Abstract
As the sole viral antigen on the HIV-1-virion surface, trimeric Env is a focus of vaccine efforts. Here we present the structure of the ligand-free HIV-1-Env trimer, fix its conformation, and determine its receptor interactions. Epitope analyses revealed trimeric ligand-free Env to be structurally compatible with broadly neutralizing antibodies, but not poorly neutralizing ones. We coupled these compatibility considerations with binding antigenicity to engineer conformationally fixed Envs, including a 201C-433C (DS) variant, specifically recognized by broadly neutralizing antibodies. DS-Env retained nanomolar affinity for the CD4 receptor, with which it formed an asymmetric intermediate: a closed trimer bound by a single CD4 without the typical antigenic hallmarks of CD4 induction. Antigenicity-guided structural design can thus be used both to delineate mechanism and to fix conformation, with DS-Env trimers in virus-like particle and soluble formats providing a new generation of vaccine antigens.
The human immunodeficiency virus type 1 (HIV-1) uses multiple mechanisms to evade the immune system, and these have stymied the development of an effective vaccine1–3. One mechanism – conformational masking4 –hides the vulnerable shape of the trimeric envelope (Env) spike recognized by broadly neutralizing antibodies via structural rearrangements that expose immunodominant epitopes recognized by non- or poorly neutralizing (‘ineffective’) antibodies5,6. The upshot is that virus infection and Env immunization both elicit abundant, Env-directed antibodies with little neutralization capacity7–9. A potential solution is to determine the structure of the vulnerable Env conformation and to use this structural information and protein design to stabilize or to fix the vulnerable shape.
Definition of the structure of trimeric HIV-1 Env has been accomplished at increasing resolution by crystallography and cryo-electron microscopy10–14. These studies have culminated in atomic-level structures of antibody-bound forms of a near-native trimer mimic, named BG505 SOSIP.664, for HIV-1 strain (BG505)15 and stabilizing mutations (SOSIP.664)16–18. Antibodies, however, can influence conformation. Structures of the Env gp120 subunit, for example, can differ substantially when ligand-free19 or bound to different antibodies3,6,20–24. HIV-1 Env, moreover, is a type 1 fusion machine, which utilizes structural rearrangements to drive the merging of virus and host-cell membranes during entry (reviewed in 25). Not only do substantial pre-fusion to post-fusion conformational changes accompany this process14,26,27, but single molecule-fluorescence resonance transfer (sm-FRET) analysis indicates that pre-fusion ligand-free Env on infectious virions undergoes transitions between at least three different conformations28.
When a viral antigen can assume multiple conformations, which is the “right” conformation to fix? Clues from smFRET28 and hydrogen-deuterium exchange (HDX) experiments29 suggest that a single dominant conformation, the mature pre-fusion closed state, is recognized by broadly neutralizing antibodies. Here we set out not only to fix HIV-1 Env in its vulnerable shape, but to determine the appropriate conformation to fix. We layered antigenic considerations – both structural and binding – onto structure-based design. To provide a basis for the analysis, we determined the crystal structure of the ligand-free HIV-1-Env trimer, and analyzed its structural compatibility with epitopes defined in previously determined antibody-bound Env structures. We coupled structural compatibility with binding measurements to identify both an “appropriate target conformation” and an “appropriate target antigenicity”, and used antigenicity-guided structural design to fix the desired target shape. We then examined the functional and antigenic consequences of conformational fixation. Functional analysis revealed HIV-1 Env to transition through an asymmetric intermediate, and antigenic analysis indicated improved specificity for broadly neutralizing antibodies. Together, our results provide a foundation by which to understand ligand-free HIV-1-Env trimer: its structure, its entry-related mechanistic interactions, and its conformational fixation as a means to overcome conformational masking.
RESULTS
Structure and properties of ligand-free HIV-1-Env trimer
To obtain the structure of mature ligand-free HIV-1 Env, we used a sparse-matrix approach30 to crystallize an endoglycosidase H-treated BG505 SOSIP.664 trimer from a PEG 400-PEG 3,350 precipitant mixture31. Diffraction data extended to 3.3 Å, but was anisotropic with a nominal resolution of 3.7 Å (Table 1). Because of the lower resolution, we were careful with crystallographic B-factors; refinement without B-factors did not reduce Rfree to below 33%, whereas refinement32 with group B-factor and TLS yielded Rwork/Rfree of 26.6%/28.5% (Fig. 1, Table 1 and Supplementary Figs. 1 and 2). The resultant B-factors correlated strongly with real-space correlation (Supplementary Table 1) suggesting a reflection of pre-fusion coordinate mobility.
Table 1.
Data collection and refinement statistics
| Ligand-free HIV-1 BG505 SOSIP.664 |
|
|---|---|
| Data collection | |
| Space group | P63 |
| Cell constants | |
| a, b, c (Å) | 107.6, 107.6, 107.6 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) | 50.0–3.30 (3.42–3.30)a |
| Rmerge | 9.4 (42.5) |
| I/σI | 14.3 (1.29) |
| Completeness (%) | 68.3 (14.6) |
| Redundancy | 5.2 (2.2) |
| Refinement | |
| Resolution (Å) | 35.3–3.3 |
| Unique reflections | 6,434 |
| Rwork/Rfree (%) | 26.6/28.5 |
| No. atoms | |
| Protein | 4,518 |
| Ligand | 338 |
| Water | 0 |
| B-factors (Å2) | |
| Protein | 93.8 |
| Ligand | 124.7 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 1.147 |
Values in parentheses are for highest-resolution shell; the highest resolution shell for which data were 50% complete with I/sigma greater than 2 was 3.91–3.72 Å. We therefore consider this structure to have a nominal resolution of 3.72 Å. One crystal was used for data measurements.
Figure 1.
Crystal structure of ligand-free HIV-1-Env trimer, and conformational changes related to individual subunit structures and virus entry. (a) The left gp120-gp41 protomer of the ligand-free BG505 SOSIP.664 trimer is shown in B-factor putty representation, the right protomer in Cα-backbone representation colored by average Cα distance between ligand-free Env and previously determined Env subunit structures, and the third protomer in grey cartoon representation. (b) Plot of pre-fusion Env B-factors versus Cα-subunit movement (see Supplementary Table 1 for a full listing of correlations and P-values for residue-level properties). (c) Correlations and P-values for pre-fusion B-factors of type 1 fusion machines versus pre-fusion to post-fusion movement of fusion subunit (see Supplementary Fig. 3 for subunit pictorials and correlation graphs). P-values in (b) and (c) were obtained by two-tailed student’s t test.
To provide insight into the physical characteristics of ligand-free HIV-1 Env, we analyzed residue and surface properties such as sequence entropy, atomic mobility, hydrophobicity, polarity and surface accessibility (Fig. 1 and Supplementary Figs. 1 and 2). Of these, Cα differences between separate subunit structures and the ligand-free trimer showed moderate correlation with B-factor (Supplementary Tables 1 and 2). When only the gp41 subunit was considered, the correlation increased (P < 0.0001) (Fig. 1b). Thus gp41 regions of ligand-free trimeric Env that are structurally similar in the separate subunit context exhibit lower B-factors, increased hydrophobicity, reduced surface accessibility, and reduced sequence variation: these regions include the nascent α7 coiled-coil that underpins the internal coiled-coil of the post-fusion six-helix bundle. In contrast, gp41 regions that differ in the separate subunit context, such as the α8 and α9 helices, displayed higher B-factors, increased polarity, and higher surface accessible in the pre-fusion closed state.
One explanation for these results is that gp41-residue level properties associated with pre-fusion mobility presage or predict regions that move between pre-fusion and post-fusion states. Indeed, pre-fusion B-factors correlated strongly with gp41 movement between pre-fusion to post-fusion states (Fig. 1c). Relevant to this, we also observed pre-fusion B-factors of the fusion subunit from influenza virus (HA2) and respiratory syncytial virus (F1) to correlate with pre-fusion to post-fusion movement (Fig. 1c, Supplementary Fig. 3). Thus clues to viral entry-related conformational change are to be found in the ligand-free pre-fusion structures of Env, with residues of the fusion subunits in these type 1 fusion machines poised on a mobility gradient to undergo the requisite conformational rearrangements required for entry.
Structural compatibility and appropriate target conformation
In addition to virus entry, HIV-1 Env functions to evade the humoral immune response, for which glycan shielding and conformational change are critical. In the refined ligand-free Env structure, we observed electron density corresponding to single protein-proximal N-acetyl glucosamine residues at all sites of N-linked glycosylation, except at residues 197, 262, and 332 (Env numbering follows standard HX convention)33, where additional monosaccharide residues were observed, or at residue 137, which was mostly disordered (Supplementary Fig. 4). Overall, despite differences in glycosylation and lattice packing (Supplementary Fig. 4), the structure of the ligand-free trimer assumed a closed conformation, which was highly similar to antibody-bound trimers12–14, especially the PGT122-35022-bound trimer14 with which it had a root-mean-square deviation (RMSD) in Cα positions of less than 1 Å, substantially lower than observed with monomeric gp120 (Fig. 2a).
