Abstract
Pathogenicity islands (PAIs) are chromosomal clusters of pathogen-specific virulence genes often found at tRNA loci. In the Yersinia pseudotuberculosis 32777 chromosome, we characterized a 98-kb segment that has all of the characteristic features of a PAI, including insertion in a (phenylalanine) tRNA gene, the presence of a bacteriophage-like integrase-encoding gene, and direct repeats at the integration sites. The G+C content of the segment ranges from 31 to 60%, reflecting a genetic mosaic: this is consistent with the notion that the sequences were horizontally acquired. The PAI, termed YAPI (for Yersinia adhesion pathogenicity island), carries 95 open reading frames and includes (i) the previously described pil operon, encoding a type IV pilus that contributes to pathogenicity (F. Collyn et al., Infect. Immun. 70:6196-6205, 2002); (ii) a block of genes potentially involved in general metabolism; (iii) a gene cluster for a restriction-modification system; and (iv) a large number of mobile genetic elements. Furthermore, the PAI can excise itself from the chromosome at low frequency and in a precise manner, and deletion does not result in a significant decrease of bacterial virulence compared to inactivation of the fimbrial gene cluster alone. The prevalence and size of the PAI vary from one Y. pseudotuberculosis strain to another, and it can be found integrated into either of the two phe tRNA loci present on the species' chromosome. YAPI was not detected in the genome of the genetically closely related species Y. pestis, whereas a homologous PAI is harbored by the Y. enterocolitica chromosome.
Pathogenicity islands (PAIs) are DNA segments of 10 to 200 kb which are present in the genome of pathogenic strains but absent from those of nonpathogenic members of the same (or related) bacterial species and typically carry genes encoding one or more virulence factors. Since their discovery in pathogenic strains of Escherichia coli during the late 1980s (11), PAIs have been described in many other gram-negative (mostly Enterobacteriaceae) bacteria, as well as in certain gram-positive species (23). These genetic elements have common features: they are often DNA regions which (i) have a G+C content and codon usage that differ from that of the rest of the genome; (ii) are flanked by small, direct-repeat sequences; (iii) are associated with tRNA genes; (iv) harbor cryptic or functional genes that encode mobility factors such as integrases, transposases, and insertion sequence (IS) elements or parts of these elements; and (v) are unstable (8). To date, only one chromosomal PAI (called the high-pathogenicity island [HPI]) has been well characterized in the three pathogenic Yersinia species: Yersinia pestis (the causative agent of plague) and Yersinia pseudotuberculosis and Yersinia enterocolitica (both responsible for digestive tract infections) (24). The HPI ranges from 36 to 43 kb (according to the species in question) and bears genes involved in the biosynthesis, transport, and regulation of the yersiniabactin siderophore (for a review, see reference 4). Apart from the HPI, additional putative PAIs have been detected in silico within the Y. pestis whole-genome sequence (7, 10).
We recently discovered an 11-kb chromosomal type IV pilus gene cluster (pil) that contributes to bacterial virulence in Y. pseudotuberculosis serotype O:1 (6). In silico analysis (www.sanger.ac.uk/Projects/Y_pestis/ and www.genome.wisc.edu/sequencing/pestis) revealed that the pil gene cluster was absent from the genome of the genetically closely related species Y. pestis (1), whereas, in contrast, a homologous locus (83% identity at the nucleotide level) was harbored on the Y. enterocolitica chromosome (www.sanger.ac.uk/Projects/Y_enterocolitica/). We show here that the Y. pseudotuberculosis pil locus is present on a large (98-kb) fragment possessing the characteristics of a PAI.
MATERIALS AND METHODS
Plasmids, bacterial strains, and growth conditions.
pUC18 (New England Biolabs), pBeloBac11 (New England Biolabs), and pMM70413 (5) plasmids were used for DNA cloning, with pCCY41.2 used as a source of sacB and aph(3′)-IIIa genes (5). pIL31.3 is a pMM70413 derivative, with sacB and aph(3′)-IIIa genes inserted upstream of the Y. pseudotuberculosis urease locus (5).
Escherichia coli strains DH10B and DH5α were hosts for plasmids pBeloBac11 and pUC18, respectively. SY327λpir and SM10λpir strains were recipients for replication of the pMM70413 suicide plasmid and its derivatives (15). In addition to the Y. pseudotuberculosis wild-type strain 32777, the present study used 32777 derivatives MIV (lacking the pil operon [6]), pilKS, ureKS, ΔYAPI1, and ΔYAPI2 (all newly engineered in this work), as well as reference strains YPT9, 58/87, 2926, 33054, 32945, 32842, ST, 1830, 199/90, 1553, and 682/90. Y. pseudotuberculosis and E. coli strains were grown at 28 and 37°C, respectively, in Luria-Bertani broth or on agar plates. Mating experiments between E. coli and Y. pseudotuberculosis were plated on M9 minimum medium agar, as previously described (5). X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 20 μg ml−1), IPTG (isopropyl-β-d-thiogalactopyranoside; 25 μg ml−1), ampicillin (100 μg ml−1), kanamycin (50 μg ml−1), chloramphenicol (25 μg ml−1), and sucrose (10%) were added to growth medium for bacterial selection when necessary.
DNA preparation, amplification, analysis, and hybridization.
