CX5RT/S (12–17). The cysteine residue of the CX5RT/S motif participates in SN2 nucleophilic attack on the phosphate group of the substrate (18). Replacement of the active-site cysteine by serine inactivates enzymes of this class by preventing formation of a thiol-phosphate intermediate (19,20). Cysteine-to-serine mutations resulted in complete inactivation of the lipid phosphatases MTMR2 and SAC1, whose active sites are closely related to FIG4 (20,21). Cysteine-to-serine mutations of the CX5RT/S motif also inactivate protein tyrosine phosphatases (22–26) and dual substrate phosphatases (19,27,28). Most recently, the mutation p.Cys450Ser was used to inactivate fig4 in Drosophila (Bharadwaj and Lloyd, HMG co-submitted manuscript).
We generated the active-site mutation p.Cys486Ser in FIG4 and examined the effect in Fig4-deficient cells and mice. The partial rescue of the Fig4 null phenotype by the active-site mutant indicates that mammalian FIG4 has both phosphatase-dependent and phosphatase-independent roles that are essential in vivo.
Results
Fig4 with an active-site mutation rescues vacuolization of Fig4−/− fibroblasts
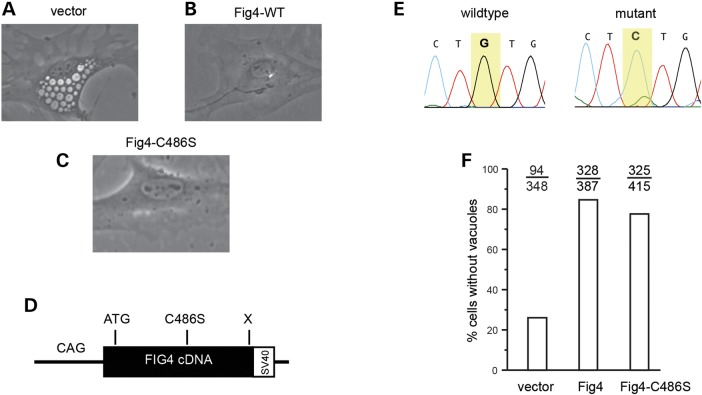
Fig4−/− fibroblasts accumulate large, cytoplasmic, fluid-filled vesicles derived from the endosome/lysosome pathway (5) (Fig. 1A). Transfection of wild-type Fig4 cDNA rescues vacuolization of null fibroblasts (Fig. 1B), and pathogenic mutations impair rescue (29,30). We introduced the p.Cys486Ser active-site mutation into the Fig4 cDNA (Fig. 1D and E). Primary fibroblasts cultured from Fig4−/− mice were transfected with the mutant and wild-type cDNA. The mutant cDNA was as effective as wild-type in correcting vacuolization of cultured cells, with >80% of null cells lacking vacuoles (Fig. 1C and F).
Figure 1.
Rescue of cell vacuolization by the Fig4 cDNA with an active-site mutation. (A) Vacuolated Fig4−/− fibroblasts in culture. (B) Representative cell after transfection of wild-type Fig4 cDNA. (C) Representative cell after transfection of Fig4Cys486Ser cDNA. (D) The structure of mutant Fig4 cDNA construct with chicken β-actin promoter. (E) Sanger sequence chromatogram of codon 486 from wild-type (TGT) and mutant (TCT) cDNAs. (F) Quantitation of rescue of vacuolization by wild-type Fig4 cDNA and cDNA containing the active-site mutation p.Cys486Ser. The number of vacuolated cells and total cells counted are indicated.
Generation of transgenic mice with neuronal expression of mutant Fig4
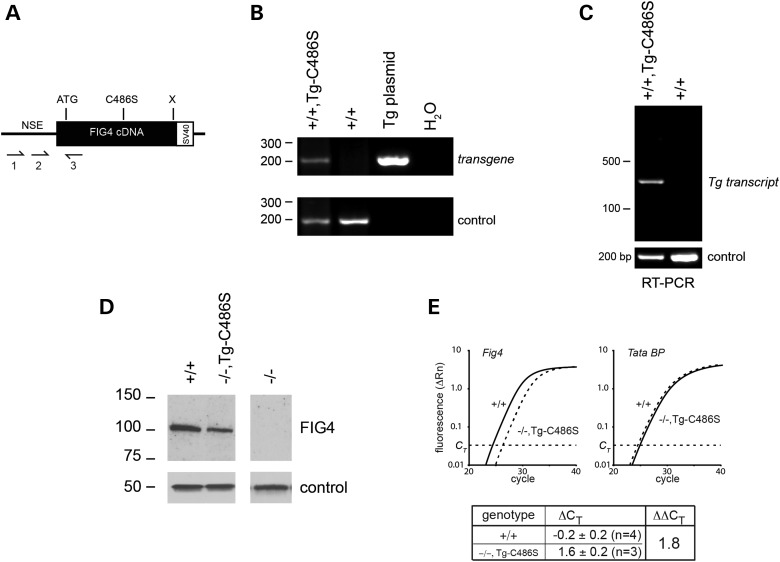
To examine the role of the mutant protein in vivo, the FIG4-Cys486Ser mutant cDNA was cloned downstream of the 4 kb NSE (neuron-specific enolase) fragment that we previously used for the expression of wild-type Fig4 (Fig. 2A) (32). The linearized cDNA construct was microinjected into fertilized eggs and founders carrying the transgene were identified by amplification of genomic DNA with transgene-specific primers (Fig. 2B). The transgene was transmitted through the germline to offspring in a cross between transgenic founders and Fig4+/− heterozygous null mice.