Figure 2.
Ligand-free HIV-1-Env trimer is structurally compatible with epitopes of broadly neutralizing, but not ineffective, antibodies. (a) Superposition of ligand-free and antibody-bound HIV-1-Env structures. (Left) Ligand-free gp120 core monomer shown in ribbon representation, with regions of less (or greater) than 2 Å RMSD upon antibody binding shown in green (or magenta), and representative antibody-bound structures in gray. (Middle and right) Ligand-free and antibody-bound HIV-1-Env trimers. In the right panel, antibodies PGT122 and 35022 are shown in gray semitransparent surface, with rear protomer removed for clarity. RMSD values are reported in Supplementary Table 7. (b) Breadth-potency of broadly neutralizing (green) and ineffective (magenta) antibodies on a diverse 170 HIV-1-isolate panel. (c) Structural compatibility of ligand-free trimer by antibody epitope: an appropriate structural compatibility. The ligand-free Env structure is displayed as Cα-ribbon, with antibody epitope residues colored green (structurally compatible) or magenta (incompatible), and grey for non-epitope regions. RMSD (solid fill) and volume overlap (striped fill) with the respective antibody-Env complexes shown as a bar graph, with two linear scales split at RMSD and antibody-antigen volume overlap cutoffs of 2 and 500 Å3, respectively; bars below the respective cutoffs are colored green, and magenta otherwise. Antibody labels are colored green if the epitope is structurally compatible, magenta if incompatible, and gray if not present in the structure. (d) Ligand-free trimer structural compatibility versus antibody breadth. Volume overlap (left), RMSD (middle) and Antigenic Structural Compatibility (ASC) score (right) are graphed versus antibody breadth on a diverse 170 HIV-1-isolate panel; pictorial representations shown in Supplemental Fig. 5. P-values for Spearman correlations provided (n = 14).
To determine the appropriateness of the ligand-free closed trimer as a vaccine template, structurally specific for broadly neutralizing antibodies and incompatible with non- or poorly neutralizing antibodies, we first sought to categorize antibodies by their functional efficacy (Fig. 2b). We defined broadly neutralizing antibodies as those of greater than 35% breadth on a diverse panel of 170 isolates, with ineffective antibodies of less than 15% breadth (for an isolate to be considered sensitive in this breath analysis, we used an IC50 cutoff for antibody of <50 ug/ml). Antibodies b1234, 35O2235 and PGT13536 were close to the cutoff for the broadly neutralizing category; and while some antibodies such as the V3-directed 447-52D37 showed clade-specific breadth (447-52D neutralizes over 20% of clade B isolates), we nonetheless classified 447-52D as ineffective because its overall breadth was only 12% (Supplementary Table 3). We then analyzed the ligand-free closed structure for its structural compatibility with antibody epitopes – most determined structurally in the context of antibody-bound subunit or antibody-bound peptide – based on two measures: antibody-volume overlap and epitope RMSD (Fig. 2c and Supplementary Fig. 5). Antibody-volume overlap, which involves the superposition of epitopes in unliganded trimer and antibody-bound context (Supplemental Fig. 5), anti-correlated strongly with neutralization breadth (P = 0.0007) (Fig. 2d). Epitope RMSD, which compares epitope structural differences in unliganded trimer and antibody-bound context, trended with breadth, but did not achieve statistical significance. An antigenic structural compatibility score (ASC), which combined both overlap and RMSD, did achieve significance (P = 0.0031) (Fig. 2d).
The ligand-free closed structure was compatible with the epitopes for all broadly neutralizing antibodies, except those of the membrane-proximal external region, which recognize epitopes C-terminal to residue 664, and those of antibodies b1221 and CH10338, with CH103 exceeding a 2 Å threshold of epitope similarity and with b12 exceeding a volume threshold of 500 Å3 (Fig. 2d). In light of the poor epitope RMSD correlation with neutralization breadth (Fig. 2d), the RMSD threshold was somewhat arbitrary. Nonetheless, the specific incompatibility of these moderately effective CD4-binding site antibodies suggests that movement of residues of the CD4-binding site could occur relative to the ligand-free closed trimer and still achieve moderate neutralization breadth; indeed, induced trimer movements have been observed for b12 (which binds poorly to the BG505 SOSIP.664 trimer18) by electron microscopy10 and hydrogen-deuterium exchange29. By contrast, none of the epitopes for non- or poorly-neutralizing antibodies were structurally compatible with the ligand-free closed structure (Fig. 2d). These results indicate the ligand-free closed trimer to be structurally specific for neutralizing antibodies and thus an “appropriate target conformation” for immunogen design.
Appropriate target antigenicity for an Env-vaccine antigen
Structural specificity as measured by epitope compatibility is only one of the requirements of an appropriate vaccine template: antigenic specificity as measured by antibody binding is also crucial. The BG505 SOSIP.664 has previously been shown to be antigenically specific for broadly neutralizing antibodies, though binding to weakly neutralizing antibodies such as those directed at the V3 loop was reported18.
Our structural compatibility analysis indicated V3 antibodies to be incompatible with the ligand-free closed state (Fig. 2c), suggesting that the binding of BG505 SOSIP.664 to V3 antibodies might not be intrinsic to the closed conformation of the SOSIP.664 construct, but an artifact of alternative folding. Indeed, we found negative selection39 by weakly neutralizing V3-directed antibodies to substantially reduce V3 antibody binding to BG505 SOSIP.664 (Fig. 3a and Supplementary Data Set 1); when we tested recognition on a panel of V3-directed antibodies, negative selection by 447-52D and by a V3-antibody cocktail reduced recognition by V3-directed antibodies to a level similar to that observed for non-cognate antibody binding (Fig. 3b, c). We also tested CD4-negative selection, but a substantial non-CD4-binding subportion of Env trimers was not observed. Together, these results indicated that an “appropriate target antigenicity” for an Env-vaccine antigen (Fig. 3d) would involve no recognition by ineffective antibodies (including those directed at the V3 region), while maintaining recognition of broadly neutralizing antibodies (except those with moderate neutralization, directed at the CD4-binding site).
Figure 3.
Structural compatibility-guided negative selection and an appropriate target antigenicity. (a) Size-exclusion chromatography profile of crystallization-grade SOSIP before negative selection (left panel), and SDS-PAGE analyses of negative selection: first with antibody 447-52D, next with a cocktail of V3 antibodies, and third by CD4 (right panels). NR: non-reducing conditions, R: reducing conditions. (b) V3-epitope exposure on BG505 SOSIP.664 quantified by SPR on a panel of V3 antibodies and displayed as percent of total V3 epitope upon CD4 triggering. *V3 epitopes for antibodies 39F and 19b have not been structurally defined. (c) V3 antigenicity for all structurally defined antibodies in b mapped onto epitope atoms in the structure of the ligand-free Env trimer. (d) Appropriate target antigenicity. Binding antigenicity is displayed on the ligand-free Env trimer with epitope atoms with tight trimer binding to broadly neutralizing antibodies in green and to ineffective antibodies in magenta.
CD4 triggering and conformational stabilization
Even after V3-antibody negative selection, CD4 triggering could efficiently expose V3 epitopes40 recognized by antibodies like 447-52D41, 307442, and others, as well as bridging sheet epitopes20 recognized by antibodies like 17b43 (Fig. 4a; Supplementary Data Sets 2 and 3). Thus, while structural compatibility (and neutralization breadth) generally correlated with antibody binding, in the presence of CD4, this correlation was lost (Fig. 4a and Supplementary Data Set 2). Notably, CD4 triggered BG505 SOSIP.664 recognition of ineffective antibodies so that their average binding was tighter than that of the broadly neutralizing ones (Fig. 4a). Such CD4 triggering makes BG505 SOSIP.664 less desirable as an immunogen: in primates, it would bind CD4 in vivo and would thus be predicted to elicit primarily ineffective antibodies against highly immunogenic CD4-induced epitopes.
Figure 4.