Genomic DNA extraction, small-scale plasmid preparation, endonuclease digestion, DNA ligation, PCR, agarose gel electrophoresis, elution of DNA fragments from agarose gels, E. coli transformation, and colony blotting were carried out by using standard procedures (22). DNA/DNA hybridization was performed by using a digoxigenin (DIG)-labeled probe and the DIG detection kit from Roche according to the manufacturer's instructions.
Synthetic oligonucleotides.
Oligonucleotide primers were custom synthesized (MWG Biotech) for PCR generation of DNA fragments. Their nucleotide sequences (5′→3′) were as follows: Af, AATTCGTCATGACCCG; Ar, CGAGTTCCACTGGT; Bf, TCCTTGCTGACCGAAGGTG; Br, AGGTACGCCACCGACCTGA; Cf, GATGGCTCAGTACGTC; Cr, GTCAGTTCGGGAGTAT; Df, AACCAAGGCGTTACACAGACA; Dr, GAACAGGCGGATAGCATCAG; Ef,GGTCGCTGTCTGTTTCTTGA; Er, AAATTACGGAACCCAGTAGCC; Ff, GTAAGTGGTACTCCAC; Fr, CCAAATGTTTAGCGGA; Gf, GCTTCACCAAGCGTAATGAC; and Gr, CCCAGAATGATCTAATGGAG (referred to below as primer sets A to G).
DNA cloning.
A bacterial artificial chromosome (BAC) library was constructed from the Y. pseudotuberculosis 32777 chromosome by using the pBeloBac11 vector as described in detail by Buchrieser et al. (3). DNA extracted from selected BAC clones was partially digested with Sau3A: 1.5- to 3-kb and 3- to 5-kb DNA fragments fractionated by agarose gel electrophoresis were randomly ligated to BamHI-restricted pUC18 (shotgun cloning).
DNA sequencing.
DNA was sequenced by using the ABI Prism dichlororhodamine dye terminator sequencing kit (Qiagen), and extension products were analyzed with the Applied Biosystems 3700 automated DNA sequencer. After assembly of the nucleotide sequence data by using Sequence Navigator (Perkin-Elmer), the remaining gaps were filled by using an Expand Long Template PCR kit (Roche) on recombinant BACs and the Y. pseudotuberculosis 32777 chromosome. Similarities to known proteins were determined by using the BLAST2N, BLAST2X, BLAST2P (2), and InterPro (16) programs, and transmembrane domains were predicted with TMHMM2.0 (12). tRNA genes were located and identified with tRNAscan (13). Finally, sequence annotation was performed with ARTEMIS software (20).
Introduction of the aph(3′)-IIIa and sacB genes into the Y. pseudotuberculosis 32777 chromosome.
A 1,353-bp fragment encompassing the pilS gene (with a SpeI restriction site created 159 bp upstream of the pilS start codon) was generated by PCR. The amplimer was cloned into a pMM70413 suicide vector to yield plasmid pMMpilS. Next, the 3,130-bp SpeI fragment harboring sacB and aph(3′)-IIIa from pCCY41.2 was inserted into the SpeI restriction site of pMMpilS, thus yielding pSACpil.1. Allelic exchange was carried out between Y. pseudotuberculosis 32777 and E. coli SM10λpir (pSACpil.1): after bacterial mating, transconjugants were plated on M9 minimum medium containing kanamycin and chloramphenicol in order to select the first recombination event. Selection of the second recombination event and elimination of the suicide plasmid was performed by using the bacteriostatic property of chloramphenicol and the bactericidal activity of cycloserine to eliminate dividing microorganisms. Similarly, the aph(3′)-IIIa and sacB genes were inserted upstream of the ureA (the first gene of the urease locus) after mating of Y. pseudotuberculosis 32777 and E. coli SM10λpir(pIL31.3), a method originally reported for Y. pseudotuberculosis AH (5).
Measurement of chromosomal deletion frequency.
Dilutions of overnight cultures of Y. pseudotuberculosis 32777 derivatives containing the aph(3′)-IIIa and sacB genes (pilKS and ureKS strains) were plated on Luria-Bertani agar in the presence or absence of 10% sucrose. Sucrose-resistant bacteria were tested for loss of kanamycin resistance, and chromosomal deletion frequencies were calculated as the ratio of sucrose-resistant and kanamycin-sensitive bacteria to the total bacterial population.
Experimental infection in mice.
Six-week-old female inbred BALB/c mice (Iffa Credo) were challenged by the intragastric or the intravenous route as previously described (6), and infected animals were monitored for 3 weeks.
Nucleotide sequence accession number.
The nucleotide sequence data reported here have been deposited in the EMBL database under accession number AJ627388.