Figure 2.
Neuronal expression of the p.Cys486Ser Fig4 cDNA transgene. (A) The structure of the neuron-specific NSE transgene, with positions of PCR primers used for genotyping and RT-PCR. (B) Genotyping of mouse genomic DNA; control = Vac14 (6). (C) RT-PCR of brain RNA from transgenic and wild-type mice; control = Scn8a (31). (D) Western blot probed with monoclonal anti-FIG4 antibody; control = α-tubulin (32). The expression of FIG4 protein in the mutant is 45 ± 5% of endogenous wild-type expression (Image J statistics; Supplementary Material, Fig. S1). (E) Quantitation of brain RNA by qRT-PCR using a TaqMan gene expression assay.
To determine whether the transgene was expressed in the Fig4+/+,Tg+ offspring, brain RNA was prepared and the transgene transcript was amplified with a forward primer in the NSE noncoding exon and a reverse primer in exon 2 of Fig4. These primers span an intron, permitting distinction between spliced transcripts and genomic DNA. Expression of spliced transcript was detected in brain (Fig. 2C), but not in other tissues of mice from line 415. This expression pattern continued through adulthood and is consistent with the predicted neuronal specificity of the NSE promoter. Fig4+/−,Tg+ mice were crossed with Fig4+/− mice to generate Fig4−/−,Tg+ mice that are predicted to express mutant FIG4 in the absence of wild-type FIG4. Expression of Fig4 protein was detected by western blotting in brain of Fig4−/−,Tg+ mice (Fig. 2D). The expression of FIG4 protein in the mutant is 45 ± 5% of endogenous wild-type expression (n = 3) (Supplementary Material, Fig. S1). Expression of the transgene transcript was quantitated by qRT-polymerase chain reaction (PCR) on brain RNA from three Fig4−/−,Tg+ mice and three wild-type mice. Expression of the transgene mRNA was 29% of the endogenous level (ΔΔCT = 1.8) (Fig. 2E).
Rescue of postnatal growth and juvenile lethality by the mutant transgene
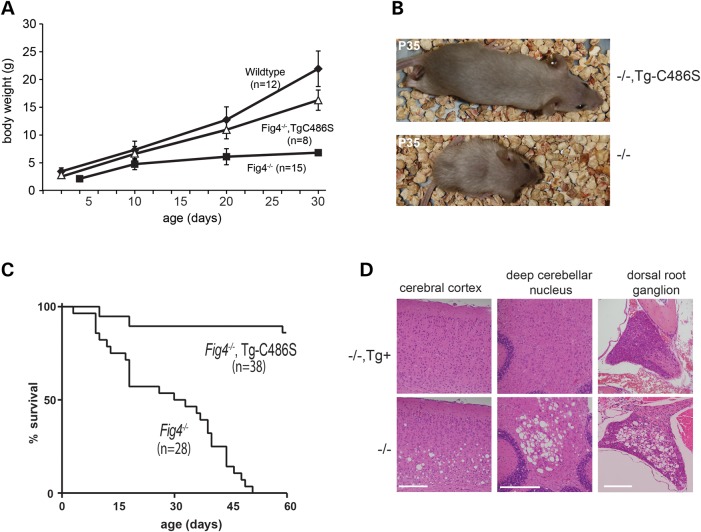
Fig4+/−,Tg+ mice were crossed with heterozygous Fig4+/− mice, and offspring were genotyped at 14 days of age. Twenty-five percent of offspring had the genotype Fig4−/− (31/115) and 50% of these carried the transgene (16/31), consistent with Mendelian predictions. The early growth impairment of Fig4−/− mice was almost completely rescued by the expression of the mutant transgene (Fig. 3A). The growth of these transgenic mice was comparable to previously described transgenic mice expressing wild-type Fig4 from the same promoter (32). At 30 days of age, Fig4−/−,Tg+ mice did not display the characteristic muscle wasting and small size observed for Fig4−/− mice (Fig. 3B). The juvenile lethality of Fig4−/− mice was also rescued by the mutant transgene (Fig. 3C). The mean survival of the control Fig4−/− non-transgenic littermates was 37 ± 11 days (n = 15), and none lived beyond 2 months of age. In contrast, 36/38 Fig4−/−,Tg+ mice expressing the mutant transgene survived beyond 2 months of age (Fig. 3C).
Figure 3.
Rescue of neurodegeneration, impaired growth and juvenile lethality by the Fig4Cys486Ser transgene. (A) Mice of the indicated genotypes were weighted daily from birth. (B) Rescue of body size at P35 by the mutant transgene. (C) Kaplan–Meier plot of survival for Fig4 null mice with and without the transgene. Open circles, currently surviving transgenic mice. (D) Rescue of neurodegeneration in CNS and PNS. Sections of brain and dorsal root ganglion were prepared at P30 from Fig4−/−, Tg+ and Fig4−/− littermates and stained with hematoxylin/eosin. The extensive in vivo vacuolization in the Fig4−/− null tissues is corrected by the mutant transgene.