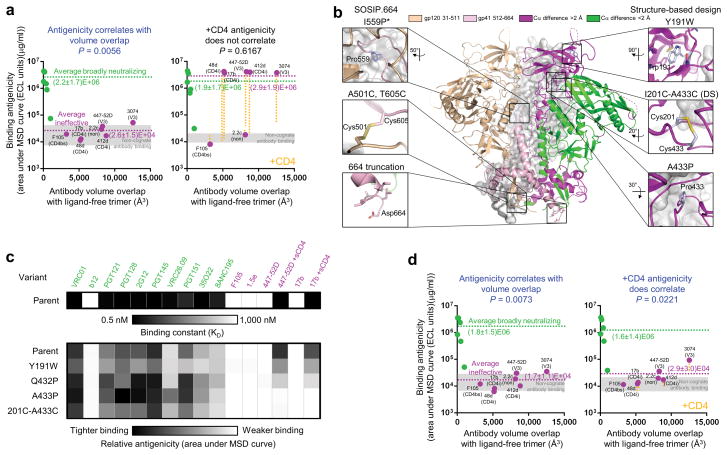
CD4-induced changes in antigenicity and conformational fixation of ligand-free HIV-1 Env. (a) BG505 SOSIP.664 ligand-free structural compatibility versus binding antigenicity, in the absence (left) and presence (right) of CD4 as measured by MSD. Antibodies are displayed in green (broadly neutralizing) and magenta (ineffective); the average binding of each is provided; and ineffective antibodies are labeled (CD4bs: CD4-binding site, CD4i: CD4-induced, non: non-neutralizing, V3: V3-loop directed). The change in binding to ineffective antibodies in the presence of CD4 is shown as a yellow dotted line. P-values for Spearman correlations are provided (n = 13). Binding levels for non-cognate antibodies lie within the gray shaded areas. (b) Conformational fixation of HIV-1-Env trimer. The central image depicts the ligand-free BG505 SOSIP.664 HIV-1-Env trimer, with two protomers shown in cartoon representation, one colored by domains (gp120 in wheat and gp41 in light pink), a second colored by RMSD distance between ligand-free trimer and subunit structures of CD4-bound gp120 and post-fusion gp41 (green if less than 2 Å, magenta if more than 2 Å). A third protomer is shown in gray. Insets: atomic-level details. *Residue 559 is disordered in the ligand-free structure. (c) Binding antigenicity of BG505 SOSIP.664 variants. Heat map showing binding of BG505 SOSIP.664 and variants that stabilized the ligand-free closed state to a panel of antibodies. (d) Ligand-free trimer structural compatibility versus BG505 SOSIP.664 201C-433C binding antigenicity in the absence (left) and presence (right) of CD4, with antibodies and average binding in green (broadly neutralizing) and magenta (ineffective) as in a. Elisa, Octet and Biacore data in Supplemental Data Set 2 and Supplementary Tables 5 and 6.
To fix the ligand-free closed state and to prevent CD4 triggering, we analyzed regions of the ligand-free closed structure that moved upon CD4 binding and identified cavity-filling hydrophobic substitutions, side-chain pairs capable of forming disulfide bonds, and positions where the introduction of a proline would be compatible with only the ligand-free closed structure of Env, but not its receptor-bound conformation (Fig. 4b). We engineered these substitutions into BG505 SOSIP.664, co-expressed them with furin in a 96-well transfection format44, and assessed supernatants on an antigenic panel comprising broadly neutralizing antibodies PGT12236 and VRC0145, quaternary-specific broadly neutralizing antibodies PGT14536 and CAP256-VRC2646, and poorly neutralizing antibodies F10547 and 17b43, with the latter tested alone and in the presence of CD4. In total, 124 constructs were tested (Supplementary Table 4). Promising constructs were purified and analyzed for gp120-gp41 cleavage and oligomeric heterogeneity (Supplementary Fig. 6) and by meso-scale discovery-electrochemiluminescence immunoassay (MSD-ECLIA) for recognition on a more comprehensive panel of HIV-1-reactive antibodies (Fig. 4c, Supplementary Data Set 2, and Supplementary Tables 5 and 6). One cavity-filling alteration, Y191W, was recognized by broadly neutralizing antibodies, but exhibited only moderately reduced binding by antibody 17b, while two proline substitutions, Q432P and A433P, showed improved antigenic specificity. A 201C-433C double cysteine mutant showed virtually no antibody 17b recognition, even in the presence of CD4, but was equivalently recognized by antibody PGT145 and was bound even better by antibody CAP256-VRC26 (Fig. 4c). While A433P was better recognized by broadly neutralizing antibodies compared to 201C-433C, the temporal stability of A433P assessed over 10 days at different temperatures (Supplementary Data Set 2b) was lower than that of both BG505 SOSIP.664 and 201C-433C, with 201C-433C exhibiting the highest temporal stability. Notably, with the 201C-433C variant, structural compatibility with ligand-free Env correlated with antibody binding, even in the presence of CD4 (Fig. 4d and Supplementary Data Set 2). Modeling indicated a 201C-433C disulfide to be incompatible with the CD4-bound state, where α-carbons (Cα) of residues 201 and 433 are 9.4 Å apart, separated by a strand of the bridging sheet20, and the V3 loop is fully exposed6 (Fig. 5a). By contrast, the 201C-433C substitutions are expected to form a disulfide in the ligand-free closed trimer, and indeed the ligand-free BG505 SOSIP.664 201C-433C exhibited a 6.1°C increase in thermostability (to 73.1° C), relative to the parent SOSIP.664 (Fig. 5b), as well as increased tolerance to other physical stresses such as pH and freeze-thaw (Fig. 5c). These results indicated the 201C-433C ‘DS’ variant of BG505 SOSIP.664 (hereon named DS-SOSIP.664) to be conformationally stabilized and not to be triggered by CD4.
Figure 5.
Atomic-level models and physical stability of ligand-free 201C-433C mutant (DS-SOSIP). (a) Atomic-level models of residues 201 and 433 in ligand-free pre-fusion closed state (grey) and CD4-bound state (yellow). Ribbon representations of the two structures are shown with residues that make-up the bridging sheet in the CD4-bound conformation colored orange, residues 201 and 433 in cyan spheres, and 201-433 Cα distance indicated. Variable loops are labeled. (Monomeric CD4-bound conformation modeled from PDB ID 2B4C6, 3U4E59 and 3JWD14). (b) Thermostability of the DS-SOSIP was assessed by differential scanning calorimetry. Raw data are shown in solid line (black for BG505 SOSIP.664 and red for DS-SOSIP), with corresponding curves from the fit shown in purple dashed lines. Tm values and error were obtained from the fit. (c) Physical stability of trimeric DS-SOSIP as determined by the quaternary-specific antibody VRC26.09 after 60 minutes of incubation at physical extremes or after 10 freeze-thaw cycles. Error bars are SEM of two technical replicates.
CD4 interaction of DS-stabilized HIV-1 Env
To define the interaction of the ligand-free conformation of DS-SOSIP.664 with CD4, we used surface plasmon resonance (SPR) (Fig. 6a). DS-SOSIP.664 recognized CD4 with a similar on-rate as the parent SOSIP.664, but with ~10-fold faster off-rate, resulting in a ~10-fold reduction in KD relative to SOSIP.664 (Fig. 6a). To test for CD4 triggering over a longer time scale, we incubated both DS-SOSIP.664 and parent SOSIP.664 for 100 h in the presence of CD4, and used SPR readout of antibody 17b or antibody 3074 to assess triggering. With the parent SOSIP.664, CD4 induced a slow transition to a state with bridging sheet formed (t1/2 of 3.3±0.7 h for antibody 17b43) and V3 loop exposed (t1/2 of 4.2±1.0 h for antibody 307442) (Fig. 6b and Supplementary Data Set 3d). With DS-SOSIP.664, we could not observe triggering by CD4 of bridging sheet or V3 regions over the entire 100 h time course (Fig. 6b and Supplementary Data Set 3d).
Figure 6.
DS-SOSIP binds a single CD4 without the typical antigenic hallmarks of CD4 triggering. (a) Binding of soluble CD4 to SOSIP.664 and mutants measured by SPR with single-cycle kinetics. Values in parenthesis report standard errors from fit of the data to a 1:1 Langmuir binding model. Note that the level of binding for the P313W mutant is roughly 3-times higher than either wild-type SOSIP or 201C-433C. (b) Time course of CD4 activation of HIV-1 Env as measured by SPR. To initiate the time course, CD4 was mixed with HIV-1 Env at time point 0, and after incubation (time shown on x-axis), assessed by SPR for interaction with antibody (y-axis). (Left) Binding to antibody 17b, which recognizes a bridging sheet epitope; (right) binding to 3074, which recognizes a V3 epitope (right). (c) Sedimentation equilibrium analytical ultracentrifugation measurements of BG505 SOSIP.664 and 201C-433C variant in presence of excess 2-domain soluble CD4. Stochiometry for both 2-domain and 4-domain CD4 as well as residual calculation are provided in Supplementary Data Set 4.
