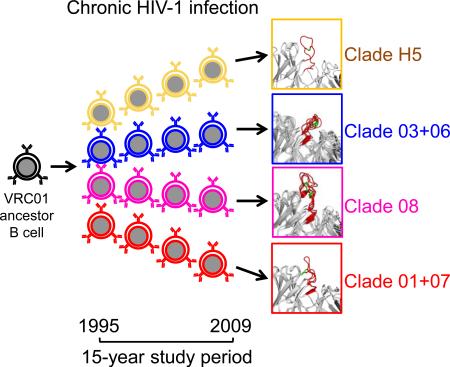
Abstract
HIV-1-neutralizing antibodies develop in most HIV-1-infected individuals, although highly effective antibodies are generally observed only after years of chronic infection. Here we characterize the rate of maturation and extent of diversity for the lineage that produced the broadly neutralizing antibody VRC01 through longitudinal sampling of peripheral B cell transcripts over 15 years and co-crystal structures of lineage members. Next-generation sequencing identified VRC01-lineage transcripts, which encompassed diverse antibodies organized into distinct phylogenetic clades. Prevalent clades maintained characteristic features of antigen recognition, though each evolved binding loops and disulfides that formed distinct recognition surfaces. Over the course of the study period, VRC01-lineage clades showed continuous evolution, with rates of ~2 substitutions per 100 nucleotides per year, comparable to that of HIV-1 evolution. This high rate of antibody evolution provides a mechanism by which antibody lineages can achieve extraordinary diversity and, over years of chronic infection, develop effective HIV-1 neutralization.
Graphical Abstract
HIV-1-neutralizing antibodies to autologous virus develop within weeks or months of infection (Albert et al., 1990; Richman et al., 2003; Wei et al., 2003), with serum neutralization generally evolving over time to gain increased potency and breadth (Gray et al., 2011; Mikell et al., 2011). In a study of ~200 chronically HIV-1-infected individuals, serum from ~50% of studied donors contained antibodies capable of neutralizing ~50% of HIV-1 strains (Hraber et al., 2014). A subset of these individuals develops potent and broadly reactive antibodies (Gray et al., 2011; Li et al., 2007; Mikell et al., 2011; Walker et al., 2010; Wu et al., 2006). Isolation and characterization reveal these broadly neutralizing antibodies to have one or more unusual features including long or protruding heavy chain third complementarity-determining regions (CDR H3s) (Burton et al., 1994; Walker et al., 2009; Zhou et al., 2007), domain swapping (Calarese et al., 2003), unusual post-translational modifications such as tyrosine sulfation (TyrS) (Huang et al., 2004; Pancera et al., 2010; Pejchal et al., 2010), poly or autoreactivity (Haynes et al., 2005), extensive somatic hypermutation (SHM) (Scheid et al., 2011; Walker et al., 2011; Wu et al., 2010; Wu et al., 2011), or dependence on framework region contacts (Klein et al., 2013). These characteristics highlight the extensive maturation process necessary for most antibodies to achieve effective HIV-1 neutralization (Burton et al., 2005; Klein et al., 2013; Mascola and Haynes, 2013; Scheid et al., 2009).
To understand how effective neutralization develops, we and others have investigated the ontogenies of neutralizing antibody lineages using monoclonal antibodies (mAbs) isolated from individual B cells (Bonsignori et al., 2012; Scheid et al., 2011; Walker et al., 2011), next-generation sequencing (NGS) of cross-sectional samples (Wu et al., 2011; Zhou et al., 2013; Zhu et al., 2012; Zhu et al., 2013), and NGS of longitudinal samples studied from time of infection (Doria-Rose et al., 2014; Liao et al., 2013). While these studies often focused on revealing the unmutated common ancestors (UCAs) of neutralizing antibody lineages and early antibody maturation, questions remain about the long-term and continuing development of antibody lineages. What is the scope of B cell development in a broadly neutralizing antibody lineage? What are the rates, compositions, extents, and continuities of lineage evolution? What biological mechanisms underlie the development of HIV-1 neutralization?
Here we investigate the VRC01-antibody lineage, which targets the site of CD4 engagement on HIV-1 (Wu et al., 2010; Zhou et al., 2010) and is a member of a class of antibodies (the VRC01 class), which share similar structural and genetic characteristics (West et al., 2012; Wu et al., 2011; Zhou et al., 2013). We identified dozens of VRC01-lineage antibodies from one donor and describe characteristics of this lineage as it evolved over the course of 15 years. Overall, these results delineate the scope of evolutionary diversity for a persistent antibody lineage. Antibody lineage characteristics identified here – such as multi-clade divergence and a high rate of evolution – may be common to effective HIV-1-neutralizing antibodies and provide insights into the immunological mechanisms that enable their development.
Results
39 probe-identified antibodies define three VRC01-lineage clades
The broadly neutralizing antibodies VRC01, VRC02 and VRC03, were previously isolated from an August 2008 sample of donor 45 peripheral blood mononuclear cells (PBMC), using the resurfaced stabilized core 3 (RSC3) probe (Wu et al., 2010). From additional 2008 samples, other probes were subsequently used to isolate 5 more VRC01-class antibodies: NIH45-46, NIH45-177, NIH45-243, VRC06 and VRC06b (Li et al., 2012; Scheid et al., 2011). All 8 of these antibodies appeared to be somatic variants from a single VRC01 lineage (Zhou et al., 2013).