Rescue of spongiform degeneration by the Fig4Cys486Ser transgene
Fig4−/− null mice exhibit spongiform degeneration in brain, spinal cord and dorsal root ganglia, with greatest severity in cortical layers 5 and 6, deep cerebellar nucleus, thalamus and dorsal root ganglia (5,10,32). To determine whether the p.Cys486Ser mutant protein could rescue these neurological phenotypes, we examined brain and dorsal root ganglia of Fig4−/−,Tg+ mice by light microscopy. At 4 weeks of age, the profound neurodegeneration seen in control non-transgenic littermates was not seen in mice expressing the Fig4Cys486Ser transgene (Fig. 3D). The extent of rescue by the mutant Fig4 was comparable to that described previously for the NSE-Fig4WT transgene (32).
Adult-onset phenotypes in Fig4−/−,TgCys486Ser mice
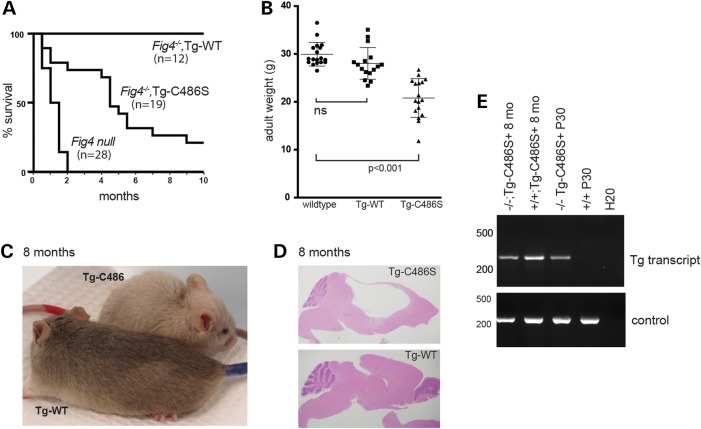
Although juvenile phenotypes were essentially rescued by the mutant transgene, adult Fig4−/−,TgCys486Ser mice exhibited reduced lifespan, with only 50% of mice surviving beyond 5 months of age (Fig. 4A). Expression of the transgene is maintained through 8 months of age (Fig. 4E). The reduced survival is in contrast to the long-term survival of mice expressing the wild-type NSE-Fig4WT transgene (Fig. 4A). Fig4−/−,TgCys486Ser adult mice were also smaller than wild-type littermates or Fig4−/−,TgWT mice (Fig. 4B). As they age, Fig4−/−,TgCys486Ser mice develop a hunched posture and domed head (Fig. 4C and Supplementary Material, Video). Dissection of the brain revealed enlarged ventricles and compression of cerebellar and cortical tissue that is indicative of high-pressure hydrocephalus (Fig. 4D). Hydrocephalus has been observed in Fig4 null mice (10) and patients with Yunis–Varon syndrome, who are also FIG4−/− (29,33,34).
Figure 4.
Shortened lifespan, hydrocephalus, and pigmentation defect in Fig4−/−,TgCys486Ser transgenic mice. (A) Reduced lifespan of transgenic mice carrying the mutant transgene. (B) Reduced body weight of Fig4−/− mice carrying the TgCys486Ser transgene at 2–3 months of age. (C) Diluted pigmentation in Fig4−/− transgenic mouse carrying Tg-C486 (red marker on tail) compared with corrected pigmentation in mouse carrying the wild-type Fig4 transgene (blue tail). (D) The saggital section of brains from Fig4−/− transgenic mice expressing mutant or wild-type FIG4 transgenes. H&E staining. (E) The expression of the mutant transgene in brain is maintained at 8 months of age (RT-PCR).
Fig4−/− mice have a diluted coat color due to clumping of melanosomes in the hair shaft (5). The NSE promoter is expressed in melanocytes in addition to neurons, and we showed previously that the NSE-Fig4WT transgene rescues the coat color dilution of Fig4 null mice (32). However, coat color was not restored in mice expressing the Fig4Cys486Ser transgene, whose diluted pigmentation mimics the Fig4−/− mouse (Fig. 4D).
Thus, the non-catalytic functions of FIG4 rescue some but not all of the abnormal phenotypes of the Fig4−/− null mouse.
Impaired myelination in Fig4−/−,TgCys486Ser mice
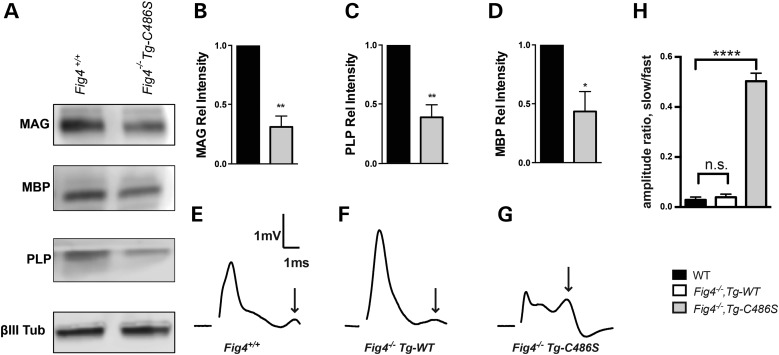
Myelination is defective in the central nervous system (CNS) and PNS of Fig4 null mice (5,34). Myelination is restored by expression of the NSE-Fig4WT transgene, which is expressed in neurons but not expressed in oligodendrocytes (32,35). To determine whether the TgCys486Ser transgene can restore CNS myelination, we examined the abundance of myelin proteins in brain membranes of Fig4−/−,TgCys486Ser mice at P21, an age when neurons appear fully rescued (Fig. 3D). The abundance of myelin-associated glycoprotein (MAG), myelin basic protein (MBP) and proteolipid protein (PLP) was reduced to <50% of wild-type levels in the Fig4−/−,TgCys486Ser mice (Fig. 5A and B).