To define the stoichiometry of CD4 interaction, we employed sedimentation equilibrium analytical ultracentrifugation of parent and DS-SOSIP.664 variants in the presence of excess CD4. Both with 2-domain CD4 (Fig. 6c) and 4-domain CD4 (Supplementary Data Set 4), we observed molecular weights consistent with the parent SOSIP.664 binding two to three CD4s and DS-SOSIP.664 binding only one CD4. DS-SOSIP.664 thus appeared to capture Env in a single CD4-bound state.
DS-stabilized HIV-1 Env in the viral context
As the single-CD4-bound state could be SOSIP.664-specific, we sought to assess DS-stabilized Envs in other contexts. When DS mutations were placed into functional virus, they ablated entry (Supplementary Data Set 5). Single molecule fluorescence energy transfer (smFRET) measurements, utilizing donor-acceptors placed in the first and fourth variable Env loops of functional JR-FL viral spikes28, revealed DS mutations to reduce transitions from the ground state. DS-viral spikes remained primarily in the closed ground state, even in the presence of dodecameric CD448 (Fig. 7a).
Figure 7.
An asymmetric intermediate in the HIV-1-entry pathway. (a) smFRET of JR-FL virions with and without 201C-433C substitution. Population FRET histograms are each paired with transition density plots, which display the relative density of observed transitions. Results for both ligand-free and dodecameric CD4 (sCD4D1D2-Igαtp), graphed in left and right pairs, respectively. The dominance of the low FRET state even in the presence of CD4 indicates DS-stabilized Env to remain in a closed state even when bound by CD4. (b) HIV-1 entry mechanism with conformation-blocking mutations, antigenicity, and interactions with functional ligands. A new mechanistic state, characterized by the binding of a single molecule of CD4 with no bridging sheet formation and reduced V3 exposure, is highlighted in red. Env density from refs. 13 and 60.
Overall, the asymmetric single CD4-bound state – with fast off-rate for CD4 – appeared to be an obligatory intermediate between the ligand-free state and a more fully CD4-triggered state capable of binding multiple CD4s and co-receptor (Fig. 7b). In this context, we note that the high off-rate of CD4 in the single CD4-bound state, coupled with the slow transition to a 3:1 CD4:trimer stoichiometry, provides a kinetics-based molecular mechanism for the ability of primary HIV-1 isolates to resist neutralization by monomeric CD449.
A new generation of DS-fixed HIV-1-Env trimeric antigens
The ligand-free Env trimer – fixed in the pre-fusion closed conformation – may be an ideal HIV-1 antigen. To obtain information on the mobility of DS-SOSIP in both ligand-free and CD4-bound states, we characterized the hydrogen-deuterium exchange (HDX) of DS-SOSIP.664 and parent SOSIP.664 with and without CD4. Without CD4, the hydrogen-deuterium exchange of DS-SOSIP.664 appeared similar to the exchange of the parent SOSIP.664 (Fig. 8a, Supplementary Fig. 7 and Supplementary Data Set 6); with CD4, the gp120 inner domain, the bridging sheet, and gp41 showed little change (Fig. 8a). The V2, V3 and the stem of V1 showed a response to CD4, consistent with the slightly increased exposure of V3 epitope observed by MSD-ECLIA (Fig. 4d), but this was substantially less than observed for the parent SOSIP.664 (Fig. 8a). The CD4-bound DS-SOSIP.664 thus differs from previously observed CD4-bound states in that the typical hallmarks of CD4-induction – such as bridging sheet formation and V3 exposure – are absent or substantially reduced.
Figure 8.
A new generation of conformationally fixed HIV-1-Env trimeric immunogens. (a) HDX characterization of conformational mobility. Changes in exchange of hydrogen-deuterium relative to wild-type BG505 SOSIP.664 are displayed, with regions that become more ordered (blue) or disordered (red) shown on one lobe of the ligand-free trimer. Full HDX profiles are shown in Supplementary Fig. 7. (b) Antigenic characteristics of SOS virus-like particles (VLPs, comprising SOS mutant without IP) from strain JR-FL (modified with E168K to allow binding of V1V2-directed broadly neutralizing antibodies) and strain BG505 (modified with T332N to allow binding of 2G12 antibody). While broadly neutralizing antibody binding is maintained between parent and 201C-433C VLPs, the 201C-433C variant showed reduced ineffective antibody binding, especially in the presence of CD4. Broadly neutralizing antibodies binding are shown in green, ineffective in magenta and binding of antibodies in presence of 2 μg/ml of sCD4 in orange. Ineffective antibodies labeled (CD4bs: CD4-binding site, CD4i: CD4-induced, V3: V3-loop directed). (c) Information flow of antigenicity-guided immunogen design: from human antibody response through structure and antigenicity-guided design to conformationally fixed immunogens.
To see if the DS substitutions might serve as a general means of reducing CD4-induced transition in other Env antigens, we placed the 201C-433C and SOS mutations into HIV-1 Env expressed on the surface of enzyme-treated pseudovirions50. We chose two types of virions: strain JR-FL, modified with E168K51, to allow binding of V1V2-directed broadly neutralizing antibodies; and strain BG505, modified with T332N18, to allow binding of the 2G1252 antibody. We observed DS-modified viral spikes to resist CD4 triggering and to retain the antigenic profile of the soluble trimer for broadly neutralizing antibodies in both JR-FL and BG505 Env backgrounds (Fig. 8b). Overall, the results indicate the disulfide-shackled 201C-433C variants of soluble SOSIP.664 and VLP SOS to be highly desirable antigens: conformationally fixed trimers in which neutralizing epitopes are almost exclusively exposed, and non- or poorly neutralizing epitopes hidden, even in the presence of CD4.
Discussion
The path to identifying the 201C-433C ‘DS’ mutant involved an information flow from broadly neutralizing antibodies, through the ligand-free Env structure and an analysis of its structural compatibility and binding antigenicity, to obtain conformationally fixed antigens of desired target antigenicity (Fig. 8c). This type of antigenicity-guided structural design may be generally applicable, and indeed an analogous design path was followed in the conformational stabilization of the fusion (F) glycoprotein from respiratory syncytial virus (RSV), to achieve high levels of RSV-neutralizing antibodies44. However, RSV F assumes primarily two conformations, pre-fusion53 and post-fusion54,55, whereas HIV-1 Env can assume a multitude of conformations, including at least three pre-fusion states28 and numerous conformations of separate gp120 and gp41 subunits. The conformational fixation of Env thus required analysis of antibody efficacy and antibody-induced conformation, and we formalized this with an in-depth analysis of structural compatibility. An understanding of the conformational complexity of the HIV-1 Env, moreover, has the potential for mechanistic dividends: fixing the conformation of a particular Env entry intermediate – in this case the ligand-free closed state – provided a means to define the mechanistic interactions of that particular state.
Unexpectedly, analysis of the DS mutant indicated an asymmetric mechanism of entry, with CD4 binding separated into two steps (Fig. 7b): first, recognition by one CD4 without the previously recognized antigenic hallmarks of CD4 binding such as bridging sheet formation; and second, recognition by more than one CD4 along with exposure or formation of characteristic CD4-induced epitopes. The restricted stoichiometry of DS-SOSIP.664 is unlikely a consequence of steric hindrance by one CD4 of the binding of additional CD4 molecules; rather the conformational change induced by the first CD4 apparently generates sufficient alterations of the Env trimer to impede the binding of a second CD4. In this context, we note that in SPR characterizations of CD4 induction (Fig. 6b), we observed a Hill co-efficient of 0.95±0.10 for the P313W mutant, which is able to rapidly engage three CD4s, and a Hill co-efficient of 0.69±0.06 for SOSIP.664, which displayed a t1/2 for CD4-induction of several hours, suggesting negative cooperativity with the binding of one CD4 impeding the binding of additional CD4s. While similar asymmetric or restricted binding for CD4 has been reported with other trimeric HIV-1-Env constructs56,57, none of these other reports observed a single CD4 to bind without the typical antigenic hallmarks induced by CD4 such as bridging sheet formation20. In general, we believe the single CD4-bound trimeric state of Env is not specific to DS-SOSIP.664, but an obligatory intermediate of an asymmetric entry pathway (Fig. 7c).
In addition to providing mechanistic insight into HIV-1 entry, antigenicity-guided conformational fixation can also improve the antigenic specificity of HIV-1-Env immunogens. We used the ligand-free HIV-1-Env trimer as a target structure, because ligand-free immunogens are typically used for vaccination. In hindsight, we could have used the PGT122-35O22 antibody-bound trimer structure14 as it is highly similar to the ligand-free structure; however, at the outset of the study, this similarity was anticipated only at the label-specific resolution of smFRET28. We nevertheless managed to achieve our goal: the creation of DS-Env antigens –conformationally fixed in the vulnerable shape– that were not recognized by ineffective antibodies (including those directed against V3) and were not triggered by CD4 to unveil epitopes for ineffective antibodies.