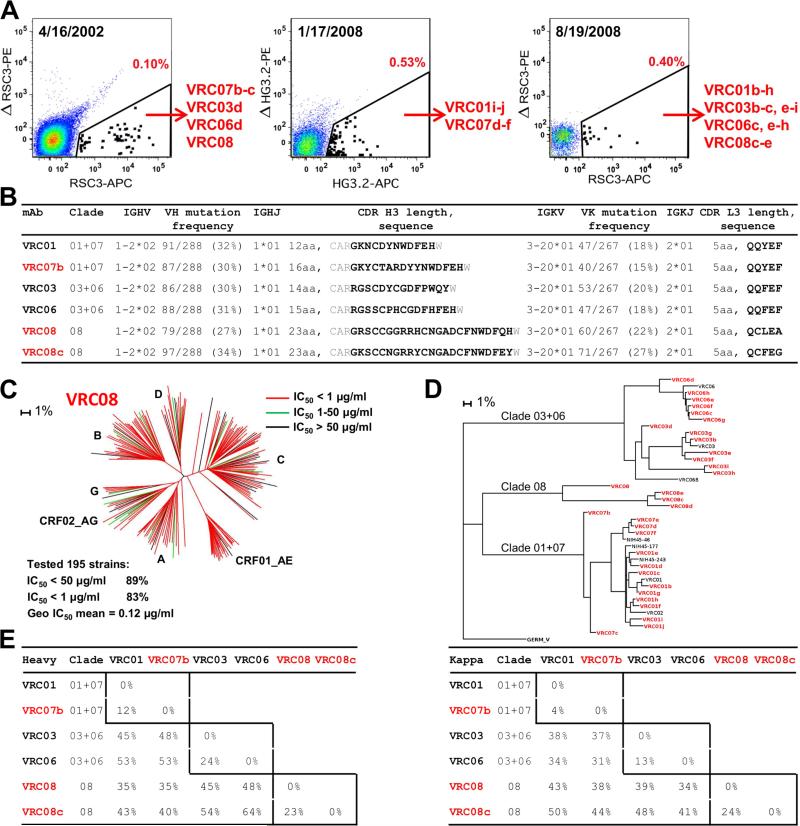
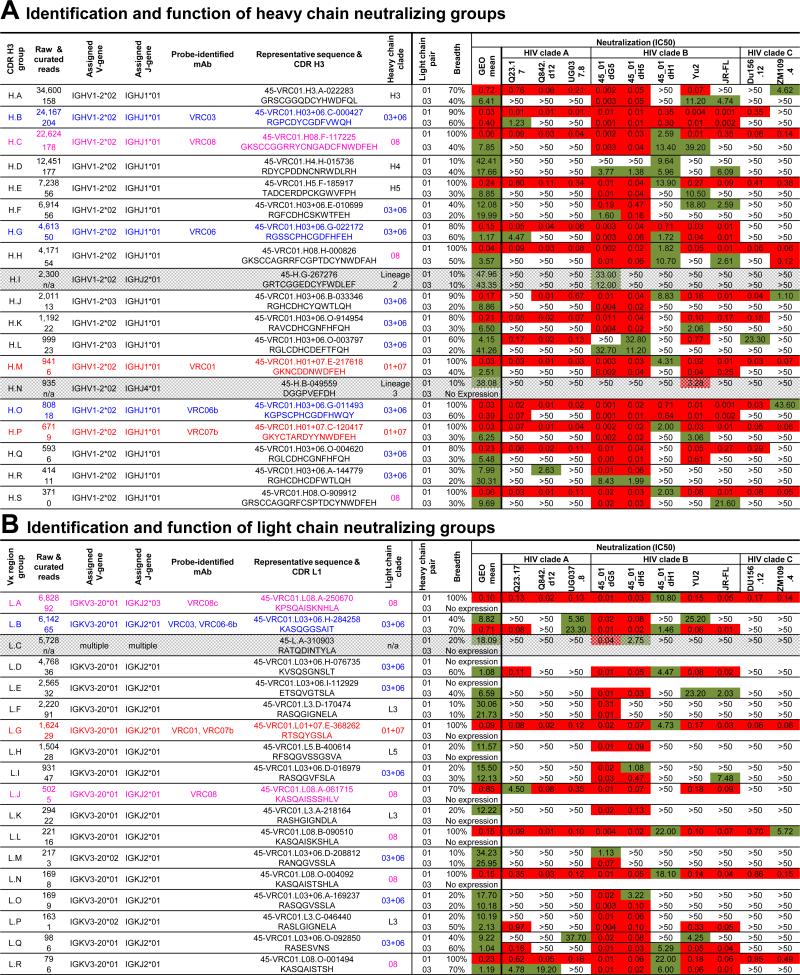
To gain insight into the scope of the VRC01 lineage, we performed RSC3-specific B cell sorting on PBMCs from April 2002 and August 2008 time points to identify additional lineage members (Figure 1A, left and right). We also performed B-cell sorting with a modified gp120 outer-domain probe (Joyce et al., 2013) on PBMCs from January 2008 (Figure 1A, middle). Five antibodies were recovered from the 2002 sample, including a new VRC01-lineage antibody, VRC08, which was substantially different from those identified previously. The CDR H3 of VRC08 was 23 amino acids in length by Kabat definition, substantially longer than those of VRC01 and VRC07 (12 and 16 amino acids, respectively) or VRC03 and VRC06 (14 and 15 amino acids, respectively) (Figure 1B). We assessed neutralization by VRC08 against 195 Env-pseudoviruses. VRC08 displayed breadth and potency similar to VRC01 (Figure 1C and Table S1). In addition to VRC08, several other closely related antibodies were identified. In all, we identified 31 new neutralizing antibodies from donor 45, all with naturally paired heavy and light chains and derived from the same origin genes (Figure 1B and Figure S1).
Figure 1. VRC01-antibody lineage: 39 probe-identified antibodies define 3 distinct clades.
(A) Isolation of antigen-specific antibodies by sorting of donor 45. B-cells were probed either with the RSC3 and its ΔI371 mutant ΔRSC3, or with a modified outer domain OD4.2 protein HG3.2 and its D368R mutant ΔHG3.2. Indicated in red is the percent of total IgG+ B-cells defined as probe-specific in each gate.
(B) Heavy and light chain sequence analysis for six representative antibodies, two from each clade. Residues flanking the CDR H3 are shown in gray.
(C) VRC08 neutralization dendrogram of 195 HIV-1 Env-pseudoviruses with branches colored by potency.
(D) Maximum-likelihood tree of VRC01-lineage antibodies from donor 45, rooted on the germline V gene sequence. Antibody clades, 01+07, 03+06, and 08 are indicated.
(E) Pairwise sequence difference of heavy (left) and light (right) chain variable domains for the six representative antibodies. Intra-clade differences are boxed.
A maximum-likelihood phylogenetic tree was constructed from the concatenated heavy and light chain nucleotide sequences of the 39 VRC01-lineage antibodies (Figure 1D); they segregated into three major clades, termed clade 01+07, clade 03+06, and clade 08. Sequence differences within clades were under 25%, while differences exceeded 50% between clades (Figure 1E), and CDR H3 lengths varied from 12 to 23 amino acids.
Tracking potential VRC01-lineage sequences in longitudinal samples
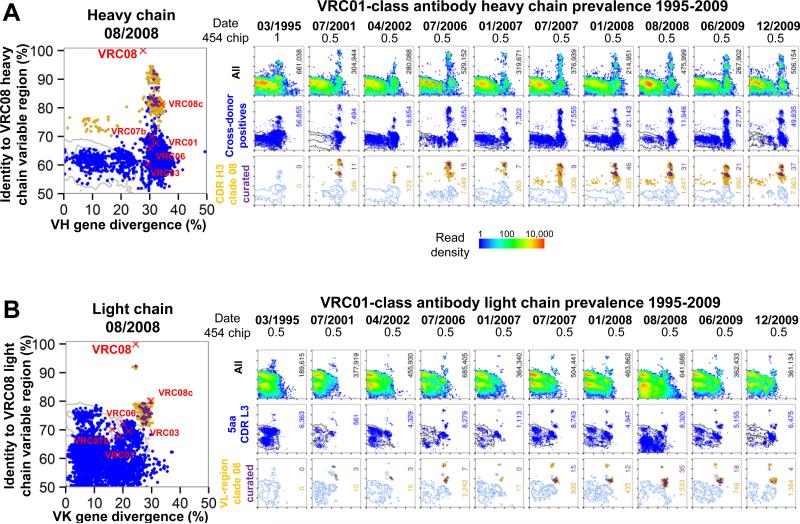
To gain a more complete understanding of the VRC01 lineage and its development, we used NGS to identify potential lineage members over 15 years. The first sample available was from March 1995, well after the diagnosis of HIV-1 infection in 1990. Ten samples were analyzed, extending to December 2009. During this 15 year period, donor 45 maintained a relatively stable plasma viral load of around 10,000 copies/ml and a CD4+ T cell count >500 cells/μl without anti-retroviral therapy (Table S2A). cDNA libraries from each time point, corresponding to the transcripts of 3-5 million PBMCs, were used as templates, and 5’ gene family-specific primers were used to amplify VH1 genes or Vκ3 genes, with 3’ primers for both IgM and IgG or Igκ constant regions. We estimate that each longitudinal sample contained 75-125,000 VH1-family-derived B cells and 100-160,000 Vκ3-family-derived B cells. 454 pyrosequencing, with either a half- or full-454 chip, generally yielded between a quarter-million to a million raw reads for each heavy or light chain reaction, for each time point (Figure 2).
Figure 2. NGS-identified VRC01-lineage transcripts in donor 45 over 15 years.
(A) Clade-specific identity-divergence plots from heavy chain longitudinal samples. Large example plot is shown at left for clade 08. Sequence divergence from the assigned germ line V gene (x axis) and sequence identity to the VRC08 heavy chain variable domain (y axis). Positions of the six representative antibodies from Figure 1D-E are shown as red X's. Identity-divergence plots for 10 longitudinal time points (right). Time points and fraction of a 454 chip used for NGS are indicated at the top. The top row shows a heat map for positions of all 454 sequences. The total number of sequences is indicated at the right borders. The middle row shows the distribution of cross-donor positive sequences as blue dots, with grey contours indicating raw sequences. The total number of cross-donor positives is displayed in blue. The bottom row shows the distribution of sequences in the same CDR H3 group as any probe-isolated antibody from that clade. Yellow dots indicate raw sequences within the CDR H3 groups, while purple dots show the subset of those sequences that survived quality control filtering. The total numbers of raw and curated sequences in the CDR H3 groups are indicated in yellow and purple, respectively; blue contours indicate cross-donor positives.