RESULTS
The Y. pseudotuberculosis 32777 pil locus is harbored on a large chromosomal segment that is missing in a pil-negative strain of the species. The type IV pilus gene cluster from Salmonella enterica serovar Typhi is harbored on a 134-kb PAI (SPI-7, previously called the major PAI) (25) and is closely related to the Y. pseudotuberculosis pil locus (6). Using the recombinant cosmids pMM2.1, pMM2.A6, and pMM3.D6 (6), we sequenced the upstream and downstream regions flanking the Y. pseudotuberculosis pil operon and found that they displayed similarities to the genetic environment of the S. enterica serovar Typhi pil gene cluster. This finding strongly suggested that the Y. pseudotuberculosis pil operon might be present on a large PAI. To identify the complete PAI, we constructed a BAC library from strain 32777 chromosomal DNA by using the pBeloBAC11 vector. By using colony blot hybridization assays with appropriately labeled, PCR-generated probes, two clones of interest (BAC 10G2 and BAC 3B9) were selected. Subsequently, we used the pUC18 plasmid to construct shotgun libraries from the BAC 10G2 and BAC 3B9 clones. After sequencing the recombinant plasmids and assembling the resulting nucleotide sequences, we used the Sanger website (www.sanger.ac.uk/Projects/Y_pestis/) to perform homology searches comparing the final contig with the whole genome sequence of Y. pestis CO92 (which lacks the pil locus) (17). This computer-assisted comparison revealed that (with the exception of the 5′ and 3′ ends of the contig, which were >99% identical to Y. pestis nucleotide sequences) a 98,058-bp segment was absent in the genome of the plague bacillus. It was found to be inserted in the Y. pseudotuberculosis 32777 chromosome between the Y. pestis CO92 YPO0339 and YPO0341 homologue genes and was associated with phenylalanine-specific tRNA gene, a frequent target for PAI integration (19). The 3′ end of the tRNA-encoding gene phe contains a 54-nucleotide motif that is repeated at the opposite extremity of the 98-kb segment. Interestingly, in the pil-negative Y. pseudotuberculosis strain 32953 (6), the region separating YPO0339 and YPO0341 was found to be identical (at least on the basis of PCR product sequencing) to that of Y. pestis CO92 (17).
The 98-kb Y. pseudotuberculosis 32777 chromosomal segment encompassing the pil locus exhibits the genetic features of a PAI.
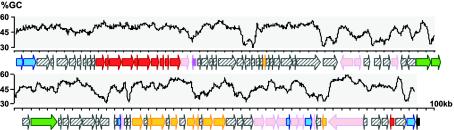
Computer analysis of the 98,058-bp sequence revealed the presence of 95 putative open reading frames (ORFs), 11 of which correspond to the previously characterized pilLMNOPQRSUVW genes (Fig. 1). The overall GC content of the fully sequenced DNA segment is 47.8%, but the base composition was found to be heterogeneous (between 31.2 and 60.6%) along the 98-kb region (Fig. 1). Of the newly identified ORFs (which we call api, for adhesion PAI), 8 encode proteins lacking significantly similar equivalents in the public databases, and 43 are similar to known protein sequences of unknown function. The latter are specified by coding sequences (CDSs) found principally on the SPI-7 PAI and, to a lesser extent, on the Ralstonia solanacearum megaplasmid. Forty-four ORFs code for proteins with putative functions (Table 1), a marked fraction of which are derived from mobile, accessory genetic elements such as IS elements, bacteriophages, and plasmids. IS elements include different subtypes (ISSod13-like, IS100, IS110-like, IS285, IS630-like, IS911-like, and IS1353-like) and can be complete or partial. One of these elements (IS100) disrupts an ORF which, at the nucleotide level, is identical to a gene previously identified in the Y. pestis genome (YPO1092) encoding a DNA-binding protein of phage origin. An intact ORF (api95) adjacent to phe-tRNA gene specifies a 326-amino-acid product displaying high homologies to recombinases of the Cre family, which includes various bacteriophage integrases. Furthermore, for two CDSs located at the other extremity of the large DNA segment, the deduced proteins are homologous to phage and plasmid DNA helicases (for api2) and ATPases involved in plasmid partitioning (for api1). Finally, one ORF (api89) codes for a product that appears to be similar to rearrangement hot spot (Rhs)-related proteins (9).
FIG. 1.
Genetic organization of YAPI from Y. pseudotuberculosis 32777. ORFs related to virulence (red), metabolism (yellow), regulation (purple), phage and plasmid function (blue), insertion elements (pink), type I restriction-modification systems (green), and unknown functions (hatched) are positioned and oriented along the 98-kb segment. The tRNA-phe gene (black) at the 3′ extremity flanks a putative integrase-encoding gene, i.e., the last gene of the PAI. Numbers on the right-hand side of the map indicate the scale in kilobases. The DNA sequence's guanine and cytosine (G+C) percentage (determined by using a 500-base window size and a 50-base window slide) is indicated above the genetic map.
TABLE 1.