Even with conformational masking disabled, HIV-1 Env is additionally protected by immune evasion involving genetic variation and glycan shielding and would thus be protected by evasion mechanisms similar to those utilized by influenza virus hemagglutinin (Supplemental Fig. 8). Immunization with conformationally fixed HIV-1 Env might therefore be expected to elicit immunological responses similar to those elicited by the seasonal flu vaccine: strain-specific responses with little neutralization breadth. Indeed, this is what initial reports of immunization with BG505 SOSIP.664 are observing58. While an improvement over what is elicited by conformationally masked Env (e.g. by gp120 subunit immunization), such strain-specific neutralization is unlikely to be generally protective. We note, however, that the immunogens in these initial reports exhibit V3-epitope exposure18, in contrast to the V3-negatively selected DS-SOSIP described here. As a result, DS-SOSIP might be expected to induce responses that are more focused on desirable neutralizing epitopes. Altogether, since the 201C-433C ‘DS’ mutation provides a means to overcome conformational masking, it should provide the basis for a new generation of vaccine antigens; however, future efforts to elicit broad HIV-1-neutralizing antibodies may require additional immunogen engineering to overcome remaining neutralization-evading mechanisms of genetic variation and glycan shielding.
Online Methods
BG505 SOSIP.664 expression, purification, and deglycosylation
BG505 SSOIP.664 trimer was produced in HEK 293 GnTI −/− cells via transient transfection of the BG505 SOSIP expressing plasmid with furin and purified as described previously over 2G12-affinity column12,14,18 (see Supplementary Note for additional details, including transient transfection in 96-well plates). The eluted protein was then dialyzed against PBS and set for deglycosylation reaction at 37 °C in the reaction buffer containing 1 mM EDTA, 150 mM NaCl, protease inhibitor cocktail (Roche), 17,000 units of Endo H/ml, and 50 mM sodium acetate, pH 5.8. The deglycosylated BG505 SOSIP was further purified with Superdex 200 16/60 (GE Healthcare) column in the buffer containing 5 mM HEPES 7.5, 150 mM NaCl, and 0.02% NaN3. The peak corresponding to trimeric HIV-1 Env was identified, pooled and concentrated to ~10 mg/ml using an Amicon Ultra-15 centrifugal filter (MWCO 50,000, Millipore) and screened for crystallization. Similarly purified “crystallization-grade” samples were also used for HDX experiments.
For most antigenicity and stability analyses, after trimers were purified by affinity chromatography and gel filtration over a Superdex 200 16/60 (GE Healthcare) column in buffer containing 5 mM HEPES 7.5, 150 mM NaCl, and 0.02% NaN3, they were subjected to negative selection39. This generally involved a V3-antibody 447-52D (PDB ID 4M1D61) affinity column to remove aberrant trimer species. However, for select antigenic analyses (e.g. Figs. 4a and 4d), an additional column comprising a cocktail of V3-directed antibodies, 1006-15D, 2219, 2557, 2558, 3074 and 50.1 (PDB IDs 3MLW42, 2B0S62, 3MLS42, 3UJI63, 3MLX42, 1GGI64, respectively), was employed (Fig. 3, Supplementary Data Set 1 and Supplementary Note).
Crystallization screening
Deglycosylated BG505 SOSIP.664 was screened for crystallization using 572 conditions from Hampton, Wizard and Precipitant Synergy31 screens using a Cartesian Honeybee crystallization robot as described previously59 and a mosquito robot using 0.1 μl of reservoir solution and 0.1 μl of protein solution. Crystals suitable for structural determination were obtained robotically in 26% PEG 400, 3.2% PEG 3350, and 0.1M sodium acetate pH 5.5. Crystals were cryoprotected in a solution containing 30% glycerol, 30% PEG 400, 4% PEG 3350, and 0.1M sodium acetate pH 5.5, and flash-frozen in liquid nitrogen. Data were collected at a wavelength of 1.00 Å at the SER-CAT beamline ID-22 (Advanced Photon Source, Argonne National Laboratory).
X-ray data collection, structure solution and model building
Diffraction data were processed with the HKL2000 suite65. The data were corrected for anisotropy using the anisotropy server http://services.mbi.ucla.edu/anisoscale/with truncations to 3.7 Å, 3.7 Å, 3.3 Å along a, b, and c axes, respectively. Structure solution was obtained with Phaser using 35O22- and PGT122-bound BG505 SOSIP.664 (PDB ID 4TVP14) as search models. Refinement was carried out with Phenix32. Model building was carried out with Coot66. Data collection and refinement statistics are shown in Table 1.
Structural analyses involving residue-specific properties
To estimate the degree of structural flexibility in the ligand-free HIV-1 trimer, we determined the average Cα RMSD distance for each residue position in the ligand-free trimer structure (Fig. 2d, e). The average Cα RMSD distance served as a proxy for structural plasticity and was computed between corresponding residues after optimal superimposition onto a set of 98 structures from the Protein Data Bank (PDB)67. Each domain of the ligand-free trimer was considered separately and superimposed onto the set of structures using either the program TM-align68 or single value decomposition (SVD). To obtain the correct registry between corresponding residues, structural superimpositions were guided by amino acid sequence alignments when necessary. A total of 63 monomeric structures were used for superimpositions involving the gp120 domain (Supplementary Table 2). To generate Figure 1a (left), we used five representative gp120 structures; ligand-free clade A/E HIV-1 gp120 coree (3TGT)19, b12-bound gp120 (2NY7)21, b13-bound gp120 (3IDX)3, F105-bound gp120 (3HI1)3, and VRC01-bound gp120 (3NGB)69 structures. For the gp41 domain we used a total of 35 structures from the PDB that included hexameric bundles as well as disordered peptides (Supplementary Table 2).
To understand the dynamic properties of ligand-free BG505 SOSIP.664, qualitative exchange profiles for observable peptides of SOSIP.664 after 3s were extracted from individual HDX -MS exchange plots70. The average exchange values (0–75%) were substituted in the B-factor field for the observed peptides of closed, ligand-free BG505 SOSIP.664 coordinates and displayed within PyMol (Supplementary Figs. 1g and 2g). Non-observable peptides in the deuterium exchange experiment as well as peptides with missing electron density were excluded from the analysis.
Other residue-specific properties were calculated and shown Supplementary Table 1. These included residue depth71, solvent-accessible surface area (SASA)72, sequence variability and hydrophobicity. Residue sequence variability was computed as the Shannon entropy for each residue position based on a representative set of 3,943 HIV-1 strains (Supplementary Figs. 1e and 2e). The electrostatic potential surfaces were generated using GRASP73. The Pearson correlation coefficient and associated P-values (computed using two tailed t-test) were computed using the statistical package R. Residue-level and surface property analyses were carried out with coordinates that differed slightly from the deposited PDB (RMSD of the analyzed coordinates differed by 0.02 Å versus the deposited PDB for regions compared, and B-factors were identical.
Assessment of antibody functionality on a panel of 170 diverse HIV-1
Neutralization was measured using single-round-of-infection HIV-1-Env pseudoviruses and TZM-bl target cells, as described previously74. Neutralization curves were fit by nonlinear regression using a 5-parameter hill slope equation. The 50% and 80% inhibitory concentrations (IC50 and IC80) were reported as the antibody concentrations required to inhibit infection by 50% and 80% respectively. The following antibodies were tested for HIV-1 neutralization efficacy- 17b20, 48D75, E5176, 35O2277, 447-52D61, 8ANC19578, VRC01 45, b12 21, b13 3, CH103 79, F105 3, PG9 36, PG16 36, PGT145 36, PGT128 36, PGT13536 and 2G1280.
Computation of antibody epitope RMSD, volume overlap, and epitope presence
HIV-1-specific antibody-antigen complex structures were compiled from the PDB, and antibodies were defined as broadly or poorly/non-neutralizing based on published or in-house neutralization data of diverse viral strains81. Antibodies that were deemed to have insufficient evidence for being classified as broadly or poorly/non-neutralizing were excluded from the analysis. A single antibody representative was included in the analysis in cases where multiple antibody clonal relatives were found. The epitope residues for each antibody were defined based on the respective antibody-antigen complex crystal structures, with an antigen residue being defined as an epitope residue if any of its heavy atoms were within 5.5 Å of any antibody heavy atom. To compute the RMSD between the epitope residues in the antibody-antigen complex structure and the same residues in the ligand-free trimer structure: (1) the epitope residues from the complex structure were aligned to the ligand-free trimer structure using the align function in PyMOL, then (2) the Cα RMSD of the epitope residues was calculated. To remove outlier residues, the top and bottom 10% of the Cα deviations were removed from the RMSD calculation. To calculate the volume overlap between a given antibody and the ligand-free trimer structure, the alignment from above was used to compute the overlap volume between the antibody from the complex structure and the ligand-free trimer structure by using the phase_volCalc utility from Schrödinger (www.schrodinger.com). An antibody epitope was considered as present in the ligand-free trimer structure if at least 70% of the epitope residues as defined by the antibody-antigen complex structure were also present in the ligand-free trimer structure. For mapping the per-residue RMSD computation onto the ligand-free trimer structure, residues part of any antibody epitope (including epitopes with less than 70% total residues present) were included in the analysis; if a given residue was part of more than one antibody epitope, the highest RMSD value for that residue among all epitopes was used. Antibody volume overlap values were mapped onto the ligand-free trimer structure for all residues part of the epitope for the given antibody; if a residue was part of more than one antibody epitope, then the lowest volume overlap for that residue among all epitopes was used. Correlations of structural properties with neutralization and/or binding data were computed using the Spearman correlation coefficient with two-tailed P-values.