(B) Clade-specific identity-divergence plots from light chain longitudinal samples. The same analyses are shown as for (A), except that the middle row shows the distribution of sequences with a 5 amino acid CDR L3. Because sequences are clustered across all 10 time points to determine the final curated sequences (Extended Experimental Procedures), groups of in-clade sequences (yellow dots) with high-identity to the referent antibody (eg in the 01/2007, 07/2007, and 01/2008 time points) may appear to have resulted in no surviving high-quality sequences (no purple dots). In actuality, the representative sequence in the final curated set has simply been chosen from a different time point (eg 07/2006).
A heavy chain-specific bioinformatics pipeline with multiple computational sieves was used to identify VRC01-lineage transcripts and to classify them into closely related groups of sequences based on CDR H3 identity (Figure S2A, bottom panels). We utilized a modified cross-donor analysis algorithm and applied several levels of data quality filtering and dereplication with a 97.25% identity threshold (Extended Experimental Procedures). These quality filters ameliorated known 454 errors and reduced the number of unique sequences. The effect of each data-filtering step on the number of retained sequences is shown in Table S2B. Similarly, a light chain-specific pipeline was used to identify and classify VRC01-lineage light chain transcripts (Figure S2B, bottom panels). As with the heavy chains, we applied data quality filters (Figure 2B, bottom panels, and Table S2B).
We were able to track the evolution of each clade of the VRC01 lineage through the positions of clusters of closely related sequences on identity/divergence (I-D) plots over time (Figure 2,and Figure S2C-F). The positions of these clusters at successive time points revealed the persistence and continued SHM of each clade, with a typical increase of 5-10% in divergence over 10 years and a maximal sequence identity around the time of isolation of the referent antibody.
Validation of functional antibody clades and definition of the VRC01 lineage
For both heavy and light chains, we found large groups of sequences that were divergent from all probe-isolated antibodies, but shared VRC01-lineage origin genes. We previously used functional complementation between heavy and light chains to confirm functionality and membership in the VRC01 class (Wu et al., 2011; Zhu et al., 2013). Importantly, reconstitution of unrelated antibody heavy chains with a VRC01 light chain, even using heavy chains with high predicted VRC01 structural compatibility, failed to neutralize HIV-1 in 10 out of 10 trials (Wu et al., 2011). To assess the thousands of divergent B cell transcripts from NGS, we clustered them into groups: heavy chains were clustered based on CDR H3 identity into CDR H3 groups; light chains, which have short CDR L3s, were clustered based on overall sequence identity into VL-region groups. We focused on the most populous groups (those containing at least 300 raw heavy chain sequences or 75 raw light chain sequences) and chose two representative sequences for testing from each heavy and light chain group (Figure 3 and Table S3A-B) (Extended Experimental Procedures). Many of these selected sequences were observed at multiple time points (Table S3C and S3D). For heavy chains, representative sequences from 19 CDR H3 groups neutralized HIV-1 when reconstituted with VRC01 or VRC03 light chain (Figure 3A and Table S3A). Two of these groups, H.I and H.N, appeared to use non-lineage matching JH genes (Extended Experimental Procedures), though the identity of the JH gene was uncertain due to the high level of SHM (~35% and ~25% diverged from VH1-2, respectively). These ambiguous groups were analyzed separately from the rest of the VRC01 lineage. For light chains, 18 VL groups neutralized HIV-1 when reconstituted with VRC01 or VRC03 heavy chain (Figure 3B, Table S3B). One group, L.C, was composed of multiple unrelated lineages using various Vκ and Jκ genes and was not included in analysis of the VRC01 lineage.
Figure 3. Many bioinformatically identified sequences not closely related to probe-identified antibodies neutralize diverse viral strains.
(A) Representative sequences from the most prevalent CDR H3 groups were synthesized, reconstituted with VRC01 and VRC03 light chains, and tested for neutralization. CDR H3 groups confirmed for neutralization were assessed for sequence identity (Figure 4A) to each other and merged into clades (center column). Two groups with non-matching J gene assignments (gray rows) were excluded from further analysis. Total sequences in each group, probe-identified representative (if any), assigned V and J genes, and the most neutralizing representative and its CDR H3 sequence (left columns) are shown. Neutralization breadth and potency for both VRC01 and VRC03 light chain pairings are provided against selected HIV-1 viruses from clades A, B, and C (right columns).
(B) The same information as shown in (A), for the most prevalent light chain-variable region groups tested for neutralization after reconstitution with the heavy chains from VRC01 and VRC03. Group L.C, shown in gray, consisted of multiple unrelated clones and was excluded from further analysis.
Although they shared the same germ line origin genes (VH1-2 and JH1), the diverse heavy chain sequences identified could derive from a single lineage or multiple lineages. We observed that the 39 probe-identified antibodies from donor 45 all contained a cysteine at position 98 (99 in some sequences due to a 1 aa insertion). This cysteine is not a required feature of the VRC01-class, as VRC01-class antibodies from other donors do not contain such a residue. We therefore used this signature cysteine to assess membership in the VRC01 lineage. Of 1041 curated NGS sequences assigned to the VRC01 lineage, 6 did not contain the cysteine while 1035 did (99.4%). By contrast, of the remaining VH1-2-derived NGS reads from this donor not assigned to the VRC01-lineage, 104,223 sequences did not have this signature cysteine, while 4,641 did (4.5%; p< 0.0001) (Figure S3B).
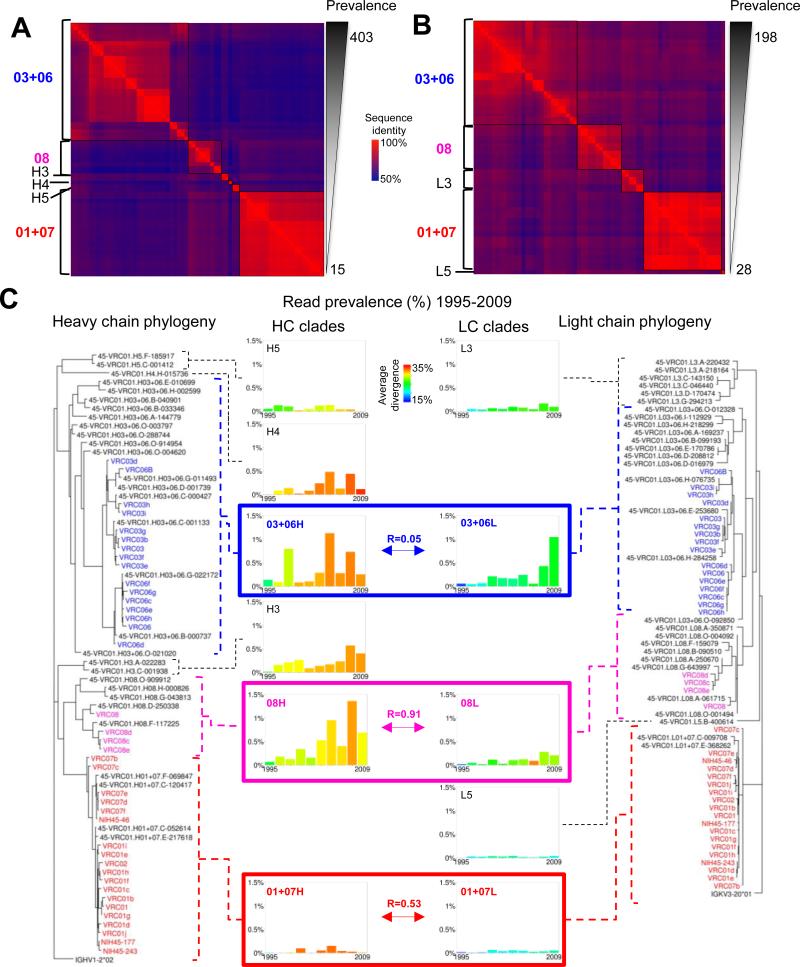
To examine the relationship among the neutralizing groups with VRC01-lineage origin genes, we calculated the full-length pairwise identities of the validated representative sequences and probe-identified antibodies. Pairwise identity matrices, grouped by similarity, are shown as heat maps for heavy and light chains in Figures 4A and B, respectively. The CDR H3 and VL groups clustered into higher-order units. Critically, correspondence with maximum-likelihood phylogenetic trees (Figure 4C) confirmed that the clusters of neutralizing groups define phylogenetic clades similar to the three originally defined by the probe-isolated antibodies (Figure S4).