Characteristics of the 95 CDSs harbored on YAPI from Y. pseudotuberculosis 32777
| CDS | Size (aa)a | Similarities (determined by BLAST) | % Identity/homology | Accession no. |
|---|---|---|---|---|
| api1 | 293 | Plasmid partitioning ATPase STY4521 (S. enterica serovar Typhi) | 61/77 | NP_458616.1 |
| api2 | 453 | DNA helicase STY4522 (S. enterica serovar Typhi) | 56/73 | NP_458617.1 |
| api3 | 560 | Hypothetical protein STY4523 (S. enterica serovar Typhi) | 47/62 | NP_458618.1 |
| api4 | 193 | Hypothetical protein STY4526 (S. enterica serovar Typhi) | 50/72 | CAD09309.1 |
| api5 | 399 | Hypothetical protein STY4528 (S. enterica serovar Typhi) | 46/62 | NP_458622.1 |
| api6 | 252 | Hypothetical protein STY4529 (S. enterica serovar Typhi) | 45/60 | NP_458623.1 |
| api7 | 180 | Hypothetical protein STY4534 (S. enterica serovar Typhi) | 64/83 | NP_458625.1 |
| api8 | 143 | ORFC8 (R. solanacearum) | 43/59 | NP_052313.1 |
| api9 | 77 | Hypothetical protein NMB1666 (Neisseria meningitidis) | 45/58 | NP_274671.1 |
| api10 | 128 | Hypothetical protein NMB1665 (N. meningitidis) | 45/65 | NP_274670.1 |
| pilL | 356 | PilL (S. enterica serovar Typhi) | 44/55 | NP_458628.1 |
| pilM | 145 | PilM (S. enterica serovar Typhi) | 42/57 | NP_458629.1 |
| pilN | 530 | PilN (S. enterica serovar Typhimurium) | 48/65 | BAA77974.1 |
| pilO | 445 | PilO (S. enterica serovar Typhi) | 33/51 | NP_458631.1 |
| pilP | 164 | PilP (S. enterica serovar Typhi) | 36/55 | NP_458632.1 |
| pilQ | 517 | PilQ (S. enterica serovar Typhi) | 55/71 | NP_458633.1 |
| pilR | 369 | PilR (S. enterica serovar Typhimurium) | 47/67 | BAA77978.1 |
| pilS | 195 | PilS (S. enterica serovar Typhimurium) | 54/70 | BAA77979.1 |
| pilU | 222 | PilU (S. enterica serovar Typhi) | 34/51 | NP_458637.1 |
| pilV | 425 | Shufflon PilV A′ (S. enterica serovar Typhimurium) | 54/68 | P09746 |
| pilW | 319 | YPO0076 transposase (Y. pestis) | 73/83 | AD0010 |
| api22 | 118 | Putative transcriptional regulator (Serratia spp.) | 56/70 | AAM47490.1 |
| api23 | 110 | Unknown | ||
| api24 | 272 | Hypothetical protein STY4558 (S. enterica serovar Typhi) | 54/72 | AE1029 |
| api25 | 212 | Hypothetical protein STY4559 (S. enterica serovar Typhi) | 59/73 | NP_458645.1 |
| api26 | 173 | Hypothetical protein STY4560 (S. enterica serovar Typhi) | 45/64 | NP_458646.1 |
| api27 | 706 | Hypothetical protein STY4562 (S. enterica serovar Typhi) | 72/85 | NP_458648.1 |
| api28 | 250 | Hypothetical protein STY4563 (S. enterica serovar Typhi) | 51/68 | NP_458649.1 |
| api29 | 230 | Unknown | ||
| api30 | 156 | Unknown | ||
| api31 | 102 | Hypothetical protein STY4564 (S. enterica serovar Typhi) | 62/76 | NP_458650.1 |
| api32 | 81 | Hypothetical protein STY4565 (S. enterica serovar Typhi) | 38/64 | NP_458651.1 |
| api33 | 116 | Putative oxidase STY4566 (S. enterica serovar Typhi) | 41/58 | NP_458652.1 |
| api34 | 124 | Hypothetical protein (Pseudomonas fluorescens) | 37/56 | ZP_00087885.1 |
| api35 | 217 | Hypothetical protein STY4568 (S. enterica serovar Typhi) | 70/82 | NP_458653.1 |
| api36 | 277 | Hypothetical protein STY4569 (S. enterica serovar Typhi) | 58/69 | NP_458654.1 |
| api37 | 505 | Hypothetical protein STY4570 (S. enterica serovar Typhi) | 51/66 | NP_458655.1 |
| api38 | 134 | Hypothetical protein STY4571 (S. enterica serovar Typhi) | 53/70 | NP_458656.1 |
| api39 | 948 | Hypothetical proteins STY4572 (S. enterica serovar Typhi) and STY4573 (S. enterica serovar Typhi) | 55/70 65/77 | NP_458657.1NP_458658.1 |
| api40 | 571 | Putative abortive infection phage resistance protein AbiR (Nitrosomonas europaea) | 20/39 | CAD84400.1 |
| api41 | 402 | IS285 transposase (Y. pestis) | 99/100 | NP_403677.1 |
| api42 | 309 | IS630-like transposase (S. enterica serovar Choleraesuis) | 70/81 | BAB20556.1 |
| api43 | 293 | Unknown | ||
| api44 | 381 | Unknown | ||
| api45 | 245 | Putative poly(A)-specific RNase subunit, C end (Schizosaccharomyces pombe) | 24/44 | CAA91128.1 |
| api46 | 289 | ISSod13-like transposase, N end (Shewanella oneidensis) | 80/89 | AAN56855.1 |
| api47 | 226 | Hypothetical protein (Streptomyces avermitilis) | 33/50 | NP_824450.1 |
| api48 | 239 | Hypothetical protein (Mesorhizobium loti) and hypothetical protein (Streptomyces avermitilis) | 40/58 | NP_824451.1 |
| 25/43 | NP_104999.1 | |||