Figure 5.
Impaired myelination and nerve conduction in Fig4−/−,TgCys486Ser transgenic mice. (A) Western blot of brain membrane proteins isolated at P21 and probed with antibodies to the myelin proteins MAG, MBP and PLP and the neuronal marker class III β-tubulin. (B–D) Quantitation of western blots normalized to βIII Tub. Results are shown as mean value ± SEM, unpaired two-tailed Student's t-test. *P < 0.03, ** < 0.01. (E–G) Representative traces of CAPs recorded at P21 reveal an increase in the population of slow conducting (non-myelinated) fibers (arrows) in optic nerve from Fig4−/−,Tg-Cys486Ser mice. (H) The amplitude ratio for slow conducting to fast conducting peaks identified in E–G. Results are shown as mean value ± SEM, unpaired Student's t-test. ****P < 0.0001.
To determine whether this reduced expression of myelin proteins results in a functional deficit in the Fig4−/−,TgCys486Ser transgenic mice, we measured nerve conduction in acutely isolated optic nerves. In Fig4−/− mice, compound action potentials (CAPs) reveal a dramatic reduction in the population of fast conducting fibers and a corresponding increase in the proportion of slowly conducting (non-myelinated) fibers in the optic nerve (35). This defect is corrected by the wild-type NSE-Fig4 transgene, which restores the normal CAP pattern seen in wild-type mice (Fig. 5E and F) (35). In contrast, the active-site mutated TgCys486Ser did not restore the normal CAP phenotype (Fig. 5G). As a result, the ratio of slow-amplitude (non-myelinated) to fast-amplitude (myelinated) conduction is much higher in mice expressing the mutant transgene (Fig. 5H), demonstrating the functional defect associated with impaired myelination of the optic nerve. Neuronal regulation of myelination in this system is thus dependent upon FIG4 catalytic activity.
Discussion
Genetic mutations affecting the generation of PI(3,5)P2, PI(4,5)P2 and PI(5)P are all deleterious to the nervous system. The work reported here provides additional support for the essential role of FIG4 in mammalian brain. Neuronal expression of Fig4 is necessary and sufficient for protection against spongiform neurodegeneration (32). We have found that this function can be provided by FIG4 protein containing an active-site mutation. The significant phenotypic rescue by the mutant protein indicates that non-catalytic functions of FIG4 are important in vivo. The stabilization of the PI(3,5)P2 biosynthetic complex by catalytically inactive FIG4 has been described in yeast (7,9), and a similar role could account for the function of catalytically inactive mammalian FIG4. By stabilizing the biosynthetic complex, the PI(3,5)P2 deficiency of Fig4 null cells may be partially ameliorated.
It is notable that a functional role for the catalytically inactive Drosophila fig4 has recently been described. Expression of the p.Cys450Ser active-site mutant was sufficient to rescue the extensive vacuolization of larval muscle in the fig4 null fly (Bharadwaj and Lloyd, HMG manuscript co-submitted). Thus in model systems including yeast, invertebrate and mouse, mutation of the active site does not completely abolish the biological activity of Fig4.
An example of the stabilization of a related enzyme by an inactive phosphatase is provided by the MTMR gene family, which dephosphorylates the third position of PI3P and PI(3,5)P2. Among the 14 MTMR paralogs in mammalian genomes, six have lost phosphatase activity (36). Mutations of either the catalytically active MTMR2 or the catalytically inactive MTMR13 can cause recessively inherited Charcot–Marie–Tooth disease type 4B (36). MTMR2 and MTMR13 interact directly on the membrane of the endolysosomal system (21,37), with MTMR13 acting as a scaffold to stabilize the activity of MTMR2.
In older mice, the essential role of the phosphatase activity of FIG4 becomes evident, as the catalytically inactive transgene does not rescue long-term survival. Myelination in particular is not rescued even in young mice, and the transgenic mice develop severe hydrocephalus like the uncorrected null mice.
The level of expression of the mutant protein is ∼45% of endogenous Fig4 protein (Supplementary Material, Fig. S1). For comparison, heterozygous Fig4+/− mice expressing 50% of wild-type protein level are unaffected (5), and transgenic mice expressing Fig4-I41T at 10% of wild-type protein level are unaffected (10). Thus, the level of expression of Cys486Ser protein is unlikely to be responsible for the incomplete rescue of late onset phenotypes.