Figure 4. Heavy and light chain of the VRC01 lineage exhibit multi-clade phylogenetic organization and concordance.
(A) Heat map showing pairwise sequence identities for heavy chains of probe-identified antibodies and validated neutralizing heavy chain sequences sorted by overall prevalence (among final curated sequences). Groups with high identity and similar prevalences were merged into clades, shown at left.
(B) Heat map for light chains of probe-identified antibodies and validated neutralizing light chain sequences, produced as in (A).
(C) Maximum-likelihood phylogenetic trees for heavy chain (left) and light chain (right) sequences. The clades described in (A) and (B) can be clearly seen in the structure of the trees, which have similar overall topology. Temporal prevalence (middle) is charted for each clade as the fraction of unique sequences with an in-frame junction and no stop codons (but without manual curation) at each time point which are assigned to that clade. For clades 01+07, 03+06, and 08, which are anchored by probe-identified antibodies, the correlation of heavy and light chain-temporal prevalence is shown (boxes). The average divergence for each clade at each time point is calculated from curated sequences.
See also Figure S4.
Known heavy and light chain pairings from the probe-identified antibodies were used to approximately align the heavy and light maximum-likelihood phylogenetic trees. Temporal prevalence was calculated from the number of non-redundant NGS reads assigned to each of the clades (Figure 4C, middle) (Extended Experimental Procedures). For each clade, prevalence of reads waxed and waned independently over time. For example, clades H5 and L3 remained at a fairly constant level, while clade 01+07 was more prevalent at intermediate time points, and clades H3, H4, and 08 increased in prevalence at later time points. For the three probe-identified clades (01+07, 03+06, and 08), heavy and light chain prevalence should correlate temporally; in practice these correlations ranged from 0.05-0.91, suggesting possible sampling issues for clade 03+06. Overall, the NGS of peripheral VRC01-lineage transcripts provided a large number of sequences (Table S2B), allowing for much greater definition of VRC01-lineage diversity than was possible with the 39 probe-identified antibodies (Figure 1). Multiple branches surround and embed the original three clades in a more diverse phylogenetic tree with 3 additional heavy chain clades and 2 additional light chain clades (Figure 4C). The greater number of NGS sequences permitted quantification of the initial and newly identified clades over time, thus illuminating the scope, diversity, and development of the lineage.
Conservation of antigen-binding mode within the 01+07 clade of the VRC01 lineage
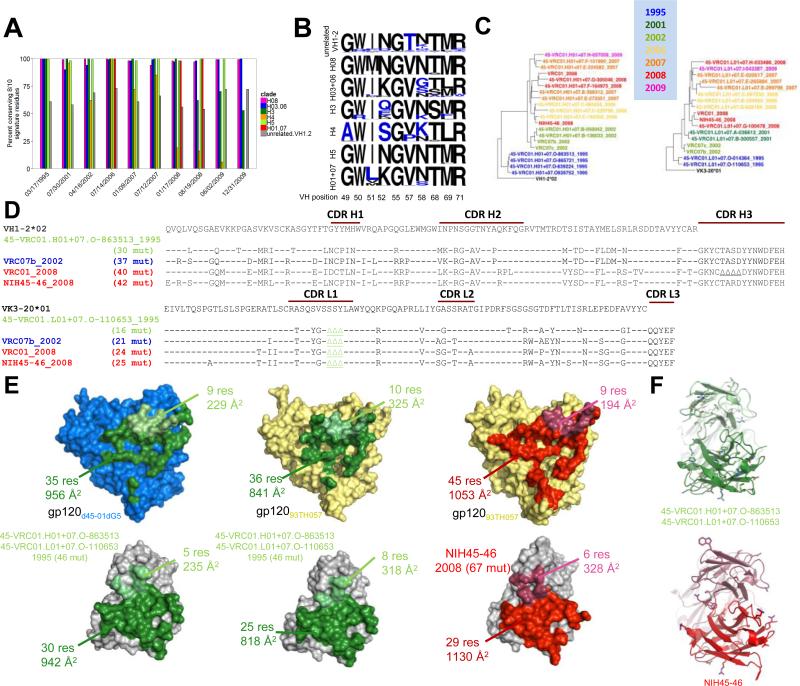
The extraordinary sequence diversity of the VRC01 lineage revealed by NGS could represent possibilities ranging from significant changes in mode of antigen recognition to sequence alteration with a conserved binding mode. To delineate between these two extremes, we evaluated the longitudinal maintenance of a previously defined set of VRC01-signature residues (Zhou et al., 2013) and the structural conservation of recognition by a clade over time. While only 60-70% of unrelated VH1-2 sequences conserved at least 8 of 10 positions in the signature, nearly all sequences from 4 of the 6 heavy chain clades did so (Figures 5A-B and S5A). For clade 01+07, we determined co-crystal structures of an antibody from 1995 and compared these to the co-crystal structure of an antibody identified from the 2008 time point (Figure 5C-F) (Diskin et al., 2011). For the 1995 antibody, co-crystal structures were determined with extended cores both from the autologous gp120 (from donor 45) and from a heterologous gp120. By contrast, the antibody from the 2008 time point was determined in complex only with a heterologous gp120 extended core (Figure 5E, Table S4). We calculated the root-mean square differences (rmsds) for the variable domains in the various crystal structures: there was a 0.38 Å rmsd on Cα of the variable domains between the two 1995 antibody structures; between the 1995 structures and 2008 structure, the rmsd was 0.45 and 0.50 Å for the same gp120 and different gp120s, respectively. Epitope recognition was highly conserved between the 1995 and 2008 antibodies, which interact with a similarly sized area of gp120 (1185 Å 2 and 1166 Å 2 for the 1995 antibody, and 1247 Å 2 for the 2008 antibody) with approximately 95 % of the contact area conserved (Figure S5B). The antibody paratope was also highly similar, but showed an increase in size for the 2008 antibody compared to the 1995 antibody (1177 Å 2 and 1136 Å 2 for the 1995 antibody, and 1458 Å 2 for the 2008 antibody). This difference in the paratope was largely due to reduced inner domain and bridging sheet interactions for the 1995 antibody compared to the 2008 antibody, and the increase in recognized surface could be attributed to SHM, especially Ser74Tyr and Ser99BArg in the heavy chain.
Figure 5. Conservation of VRC01+07 clade recognition over 15 years of chronic maturation.
(A) Percent of curated sequences in each clade conserving at least 8 of the 10-residue heavy chain signature amino acids of VRC01-class antibodies. Bars are not shown where data was unavailable for a particular clade and time point. Nearly all sequences in 4 of 6 heavy chain clades conserve at least 8 of the 10 positions, compared to only 60-70% of unrelated VH1-2-derived sequences (gray bars). Clades H3 (dark green) and H4 (orange) mutate away from the signature over the course of the study.
(B) Sequence logos of the VRC01 class signature positions for each clade. Residues colored in blue do not match the defined signature. Note, 9 of the 10 signature residues are contained in the VH1-2 germline. Thus, although in the non-VRC01-lineage sequences the dominant residue at each position matches the signature, there is variation at almost every position, and any individual read is likely to have more the 2 residues mutated (A). For VRC01 sequences, by contrast, there is strong conservation at any given position (even those which are mutated away from the signature), and relatively few positions have any variation at all.
(C) Longitudinal phylogenetic trees of clade 01+07 sequences for heavy chain (left) and light chain (right). The color of each sequence corresponds to the date at which it is first identified in the NGS data.
(D) Sequence alignment of heavy chain (top) and light chain (bottom) sequences from temporally diverse members of the 01+07 clade showing mutation from the germ line V gene.
(E) Crystal structures of autologous (blue, top left) or heterologous (pale yellow, top middle and top right) gp120s determined in complex with clade 01+07 antibodies (bottom). The complexes have been separated to show the interacting surfaces on each molecule. Contacts made by 45-VRC01.H01+07.O-863513/45-VRC01.L01+07.O-110653 (NGS-derived sequences from 1995, bottom left and bottom middle) are colored in green, while those made by NIH45-46 (isolated from 2008, bottom right) are shown in red. Heavy chain contacts are in the darker shade. These structures show a common binding mode, with a modest increase in buried surface area from the 1995 to the 2008 antibodies when bound to heterologous gp120.
