Abstract
Macrophages located in airways and the alveolar space are continually exposed to different signals from the respiratory mucosa. In this respect, epithelial cells represent an important source of cytokines and mediators modulating the state of activation and/or differentiation of mononuclear phagocytes. Many of the proinflammatory genes induced in macrophages during immune and immunopathological reactions are regulated by transcription factor NF kappa B. The aim of our study was to characterize changes in the expression of genes associated with NF kappa B activation and signalling in THP-1 human macrophages co-cultured with A549 respiratory epithelial cells. At least 4-fold upregulation of mRNA level was found in 29 of 84 tested genes including genes for multiple cytokines and chemokines, membrane antigens and receptors, and molecules associated with NF kappa B signalling. The mRNA induction was confirmed at the level of protein expression by evaluating the release of IL-6 and IL-8 and by ICAM-1 expression. Blocking of one NFκB subunit by p65 siRNA inhibited the production of IL-6 in both cell types while IL-8 release from THP-1 cells did not seem to be affected.
We conclude from our data that unstimulated respiratory epithelial cells regulate genes associated with NF kappa B dependent immune responses in human macrophages and that these interactions may play a key role in immediate responses in the respiratory mucosa.
Keywords: Co-cultures, Cytokines, Epithelial cells, Macrophages, NF kappa B
Introduction
Mononuclear phagocytes represent highly efficient effector cells of innate immunity mechanisms and in the case of dendritic cells are responsible for the initiation of adaptive immune responses. Traditionally, peripheral blood monocytes are recruited during their development (more extensively in inflammation) into tissue compartments and consequently mature into macrophages or dendritic cells. There are several lines of evidence showing that tissue macrophages are not a homogeneous population of cells and different subpopulations can be defined with respect to phenotype or cell function (Gordon and Martinez, 2010 and Mantovani et al., 2007). The level of maturation is probably a key factor in macrophage heterogeneity. For example, human alveolar macrophages sampled by bronchoalveolar lavage can reflect either a phenotype of mature tissue macrophages or less mature monocyte-like cells (Striz et al. 1993). Moreover, the phenotypic pattern of mononuclear phagocytes undergoes prominent changes in the course of immune a immunopathological reactions.
Macrophages located in the airways and the alveolar space are continually exposed to different signals from the respiratory mucosa which is known to regulate a number of immune responses (Bulek et al. 2010). In this respect, epithelial cells represent an important source of cytokines and mediators modulating the state of activation and/or differentiation of mononuclear phagocytes. We have shown previously that human monocytes co-cultured in a direct cell–cell contact with primary bronchial epithelial cells change their phenotype into a pattern resembling mature alveolar macrophages. Some of the changes, such as the induction of ICAM-1 (CD54) also were inducible by soluble factors. Moreover, similar results were obtained when using cell lines (Striz et al. 2001).
Many of the proinflammatory genes induced in macrophages during their activation are regulated by transcription factor NF kappa B, which functions as a homo or heterodimer of five constituent proteins: c-REL, RelA (p65), RelB, p50, and p52 (Wang et al. 2007). This transcription factor regulates genes involved in multiple stages of immune responses including activation of both cells participating in innate immunity cells and acquired immunity mediated by lymphocytes and dendritic cells (Wan and Lenardo 2010).
The aim of our study was to characterize changes in the expression of genes associated with NF kappa B activation and signalling in human macrophages co-cultured with respiratory epithelial cells. Furthermore, the potential role of transcription factor NFκB in cell–cell interactions was evaluated using p65-siRNA.
Methods
Culture conditions
The A549 alveolar type II-like cell line (from ATCC) was cultured in Iscove's modified Dulbecco's medium (IMDM) with 10% fetal calf serum (FCS) and l-glutamine, penicillin, streptomycin (Sigma–Aldrich) until confluency in 6-well tissue culture plates. The medium was removed and the cells were washed twice with Earle's Balanced Salt Solution (EBS). THP-1 cells (monocyte/macrophage cell line) were cultured in the same medium as A549 cells and in co-culture experiments, the cells were physically separated with 0,4 µm pore-filters. At different times (0, 4, 8, 12 and 24 h) we harvested the cells separately to evaluate gene and protein expression.
Quantitative RT-PCR (qRT-PCR)
The qRT-PCR analysis was performed using RT2 Profiler PCR Arrays (Common Cytokines Array Kit) from SABiosciences (USA) on the instrument ABI 7900HT. This kit contains 84 different specific primers for NFκB related genes, plus controls for genomic contamination and reverse transcription and positive PCR controls (Table 1). Data were evaluated with a web-based program provided by the manufacturer.
Table 1.
Abbreviations of genes evaluated.
| Symbol | Description |
|---|---|
| AGT | Angiotensinogen (serpin peptidase inhibitor, clade A, member 8) |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 |
| ATF1 | Activating transcription factor 1 |
| BCL10 | B-cell CLL/lymphoma 10 |
| BCL3 | B-cell CLL/lymphoma 3 |
| CFB | Complement factor B |
| BIRC2 | Baculoviral IAP repeat-containing 2 |
| NOD1 | Nucleotide-binding oligomerization domain containing 1 |
| CASP1 | Caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) |
| CASP8 | Caspase 8, apoptosis-related cysteine peptidase |
| CCL2 | Chemokine (C-C motif) ligand 2 |
| CD40 | CD40 molecule, TNF receptor superfamily member 5 |
| CFLAR | CASP8 and FADD-like apoptosis regulator |
| CHUK | Conserved helix-loop-helix ubiquitous kinase |
| CSF2 | Colony stimulating factor 2 (granulocyte-macrophage) |
| CSF3 | Colony stimulating factor 3 (granulocyte) |
| SLC44A2 | Solute carrier family 44, member 2 |
| EDARADD | EDAR-associated death domain |
| LPAR1 | Lysophosphatidic acid receptor 1 |
| EGR1 | Early growth response 1 |
| ELK1 | ELK1, member of ETS oncogene family |
| F2R | Coagulation factor II (thrombin) receptor |
| FADD | Fas (TNFRSF6)-associated via death domain |
| FASLG | Fas ligand (TNF superfamily, member 6) |