| api49 | 568 | Type I restriction-modification system, methylation subunit HsdM (Methanosarcina mazei) | 74/83 | NP_632453.1 |
| api50 | 449 | Type I restriction-modification system, specificity subunit HsdS (Klebsiella pneumoniae) | 40/55 | AAB70708.1 |
| api51 | 395 | Hypothetical protein (Haemophilus somnus) | 41/61 | ZP_00122800.1 |
| api52 | 1,043 | Type I restriction-modification system, restriction subunit HsdR (Methanosarcina mazei) | 69/82 | NP_632455.1 |
| api53 | 134 | Hypothetical protein STY4575 (S. enterica serovar Typhi) | 49/65 | NP_458660.1 |
| api54 | 317 | Hypothetical protein STY4576 (S. enterica serovar Typhi) | 63/78 | NP_458661.1 |
| api55 | 468 | Hypothetical protein STY4577 (S. enterica serovar Typhi) | 60/78 | NP_458662.1 |
| api56 | 507 | Hypothetical protein STY4579 (S. enterica serovar Typhi) | 45/65 | NP_458664.1 |
| api57 | 98 | Unknown | ||
| api58 | 419 | Hypothetical protein BPP0992 (Bordetella parapertussis) and hypothetical protein BPP0991 (B. parapertussis) | 54/76 48/71 | NP_883312.1NP_883311.1 |
| api59 | 65 | Unknown | ||
| api60 | 55 | DNA-binding phagic protein YPO1092, N end (Y. pestis) | 51/76 | AE0134 |
| api61 | 80 | IS1353-like transposase, C end (Shigella flexneri) | 55/63 | AAL72503.1 |
| api62 | 63 | Unknown | ||
| api63 | 83 | Hypothetical protein c4523, C end (E. coli CFT073) | 60/71 | NP_756383.1 |
| api64 | 486 | Probable tRNA synthetase RSp1438 (R. solanacearum) | 48/64 | NP_522997.1 |
| api65 | 154 | Hypothetical protein RSp1437 (R. solanacearum) | 35/46 | NP_522996.1 |
| api66 | 157 | Putative acetyltransferase RSp1436 (R. solanacearum) | 47/61 | NP_522995.1 |
| api67 | 542 | Putative AMP-binding enzyme RSp1434 (R. solanacearum) | 50/62 | NP_522993.1 |
| api68 | 219 | Hypothetical proteins RSp1433 (R. solanacearum) and RSp1432 (R. solanacearum) | 54/70 | CAD18584.1 |
| 42/60 | CAD18583.1 | |||
| api69 | 348 | Putative oxidoreductase signal peptide RSp1431 (R. solanacearum) | 39/54 | NP_522990.1 |
| api70 | 264 | Hypothetical protein RSp1430 (R. solanacearum)b | 42/58 | NP_522989.1 |
| api71 | 298 | Putative esterase RSp1429 (R. solanacearum)b | 36/49 | NP_522988.1 |
| api72 | 132 | Rhodanese-like protein RSp1428 (R. solanacearum)b | 66/84 | NP_522987.1 |
| api73 | 121 | Hypothetical protein RSp1426 (R. solanacearum) | 45/62 | NP_522985.1 |
| api74 | 443 | Probable l-ornithine 5-monooxygenase oxidoreductase protein PvdA (R. solanacearum) | 57/71 | NP_522984.1 |
| api75 | 443 | Probable diaminobutyrate-pyruvate aminotransferase protein EctB (R. solanacearum) | 60/73 | NP_522983.1 |
| api76 | 410 | Putative transmembrane protein RSp1423 (R. solanacearum) | 44/60 | NP_522982.1 |
| api77 | 319 | Putative transmembrane protein RSp1427 (R. solanacearum) | 28/51 | NP_522986.1 |
| api78 | 305 | Hypothetical protein BtrH (Bacillus circulans) | 25/39 | BAC41215.1 |
| api79 | 344 | IS110-like transposase (Shewanella oneidensis) | 47/66 | NP_719476.1 |
| api80 | 88 | IS911-like inactive transposase (E. coli) | 95/98 | CAD48417.1 |
| api81 | 344 | IS110-like transposase (Shewanella oneidensis) | 45/68 | NP_719476.1 |
| api82 | 102 | DNA-binding phagic protein YPO1092, N end (Y. pestis) | 50/72 | NP_404706.1 |
| api83 | 340 | IS100 transposase (Y. pestis) | 100 | NP_403697.1 |
| api84 | 259 | orfB of IS100 (Y. pestis) | 100 | NP_395139.1 |
| api85 | 236 | DNA-binding phagic protein YPO1092, C end (Y. pestis) | 47/67 | NP_404706.1 |
| api86 | 17 | Transposase, fragment (Shewanella oneidensis) | 77/94 | NP_858158.1 |
| api87 | 206 | Hypothetical protein (Photorhabdus luminescens) | 66/82 | NP_931453.1 |
| api88 | 157 | Hypothetical protein SciY (S. enterica serovar Typhimurium) | 40/57 | NP_459292.1 |
| api89 | 1,423 | Hypothetical protein similar to Rhs family Plu4280 (P. luminescens) | 54/67 | NP_931456.1 |
| api90 | 143 | Hypothetical protein STY0320 (S. enterica serovar Typhi) | 33/53 | AF0538 |
| api91 | 323 | Hypothetical protein (P. fluorescens) | 47/60 | ZP_00087873.1 |
| api92 | 301 | Putative membrane protein YccB (S. enterica serovar Typhimurium) | 32/52 | NP_052478.1 |
| api93 | 215 | pilK (S. enterica serovar Typhimurium) | 24/40 | BAA77971.1 |
| api94 | 398 | Hypothetical protein STY 4665 (S. enterica serovar Typhi) | 26/40 | NP_928477.1 |
| api95 | 326 | Probable phage integrase STY 4666 (S. enterica serovar Typhi) | 54/70 | NP_928478.1 |
aa, amino acids.