Cysteine to serine mutations of the CX5RT/S phosphatase motif have been widely used to eliminate enzymatic activity (19–28). We were unable to directly evaluate the activity of the FIG4-Cys486Ser mutant because of the instability of recombinant FIG4 protein and the difficulty of purifying the protein from mammalian cells. Quantitation of the impact of the mutation on the cellular abundance of PI(3,5)P2 and PI5P would provide additional insight into the phenotype of the mutant mice described here.
Mutations of Fig4 and the scaffold protein Vac14 are proposed to reduce synaptic activity of neurons via altered post-synaptic trafficking of AMPA receptors between endosomes and the surface of dendritic spines (38). Pharmacological inhibition of PIKfyve also reduces synaptic strength (39). Protein effectors of PI(3,5)P2 that could mediate these synaptic effects include WD40 domain proteins, sorting nexins and class II formins (4). The lysosomal cation channels TRPML1, TPC1 and TPC2 are also directly regulated by PI(3,5)P2 (40–42). Reduced activity of the lysosomal cation channels secondary to reduced PI(3,5)P2 could lead to osmotic enlargement of the lysosome compartment that may underlie vacuolization of Fig4 null cells (43). In support of this model, rescue of vacuolization by overexpression of TRPML1 has been reported in Vac14 and Fig4 mutant cells (40,44). Similarly, mutation of drosophila TRPML1 generates a muscle vacuolization phenotype similar to that of the FIG4-deficient mutant (Bharadwaj and Lloyd, HMG manuscript co-submitted).
The work described here demonstrates that the defects leading to juvenile lethality of the Fig4 null mouse are substantially rescued by the Fig4Cys486Ser protein. Nonetheless, survival of adult mice was greatly impaired. Thus, both catalytic and non-catalytic roles of FIG4 are essential. The mechanisms underlying the catalytic and non-catalytic functions of FIG4 remain to be fully understood. The mouse model described here will be a useful system to better understand normal protein function and pathophysiology of human disorders caused by FIG4 deficiency.
Materials and Methods
Mutagenesis of Fig4 cDNA and generation of expression constructs
The full-length Fig4 cDNA was previously cloned downstream of the chicken β-actin (CAG) promoter (10). We mutated the phosphatase active-site motif CX5RT by the PCR extension method. A fragment containing the substitution c.1458G>C encoding the missense mutation p.Cys486Ser was amplified from the CAG-Fig4 construct using the forward primer 5′ GTGAT GCTTC TGTGA TGTCT TTTAC and a reverse primer containing the mutation (underlined): 5′ CCGTA GGAAG CTTGG TTGAG ACACC TGACA AACC. The CAG-Fig4 plasmid was digested with BstXI (New England Biolabs, Ipswich, MA, USA) and the products of 7.2 kb and 420 bp were gel purified. The PCR product containing the mutation was digested with BstX1 and the resulting mutant 420 bp fragment was ligated to the 7.2 kb fragment from the wild-type clone to generate the CAG-Fig4-Cys486Ser construct. For neuron-specific expression in transgenic mice, the mutant cDNA was PCR amplified from the CAG clone and cloned downstream of the 4 kb enolase (NSE) promoter as previously described (32). Promoters, coding regions and UTRs of cDNA clones were subjected to Sanger sequencing for sequence verification.
Cell vacuolization
Mouse embryonic fibroblasts were isolated from plt/plt homozygous null embryos at E13 (5). Cells were cultured to Passage 3 and stored frozen in Recovery Cell Culture Freezing Medium #12648 (Invitrogen, Carlsbad, CA, USA). After co-transfection with enhanced green fluorescent protein, cells were cultured and vacuolization was evaluated by light microscopy as previously described (29,30).
Transgenic mice
The NSE-Fig4 cDNA was microinjected into (C57BL/6 × SJL)F2 fertilized mouse eggs in the University of Michigan Transgenic Animal Core (www.med.umich.edu/tamc/) as previously described (32). Transgenic founders were identified by PCR of genomic DNA as described in the text. Founders were crossed with congenic C3H. Fig4+/− heterozygous mice (N23) carrying the plt null allele (43). Fig4+/− offspring inheriting the transgene were crossed again with C3H.Fig4+/− mice to generate Fig4−/−, transgene positive (Tg+) mice and non-transgenic littermate controls for experiments. Wildtype mice expressing the transgene did not exhibit abnormal phenotypes or reduced fertility. Animals were housed and cared for in accordance with NIH guidelines. Experiments were approved by the University of Michigan Committee on the Use and Care of Animals.
Reverse transcriptase-polymerase chain reaction
Transgene transcripts were detected with the forward primer in NSE exon 1, CACCG GCTGA GTCTG CAGTC CTCG, and the reverse in Fig4 exon 2, TGATT GCTCC CAACT AGAAA GTATC, with 30 cycles of amplification. qRT-PCR was carried out as previously described (10). RNA was prepared from whole brain using the Trizol reagent (Invitrogen, Carlsbad, CA, USA). cDNA was generated with the first-strand cDNA synthesis kit (Invitrogen). Transgene expression on the Fig4plt/plt null background was detected with the pre-designed Taqman assay spanning Fig4 exons 18 and 19 (Catalog # Mm 01189585_m1) (Applied Biosystems, Foster City, CA, USA) that does not detect the plt transcript due to the retrotransposon insert in intron 18 (5,32).