(F) Antibody structures from (C) shown in ribbon format, with residues mutated from the germ line V gene (top) or the 1995 sequence (bottom) shown in stick representation.
We assessed gp120 binding of a VRC01 variant with the germline sequence in the heavy and light chain variable regions, but mature CDR3 regions (since the original unmutated versions of the CDR3 junctions cannot be accurately inferred). Such germline-reverted variants of VRC01-class antibodies have been reported to lack gp120 binding activity (Jardine et al., 2013; McGuire et al., 2013; Zhou et al., 2010). Nonetheless, here we show binding to the early autologous gp120 molecule, d45-01dG5 (GenBank accession number JQ609687) (Wu et al., 2012) (Supplemental Figure S5C) with affinity in the μM range. The affinity of the VRC01 lineage for this gp120 improved to sub-nanomolar levels for the 1995 and 2008 antibodies. These later antibodies were also able to interact with many more autologous and heterologous gp120 strains. Overall, the structures indicate that the 20 changes in amino acid sequence due to SHM between 1995 and 2008 antibodies enhance antibody-antigen interactions. However, antibody recognition over the 13 year time period remains highly similar, with core interactive residues mostly unchanged. Thus, despite extraordinary sequence variation, the VRC01 lineage maintains a largely conserved binding mode.
Rate of VRC01-lineage evolution
Since somatic variants of the VRC01 lineage derived from a common precursor B cell, it is possible to determine evolutionary rates for the maturation of the lineage as well as for each clade. To quantify the rate of lineage evolution, we used BEAST v1.8 (Drummond and Rambaut, 2007) which has been used previously to estimate evolutionary rates for sequences from various population types, including HIV (Alizon and Fraser, 2013; Vrancken et al., 2014). It is important to note that evolutionary rate is not equivalent to the increase in germline divergence, as it describes the overall rate of substitutions, not all of which result in increased germline divergence. For example, the same site can mutate multiple times, including reverting to the original nucleotide.
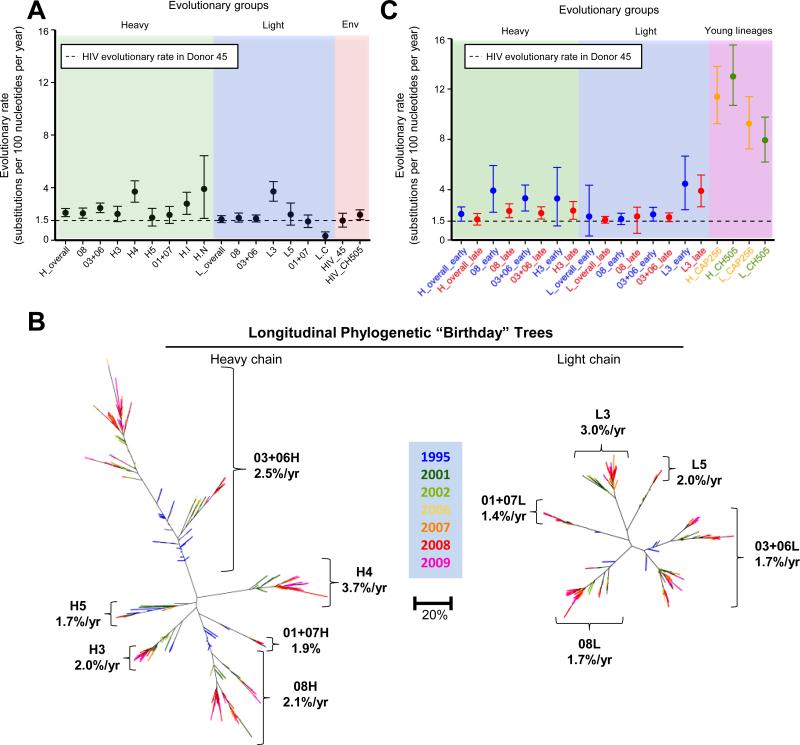
Over the entire time period of the study, VRC01-lineage heavy and light chains had similar evolutionary rates of 2.1 and 1.6 substitutions per 100 nucleotides per year, respectively (Figure 6A, green and blue panels). This rate of mutation was similar to the rate of 1.5 substitutions per 100 nucleotides per year that we determined for the Env gene in this donor (Figure 6A, red panel) based on previously determined viral genome sequences (Wu et al., 2012), and the rate of 1.9 substitutions per 100 nucleotides per year determined for previously reported Env sequences from donor CH505 (Liao et al., 2013). These results are also consistent with previous estimations of Env evolution rates, which have ranged from 0.69 to 1.4 substitutions per 100 nucleotides per year (Alizon and Fraser, 2013; Chaillon et al., 2012; Vrancken et al., 2014). We also calculated evolutionary rates for each clade independently (Figure 6A, green and blue panels). Although clades H4 and L3 showed rates somewhat higher than the other clades, evolutionary rates for all of the VRC01 clades were similar at ~2 substitutions per 100 nucleotides per year.
Figure 6. Rates of evolution and extents of divergence for clades of the VRC01 lineage over 15 years of chronic infection.
(A) Point estimates and 95% highest probability density of evolutionary rates for heavy (green panel) and light chain (blue panel) transcripts of the VRC01 lineage, shown for the lineage overall and individual clades, calculated from subsets of the curated sequence set. Dashed line represents the rate calculated for HIV-1 Env in Donor 45. Evolutionary rates for Env for HIV genomes isolated from both donor 45 and donor CH505 (red panel) are comparable to the evolutionary rate of the VRC01 lineage, calculated from deposited sequences for those data sets.
(B) Heavy (left) and light chain (right) phylogenetic trees for curated sequences from the VRC01 lineage annotated to show point estimates of evolutionary rate for each clade. Trees are shown as “birthday” trees, with each sequence colored to show the time point at which it was first observed. Later observation times (warmer colors) and greater maturation (larger radial distances) were consistent.
(C) Point estimates and 95% confidence intervals of evolutionary rate for VRC01 lineage transcripts calculated separately for the beginning (1995-2002) and end (2006-2009) of the study period (green and blue panels). Sequences used are subsets of those described for panel A. In each case, the earlier rate is faster than the later one, though the differences do not reach statistical significance. For comparison, evolutionary rates of early HIV-1-neutralizing antibody lineages from donors CH505 and CAP256 (purple panel) are ~5-fold higher, as calculated from deposited sequences of those. Additionally estimation of least common ancestors for each lineage are consistently earlier than known or plausible dates (Figures S6), suggesting that evolutionary rates were faster earlier in the history of each lineage.
See also Figure S6.
To provide an overview of the VRC01 lineage and its development, we produced maximum-likelihood phylogenetic trees with curated sequences, annotated with evolutionary rates for the VRC01-lineage clades (Figure 6B). Each sequence is colored based on its apparent “birthday” – the time point from which it was first identified in the NGS data. Importantly, advancing time (indicated by the progression of colors) and advancing maturation (indicated by branch positions) were consistent, in that later transcripts appeared at greater radial distances on the tree.
Evolutionary rate of the VRC01 lineage appears to slow
To compare the evolutionary rates of the VRC01 lineage with other broadly HIV-1-neutralizing antibody lineages, we retrieved CAP256-VRC26 (Doria-Rose et al., 2014) and CH103 (Liao et al., 2013) lineage NGS sequences from GenBank. Average germline divergence for these lineages increased more rapidly than for the VRC01 lineage (Figure S6A). We also found an evolutionary rate of 11 and 9.3 substitutions per 100 nucleotides per year, respectively, for the CAP256-VRC26 lineage heavy and light chains, and 13 and 8 substitutions per 100 nucleotides per year, respectively, for the CH103 lineage (from donor CH505) heavy and light chains (Figure 6C, purple panel). Each of these early rates was ~5-fold higher than that observed for the VRC01 lineage, which was at a much later stage in its development.