| FOS | V-fos FBJ murine osteosarcoma viral oncogene homolog |
| GJA1 | Gap junction protein, alpha 1, 43kDa |
| HMOX1 | Heme oxygenase (decycling) 1 |
| HTR2B | 5-hydroxytryptamine (serotonin) receptor 2B |
| ICAM1 | Intercellular adhesion molecule 1 |
| IFNA1 | Interferon, alpha 1 |
| IFNB1 | Interferon, beta 1, fibroblast |
| IFNG | Interferon, gamma |
| IKBKB | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| IKBKE | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| IKBKG | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| IL10 | Interleukin 10 |
| IL1A | Interleukin 1, alpha |
| IL1B | Interleukin 1, beta |
| IL1R1 | Interleukin 1 receptor, type I |
| IL6 | Interleukin 6 (interferon, beta 2) |
| IL8 | Interleukin 8 |
| IRAK1 | Interleukin-1 receptor-associated kinase 1 |
| IRAK2 | Interleukin-1 receptor-associated kinase 2 |
| JUN | Jun oncogene |
| LTA | Lymphotoxin alpha (TNF superfamily, member 1) |
| LTBR | Lymphotoxin beta receptor (TNFR superfamily, member 3) |
| MALT1 | Mucosa associated lymphoid tissue lymphoma translocation gene 1 |
| MAP3K1 | Mitogen-activated protein kinase kinase kinase 1 |
| MYD88 | Myeloid differentiation primary response gene (88) |
| NLRP12 | NLR family, pyrin domain containing 12 |
| NFκB1 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| NFκB2 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| NFκBIA | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| PPM1A | Protein phosphatase 1A (formerly 2C), magnesium-dependent, alpha isoform |
| RAF1 | V-raf-1 murine leukemia viral oncogene homolog 1 |
| REL | V-rel reticuloendotheliosis viral oncogene homolog (avian) |
| RELA | V-rel reticuloendotheliosis viral oncogene homolog A (avian) |
| RELB | V-rel reticuloendotheliosis viral oncogene homolog B |
| TRIM13 | Tripartite motif-containing 13 |
| RHOA | Ras homolog gene family, member A |
| RIPK1 | Receptor (TNFRSF)-interacting serine-threonine kinase 1 |
| SLC20A1 | Solute carrier family 20 (phosphate transporter), member 1 |
| STAT1 | Signal transducer and activator of transcription 1, 91kDa |
| TBK1 | TANK-binding kinase 1 |
| TICAM2 | Toll-like receptor adaptor molecule 2 |
| TLR1 | Toll-like receptor 1 |
| TLR2 | Toll-like receptor 2 |
| TLR3 | Toll-like receptor 3 |
| TLR4 | Toll-like receptor 4 |
| TLR6 | Toll-like receptor 6 |
| TLR7 | Toll-like receptor 7 |
| TLR8 | Toll-like receptor 8 |
| TLR9 | Toll-like receptor 9 |
| TMED4 | Transmembrane emp24 protein transport domain containing 4 |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) |
| TNFAIP3 | Tumor necrosis factor, alpha-induced protein 3 |
| TNFRSF10A | Tumor necrosis factor receptor superfamily, member 10a |
| TNFRSF10B | Tumor necrosis factor receptor superfamily, member 10b |
| TNFRSF1A | Tumor necrosis factor receptor superfamily, member 1A |
| CD27 | CD27 molecule |
| TNFSF10 | Tumor necrosis factor (ligand) superfamily, member 10 |
| TNFSF14 | Tumor necrosis factor (ligand) superfamily, member 14 |
| TRADD | TNFRSF1A-associated via death domain |
| TICAM1 | Toll-like receptor adaptor molecule 1 |
Flow cytometry
The expression of CD54 (ICAM-1) was measured by flow cytometry (Cytomics FC 500, Beckman Coulter) using PE labeled monoclonal antibody (Beckman Coulter). Isotype specific IgG1 mouse immunoglobulin (Beckman Coulter) was used as a control. The viability of the cells was detected by 7-amino-actinomycin D (7-AAD Viability Dye, Beckman Coulter). Non-viable cells can be characterized as they are stained by 7-AAD, while living cells, retaining their membrane integrity, are impermeable to 7-AAD, are unstained and 7-AAD negative.
Cytokine levels
Simultaneous determination of the concentrations of IL-6 and IL-8 released from co-cultured cells was assessed by the immunoluminescent x-MAP technology, Luminex100 System (Luminex Corporation, Austin, Texas, USA), based on the laser analysis of analyte-specific antibodies that are coated onto microparticles. In the first step, 50 µl of the samples/standards was incubated with 50 µl of microparticles for 3 h at RT on a horizontal orbital microplate shaker. After washing the beads to remove unbound substances, we added 50 µl of the secondary antibodies conjugated with biotin to each well and incubated the samples/standards for another 1 h. After the incubation, we washed the beads to remove unbound secondary antibodies and added 50 µl of the Streptavidin-PE. After 30 min incubation and another washing, the microparticles were resuspended in 100 µl of wash buffer and read using a Luminex analyzer.
RNA interference by p65-siRNA suppression of NFκB
Targeting p65 siRNA and non-targeting control siRNA (Dharmcon, Inc, Lafayette, CO) were introduced into the cells using a method previously described with modification (Liu et al. 2008). Briefly, A549 cells were cultured in 1% FCS–DMEM and THP-1 cells were cultured in 10% FCS–RPMI 1640 without antibiotics and after washing with PBS, the cells were treated with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) containing p65-siRNA or non-targeting control siRNA (final concentration 100 nM in Opti-MEM) for 6 h. Then, the cells were re-fed with 0.5% FCS–RPMI, and THP-1 cells plated on top of confluent monolayer of A549 cells. Control cultures with isolated cells were prepared. The levels of IL-6 and IL-8 released into culture media under different culture conditions were evaluated by ELISA.
IL-6 ELISA
IL-6 levels in culture supernatants were quantified using a sandwich ELISA. Briefly, 96 well flat bottomed microtiter plates (Immulon, Chantilly, VA) were coated with 200 µl/well of purified goat anti-hIL-6 antibody (R&D, Minneapolis, MN) diluted 1:1000 in Voller's buffer for 24 h at 4 °C. After 3× washing in PBS, 0.25% Tween-20 (washing solution), undiluted culture supernatants and serial dilutions of standard rIL-6 (Sigma, St. Louis) were incubated at room temperature for 90 min. Plates were rinsed 3 times with PBS–Tween followed by the addition of rabbit anti-human IL-6 antibody (Calbiochem, LaJolla, CA) diluted 1:1000 in washing solution. After 1 h incubation and washing 3 times, human serum absorbed peroxidase conjugated goat anti-rabbit antibody (ICN Biomedicals, Costa Mesa, CA) was added at a 1:2000 dilution for a 1 h incubation. The plates were then washed again three times with PBS–Tween and the substrate solution orthophenyldiamine (Sigma, St. Louis) at 10 ng/ml in a solution of 0.01% H2O2 was added. The reaction was terminated with 27.5 µl of 3 M sulfuric acid and plates were read at 490 nm in an automated ELISA reader.