CDS (entire or not) present in the Y. pestis chromosome.
The 98,058-bp segment harbors the previously described 11-kb pil locus (6). A striking finding is the presence of an ORF (api93) near the 3′ end of the island; it is homologous to the Salmonella enterica pilK gene. Expression of the pil operon was found to be transcriptionally regulated (6), and a putative transcriptional regulator gene (api22) is contiguous to pilW (the last gene of the pil operon) but with opposite polarity. Another notable feature is the presence of a gene cluster (api49, api50, and api52) that is predicted to encode enzymatic subunits of a type I restriction-modification system (18). Finally, the downstream region of the 98-kb chromosomal fragment comprises a 13-kb region (api64 to api77), the gene organization and products of which are similar to an R. solanacearum megaplasmid segment containing 15 CDSs (rsp1423 to rsp1438), mostly involved in general metabolism (21).
Deletion of the 98-kb fragment occurs at low frequency in Y. pseudotuberculosis 32777.
PAIs tend to display deletion of specific sequences or even the whole genetic element (23). Except for type IV pili production, no other phenotypic traits were known to be associated with the presence of the 98-kb fragment in Y. pseudotuberculosis 32777. Therefore, since we failed to detect pil deletion phenotypically, we screened spontaneous deletion mutants after insertion of a selection gene [kanamycin resistance aph(3′)-IIIa] and a counterselectable marker (the sacB levane sucrase-encoding gene) into the pil gene of Y. pseudotuberculosis 32777 (14). We placed the two reporter genes, as a control, upstream of the Y. pseudotuberculosis urease operon, a chromosomal region known to be stable (5). Since the product of the sacB gene is toxic for gram-negative bacteria in the presence of sucrose, only clones from which sacB is deleted will grow on agar containing this sugar. Selection on 10% sucrose and then on kanamycin medium revealed that when the reporter genes were located within pil, the occurrence of sucrose-resistant, kanamycin-sensitive clones was as frequent as when they are inserted upstream of the urease locus (mean value of four separate experiments ± the standard deviation = 1.7 ± 2.5 × 10−7 versus 0.6 ± 0.3 × 10−7, respectively). Two independent, sucrose-resistant, kanamycin-sensitive clones (ΔYAPI1and ΔYAPI2) were further characterized. PCR analysis and sequencing of PCR products revealed perfect excision of the 98,058-bp segment in these deletion mutants; they carried an intact phe-tRNA gene.
Apart from the pil locus, the 98-kb fragment does not contain any other genes for virulence in the mouse.
We previously reported that the Y. pseudotuberculosis type IV pilus gene cluster contributes to bacterial virulence in the mouse (6). PAIs often contain several pathogenicity genes (23). To determine whether any other genes on the 98-kb segment (i.e., apart from pil) are potentially involved in bacterial virulence (especially those encoding products without any known function), we compared rates of survival of BALB/c mice inoculated orally or intravenously with Y. pseudotuberculosis 32777, from which either the 98,058-bp DNA segment (strain ΔYAPI1) or the pil locus (strain MIV) had been deleted. When given orally, all mutants were found to be less virulent in the mouse than was the wild-type strain 32777. However, deletion of the entire PAI did not result in a significant decrease of bacterial virulence compared to inactivation of the pil gene cluster alone (the 50% lethal dose [LD50] for the wild type was ∼107 cells; the LD50s for MIV and ΔYAPI were ∼108 cells). Bearing in mind the limits of our model of oral infection, it appears that no virulence genes other than the fimbrial gene cluster pil are harbored on the 98-kb chromosomal segment. Concerning the intravenous route, the LD50 values for the three strains were all <102 cells, indicating that no virulence factors required for systemic infection are encoded within YAPI.
YAPI is not complete in all Y. pseudotuberculosis strains and can be situated in either of the two phe-tRNA genes.
Detection of YAPI in 11 Y. pseudotuberculosis strains (originating from various geographical areas and belonging to six different O serotypes) was screened by PCR with primers located outside mobile genetic elements. Three primer sets (B, A, and C), respectively, amplified fragments of the conserved pilin-encoding gene pilS (6) and the CDSs located upstream (api5) and downstream (api24 to api26) in the pil operon; two others (D and E) amplified the Ralstonia megaplasmid homologues api64 and api74 (Table 1). As shown in Table 2, YAPI was not present in all strains tested, and its distribution was found to be independent of the O serotype. It is interesting that for three of the six YAPI-positive strains (32945, ST, and 1553), amplification products were yielded with primer sets A, B, and C but not with primer sets D and E. Our data thus indicate that these strains lacked the YAPI fragment homologous to that in Ralstonia (which we called the metabolic segment).