Histology
Brain and spinal cord were fixed for 24 h at 4°C in phosphate buffered 10% formalin and dehydrated in 70% ethanol for 24 h at 4°C prior to processing. Paraffin embedding and hematoxylin and eosin (H&E) staining were carried out at Histoserv, Inc. (Bethesda, MD, USA). Images were obtained with an Olympus BX51 microscope and DP50 camera.
Western blotting was carried out for FIG4 (30), and myelin proteins MAG, MBP and PLP as previously described (45). Brain membrane proteins were prepared from three mice of each genotype at P20-P22 as described (35). Primary antibodies included mouse anti-βIII tubulin (1:20 000; Promega), rabbit anti-MAG (1:1000) (34), rat anti-MBP (1:1000; Millipore), anti-PLP (1:1000, Abcam) and mouse anti-Fig4 (NeuroMab, 1:200). Primary antibodies were detected using either horseradish peroxidase-conjugated secondary antibodies (1:2000–15 000; Millipore Bioscience Research Reagents) or Alexa-conjugated secondary antibodies (1:20 000, Molecular Probes). The Licor C-DiGit and Odyssey imaging systems and software were used for visualization and quantification of myelin proteins. For quantitation of FIG4 protein, the ratio of FIG4 protein to loading control was determined using the ImageJ software (46) (Supplementary Material, Fig. S1).
Optic nerve conduction
CAP recordings of acutely isolated optic nerves from P20-P22 from mice of genotype Fig4++ (n = 8 nerves), Fig4−/−,NSE-Fig4TG (n = 4 nerves) and Fig4−/−,NSE-Fig4Cys486Ser (n = 6 nerves) were performed and analyzed as previously described (35,47).
Supplementary Material
Authors’ Contributions
Conception and design of experiments: G.M.L., M.H.M.; execution of experiments: G.M.L., C.M.F., A.C.M., R.C.W. and Y.A.M.; interpretation of data: G.M.L., C.M.F., A.C.M., R.C.W., Y.A.M., R.J.G. and M.H.M.; manuscript preparation: G.M.L., M.H.M.
Funding
Funded by National Institutes of Health research grants R01 GM24872 (M.H.M.), R01 NS81281 (R.J.G. and M.H.M.) and the Dr Miriam and Sheldon G. Adelson Medical Foundation on Neural Repair and Rehabilitation (R.J.G.). Y.A.M. acknowledges support from the National Institutes of Health Cellular and Molecular Biology Training grant T32-GM007315.
Supplementary Material
Acknowledgements
We thank A. Antonellis for critical reading of the manuscript.
Conflict of Interest statement. None declared.
References
- 1.Ho C.Y., Alghamdi T.A., Botelho R.J. (2012) Phosphatidylinositol-3,5-bisphosphate: no longer the poor PIP2. Traffic, 13, 1–8. [DOI] [PubMed] [Google Scholar]
- 2.Takasuga S., Sasaki T. (2013) Phosphatidylinositol-3,5-bisphosphate: metabolism and physiological functions. J. Biochem., 154, 211–218. [DOI] [PubMed] [Google Scholar]
- 3.Michell R.H. (2013) Inositol lipids: from an archaeal origin to phosphatidylinositol 3,5-bisphosphate faults in human disease. FEBS J., 280, 6281–6294. [DOI] [PubMed] [Google Scholar]
- 4.McCartney A.J., Zhang Y., Weisman L.S. (2014) Phosphatidylinositol 3,5-bisphosphate: low abundance, high significance. Bioessays, 36, 52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow C.Y., Zhang Y., Dowling J.J., Jin N., Adamska M., Shiga K., Szigeti K., Shy M.E., Li J., Zhang X. et al. (2007) Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature, 448, 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin N., Chow C.Y., Liu L., Zolov S.N., Bronson R., Davisson M., Petersen J.L., Zhang Y., Park S., Duex J.E. et al. (2008) VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J., 27, 3221–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Botelho R.J., Efe J.A., Teis D., Emr S.D. (2008) Assembly of a Fab1 phosphoinositide kinase signaling complex requires the Fig4 phosphoinositide phosphatase. Mol. Biol. Cell, 19, 4273–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sbrissa D., Ikonomov O.C., Fenner H., Shisheva A. (2008) ArPIKfyve homomeric and heteromeric interactions scaffold PIKfyve and Sac3 in a complex to promote PIKfyve activity and functionality. J. Mol. Biol., 384, 766–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudge S.A., Anderson D.M., Emr S.D. (2004) Vacuole size control: regulation of PtdIns(3,5)P2 levels by the vacuole-associated Vac14-Fig4 complex, a PtdIns(3,5)P2-specific phosphatase. Mol. Biol. Cell, 15, 24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lenk G.M., Ferguson C.J., Chow C.Y., Jin N., Jones J.M., Grant A.E., Zolov S.N., Winters J.J., Giger R.J., Dowling J.J. et al. (2011) Pathogenic mechanism of the FIG4 mutation responsible for Charcot-Marie-Tooth disease CMT4J. PLoS Genet., 7, e1002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zolov S.N., Bridges D., Zhang Y., Lee W.W., Riehle E., Verma R., Lenk G.M., Converso-Baran K., Weide T., Albin R.L. et al. (2012) In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc. Natl Acad. Sci. USA, 109, 17472–17477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo S., Stolz L.E., Lemrow S.M., York J.D. (1999) SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J. Biol. Chem., 274, 12990–12995. [DOI] [PubMed] [Google Scholar]