Since these rate differences in principle could arise from differences between the donors, we undertook direct rate calculations on donor 45 datasets comprising the beginning (1995-2002) and end (2006-2009) of the study period (Figure 6C, green and blue panels). For aggregate data, including all clades, the evolutionary rate was higher for the early time period than the later one (2.1 versus 1.6 substitutions per 100 nucleotides per year). Although this difference was not statistically significant, it is consistent with the hypothesis that antibody SHM occurs at a faster rate during the early phase of lineage development. Similarly, when data from each clade was considered separately, the calculated rate was in all cases higher for the early time period than the later one, further supporting the idea that evolutionary rates slowed over time.
As a third test for the slowing of evolutionary rates, we used the BEAST package to infer the date of the most recent common ancestor sequence for each lineage. If evolutionary rate were stable over time, we would expect this extrapolation to give a reasonable estimate (Smith et al., 2009). For the VRC01 lineage, the most recent common ancestor was estimated to have occurred for heavy chain in 1971 and for light chain in 1979 (Figure S6B). Although the exact date at which donor 45 was infected is unknown, the inferred dates are implausible, as the AIDS epidemic began in the early 1980s. Similarly, the most recent common ancestor of the CAP256-VRC26 lineage was calculated to have occurred in early 2005 (Figure S6C), although the UCA of that lineage is known to have appeared sometime in March 2006 (Doria-Rose et al., 2014). The same inconsistency is also observed for the CH103 lineage (Liao et al., 2013) (Figure S6D). The consistent prediction of common ancestors at impossibly or implausibly early dates suggests that the evolutionary rate was faster earlier in lineage development. The fact that this was observed for the younger CAP256-VRC26 and CH103 lineages, as well, implies that this slowing begins almost immediately.
Thus, three distinct lines of evidence support the idea that SHM persists over an extended period of time, but evolutionary rate slows as an antibody lineage matures. Overall, clades within the VRC01 lineage had similar evolutionary rates. Rates for other lineages are not similar, and varied over at least a factor of 5 between fast-early and slower-late lineage development. Studies of the early lineages CAP256-VRC26 and CH103 suggested that this slowing begins very early in lineage development.
VRC01-lineage recognition: epitope variation, disulfide patterns, and probed surfaces
The continuous mutation observed over years of chronic infection provides a means to achieve the extraordinary levels of SHM observed with many of the anti-HIV-1 broadly neutralizing antibodies. For the VRC01 lineage, this divergence reaches over 30% for individual lineage members relative to the germline genes, although the variance between individual members of the lineage is considerably greater, with multiple clades evolving in a multiplexed manner (Figure 6B).
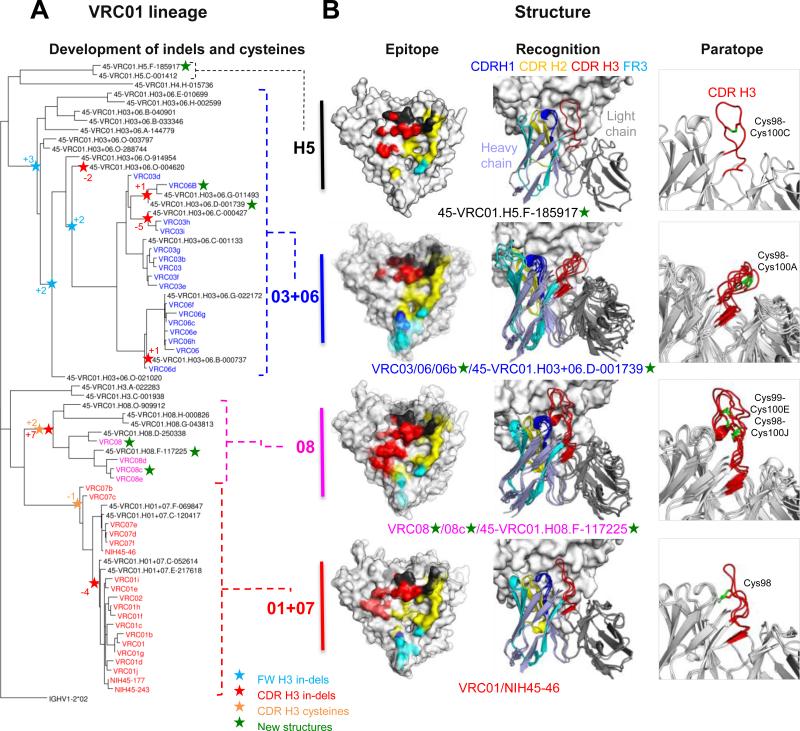
Several of the newly identified VRC01-lineage clades showed remarkable differences. For example, the VRC08 CDR H3 is 11 residues longer than that of VRC01 and contains 3 additional cysteines. To understand the variation of structural recognition of VRC01-lineage antibodies between different clades, we crystallized the antigen-binding fragments (Fabs) of representative antibodies from each clade in complex with HIV-1 gp120. Representative antibodies were chosen based on their expression level (> 5 mg/L), neutralization potency (Figure 3), and phylogenetic placement (Figure 7A). From a total of 10 antibodies, 6 formed crystals with extended core gp120 which diffracted beyond 3.5Å; structures of these were solved by molecular replacement, and structural features of their recognition analyzed.
Figure 7. The VRC01 lineage evolves divergent recognition loops and CDR H3 disulfides.
(A) Heavy chain phylogenetic tree for probe-identified antibodies and validated neutralizing sequences of the VRC01 lineage annotated for the acquisition of molecular features.
(B) Epitope, recognition, and paratope for representative VRC01 clades. Structures of VRC01-lineage antibodies in complex with gp120 are shown with CDR H1 in blue, CDR H2 in yellow, CDR H3 in red, FR3 in cyan, and light chain in dark gray. These are displayed on the gp120 surface for epitope (left column), as colored regions of a ribbon diagram for recognition (middle column), and the CDR H3 and associated cysteines are highlighted in paratope (right column). For clade H5 (top row), the NGS-identified heavy chain 45-VRC01.H5.F-185917 paired with the light chain from VRC01 is shown. NGS-derived heavy chains in clades 03+06 and 08 were paired with VRC03 and VRC08 light chains, respectively.
New structures included two from the 03+06 heavy chain clade (VRC06b and 45-VRC01.H03+06.D-001739; for NGS-defined antibodies we use the nomenclature “donor-lineage.[H/L]clade.timepoint-read_number” for heavy and light chains), three from the 08 heavy chain clade (VRC08, VRC08c, and 45-VRC01.H08.F-117225), and one from the H5 heavy chain clade (45-VRC01.H5.F-185917). We analyzed these structures along with previously determined antibody-gp120 structures from donor 45 comprising those of VRC01, VRC03, NIH45-46 and VRC06 (Diskin et al., 2011; Georgiev et al., 2013; Wu et al., 2011; Zhou et al., 2010). When gp120s were superimposed, modes of gp120 recognition by the variable domains of the donor 45 antibodies resembled each other, with the consensus/shared framework region of heavy chains (rmsds=0.46-2.41Å) (Table S5) showing moderately greater diversity than that of light chain-variable domains (rmsds =0.36-1.65Å) (Table S5). While intra-donor differences between common/consensus framework regions were not significantly different from the inter-donor differences for VRC01-class antibodies from different donors, two antibody heavy chain regions showed substantial clade diversity: the CDR H3 and the heavy chain-framework region 3 (FR3) (Figure 7B). The clade 08 CDR H3 loop (23 amino acids) extends toward the inner domain of gp120 and contributes ~35% and ~70% more binding surfaces on gp120 than CDR H3 loops of 01+07 and 03+06 clades, respectively (Figure 7B and Table S5). Unlike the CDR H3 of clade 08, the CDR H3s of clade 03+06 and clade H5 make substantially less contact with the inner domain of gp120 (~130Å2 for clade 03+06, ~200 Å2 for clade H5, and ~450Å2 for clade 08). Meanwhile, clade 03+06 antibodies are unique among antibodies of the lineage for their extended FR3s, the result of a seven amino acid insertion. These extended FR3s provide ~1.5-, ~2.5-, and ~4.5-fold more binding surface on gp120 V1/V2 stem region than FR3s of clades 01+07, 08, and H5, respectively (Table S5), and induce conformation of the V1V2 region different from that of other antibody-gp120 structures (Figure S7A). Because antibody-Env complexes determined here were in the monomeric gp120 core context, we docked antibodies in the recently determined structure of trimeric BG505 SOSIP (Julien et al., 2013; Lyumkis et al., 2013; Pancera et al., 2014). The extended FR3s were spatially proximal in the trimeric context to V1V2 (residues 203-206) and V3 (residues 314-318) of the neighboring protomer (Figure S7B). Small rearrangements could potentially allow for energetically favorable contact, with positive V3 interacting electrostatically with negatively charged FR3 insertions. In terms of the large CDR H3 alterations, these projected onto the inner domain and did not interact with neighboring protomers. However, the CDR H3 of VRC08c makes potential contacts with helix-α0 of the neighboring gp120 (Figure S7C).