Structural compatibility analysis
For a given antibody, the Antigenic Structural Compatibility (ASC) score with the HIV-1-Env ligand-free pre-fusion trimer structure was computed based on a comparison to a structure of the antibody bound to an Env-derived antigen (e.g., gp120 core or V3 peptide). ASC scores were computed on a 0–1 scale using the following variables: (i) The fraction f of epitope residues (as defined by the structure of the antibody complex) exposed to solvent in the ligand-free trimer structure was computed, with a residue considered accessible to solvent if its solvent-accessible surface area (SASA) was at least half its SASA in the respective antibody complex structure; f was set to 0 if less than 70% of epitope residues were present in the antigen. (ii) A resolution estimate r was used such that Cα RMSDs d below r=2 were not penalized in the scores. (iii) The volume overlap values were used to define a volume overlap factor v that is equal to 1 for overlap below 200 Å3, is equal to 0 for overlap over 1000 Å3, and decays linearly in between. Intuitively, the ligand-free trimer structure is expected to be structurally compatible with an antibody if f and v are high and if the RMSD d is low, since such conditions would indicate similarity between the ligand-free trimer structure and the Env conformation in the antibody complex. Thus, the ASC score for each antibody with the ligand-free trimer was defined by the formula: f v exp(−0.5 max(0, d − r)).
Antigenic analysis of BG505 SOSIP.664 and mutants by MSD-ECLIA and ELISA
Standard 96-well bare MULTI-ARRAY Meso Scale Discovery (MSD) Plates (MSD, cat #L15XA-3) were coated with a panel of HIV neutralizing (VRC0145, b1221, PGT12136, PGT12836, 2G1280, PGT14536, VRC26.0982, 35O2277 and 8ANC19578), non-neutralizing monoclonal antibodies (F1053, 1.5e83, 17b20 and 447-52D61) and non-cognate antibodies (anti-influenza antibodies CR911484 and CR802085, and anti-RSV antibodies palivizumab86, D2553 and 5C453) in duplicates (30 μL/well) at a concentration of 10 μg/mL, diluted in 1X PBS and the plates were incubated overnight at 4°C. The following day, plates were washed (wash buffer: 0.05% Tween-20 +1 X PBS) and blocked with 150 μL of blocking buffer [5% [W/V] MSD Blocker A (MSD, Cat # R93BA-4)] and incubated for 1 hr on a vibrational shaker (Heidolph TITRAMAX 100 ;CAT # P/N: 544-11200-00) at 650 rpm. All the incubations were performed at room temperature, except the coating step. During the incubation, BG505 SOSIP trimer was titrated down in a serial 2 fold dilutions starting at 4 μg/mL concentration of the trimer in assay diluent (1% [W/V] MSD blocker A + 0.05 % Tween-20). After the incubation with blocking buffer was complete, the plates were washed and the diluted trimer was transferred (25 μl/well) to the MSD plates and incubated for 2 hrs on the vibrational shaker at 650 rpm. For soluble CD4 (sCD4) induction, trimer was pre-incubated with sCD4 at a constant molar concentration of 1uM for 1 hour before adding to the MSD plate. After the 2 hr incubation with trimer, the plates were washed again and secondary detection MSD Sulfotag labeled D7324 antibody (prior to running the assay D7324 antibody was labeled with MSD Sulfotag (MSD; Cat #R91AN-1) at a conjugation ratio of 1:15 [D7324: Sulfotag]), which was diluted in assay diluent at 5 ug/mL and was added to the plates (25 μL/well) and incubated for 1 hr on the vibrational shaker at 650 rpm. The plates were washed and read using the 1X read buffer (MSD Read Buffer T (4x); Cat# R92TC-2) on MSD Sector Imager 2400.
See Supplementary Note for ELISA methods.
Surface plasmon resonance analysis
Affinities and kinetics of binding to BG505 SOSIP.664 soluble trimer and its mutants were assessed by surface plasmon resonance on a Biacore T-200 (GE Healthcare) at 20° C with buffer HBS-EP+ (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.05% surfactant P-20). Affinities for mAbs PGT121 and 2G12 were taken from published values18.
For assessing binding of trimer to sCD4, single-cycle kinetics analyses were carried out. First, ~2000RU of antibody 2G12 were immobilized on two flow cells. Next, 200 nM of trimer was injected on the sample flow cell. Finally, 5 concentrations of sCD4 (100 nM, 50 nM, 25 nM, 12.5 nM, 6.25 nM) were injected incrementally in a single cycle, starting from the lowest concentration, followed by a dissociation phase of 30 min. Blank sensorgrams were obtained by injection of same volume of HBS-EP+ buffer in place of sCD4. Sensorgrams of the concentration series were corrected with corresponding blank curves and fitted globally with Biacore T200 evaluation software using a 1:1 Langmuir model of binding.
For determining the time-course of CD4 activation of the soluble trimers, 17b IgG, 3074 IgG and 2G12 IgG were captured on three separate flow cells of a CM5 chip immobilized with ~10,000 RU of mouse anti-human Fc antibody. Trimers were incubated in 4-fold molar excess of sCD4 and samples were injected at different time-points. Blank sensorgrams were obtained by injection of same volume of HBS-EP+ buffer in place of trimer. To measure any change in the trimer samples on incubation, ligand-free trimers were injected before and 72 hours after start of the experiment. (See Supplementary Note for additional SPR assay methods).
Biolayer interferometry analysis
A fortéBio HTX instrument was used to measure affinities of SOSIP.664 and DS-SOSIP.664 to a panel of HIV-1-Env reactive antibodies at 30° C. (See Supplementary Note for details).
Negative-stain electron microscopy
Negative-stain electron microscopy samples were diluted to about 0.03 mg/ml, adsorbed to a freshly glow-discharged carbon-film grid for 15s, and stained with 0.7% uranyl formate. Images were collected semi-automatically using SerialEM87 on a FEI Tecnai T20 with a 2k x 2k Eagle CCD camera at a pixel size of 0.22 nm/px. Particles were picked automatically and reference-free 2D classification was performed in EMAN288.
Differential scanning calorimetry
The heat capacity of BG505 SOSIP.664 and DS-SOSIP.664 was measured as a function of temperature using a high-precision differential scanning VP-DSC microcalorimeter (GE Healthcare/Microcal, Northampton, MA). The samples were extensively dialyzed against PBS, pH 7.4, and then degassed to avoid the formation of bubbles in the calorimetric cells. Thermal denaturation scans were conducted from 10 to 100 °C at a rate of 1 °C/min. The protein concentration was about 0.3 mg/mL.
Analytical ultracentrifugation equilibrium measurements
Analytical ultracentrifugation (AUC) equilibrium experiments were performed at 15°C, using a Beckman XL-A/I ultracentrifuge equipped with a Ti60An rotor (See Supplementary Note for additional details).
Hydrogen Deuterium Exchange (HDX)
The hydrogen-deuterium exchange rates for BG505 SOSIP.664 and the DS-SOSIP.664 both alone and in the presence of CD4 were assessed. Complexes with soluble CD4 (D1D2) were formed by overnight incubation with a nine-fold molar excess of ligand (relative to trimer). Proteins (10 μg) were diluted 10-fold into deuterated PBS buffer and incubated at room temperature. Aliquots removed after 3 s, 1 min, 30 min and 20 h were quenched by mixing with an equal volume of cold 200 mM Tris-2-carboxyethyl phosphine (TCEP), 0.2% formic acid (final pH 2.5). The samples were subsequently digested with pepsin (at 0.15 mg/mL) for 5 min on ice, flash frozen in liquid nitrogen, and stored at −80° C. For LC-MS analysis, samples were thawed on ice for 5 minutes and manually injected onto a Waters BEH 1.7 μm 1.2×5 mm trap column (Waters) flowing 0.1% TFA at 200 μL/min. After 3 minutes of washing the peptides were resolved over a Hypersil 1×50 mm 2.1 μm C18 column (Thermo Scientific) using a gradient of 15 to 40% B in 8 minutes (A: 0.05% TFA 5% ACN; B: 0.05% TFA 80% ACN). Eluted peptides were analyzed with a Waters Synapt Q-TOF mass spectrometer. Peptide identification and exchange analysis were as described previously70.