- 13.Hughes W.E., Cooke F.T., Parker P.J. (2000) Sac phosphatase domain proteins. Biochem. J., 350(Pt 2), 337–352. [PMC free article] [PubMed] [Google Scholar]
- 14.Gary J.D., Sato T.K., Stefan C.J., Bonangelino C.J., Weisman L.S., Emr S.D. (2002) Regulation of Fab1 phosphatidylinositol 3-phosphate 5-kinase pathway by Vac7 protein and Fig4, a polyphosphoinositide phosphatase family member. Mol. Biol. Cell, 13, 1238–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sbrissa D., Ikonomov O.C., Fu Z., Ijuin T., Gruenberg J., Takenawa T., Shisheva A. (2007) Core protein machinery for mammalian phosphatidylinositol 3,5-bisphosphate synthesis and turnover that regulates the progression of endosomal transport. Novel Sac phosphatase joins the ArPIKfyve-PIKfyve complex. J. Biol. Chem., 282, 23878–23891. [DOI] [PubMed] [Google Scholar]
- 16.Manford A., Xia T., Saxena A.K., Stefan C., Hu F., Emr S.D., Mao Y. (2010) Crystal structure of the yeast Sac1: implications for its phosphoinositide phosphatase function. EMBO J., 29, 1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okamura Y., Dixon J.E. (2011) Voltage-sensing phosphatase: its molecular relationship with PTEN. Physiology (Bethesda), 26, 6–13. [DOI] [PubMed] [Google Scholar]
- 18.Jackson M.D., Denu J.M. (2001) Molecular reactions of protein phosphatases—insights from structure and chemistry. Chem. Rev., 101, 2313–2340. [DOI] [PubMed] [Google Scholar]
- 19.Zhou G., Denu J.M., Wu L., Dixon J.E. (1994) The catalytic role of Cys124 in the dual specificity phosphatase VHR. J. Biol. Chem., 269, 28084–28090. [PubMed] [Google Scholar]
- 20.Rohde H.M., Cheong F.Y., Konrad G., Paiha K., Mayinger P., Boehmelt G. (2003) The human phosphatidylinositol phosphatase SAC1 interacts with the coatomer I complex. J. Biol. Chem., 278, 52689–52699. [DOI] [PubMed] [Google Scholar]
- 21.Berger P., Berger I., Schaffitzel C., Tersar K., Volkmer B., Suter U. (2006) Multi-level regulation of myotubularin-related protein-2 phosphatase activity by myotubularin-related protein-13/set-binding factor-2. Hum. Mol. Genet., 15, 569–579. [DOI] [PubMed] [Google Scholar]
- 22.Guan K.L., Dixon J.E. (1990) Protein tyrosine phosphatase activity of an essential virulence determinant in Yersinia. Science, 249, 553–556. [DOI] [PubMed] [Google Scholar]
- 23.Milarski K.L., Zhu G., Pearl C.G., McNamara D.J., Dobrusin E.M., MacLean D., Thieme-Sefler A., Zhang Z.Y., Sawyer T., Decker S.J. et al. (1993) Sequence specificity in recognition of the epidermal growth factor receptor by protein tyrosine phosphatase 1B. J. Biol. Chem., 268, 23634–23639. [PubMed] [Google Scholar]
- 24.Milarski K.L., Saltiel A.R. (1994) Expression of catalytically inactive Syp phosphatase in 3T3 cells blocks stimulation of mitogen-activated protein kinase by insulin. J. Biol. Chem., 269, 21239–21243. [PubMed] [Google Scholar]
- 25.Noguchi T., Matozaki T., Horita K., Fujioka Y., Kasuga M. (1994) Role of SH-PTP2, a protein-tyrosine phosphatase with Src homology 2 domains, in insulin-stimulated Ras activation. Mol. Cell. Biol., 14, 6674–6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pei D., Lorenz U., Klingmuller U., Neel B.G., Walsh C.T. (1994) Intramolecular regulation of protein tyrosine phosphatase SH-PTP1: a new function for Src homology 2 domains. Biochemistry, 33, 15483–15493. [DOI] [PubMed] [Google Scholar]
- 27.Sun H., Charles C.H., Lau L.F., Tonks N.K. (1993) MKP-1 (3CH134), an immediate early gene product, is a dual specificity phosphatase that dephosphorylates MAP kinase in vivo. Cell, 75, 487–493. [DOI] [PubMed] [Google Scholar]
- 28.Ward Y., Gupta S., Jensen P., Wartmann M., Davis R.J., Kelly K. (1994) Control of MAP kinase activation by the mitogen-induced threonine/tyrosine phosphatase PAC1. Nature, 367, 651–654. [DOI] [PubMed] [Google Scholar]