TABLE 2.
Distribution of YAPI among different Y. pseudotuberculosis serotypesa
| Strain | Serotype | Presence (+) or absence (−) of amplimers yielded with primer set:
|
||||
|---|---|---|---|---|---|---|
| A | B | C | D | E | ||
| YPT9 | O:1 | + | + | + | + | + |
| 58/87 | O:1 | − | − | − | − | − |
| 2926 | O:2 | + | + | + | + | + |
| 33054 | O:2 | − | − | − | − | − |
| 32945 | O:3 | + | + | + | − | − |
| 32842 | O:3 | − | − | − | − | − |
| ST | O:4 | + | + | + | − | − |
| 1830 | O:4 | − | − | − | − | − |
| 199/90 | O:5 | + | + | + | + | + |
| 1553 | O:6 | + | + | + | − | − |
| 682/90 | O:6 | − | − | − | − | − |
The first three primer sets and the two last ones amplified portions of the adhesion and metabolic segment, respectively. Amplification products generated with primer sets A (605 bp), B (523 bp), C (1,017 bp), D (888 bp), and E (941 bp) were analyzed by agarose gel electrophoresis.
Two copies of the phe-tRNA gene have been detected in the chromosome of Y. pestis and Y. pseudotuberculosis 32777, and their nucleotide sequences were identical in each of the two species (7, 17; F. Collyn et al., unpublished data). To determine into which allele YAPI is inserted, PCRs were carried out with primer sets F and G, i.e., flanking each phe-tRNA gene. In the six YAPI-positive strains, the PAI was found to be integrated into either tRNA copy.
DISCUSSION
We describe here a large (98-kb) Y. pseudotuberculosis chromosomal segment, YAPI, with all of the characteristics of a PAI: (i) it bears at least one gene cluster involved in bacterial pathogenicity (the pil locus, encoding a type IV pilus); (ii) it is associated with a tRNA locus (phe-tRNA); (iii) it is flanked by direct-repeat sequences; (iv) it comprises a variety of mobility genes (whether intact or not), notably an ORF encoding a prophage integrase and situated upstream of the flanking tRNA gene; and, finally, (v) its sequence has a heterogeneous GC content, reflecting a genetic mosaic (Fig. 1). Deletion at high frequency is a common feature of PAIs. This is not the case for YAPI, which excises from the Y. pseudotuberculosis chromosome in only 1 per 107 cells. However, high stability of these genetic elements (although infrequent) has indeed been reported elsewhere (23).
Like the HPI, YAPI is not uniformly distributed within the species (Table 2). However, whereas the HPI is recovered only in O:1 and O:3 strains (4), YAPI seems to be spread across the species independently of the O serotype. This is consistent with a previous study in which we showed that the pil locus was detected in various Y. pseudotuberculosis serotypes (6). However, this finding needs to be confirmed by an extensive study on a broader strain collection (such studies are in progress). This strongly suggests that YAPI has emerged earlier in Y. pseudotuberculosis than did the HPI. Another feature shared by the HPI and YAPI is the insertion of the two PAIs into either copy of their target tRNA gene (asn for HPI and phe for YAPI) (4). YAPI can spontaneously and perfectly excise itself from the chromosome and comprises an intact putative integrase-encoding gene. Although we failed to detect an excised, circular form of YAPI (most probably due to the relative infrequency of this type of genetic event), intracellular mobility of the PAI is an alternative to a random phe-tRNA insertion or homologous recombination between two copies of the same gene.
In contrast to the HPI, YAPI was not detected in Y. pestis. This finding was established both by in silico analysis of the sequenced genomes of strain KIM (7) and strain CO92 (17) and by PCR experiments with primer sets A and B on several other strains of the Medievalis and Orientalis biovars (strains 518 and 556 and strains 573, 610, and 612, respectively), as well as in biovar Antiqua strains (strains 536, 537, and 539) (data not shown). However, a DNA segment homologous to YAPI (although shorter in length [66 kb]) was detected in silico in the genome-sequenced strain 8081 (biotype 1B [O:8]) of Y. enterocolitica. Also associated with a phe-specific tRNA locus, this fragment shared 41 of its 61 predicted ORFs (64 to 97% identity at the protein level) with the YAPI from Y. pseudotuberculosis 32777 (unpublished data).
The right part of Y. pseudotuberculosis YAPI (which notably encompasses metabolic genes [see Table 1]) was missing in some strains of this species, whereas the left part (which includes the type IV pilus gene cluster and was designated the adhesion segment) was never seen to be lacking in PAI-positive strains (Table 2). The pil operon, which is highly conserved in distantly related strains on the basis of PCR-restriction fragment length polymorphism (data not shown), therefore represents the best probe for detecting YAPI in Y. pseudotuberculosis. Considering the PCR results presented in Table 2 raises the question of whether YAPI was generated in one or two steps. Would the PAI have emerged in one block of genes (which would then have sustained deletion of its right part) or, in contrast, would the latter have been added once the left part had been laterally acquired by the bacterium? In Y. enterocolitica, the presence of a gene cluster encoding putative proteins involved in arsenic resistance (instead of the homologous Ralstonia megaplasmid segment in the corresponding region of Y. pseudotuberculosis YAPI [Table 1 and unpublished data]) argues in favor of the second hypothesis. Comparative analysis of PAI gene composition in a large collection of YAPI-positive strains from various animal and environmental sources (work in progress) should provide insight not only into the pathogenesis and ecological fitness of Y. pseudotuberculosis but also into the PAI's evolution within the species.