Notably, the disulfide bonding patterns in the CDR H3 loops vary between clades (Figure 7B). 01+07 clades have an interloop disulfide bond between Cys98 (CDR H3) and Cys32 (CDR H1). Clade 03+06 and clade H5 have a single disulfide bond between Cys98 and Cys100A, Cys98 and Cys100C, respectively. Clade 08 has two pairs of intra disulfide bonds (Cys98-Cys100J and Cys99-Cys100E) within the CDR H3 loop, which likely adds structural rigidity to this CDR H3, which is ~10 residues longer than most of the other clades. Although CDR H3 cysteine diversity has been observed before with bovine antibodies (Wang et al., 2013), the bovine mechanism depends on specific D genes optimized for cysteine pairing, whereas, in the current case, cysteine diversity derives from SHM within a single lineage.
To investigate the functional consequences of the clade diversity, we analyzed neutralization fingerprints for a representative selection of VRC01-lineage antibodies. Remarkably, a dendrogram built to represent the neutralization (functional) features mirrors dendrograms based on sequence phylogeny, and in particular replicated the overall clade structure (Supplemental Figure S7D). Thus, the diverse clades do have functional differences that reflect their divergent sequences and the clade organization.
Despite originating from a single ancestor B cell, different clades of the VRC01 lineage have evolved substantial differences in disulfide-bonding patterns and sequence lengths for some of their antigen-binding loops. The CDR H2 – the central contact surface of the antibody lineage with gp120 – does not vary in this manner, nor do other loops of the variable domains. The peripheral role of both the CDR H3 and the FR3 region in gp120 recognition may allow for extraordinary variation as different antibody variants evolve to probe the gp120 surface in distinctive ways.
Discussion
It has been unclear how antibody lineages achieve the unusually high levels of mutation commonly found for broadly HIV-1-neutralizing antibodies. This led us to investigate the evolutionary history of B cell lineages that produce such antibodies. SHM and B cell selection are a form of “accelerated evolution” that generates high-affinity antibodies. In most cases, this process takes a few weeks and involves only a handful of mutations, averaging about 5% nucleotide difference from the originating antibody genes. However, some broadly HIV-1-neutralizing antibodies are characterized by extraordinary levels of SHM where >30% of nucleotides in the antibody variable region differ from the germline-encoded sequence (Burton et al., 2012; Kwong and Mascola, 2012; Mascola and Haynes, 2013). One such antibody, VRC01 (Wu et al., 2010), was isolated approximately two decades after the donor was diagnosed with HIV-1 infection. Here we used antibody isolation, NGS and crystal structures to characterize the VRC01 lineage from 1995-2009. We observed extraordinary diversity, a result of a high rate of SHM over years of chronic infection. Of note, several prior studies indicated germline-reverted variants of VRC01-class antibodies not to bind HIV-1 gp120, raising the question of how they arise (Jardine et al., 2013; McGuire et al., 2013; Zhou et al., 2010); our results indicate germline-reverted VRC01 to bind an early autologous Env (d45-01dG5) (Wu et al., 2012), suggesting the VRC01-lineage to have been initiated by interaction with a specific autologous Env sequence (Supplemental Figure S5C).
The NGS-derived sequences determined here are expected to contain errors arising from cDNA preparation, PCR amplification and 454 sequencing platform. We used the 454 pyrosequencing platform because its read lengths allowed identification of full variable region sequences. Unfortunately, the sequencing reactions were error-prone. We implemented quality control procedures to reduce errors (Extended Experimental Procedures) and to parse the NGS-derived data into three levels of experimental certainty. First, “raw” VRC01-lineage sequences, from all ten time points, comprise 124,834 heavy chain and 28,500 light chain sequences. Based on prior published data, these sequences are expected to have an RMS error in sequence of 1.38% (Zhu et al., 2012). Second, “curated” sequences comprise 1,041 heavy and 492 light chain sequences, and of these, 162 heavy chain and 119 light chain sequences were observed to have biological replicates at the 97.25% level in other time points. Third, “confirmed neutralizers” comprise the closest to consensus or most highly represented sequences, and these are expected to have median accuracy in sequence of 98.9% identity based on the accuracy of VRC01-class plasmids (Extended Experimental Procedures). Additionally, 36% of the confirmed neutralizing sequences had temporal biological replicates at >97.25% identity. Finally, we note that evolutionary rate calculations were performed with curated sequences, and the results were similar over multiple bootstraps, suggesting that these calculated rates were not substantially affected by NGS-sequence errors. Thus the “leaves” of the VRC01-lineage phylogenetic tree and raw data (Figure 2) contain substantial uncertainty, while the overall clade structure (Figure 4, 7) (which is defined by the confirmed neutralizers and probe-identified antibodies and consists of clades that differ on average by 40-50% in sequence) and overall rate of evolution (Figure 6) should not be substantially affected by errors in the NGS data.
Despite the substantial differences between clades of the VRC01 lineage, their average divergences were similar at each time point, and they also showed similar evolutionary rates (Figure 6A). Antibody lineages unrelated to the VRC01 lineage, however, showed substantially different evolutionary rates. Thus, each member of a lineage appeared to share common rate characteristics. Notably, this rate does not appear to be influenced by the differing neutralization of the individual clades on autologous virus (Wu et al., 2012), as might be expected from the co-evolution of virus and neutralizing antibody (Richman et al., 2003; Wei et al., 2003). These finding suggest that the criteria for continued maturation may be less stringent than the criteria for effective neutralization and that the rate of antibody evolution may depend on the number of cell divisions from the single originating common ancestor B cell.
Our data suggests that the evolutionary rate of an antibody lineage slows as it matures. Importantly, even the lowest rates of antibody evolution that we observed were comparable to the evolutionary rate of HIV-1 Env. For the 01+07 clade, heavy chains had already mutated to 20% divergence at the earliest study time point, and we observed continuous SHM to >30% at the end study point (Figure S6A). Likewise, the levels of SHM for other heavy chain and light chain clades of the lineage also increased over time, despite high initial divergence. These data suggest that SHM may continue for as long as antigen persists in long-term chronic infection. The rapid evolutionary rates described here, combined with persistent antigen due to chronic infection over years, provide a mechanism to explain how antibodies can develop the high levels of SHM and lineage diversity required for broad and potent neutralization of HIV-1.
Previously described antibodies from the VRC01 lineage of donor 45 were diverse enough that they appeared to represent several separate lineages (Wu et al., 2010; Wu et al., 2011). Here we show that the diversity of the VRC01 lineage extends far beyond what has generally been thought possible, comprising at least six distinct heavy chain clades and five light chain clades. The curated deep-sequencing data fills in details of this clade structure, without deviating from the outlines provided by functionally validated sequences and antibodies (Figure S7E). Each clade had distinctive sequence and structure characteristics. CDR H3 length within the lineage ranged from 9 to 23 residues, and CDR H3 cysteines ranged from a single cysteine, which formed an interloop disulfide with another CDR, to four cysteines, which formed two intraloop disulfides (Figure 7B). This diversity and its continued evolution present a picture of antibody immunity in which extraordinary variation within just a few antibody lineages – or even a single lineage – may be of critical importance for opposing HIV-1.
EXPERIMENTAL PROCEDURES
Human Specimens
Recovery of PBMCs from donor 45 (Li et al., 2007; Wu et al., 2010) has been described previously. Donor 45 samples from different time points were collected with informed consent under clinical protocols approved by the appropriate institutional review board (IRB).
Isolation of Donor 45 Antibodies
Fluorescence-activated cell sorting of antigen-specific IgG+ B cells from donor 45 PBMC and the amplification and cloning of immunoglobulin genes were carried out using previously described protocols (Wu et al., 2010).