- 29.Campeau P.M., Lenk G.M., Lu J.T., Bae Y., Burrage L., Turnpenny P., Roman Corona-Rivera J., Morandi L., Mora M., Reutter H. et al. (2013) Yunis-Varon syndrome is caused by mutations in FIG4, encoding a phosphoinositide phosphatase. Am. J. Hum. Genet., 92, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baulac S., Lenk G.M., Dufresnois B., Ouled Amar Bencheikh B., Couarch P., Renard J., Larson P.A., Ferguson C.J., Noe E., Poirier K. et al. (2014) Role of the phosphoinositide phosphatase FIG4 gene in familial epilepsy with polymicrogyria. Neurology, 82, 1068–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levin S.I., Meisler M.H. (2004) Floxed allele for conditional inactivation of the voltage-gated sodium channel Scn8a (NaV1.6). Genesis, 39, 234–239. [DOI] [PubMed] [Google Scholar]
- 32.Ferguson C.J., Lenk G.M., Jones J.M., Grant A.E., Winters J.J., Dowling J.J., Giger R.J., Meisler M.H. (2012) Neuronal expression of Fig4 is both necessary and sufficient to prevent spongiform neurodegeneration. Hum. Mol. Genet., 21, 3525–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dworzak F., Mora M., Borroni C., Cornelio F., Blasevich F., Cappellini A., Tagliavini F., Bertagnolio B. (1995) Generalized lysosomal storage in Yunis Varon syndrome. Neuromuscul. Disord., 5, 423–428. [DOI] [PubMed] [Google Scholar]
- 34.Walch E., Schmidt M., Brenner R.E., Emons D., Dame C., Pontz B., Wiestler O.D., Bartmann P. (2000) Yunis-Varon syndrome: evidence for a lysosomal storage disease. Am. J. Med. Genet., 95, 157–160. [DOI] [PubMed] [Google Scholar]
- 35.Winters J.J., Ferguson C.J., Lenk G.M., Giger-Mateeva V.I., Shrager P., Meisler M.H., Giger R.J. (2011) Congenital CNS hypomyelination in the Fig4 null mouse is rescued by neuronal expression of the PI(3,5)P(2) phosphatase Fig4. J. Neurosci., 31, 17736–17751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolis A., Zordan P., Coviello S., Bolino A. (2007) Myotubularin-related (MTMR) phospholipid phosphatase proteins in the peripheral nervous system. Mol. Neurobiol., 35, 308–316. [DOI] [PubMed] [Google Scholar]
- 37.Ng A.A., Logan A.M., Schmidt E.J., Robinson F.L. (2013) The CMT4B disease-causing phosphatases Mtmr2 and Mtmr13 localize to the Schwann cell cytoplasm and endomembrane compartments, where they depend upon each other to achieve wild-type levels of protein expression. Hum. Mol. Genet., 22, 1493–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y., McCartney A.J., Zolov S.N., Ferguson C.J., Meisler M.H., Sutton M.A., Weisman L.S. (2012) Modulation of synaptic function by VAC14, a protein that regulates the phosphoinositides PI(3,5)P(2) and PI(5)P. EMBO J., 31, 3442–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCartney A.J., Zolov S.N., Kauffman E.J., Zhang Y., Strunk B.S., Weisman L.S., Sutton M.A. (2014) Activity-dependent PI(3,5)P2 synthesis controls AMPA receptor trafficking during synaptic depression. Proc. Natl Acad. Sci. USA, 111, E4896–E4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong X.P., Shen D., Wang X., Dawson T., Li X., Zhang Q., Cheng X., Zhang Y., Weisman L.S., Delling M. et al. (2010) PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca(2+) release channels in the endolysosome. Nat. Commun., 1, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X., Zhang X., Dong X.P., Samie M., Li X., Cheng X., Goschka A., Shen D., Zhou Y., Harlow J. et al. (2012) TPC proteins are phosphoinositide-activated sodium-selective ion channels in endosomes and lysosomes. Cell, 151, 372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang W., Zhang X., Gao Q., Xu H. (2014) TRPML1: an ion channel in the lysosome. Handb. Exp. Pharmacol., 222, 631–645. [DOI] [PubMed] [Google Scholar]
- 43.Lenk G.M., Meisler M.H. (2014) Mouse models of PI(3,5)P2 deficiency with impaired lysosome function. Methods Enzymol., 534, 245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zou J., Hu B., Arpag S., Yan Q., Hamilton A., Zeng Y.S., Vanoye C.G., Li J. (2015) Reactivation of lysosomal Ca2+ efflux rescues abnormal lysosomal storage in FIG4-deficient cells. J. Neurosci., 35, 6801–6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baldwin K.T., Carbajal K.S., Segal B.M., Giger R.J. (2015) Neuroinflammation triggered by beta-glucan/dectin-1 signaling enables CNS axon regeneration. Proc. Natl Acad. Sci. USA, 112, 2581–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider C.A., Rasband W.S., Eliceiri K.W. (2012) NIH image to ImageJ: 25 years of image analysis. Nat. Methods, 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carbajal K.S., Mironova Y., Ulrich-Lewis J.T., Kulkarni D., Grifka-Walk H.M., Huber A.K., Shrager P., Giger R.J., Segal B.M. (2015) Th cell diversity in experimental autoimmune encephalomyelitis and multiple sclerosis. J. Immunol., 195, 2552–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.