Acknowledgments
We gratefully acknowledge J. Parkhill for his authorization to use Y. enterocolitica sequence data produced by the Yersinia enterocolitica Sequencing Group at the Sanger Institute (ftp://ftp.sanger.ac.uk/pub/pathogens/ye/). We also thank S. Aleksić, E. Carniel, J. Sundar, R. Van Noyen, and N. Takeda for supplying reference Yersinia strains.
F. Collyn received a doctoral studentship from the Ministère de l'Enseignement Supérieur, de la Recherche et de la Technologie and from the Fondation pour la Recherche Médicale. This study was supported in part by the European Regional Development Fund.
Editor: J. B. Bliska
REFERENCES
- 1.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchrieser, C., C. Rusniok, L. Frangeul, E. Couve, A. Billault, F. Kunst, E. Carniel, and P. Glaser. 1999. The 102-kilobase pgm locus of Yersinia pestis: sequence analysis and comparison of selected regions among different Yersinia pestis and Yersinia pseudotuberculosis strains. Infect. Immun. 67:4851-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carniel, E. 2001. The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 3:561-569. [DOI] [PubMed] [Google Scholar]
- 5.Carnoy, C., S. Floquet, M. Marceau, F. Sebbane, S. Haentjens-Herwegh, A. Devalckenaere, and M. Simonet. 2002. The superantigen gene ypm is located in an unstable chromosomal locus of Yersinia pseudotuberculosis. J. Bacteriol. 184:4489-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collyn, F., M. A. Léty, S. Nair, V. Escuyer, A. Ben Younes, M. Simonet, and M. Marceau. 2002. Yersinia pseudotuberculosis harbors a type IV pilus gene cluster that contributes to pathogenicity. Infect. Immun. 70:6196-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 9.Hill, C. W. 1999. Large genomic sequence repetitions in bacteria: lessons from rRNA operons and Rhs elements. Res. Microbiol. 150:665-674. [DOI] [PubMed] [Google Scholar]
- 10.Hinchliffe, S. J., K. E. Isherwood, R. A. Stabler, M. B. Prentice, A. Rakin, R. A. Nichols, P. C. Oyston, J. Hinds, R. W. Titball, and B. W. Wren. 2003. Application of DNA microarrays to study the evolutionary genomics of Yersinia pestis and Yersinia pseudotuberculosis. Genome Res. 13:2018-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knapp, S., J. Hacker, I. Then, D. Muller, and W. Goebel. 1984. Multiple copies of hemolysin genes and associated sequences in the chromosomes of uropathogenic Escherichia coli strains. J. Bacteriol. 159:1027-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 13.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Middendorf, B., G. Blum-Oehler, U. Dobrindt, I. Muhldorfer, S. Salge, and J. Hacker. 2001. The pathogenicity islands (PAIs) of the uropathogenic Escherichia coli strain 536: island probing of PAI II536. J. Infect. Dis. 183(Suppl. 1):S17-S20. [DOI] [PubMed] [Google Scholar]
- 15.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires. toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulder, N. J., R. Apweiler, T. K. Attwood, A. Bairoch, D. Barrell, A. Bateman, D. Binns, M. Biswas, P. Bradley, P. Bork, P. Bucher, R. R. Copley, E. Courcelle, U. Das, R. Durbin, L. Falquet, W. Fleischmann, S. Griffiths-Jones, D. Haft, N. Harte, N. Hulo, D. Kahn, A. Kanapin, M. Krestyaninova, R. Lopez, I. Letunic, D. Lonsdale, V. Silventoinen, S. E. Orchard, M. Pagni, D. Peyruc, C. P. Ponting, J. D. Selengut, F. Servant, C. J. Sigrist, R. Vaughan, and E. M. Zdobnov. 2003. The InterPro Database 2003 brings increased coverage and new features. Nucleic Acids Res. 31:315-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 18.Piekarowicz, A., A. Klyz, A. Kwiatek, and D. C. Stein. 2001. Analysis of type I restriction-modification systems in the Neisseriaceae: genetic organization and properties of the gene products. Mol. Microbiol. 41:1199-1210. [DOI] [PubMed] [Google Scholar]
- 19.Reiter, W. D., P. Palm, and S. Yeats. 1989. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 17:1907-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 21.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Schmidt, H., and M. Hensel. 2004. Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 17:14-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smego, R. A., J. Frean, and H. J. Koornhof. 1999. Yersiniosis I: microbiological and clinicoepidemiological aspects of plague and non-plague Yersinia infections. Eur. J. Clin. Microbiol. Infect. Dis. 18:1-15. [DOI] [PubMed] [Google Scholar]
- 25.Zhang, X. L., I. S. Tsui, C. M. Yip, A. W. Fung, D. K. Wong, X. Dai, Y. Yang, J. Hackett, and C. Morris. 2000. Salmonella enterica serovar Typhi uses type IVB pili to enter human intestinal epithelial cells. Infect. Immun. 68:3067-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]