IL-8 ELISA
IL-8 levels in culture supernatants were quantified using a sandwich ELISA. The 96 well flat bottomed microtiter plates (Dynatech, Chantilly, VA) were coated with 200 µl/well of goat anti-human IL-8 antibody (R&D, Minneapolis, MN) diluted 1:2000 in Voller‘s buffer for 24 h at 4 °C. After 3× washing in PBS–Tween, undiluted culture supernatants and serial dilutions of standard human recombinant rIL-8 (Sigma, St. Louis, MO) were incubated at room temperature for 90 min. Plates were rinsed 3 times with PBS–Tween followed by the addition of rabbit anti-human IL-8 antibody (Upstate Biotechnology Inc., Lake Placid, NY) diluted 1:2000 in washing solution. After a 1 h incubation, dishes were washed 3 times, and peroxidase conjugated goat anti-rabbit antibody (Sigma, St. Louis, MO) was added at a 1:2000 dilution for a 1 h incubation. The plates were then washed again and orthophenyldiamine (Sigma, St. Louis, MO) dissolved in methanol (1 mg/ml) and diluted (10 ng/ml) in distilled water containing 0.01% H2O2 was added. The subsequent reaction was terminated with 27.5 l of 3 M sulfuric acid and plates were read at 490 nm at an ELISA reader.
Statistical analysis
Statistical analysis was performed using the Mann–Whitney U test and Student's t-test, for qRT-PCR data, a web-based program provided by the manufacturer (SABiosciences) was used.
Results
Genes induced in THP-1 monocytes/macrophages after a co-culture with A549 epithelial cells
At least 4-fold upregulation of mRNA level was found in 29 out of 84 tested genes. In the early phases of co-culture (4 h), upregulation of proinflammatory cytokine genes IL-1 beta and TNF alpha (more than 40-fold increase in mRNA level compared to basal expression) was detected together with increased expression of IL-8/CXCL8 chemokine (the highest mRNA expression), EGR1, HMOX1, CCL2, JUN, LTA, IRAK2, FOS, IFN beta 1, and ICAM-1 (Table 2). Macrophages isolated after 8 h of co-culture upregulated gene expression of IL-1R-type I 1 and CD40. After 12 h of co-culture with epithelial monolayers, other genes associated with NF kappa B activation are upregulated including REL, transcription co-activator BCL3, MALT gene, NFκB1 subunit, TNFRSF member 10b, IFN alpha 1, IL-6 and IL-10. After 24 h, mRNA for CD27, Fas ligand (CD178), TLR3, CSF2, CSF3, IL-1 alpha and IFN gamma are induced.
Table 2.
mRNA expression in THP-1 cells after a co-culture with A549 epithelial monolayers. THP-1 cells were co-cultured with a monolayer of A549 cells separated by a filter insert. After 4 h, 8 h, 12 h, and 24 h, the THP-1 cells were aspirated, mRNA extracted and the expression of mRNA evaluated by qRT-PCR. The experiment has been repeated three times with consistent results. Data were correlated with mRNA expression of unstimulated THP-1 cells and statistically evaluated with a web-based program provided by the manufacturer. Bold are the values of at least 4-fold change of mRNA expression.
| Upregulated | ||||||||||||
| Symbol | 95% CI | Fold reg. | p-Value | 95% CI | Fold reg. | p-Value | 95% CI | Fold reg. | p-Value | 95% CI | Fold reg. | p-Value |
| IL8 | (147.30, 296.04) | 221.67 | 0 | (174.85, 197.32) | 186.08 | 0 | (207.99, 245.93) | 226.96 | 0 | (27.32, 30.74) | 29.03 | 0 |
| EGR1 | (82.59, 111.12) | 96.85 | 0 | (115.54, 124.96) | 120.25 | 0 | (45.21, 65.48) | 55.34 | 0 | (7.73, 18.74) | 13.23 | 0.01 |
| TNF | (58.66, 69.62) | 64.14 | 0 | (41.25, 48.81) | 45.03 | 0 | (18.15, 22.35) | 20.25 | 0 | (22.34, 29.40) | 25.87 | 0 |
| IL1B | (29.80, 55.01) | 42.41 | 0 | (32.09, 59.42) | 45.75 | 0 | (286.84, 483.44) | 385.14 | 0 | (1.43, 3.28) | 2.35 | 0.02 |
| HMOX1 | (31.26, 39.29) | 35.28 | 0 | (26.57, 30.66) | 28.61 | 0 | (25.31, 29.56) | 27.43 | 0 | (3.32, 8.68) | 6 | 0.02 |