Neutralization of viral entry
The point mutations were introduced into full-length Env clone BG505.W6M.C215 in expression vector pcDNA3.1/V5-His-TOPO (Invitrogen). Pseudotyped, single round of entry virus was produced as described in Shu et al.89. Briefly, plasmid DNA was used to transfect 293T cells along with an envelope-deficient HIV-1 subtype A proviral plasmid, SG3dEnv90 to generate pseudotyped viral particles. Serial dilutions of the pseudovirus stocks were added to TZMbl reporter cells, and two days later the activity of the luciferase reporter gene in infected cells was assessed with a Luciferase Assay kit (Promega) and measured in a luminometer; activity was reported as Relative Light Units (RLU).
smFRET on JR-FL viral spikes
smFRET experiments were performed as described28. Briefly, HEK293 cells were transfected at a 40:1 ratio of wild-type HIV-1JR-FL or HIV-1JR-FL 201C-433C Env to dually V1-Q3/V4-A1 tagged Env and the additional presence of pNL4-3 Δenv ΔRT (See Supplementary Note for details).
Assessment of physical stability
To assess physical stability of the pre-fusion, closed conformation of SOSIP.664 and DS-SOSIP.664, we subjected the proteins to a variety of pharmaceutically relevant stresses such as extreme pH, high temperature, low and high osmolarity, as well as repeated freeze/thaw cycles. The physical stability of treated BG505 SOSIP and 201C-433C proteins was reported as the fraction of binding retained to the quaternary-specific V1V2-directed antibody CAP256-VRC26.09 by using a fortéBio Octet HTX instrument. To assess physical stability over time, the trimers were incubated at 4°C, 20°C, 37°C and 42°C, and aliquots were taken at different time-points and binding to CAP256-VRC26.09 was measured. (See Supplementary Note for details).
Virus-like particles
Virus-like particles containing protease-purified Env trimers were prepared as previously described50,51,91, and ELISAs on these VLPs were performed as described51. Briefly, Immulon II plates were coated overnight at 4°C with VLPs at 20 times their concentration in transfection supernatants. Wells were washed with PBS and then blocked with 4% bovine serum albumin/10% fetal bovine serum in PBS. Various biotinylated monoclonal antibodies (biotinylated using sulfo-NHS-Xbiotin, Thermo), and CD4-IgG2 were then titrated in the presence or absence of a fixed concentration of 2μg/ml soluble CD4. Alkaline phosphatase conjugated to streptavidin (Vector Laboratories, Burlingame, CA; to detect biotinylated mAbs) or anti-Fc (Accurate, Westbury, NY; to detect CD4-IgG2) and SigmaFAST p-nitrophenyl phosphate tablets (Sigma) were then used to detect binding. Plates were read at 405 nm.
Supplementary Material
Acknowledgments
We thank Y. Dai for ERV MuLV Gag plasmid, M. Murphy for SPR discussions, B. Whalen for the Rev plasmid, members of the Structural Biology Section and Structural Bioinformatics Core, Vaccine Research Center for discussions and comments on the manuscript, and the WCMC/AMC/TSRI HIVRAD team for their contributions to the design and validation of near-native mimicry for soluble BG505 SOSIP.664 trimers. We thank J. Baalwa, D. Ellenberger, F. Gao, B. Hahn, K. Hong, J. Kim, F. McCutchan, D. Montefiori, L. Morris, J. Overbaugh, E. Sanders-Buell, G. Shaw, R. Swanstrom, M. Thomson, S. Tovanabutra, C. Williamson, and L. Zhang for contributing the HIV-1-Env plasmids used in our neutralization panel. Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (to J.A., A.B.M, J.R.M, and P.D.K), and by grants from the Division of AIDS, NIAID, National Institutes of Health (P01-AI100151 to S.Z.P., P01-AI104722 to L.S., R01-AI93278 to J.M.B., R21-AI100696 to W.M. and S.C.B, R21-AI112389 to K.K.L., and R33-AI84714 to J.M.B.), from the National Institutes of General Medical Sciences (P01-GM56550 to E.F., S.C.B and W.M., R01-GM78031 to B.R.D., and R01-GM98859 to S.C.B.), from the National Institute Heart, Lung and Blood (PO1-HL59725 to S.Z.P.), from the National Science Foundation (MCB-1157506 to E.F.), from the Bill and Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery (OPP1033102 to K.K.L.), from the Australian Research Council (DP130102219 to L.K.L.), from the Irvington Fellows Program of the Cancer Research Program to J.B.M., by funds from the Department of Veterans Affairs to S.Z.P., and by a fellowship from the China Scholarship Council-Yale World Scholars to X.M. This project was funded in part with Federal funds to U.B. from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. Use of sector 22 (Southeast Region Collaborative Access team) at the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract number W-31-109-Eng-38.
Footnotes
Accession Codes. Coordinates and structure factors for the ligand-free BG505 SOSIP.664 trimer have been deposited with the Protein Data Bank under accession code 4ZMJ.
Author Contributions YDK headed the ligand-free trimer structure determination; MP co-headed the conformational fixation, leading atomic-level investigations; PA co-headed the conformational fixation, leading antigenic assessments; and ISG headed the structural compatibility bioinformatics and designed the DS mutation. MP, TZ, AD and PDK contributed to structure determination, YDK and CS performed structural analysis, RTB and MKL assessed neutralization breadth, ISG, GYC, MAH, TK, BRD and LKL performed structural compatibility bioinformatics, MP, PA, MGJ, SN, MC, GO, MP, MS, TT, CW, SZP, and ABM performed antigenic analyses, JG, GSJ, YY, BZ, and JRM contributed to conformational fixation, AH and UB performed electron microscopy, MP, PA, AS and EF performed calorimetry, YDK, GA and LS performed ultracentrifugation, YDK, MG and KKL performed and analyzed HDX-MS, NAD, SOD and JRM created and analyzed mutant virus, JG, XM, DST, HZ, ZZ, JA, JBM, SCB and WM performed smFRET, PA, MGJ, and PVT assessed physical and temporal stability, MP, ETC, KO and JMB contributed VLP analysis, and ISG, JS and PDK evaluated information flow. YDK, MP, PA, ISG, and PDK assembled and wrote the paper, on which all principal investigators commented.
Competing financial interests The authors declare no competing financial interests.