| CCL2 | (6.93, 11.50) | 9.21 | 0 | (9.83, 10.96) | 10.4 | 0 | (31.39, 33.10) | 32.24 | 0 | (4.05, 5.20) | 4.62 | 0 |
| JUN | (6.24, 12.00) | 9.12 | 0 | (5.80, 7.05) | 6.43 | 0 | (7.27, 13.11) | 10.19 | 0 | (2.48, 4.01) | 3.25 | 0 |
| LTA | (6.94, 8.66) | 7.8 | 0 | (7.91, 9.49) | 8.7 | 0 | (5.49, 6.90) | 6.2 | 0 | (5.44, 10.86) | 8.15 | 0.01 |
| IRAK2 | (3.50, 7.43) | 5.46 | 0.01 | (4.30, 6.67) | 5.49 | 0 | (17.13, 23.24) | 20.19 | 0 | (1.38, 2.61) | 1.99 | 0.03 |
| FOS | (4.30, 6.28) | 5.29 | 0 | (5.10, 7.15) | 6.13 | 0 | (9.26, 11.60) | 10.43 | 0 | (5.49, 8.71) | 7.1 | 0 |
| IFNB1 | (2.11, 8.17) | 5.14 | 0 | (1.81, 6.41) | 4.11 | 0 | (0.00001, 9.74) | 4.77 | 0.14 | (2.12, 8.58) | 5.35 | 0 |
| ICAM1 | (3.38, 5.53) | 4.45 | 0 | (5.01, 7.21) | 6.11 | 0 | (11.25, 18.77) | 15.01 | 0 | (0.86, 1.88) | 1.37 | 0.18 |
| IL1R1 | (3.05, 3.91) | 3.48 | 0 | (3.74, 4.41) | 4.08 | 0 | (5.51, 7.17) | 6.34 | 0 | (1.73, 2.57) | 2.15 | 0 |
| CD40 | (2.08, 3.54) | 2.81 | 0 | (4.63, 7.59) | 6.11 | 0 | (3.54, 7.86) | 5.7 | 0.01 | (1.65, 2.45) | 2.05 | 0 |
| BCL3 | (2.42, 3.07) | 2.75 | 0 | (2.24, 3.24) | 2.74 | 0 | (11.64, 14.65) | 13.15 | 0 | (0.47, 0.79) | 1.59 | 0.02 |
| CD27 | (1.79, 3.54) | 2.66 | 0 | (1.10, 3.01) | 2.06 | 0.05 | (1.21, 2.68) | 1.94 | 0.02 | (2.98, 8.06) | 5.52 | 0.01 |
| NFκB1 | (2.02, 2.52) | 2.27 | 0 | (1.63, 2.04) | 1.84 | 0 | (4.10, 5.20) | 4.65 | 0 | (0.77, 1.24) | 1.01 | 0.92 |
| IL6 | (1.42, 2.98) | 2.2 | 0.04 | (2.95, 3.61) | 3.28 | 0 | (2.79, 7.21) | 5 | 0.03 | (6.43, 10.26) | 8.34 | 0 |
| MALT1 | (1.04, 2.70) | 1.87 | 0.09 | (2.11, 2.34) | 2.23 | 0 | (9.36, 10.57) | 9.96 | 0 | (1.02, 1.54) | 1.28 | 0.1 |
| TNFRSF10B | (1.61, 1.95) | 1.78 | 0 | (2.23, 2.69) | 2.46 | 0 | (4.46, 7.40) | 5.93 | 0 | (1.07, 1.38) | 1.22 | 0.04 |
| IL10 | (1.41, 1.65) | 1.53 | 0 | (1.45, 2.53) | 1.99 | 0.02 | (3.31, 7.95) | 5.63 | 0.01 | (6.43, 10.26) | 8.34 | 0 |
| IFNA1 | (1.41, 1.65) | 1.53 | 0 | (0.94, 0.96) | 1.05 | 0 | (1.40, 1.77) | 1.58 | 0 | (6.43, 10.26) | 8.34 | 0 |
| CSF2 | (1.41, 1.65) | 1.53 | 0 | (0.39, 2.57) | 1.48 | 0.36 | (1.13, 1.41) | 1.27 | 0.02 | (6.43, 10.26) | 8.34 | 0 |
| CSF3 | (1.41, 1.65) | 1.53 | 0 | (0.94, 0.96) | 1.05 | 0 | (1.13, 1.41) | 1.27 | 0.02 | (6.43, 10.26) | 8.34 | 0 |
| FASLG | (1.41, 1.65) | 1.53 | 0 | (0.94, 0.96) | 1.05 | 0 | (1.13, 1.41) | 1.27 | 0.02 | (6.43, 10.26) | 8.34 | 0 |
| IFNG | (1.41, 1.65) | 1.53 | 0 | (0.94, 0.96) | 1.05 | 0 | (1.13, 1.41) | 1.27 | 0.02 | (6.43, 10.26) | 8.34 | 0 |
| TLR3 | (1.41, 1.65) | 1.53 | 0 | (1.07, 1.15) | 1.11 | 0 | (1.13, 1.41) | 1.27 | 0.02 | (6.43, 10.26) | 8.34 | 0 |
| IL1A | (1.19, 1.29) | 1.24 | 0 | (1.45, 2.97) | 2.21 | 0.03 | (4.49, 8.15) | 6.32 | 0.01 | (0.54, 0.77) | 1.53 | 0.01 |
| REL | (1.11, 1.19) | 1.15 | 0 | (0.93, 1.08) | 1.01 | 0.83 | (4.25, 5.27) | 4.76 | 0 | (0.52, 0.67) | 1.68 | 0 |
| Downregulated | ||||||||||||
| F2R | (0.13, 0.37) | 3.99 | 0.02 | (0.14, 0.35) | 4.14 | 0.02 | (0.09, 0.31) | 4.99 | 0.02 | (0.19, 0.63) | 2.45 | 0.06 |
| MAP3K1 | (0.25, 0.52) | 2.58 | 0 | (0.62, 0.68) | 1.54 | 0 | (0.43, 0.52) | 2.11 | 0 | (0.22, 0.27) | 4.08 | 0 |
| GJA1 | (0.40, 0.70) | 1.81 | 0.02 | (0.78, 1.34) | 1.06 | 0.75 | (3.35, 6.17) | 4.76 | 0 | (0.39, 1.01) | 1.42 | 0.21 |
In most of the genes evaluated, the increase in mRNA expression was followed by their sequential down-regulation, on the other hand, IL-6 and IL-10 gradually increased the expression in 24 h period.
During co-culture, only a limited number of genes showed transient down-regulation compared to basal levels; these included Gap junction protein alpha 1, MAP3 kinase 1, and coagulation factor II (thrombin) receptor.
CD54 expression and cytokine release induced by co-culture of THP-1 monocytes/macrophages with A549 epithelial cells
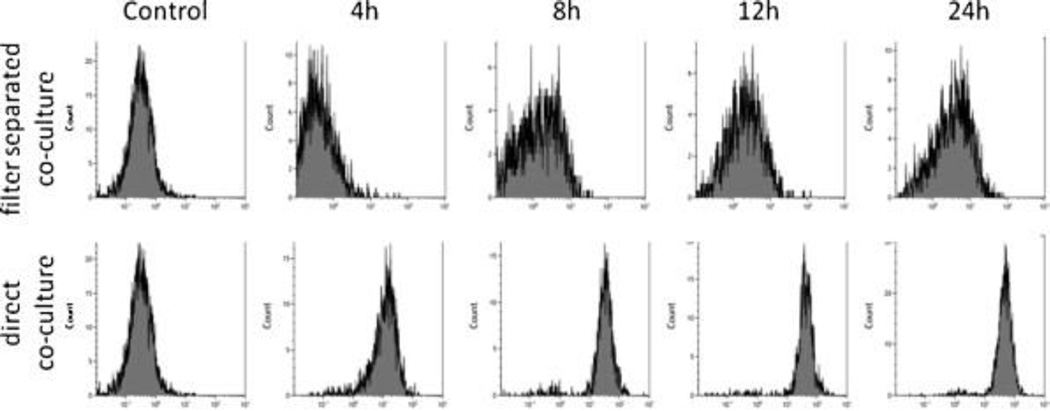
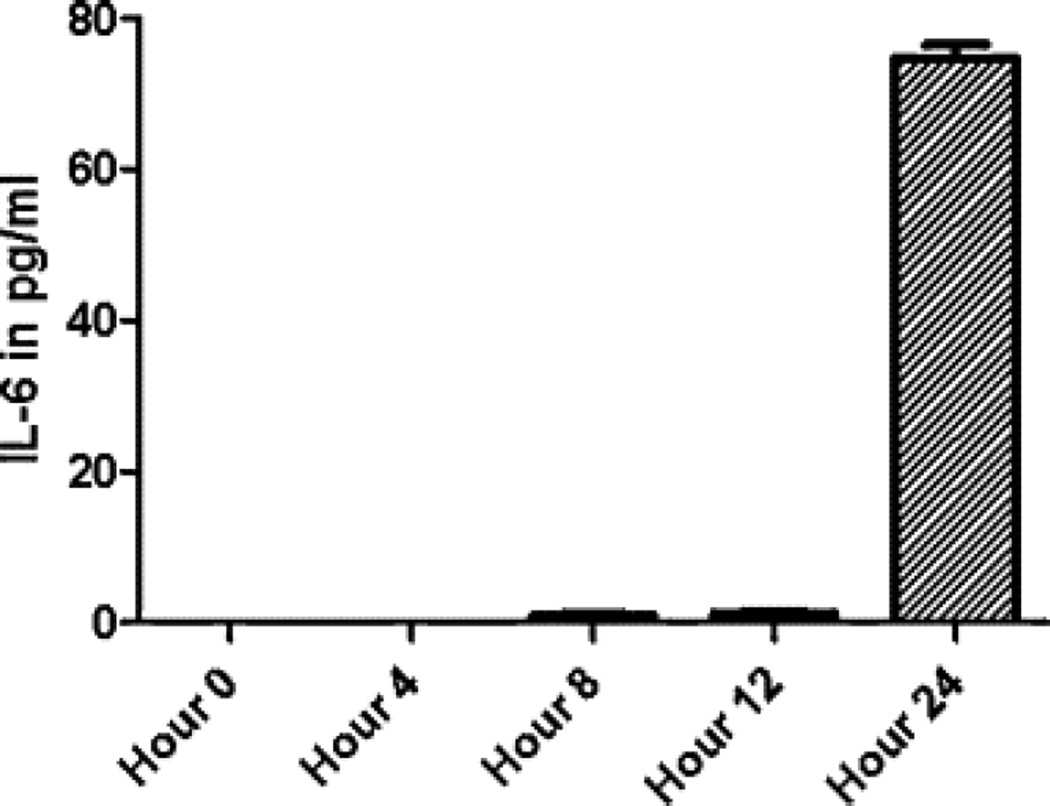
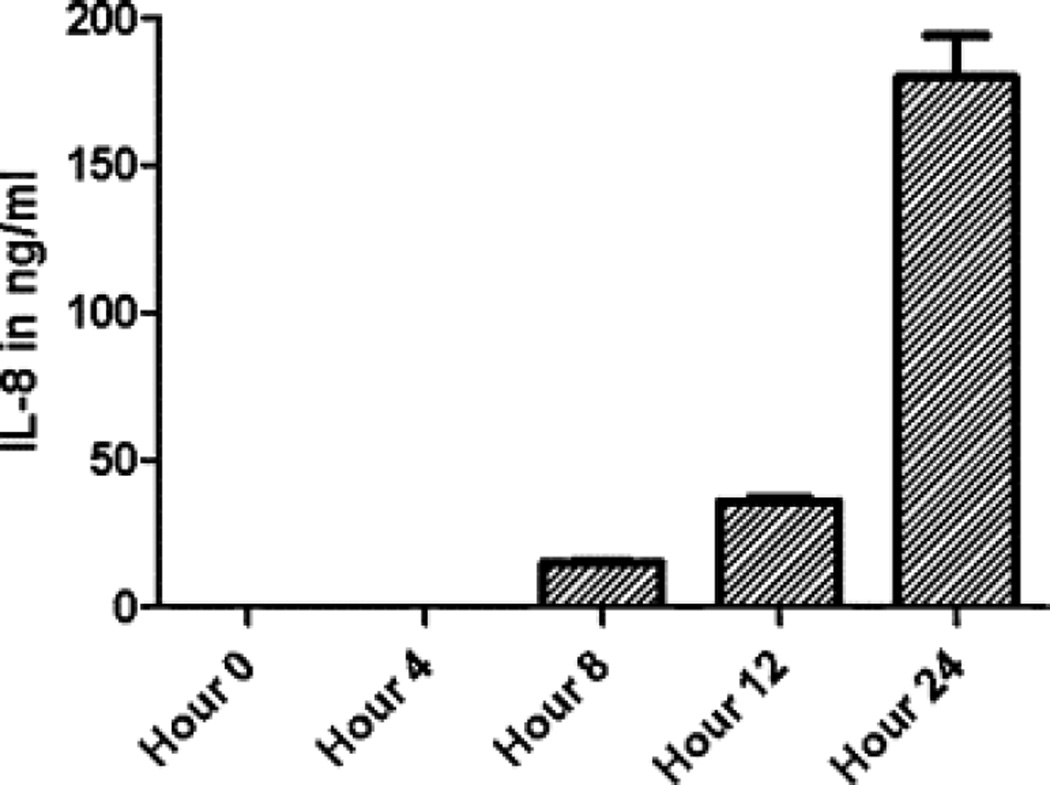
Co-culture with a monolayer of A549 cells induced the expression of ICAM-1 (CD54) on the surface of THP-1 cells (Fig. 1). The changes were more pronounced in a direct co-culture without separation of cells by filter insert. When looking at the cytokine levels in culture media, IL-8 was released earlier than IL-6 (Fig. 2 and Fig. 3).
Fig. 1.
ICAM-1 (CD54) expression on THP-1 cells co-cultured with A549 epithelial monolayers. THP-1 cells were either directly co-cultured with confluent A549 cells in the same well or separated by a filter insert. At different time points (4, 8, 12, 24 h) THP-…
Fig. 2.
IL-6 induction by a co-culture of THP-1 monocytes/macrophages with A549 epithelial cell line. Both cell lines were co-cultured for 24 h and at different time points (4, 8, 12, 24 h) the supernatants with floating THP-1 cells were aspirated, centrifuged…
Fig. 3.