References
- 1.Starcich BR, et al. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell. 1986;45:637–48. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- 2.Wei X, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–12. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 3.Chen L, et al. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science. 2009;326:1123–7. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwong PD, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–82. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 5.Fouts TR, Binley JM, Trkola A, Robinson JE, Moore JP. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J Virol. 1997;71:2779–85. doi: 10.1128/jvi.71.4.2779-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang CC, et al. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–8. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss RA, et al. Neutralization of human T-lymphotropic virus type III by sera of AIDS and AIDS-risk patients. Nature. 1985;316:69–72. doi: 10.1038/316069a0. [DOI] [PubMed] [Google Scholar]
- 8.Bures R, et al. Immunization with recombinant canarypox vectors expressing membrane- anchored glycoprotein 120 followed by glycoprotein 160 boosting fails to generate antibodies that neutralize R5 primary isolates of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2000;16:2019–35. doi: 10.1089/088922200750054756. [DOI] [PubMed] [Google Scholar]
- 9.Flynn NM, et al. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis. 2005;191:654–65. doi: 10.1086/428404. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–13. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartesaghi A, Merk A, Borgnia MJ, Milne JL, Subramaniam S. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol. 2013;20:1352–7. doi: 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Julien JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–83. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyumkis D, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–90. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pancera M, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–61. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu X, et al. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J Virol. 2006;80:835–844. doi: 10.1128/JVI.80.2.835-844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Binley JM, et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion- associated structure. J Virol. 2000;74:627–43. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanders RW, et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2002;76:8875–89. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders RW, et al. A next-generation cleaved, soluble HIV-1 Env Trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon YD, et al. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc Natl Acad Sci U S A. 2012;109:5663–8. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–59. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou T, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–7. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou T, et al. Structural Basis for Broad and Potent Neutralization of HIV-1 by Antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pejchal R, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scharf L, et al. Antibody 8ANC195 Reveals a Site of Broad Vulnerability on the HIV-1 Envelope Spike. Cell Rep. 2014;7:785–95. doi: 10.1016/j.celrep.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–8. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 26.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–73. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 27.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–30. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 28.Munro JB, et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346:759–63. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guttman M, et al. Antibody potency relates to the ability to recognize the closed, pre-fusion form of HIV Env. Nat Commun. 2015;6:6144. doi: 10.1038/ncomms7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jancarik JaKS-H. Sparse matrix sampling: a screening method for crystallization of proteins. J Appl Cryst. 1991;24:409–411. [Google Scholar]
- 31.Majeed S, et al. Enhancing protein crystallization through precipitant synergy. Structure. 2003;11:1061–70. doi: 10.1016/s0969-2126(03)00185-0. [DOI] [PubMed] [Google Scholar]
- 32.Adams PD, et al. Recent developments in the PHENIX software for automated crystallographic structure determination. J Synchrotron Radiat. 2004;11:53–5. doi: 10.1107/s0909049503024130. [DOI] [PubMed] [Google Scholar]
- 33.Korber BTF, BT, Kuiken CL, Pillai SK, Sodroski JG. Numbering Positions in HIV Relative to HXB2CG. 1998. [Google Scholar]
- 34.Burton DR, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–7. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- 35.Huang J, et al. Broad and potent neutralization of HIV-1 by a human antibody that recognizes an intersubunit site on the envelope glycoprotein. Nature. 2014 In Press. [Google Scholar]
- 36.Walker LM, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–70. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zolla-Pazner S, et al. The cross-clade neutralizing activity of a human monoclonal antibody is determined by the GPGR V3 motif of HIV type 1. AIDS Res Hum Retroviruses. 2004;20:1254–8. doi: 10.1089/aid.2004.20.1254. [DOI] [PubMed] [Google Scholar]
- 38.Liao HX, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guenaga J, et al. Well-ordered trimeric HIV-1 subtype B and C soluble spike mimetics generated by negative selection display native-like properties. PLoS Pathog. 2015;11:e1004570. doi: 10.1371/journal.ppat.1004570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mbah HA, et al. Effect of soluble CD4 on exposure of epitopes on primary, intact, native human immunodeficiency virus type 1 virions of different genetic clades. J Virol. 2001;75:7785–8. doi: 10.1128/JVI.75.16.7785-7788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stanfield RL, Gorny MK, Williams C, Zolla-Pazner S, Wilson IA. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D. Structure (Camb) 2004;12:193–204. doi: 10.1016/j.str.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Jiang X, et al. Conserved structural elements in the V3 crown of HIV-1 gp120. Nat Struct Mol Biol. 2010;17:955–61. doi: 10.1038/nsmb.1861. [DOI] [PubMed] [Google Scholar]
- 43.Thali M, et al. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–88. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLellan JS, et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–8. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu X, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–61. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doria-Rose NA, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Posner MR, Cavacini LA, Emes CL, Power J, Byrn R. Neutralization of HIV-1 by F105, a human monoclonal antibody to the CD4 binding site of gp120. J Acquir Immune Defic Syndr. 1993;6:7–14. [PubMed] [Google Scholar]
- 48.Arthos J, et al. Biochemical and biological characterization of a dodecameric CD4-Ig fusion protein: implications for therapeutic and vaccine strategies. J Biol Chem. 2002;277:11456–64. doi: 10.1074/jbc.M111191200. [DOI] [PubMed] [Google Scholar]
- 49.Daar ES, Ho DD. Relative resistance of primary HIV-1 isolates to neutralization by soluble CD4. Am J Med. 1991;90:22S–26S. doi: 10.1016/0002-9343(91)90407-o. [DOI] [PubMed] [Google Scholar]
- 50.Crooks ET, Tong T, Osawa K, Binley JM. Enzyme digests eliminate nonfunctional Env from HIV-1 particle surfaces, leaving native Env trimers intact and viral infectivity unaffected. J Virol. 2011;85:5825–39. doi: 10.1128/JVI.00154-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tong T, Crooks ET, Osawa K, Binley JM. HIV-1 virus-like particles bearing pure env trimers expose neutralizing epitopes but occlude nonneutralizing epitopes. J Virol. 2012;86:3574–87. doi: 10.1128/JVI.06938-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trkola A, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–8. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McLellan JS, et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013;340:1113–7. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McLellan JS, Yang Y, Graham BS, Kwong PD. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol. 2011;85:7788–96. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swanson KA, et al. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc Natl Acad Sci U S A. 2011;108:9619–24. doi: 10.1073/pnas.1106536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kovacs JM, et al. Stable, uncleaved HIV-1 envelope glycoprotein gp140 forms a tightly folded trimer with a native-like structure. Proc Natl Acad Sci U S A. 2014;111:18542–7. doi: 10.1073/pnas.1422269112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pancera M, et al. Soluble mimetics of human immunodeficiency virus type 1 viral spikes produced by replacement of the native trimerization domain with a heterologous trimerization motif: characterization and ligand binding analysis. J Virol. 2005;79:9954–69. doi: 10.1128/JVI.79.15.9954-9969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moore JP. Native-like BG505 SOSIP.664 Trimers Induce Autologous Tier-2 NAbs against Complex Epitopes in Rabbits and Macaques. AIDS Res Hum Retroviruses. 2014;30:A67–A67. [Google Scholar]
- 59.McLellan JS, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–43. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tran EE, et al. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 2012;8:e1002797. doi: 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Killikelly A, et al. Thermodynamic signatures of the antigen binding site of mAb 447-52D targeting the third variable region of HIV-1 gp120. Biochemistry. 2013;52:6249–57. doi: 10.1021/bi400645e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stanfield RL, Gorny MK, Zolla-Pazner S, Wilson IA. Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity. J Virol. 2006;80:6093–105. doi: 10.1128/JVI.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gorny MK, et al. Human anti-V3 HIV-1 monoclonal antibodies encoded by the VH5-51/VL lambda genes define a conserved antigenic structure. PLoS One. 2011;6:e27780. doi: 10.1371/journal.pone.0027780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rini JM, et al. Crystal structure of a human immunodeficiency virus type 1 neutralizing antibody, 50.1, in complex with its V3 loop peptide antigen. Proc Natl Acad Sci U S A. 1993;90:6325–9. doi: 10.1073/pnas.90.13.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 66.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 67.Bernstein FC, et al. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–42. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–9. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pancera M, et al. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci U S A. 2010;107:1166–71. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guttman M, et al. CD4-Induced Activation in a Soluble HIV-1 Env Trimer. Structure. 2014;22:974–84. doi: 10.1016/j.str.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanner MF, Olson AJ, Spehner JC. Reduced surface: an efficient way to compute molecular surfaces. Biopolymers. 1996;38:305–20. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 72.Hubbard SJaT, JM . “NACCESS”, Computer Program. Department of Biochemistry and Molecular Biology, University College London; 1993. [Google Scholar]
- 73.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–96. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 74.Sarzotti-Kelsoe M, et al. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J Immunol Methods. 2014;409:131–46. doi: 10.1016/j.jim.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang CC, et al. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci U S A. 2004;101:2706–11. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xiang SH, et al. Epitope mapping and characterization of a novel CD4-induced human monoclonal antibody capable of neutralizing primary HIV-1 strains. Virology. 2003;315:124–34. doi: 10.1016/s0042-6822(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 77.Huang J, et al. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature. 2014;515:138–42. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scheid JF, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–40. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 79.Liao HX, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–76. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Calarese DA, et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–71. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 81.Georgiev IS, et al. Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science. 2013;340:751–6. doi: 10.1126/science.1233989. [DOI] [PubMed] [Google Scholar]
- 82.Doria-Rose NA, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seaman MS, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol. 2010;84:1439–52. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dreyfus C, et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343–8. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tharakaraman K, Subramanian V, Cain D, Sasisekharan V, Sasisekharan R. Broadly neutralizing influenza hemagglutinin stem-specific antibody CR8020 targets residues that are prone to escape due to host selection pressure. Cell Host Microbe. 2014;15:644–51. doi: 10.1016/j.chom.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Palivizumab a humanized respiratory syncytial virus monoclonal antibody reduces hospitalization from respiratory syncytial virus infection in high-risk infants. The IMpact-RSV Study Group. Pediatrics. 1998;102:531–7. [PubMed] [Google Scholar]
- 87.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 88.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 89.Shu Y, et al. Efficient protein boosting after plasmid DNA or recombinant adenovirus immunization with HIV-1 vaccine constructs. Vaccine. 2007;25:1398–408. doi: 10.1016/j.vaccine.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wei X, et al. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 2002;46:1896–905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tong T, Osawa K, Robinson JE, Crooks ET, Binley JM. Topological analysis of HIV-1 glycoproteins expressed in situ on virus surfaces reveals tighter packing but greater conformational flexibility than for soluble gp120. J Virol. 2013;87:9233–49. doi: 10.1128/JVI.01145-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.