IL-8 induction by a co-culture of THP-1 monocytes/macrophages with A549 epithelial cell line. Both cell lines were co-cultured for 24 h and at different time points (4, 8, 12, 24 h) the supernatants with floating THP-1 cells were aspirated, centrifuged…
Effect of p65 suppression on IL-6 release by A549/THP-1 co-culture
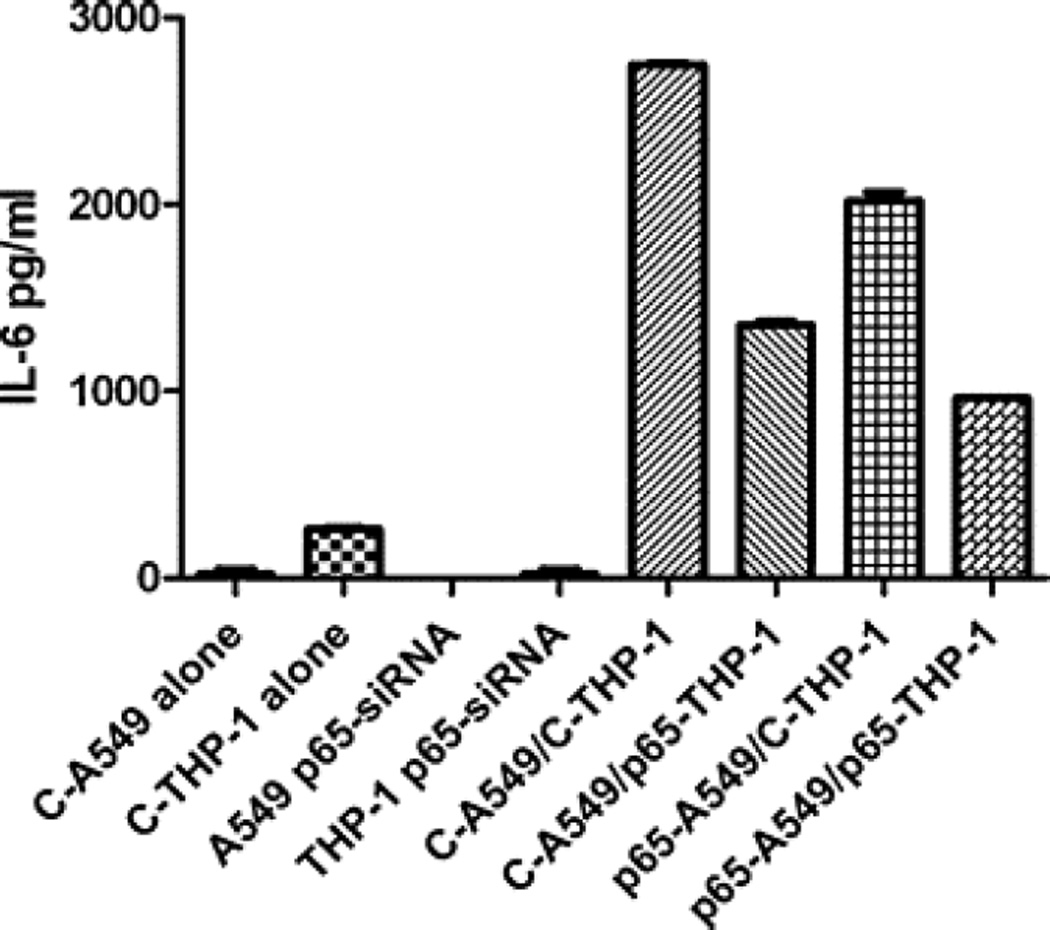
Suppression of the NFκB subunit by p65 demonstrated that this pathway is involved in the regulation of IL-6 in both cell types (Fig. 4). The release of IL-6 was inhibited in a co-culture of p65 siRNA transfected THP-1 cells with control A549 cells (p < 0.0001) and similarly in a co-culture of control THP-1 cells with siRNA p65 transfected A549 cells (p < 0.0001) as compared with control A549/THP-1 co-culture.
Fig. 4.
P65-siRNA effect on IL-6 release by A549/THP-1 co-culture. A549 and THP-1 cells were transfected with p65-siRNA or non-targeting control siRNA. Then, the THP-1 cells were plated on top of confluent monolayer of A549 cells with relevant control culture…
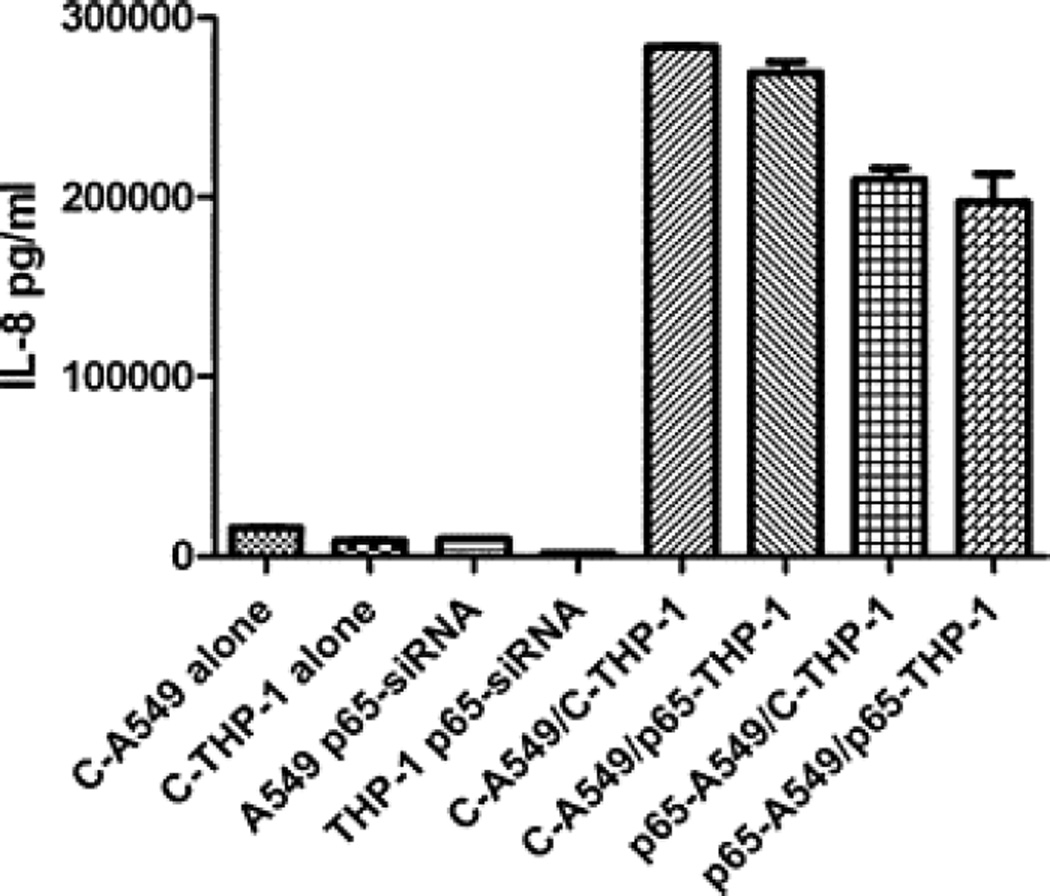
Effect of p65 suppression on IL-8 release by A549/THP-1 co-culture
The release of IL-8 was inhibited only in a co-culture of p65 transfected A549 cells with control THP-1 cells (p < 0.0003) or p65 transfected THP-1 cells (p < 0.0053). Isolated suppression of p65 only in THP-1 did not affect IL-8 production (Fig. 5).
Fig. 5.
P65-siRNA effect on IL-8 release by A549/THP-1 co-culture. After the transfection of A549 and THP-1 cells with p65-siRNA or non-targeting control siRNA, the cells were co-cultured for 24 h with relevant control culture conditions of isolated cells. The…
Discussion
Coordinated efforts of different cell populations present in the lung are important for rapid and effective responses to stimuli inhaled from the environment. Epithelial cells in both the airways and alveoli are able to release multiple chemokines that regulate the influx of different types of immune cells (Mercer et al., 2009, Sachse et al., 2006 and Vroling et al., 2007), proinflammatory cytokines that amplify immune responses (Ge et al., 2010 and Hashimoto et al., 2000), and regulate functions of T lymphocytes (Regamey et al. 2007), as well as growth factors (Leigh et al. 2008) and mediators of repair (Howat et al. 2002). In the lung, macrophages are the predominant population of “professional” immune cells and are thus continuously affected by mediators released from the respiratory epithelium. It is likely that both soluble factors and direct cell–cell contacts can play a role in this communication. We have shown, using an in vitro model, that THP-1 macrophages co-cultured with filter separated A549 epithelial cells upregulate the transcription of numerous proinflammatory genes of cytokines, chemokines and signalling molecules within hours. The highest mRNA expression at 4 h of co-culture was found for IL-8/CXCL8, a major chemokine regulating the influx of neutrophils. This was followed by early growth response protein 1 which induces activation and differentiation of lymphocytes (Gomez-Martin et al. 2010). The induction of macrophage-related proinflammatory cytokines TNF alpha and IL-1 beta was also very early and robust. On the other hand, slightly delayed induction of the anti-inflammatory cytokine IL-10 mRNA was detected, which increased gradually during the 24 h period. Delayed release of IL-10 could function as a control mechanism to dampen and/or terminate inflammation induced by earlier expression of pro-inflammatory genes. Suppression of inflammation by delayed induction of some NF kappa B related genes is also consistent with recent findings that this transcription factor is associated not only with inflammatory responses but also mediates pathways leading to resolution of inflammation (Han et al., 2009 and Lawrence and Fong, 2010). This is also supported by the observation that A549 cells reduce NO production by alveolar macrophages (Rubovitch et al. 2007).
Findings at the protein level, consistent data with the mRNA induction, were found in the release of IL-6 and IL-8 and ICAM-1 expression. These proteins were selected as we have previously studied their regulation in primary cultures of human bronchial epithelial cells and human blood monocytes and know that similar regulatory mechanisms are transferable to studies of cell lines. Both macrophages and epithelial cells might contribute as a source of IL-6 and IL-8. It has been found in previous study, that a co-culture of A549 cells with mononuclear phagocytes modulates the release of cytokines and chemokines in response to endotoxin and staphylococcal exotoxins (Krakauer 2002), hyperoxia (Hjort et al. 2003), and microparticles (Alfaro-Moreno et al. 2008).
Membrane expression of ICAM-1 on THP-1 macrophages was induced to a higher degree when the cells were in a direct contact with epithelial cells as compared to filter-separated conditions. This observation is consistent with our data using primary cells and the interaction probably involves multiple ligand signalling since our preliminary attempts to block it with a large panel of monoclonal antibodies against major contact and co-stimulatory molecules have not shown any specific ligand dependence (data not shown).
NF kappa B regulates multiple genes involved in different stages of immune responses and controls also many functions of macrophages and epithelial cells. This transcription factor plays a key role in the regulation of proinflammatory cytokine genes (TNF alpha, IL-1beta, IL-6), Toll-like receptors, IL-1 and TNF receptors. NF kappa B triggers activation of MAPK and STAT proteins and downstream leads to induction of interferons (Biswas and Lewis 2010). RNA interference targeting the p65 (RelA) subunit of NF kappa B has been shown to modulate functions of primary epithelial cells in our previous study (Liu et al. 2008) and we thus decided to use this approach in the current co-culture model. We have found that THP-1 cells lacking p65 are defective in IL-6 release after co-culture with epithelial cells. In contrast, IL-8 concentrations were not significantly inhibited by blocking THP-1 p65 using siRNA in the same set of experiments, suggesting regulation through different pathways. A role for NF kappa B is not excluded, however, as atypical NF kappa B pathways, involving p105, p100, and RelB (Sun and Ley 2008) are responsible for regulation of IL-8 gene transcription. Negative results have been obtained in experiments targeting p65 to modulate VEGF release from THP-1 cells (data not shown).
We conclude that unstimulated respiratory epithelial cells regulate the expression of multiple genes in human macrophages, that this is, at least partially due to NF kappa B dependent signaling and that these interactions might play a key role in immediate responses in the respiratory epithelium.
Acknowledgements
The study was supported by the MSMT grant No. ME 906, IGA MZCR grant No. 10524-3 and by the Institute for Clinical and Experimental Medicine (MZO 00023001).
Abbreviations
- NF
nuclear factor
- mRNA
messenger ribonucleic acid
- ICAM
intercellular adhesion molecule
- IL
interleukin
- hIL
human interleukin
- rIL
recombinant interleukin
- siRNA
small interfering ribonucleic acid
- qRT-PCR
quantitative reverse transcription real-time polymerase chain reaction
- Ig
immunoglobulin
- ELISA
enzyme-linked immunosorbent assay
References
- Alfaro-Moreno E, Nawrot TS, Vanaudenaerde BM, Hoylaerts MF, Vanoirbeek JA, Nemery B, Hoet PH. Co-cultures of multiple cell types mimic pulmonary cell communication in response to urban PM10. Eur. Respir. J. 2008;32:1184–1194. doi: 10.1183/09031936.00044008. [DOI] [PubMed] [Google Scholar]
- Biswas SK, Lewis CE. NF-{kappa}B as a central regulator of macrophage function in tumors. J. Leukoc. Biol. 2010;88:877–884. doi: 10.1189/jlb.0310153. [DOI] [PubMed] [Google Scholar]
- Bulek K, Swaidani S, Aronica M, Li X. Epithelium: the interplay between innate and Th2 immunity. Immunol. Cell Biol. 2010;88:257–268. doi: 10.1038/icb.2009.113. [DOI] [PubMed] [Google Scholar]
- Ge Q, Moir LM, Black JL, Oliver BG, Burgess JK. TGFbeta1 induces IL-6 and inhibits IL-8 release in human bronchial epithelial cells-the role of SMAD2/3. J. Cell Physiol. 2010;225:846–854. doi: 10.1002/jcp.22295. [DOI] [PubMed] [Google Scholar]
- Gomez-Martin D, Diaz-Zamudio M, Galindo-Campos M, Alcocer-Varela J. Early growth response transcription factors and the modulation of immune response: implications towards autoimmunity. Autoimmun. Rev. 2010;9:454–458. doi: 10.1016/j.autrev.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Han W, Joo M, Everhart MB, Christman JW, Yull FE, Blackwell TS. Myeloid cells control termination of lung inflammation through the NF-kappaB pathway. Am. J. Physiol. Lung Cell Mol. Physiol. 2009;296:L320–L327. doi: 10.1152/ajplung.90485.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto S, Matsumoto K, Gon Y, Maruoka S, Kujime K, Hayashi S, Takeshita I, Horie T. p38 MAP kinase regulates TNF alpha-, IL-1 alpha- and PAF-induced RANTES and GM-CSF production by human bronchial epithelial cells. Clin. Exp. Allergy. 2000;30:48–55. doi: 10.1046/j.1365-2222.2000.00641.x. [DOI] [PubMed] [Google Scholar]
- Hjort MR, Brenyo AJ, Finkelstein JN, Frampton MW, LoMonaco MB, Stewart JC, Johnston CJ, D’Angio CT. Alveolar epithelial cell–macrophage interactions affect oxygen-stimulated interleukin-8 release. Inflammation. 2003;27:137–145. doi: 10.1023/a:1023817811850. [DOI] [PubMed] [Google Scholar]
- Howat WJ, Holgate ST, Lackie PM. TGF-beta isoform release and activation during in vitro bronchial epithelial wound repair. Am. J. Physiol. Lung Cell Mol. Physiol. 2002;282:L115–L123. doi: 10.1152/ajplung.2002.282.1.L115. [DOI] [PubMed] [Google Scholar]
- Krakauer T. Stimulant-dependent modulation of cytokines and chemokines by airway epithelial cells: cross talk between pulmonary epithelial and peripheral blood mononuclear cells. Clin. Diagn. Lab. Immunol. 2002;9:126–131. doi: 10.1128/CDLI.9.1.126-131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T, Fong C. The resolution of inflammation: anti-inflammatory roles for NF-kappaB. Int. J. Biochem. Cell Biol. 2010;42:519–523. doi: 10.1016/j.biocel.2009.12.016. [DOI] [PubMed] [Google Scholar]
- Leigh R, Oyelusi W, Wiehler S, Koetzler R, Zaheer RS, Newton R, Proud D. Human rhinovirus infection enhances airway epithelial cell production of growth factors involved in airway remodeling. J. Allergy Clin. Immunol. 2008;121:1238–1245. e1234. doi: 10.1016/j.jaci.2008.01.067. [DOI] [PubMed] [Google Scholar]
- Liu X, Togo S, Al-Mugotir M, Kim H, Fang Q, Kobayashi T, Wang X, Mao L, Bitterman P, Rennard S. NF-kappaB mediates the survival of human bronchial epithelial cells exposed to cigarette smoke extract. Respir. Res. 2008;9:66. doi: 10.1186/1465-9921-9-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A, Sica A, Locati M. New vistas on macrophage differentiation and activation. Eur. J. Immunol. 2007;37:14–16. doi: 10.1002/eji.200636910. [DOI] [PubMed] [Google Scholar]
- Mercer PF, Johns RH, Scotton CJ, Krupiczojc MA, Konigshoff M, Howell DC, McAnulty RJ, Das A, Thorley AJ, Tetley TD, et al. Pulmonary epithelium is a prominent source of proteinase-activated receptor-1-inducible CCL2 in pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2009;179:414–425. doi: 10.1164/rccm.200712-1827OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regamey N, Obregon C, Ferrari-Lacraz S, van Leer C, Chanson M, Nicod LP, Geiser T. Airway epithelial IL-15 transforms monocytes into dendritic cells. Am. J. Respir. Cell Mol. Biol. 2007;37:75–84. doi: 10.1165/rcmb.2006-0235OC. [DOI] [PubMed] [Google Scholar]
- Rubovitch V, Gershnabel S, Kalina M. Lung epithelial cells modulate the inflammatory response of alveolar macrophages. Inflammation. 2007;30:236–243. doi: 10.1007/s10753-007-9042-2. [DOI] [PubMed] [Google Scholar]
- Sachse F, von Eiff C, Stoll W, Becker K, Rudack C. Induction of CXC chemokines in A549 airway epithelial cells by trypsin and staphylococcal proteases – a possible route for neutrophilic inflammation in chronic rhinosinusitis. Clin. Exp. Immunol. 2006;144:534–542. doi: 10.1111/j.1365-2249.2006.03089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striz I, Slavcev A, Kalanin J, Jaresova M, Rennard SI. Cell–cell contacts with epithelial cells modulate the phenotype of human macrophages. Inflammation. 2001;25:241–246. doi: 10.1023/a:1010975804179. [DOI] [PubMed] [Google Scholar]
- Striz I, Wang YM, Svarcova I, Trnka L, Sorg C, Costabel U. The phenotype of alveolar macrophages and its correlation with immune cells in bronchoalveolar lavage. Eur. Respir. J. 1993;6:1287–1294. [PubMed] [Google Scholar]
- Sun SC, Ley SC. New insights into NF-kappaB regulation and function. Trends Immunol. 2008;29:469–478. doi: 10.1016/j.it.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroling AB, Duinsbergen D, Fokkens WJ, van Drunen CM. Allergen induced gene expression of airway epithelial cells shows a possible role for TNF-alpha. Allergy. 2007;62:1310–1319. doi: 10.1111/j.1398-9995.2007.01495.x. [DOI] [PubMed] [Google Scholar]
- Wan F, Lenardo MJ. The nuclear signaling of NF-kappaB: current knowledge, new insights, and future perspectives. Cell Res. 2010;20:24–33. doi: 10.1038/cr.2009.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang X, Hussain S, Zheng Y, Sanjabi S, Ouaaz F, Beg AA. Distinct roles of different NF-kappa B subunits in regulating inflammatory and T cell stimulatory gene expression in dendritic cells. J. Immunol. 2007;178:6777–6788. doi: 10.4049/jimmunol.178.11.6777. [DOI] [PubMed] [Google Scholar]