Expression and Purification of Antibodies and Fab Fragments
Expression plasmids for heavy and kappa chains were constructed as described previously (Zhou et al., 2010). The expression and purification of antibody IgGs and preparation of Fab fragments were carried out as described in Extended Experimental Procedures.
Neutralization Assessment
Neutralization donor 45 antibodies were measured using single-round-of-infection HIV-1 Env-pseudoviruses and TZM-bl target cells using protocols described in Extended Experimental Procedures.
Crystallization, X-ray Data Collection, Structure Determination and Refinement of Donor 45 Antibodies in Complex with HIV-1 gp120
Purification, crystallization of antibody-gp120 complexes and data collection are described in Extended Experimental Procedures. All diffraction data were integrated and scaled with the HKL2000 suite (Otwinowski and Minor, 1997). Structure solution, refinement, and analysis are described in Extended Experimental Procedures.
454 Pyrosequencing
454 pyrosequencing libraries from donor 45 were prepared and 454 pyrosequencing of the PCR products were performed with modifications to those described previously (Wu et al., 2011) and in Extended Experimental Procedures.
Bioinformatics Analysis
Bioinformatics analyses of the longitudinal 454 data were performed using algorithms similar to those described previously (Zhu et al., 2013), implemented in a new Python code base. Data quality filtering and other new bioinformatics methods are described in Extended Experimental Procedures.
Supplementary Material
Acknowledgments
We thank E. Turk and C,-L, Lin for technical assistance with large neutralization panels. We thank members of the Structural Biology Section, Structural Bioinformatics Core Section, and Humoral Immunology Core Section, Vaccine Research Center (VRC), NIAID, NIH, for comments and suggestions on the manuscript. We thank J. Baalwa, D. Ellenberger, F. Gao, B. Hahn, K. Hong, J. Kim, F. McCutchan, D. Montefiori, L. Morris, J. Overbaugh, E. Sanders-Buell, G. Shaw, R. Swanstrom, M. Thomson, S. Tovanabutra, C. Williamson, and L. Zhang for contributing the HIV-1 Envelope plasmids used in our neutralization panel. The sequences of NIH45-177 and NIH45-243 were kindly provided by J. Scheid and M. Nussenzweig. We thank Fred Alt for helpful discussions on antibody lineage development and rates. Support for this work was provided by the Intramural Research Program of the VRC and the NHGRI. Use of sector 22 at the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract number W-31-109-Eng-38.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
NISC Comparative Sequencing Program
Betty Benjamin5, Robert Blakesley5, Gerry Bouffard5, Shelise Brooks5, Holly Coleman5, Mila Dekhtyar5, Michael Gregory5, Xiaobin Guan5, Jyoti Gupta5, Joel Han5, April Hargrove5, Shi-ling Ho5, Richelle Legaspi5, Quino Maduro5, Cathy Masiello5, Baishali Maskeri5, Jenny McDowell5, Casandra Montemayor5, Morgan Park5, Nancy Riebow5, Karen Schandler5, Brian Schmidt5, Christina Sison5, Mal Stantripop5, James Thomas5, Pam Thomas5, Meg Vemulapalli5, Alice Young5
5 NISC Comparative Sequencing Program, NIH, Bethesda, Maryland 20892, USA.
Author Contributions
X.W., Z.Z., C.A.S., M.G.J., Y.D.K., T.Z., Z.S., P.D.K., J.R.M., and L.S. designed research, analyzed data, and wrote and edited the paper. X.W. isolated and characterized antibodies, prepared samples for 454 pyrosequencing, and performed functional characterization. Z.Z. and C.A.S. designed and carried out bioinformatics analyses. M.G.J., Y.D.K., and T.Z. determined and analyzed structures of VRC01-lineage antibodies with gp120. Z.S. performed the evolutionary rate analysis. B.Z. produced recombinant antibodies and gp120 core proteins. S.O., K.M., and R.T.B. performed neutralization assays. I.S.G. conducted computational analyses of neutralization fingerprinting and G.-Y.C. conducted statistical analyses of lineage membership. C.S. and N.S.L. contributed additional bioinformatics analyses. R.M.L. tested germline reverted antibody variants. K.O.S. isolated antibodies, S.S. assisted with crystallization, Y.Y. expressed proteins for crystallization, R.T.B. and M.K.L. defined neutralization on 195-isolate panel, J.C.M and NISC performed next-generation sequencing and bioinformatics analysis, and M.C. contributed donor 45 samples.
Accession Numbers Coordinates and structure factors for the eight antibody-HIV-1 gp120 complex structures have been deposited in the Protein Data Bank with accession numbers 4XVS, 4XVT, 4S1Q, 4S1R, 4S1S, 4XNZ, 4XMP, and 4XNY. Raw 454 data has been deposited in the NCBI Short Reads Archive with accession number SRP052625. In addition, 1,041 curated heavy chain sequences, 33 functionally validated NGS-derived heavy chain sequences, 492 curated light chain sequences, 32 functionally validated NGS-derived light chain sequences, and 31 new probe-identified antibodies with both heavy and light chain sequences have been deposited in GenBank.
Consortia
The NISC Comparative Sequencing Program includes Betty Benjamin, Robert Blakesley, Gerry Bouffard, Shelise Brooks, Holly Coleman, Mila Dekhtyar, Michael Gregory, Xiaobin Guan, Jyoti Gupta, Joel Han, April Hargrove, Shi-ling Ho, Richelle Legaspi, Quino Maduro, Cathy Masiello, Baishali Maskeri, Jenny McDowell, Casandra Montemayor, James Mullikin, Morgan Park, Nancy Riebow, Karen Schandler, Brian Schmidt, Christina Sison, Mal Stantripop, James Thomas, Pam Thomas, Meg Vemulapalli, and Alice Young.
Supplementary information is provided.
REFERENCES
- Albert J, Abrahamsson B, Nagy K, Aurelius E, Gaines H, Nystrom G, Fenyo EM. Rapid development of isolate-specific neutralizing antibodies after primary HIV-1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. Aids. 1990;4:107–112. doi: 10.1097/00002030-199002000-00002. [DOI] [PubMed] [Google Scholar]
- Alizon S, Fraser C. Within-host and between-host evolutionary rates across the HIV-1 genome. Retrovirology. 2013;10:49. doi: 10.1186/1742-4690-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Montefiori DC, Wu X, Chen X, Hwang KK, Tsao CY, Kozink DM, Parks RJ, Tomaras GD, Crump JA, et al. Two distinct broadly neutralizing antibody specificities of different clonal lineages in a single HIV-1-infected donor: implications for vaccine design. J Virol. 2012;86:4688–4692. doi: 10.1128/JVI.07163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- Burton DR, Stanfield RL, Wilson IA. Antibody vs. HIV in a clash of evolutionary titans. Proc Natl Acad Sci U S A. 2005;102:14943–14948. doi: 10.1073/pnas.0505126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH, et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- Chaillon A, Braibant M, Hue S, Bencharif S, Enard D, Moreau A, Samri A, Agut H, Barin F. Human immunodeficiency virus type-1 (HIV-1) continues to evolve in presence of broadly neutralizing antibodies more than ten years after infection. PloS one. 2012;7:e44163. doi: 10.1371/journal.pone.0044163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin R, Scheid JF, Marcovecchio PM, West AP, Jr., Klein F, Gao H, Gnanapragasam PN, Abadir A, Seaman MS, Nussenzweig MC, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC evolutionary biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev IS, Doria-Rose NA, Zhou T, Kwon YD, Staupe RP, Moquin S, Chuang GY, Louder MK, Schmidt SD, Altae-Tran HR, et al. Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science. 2013;340:751–756. doi: 10.1126/science.1233989. [DOI] [PubMed] [Google Scholar]
- Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, et al. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J Virol. 2011;85:4828–4840. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, Robinson J, Scearce RM, Plonk K, Staats HF, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS. 2014;28:163–169. doi: 10.1097/QAD.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, Dimitrov DS, Hendrickson WA, Robinson J, Sodroski J, et al. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci U S A. 2004;101:2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce MG, Kanekiyo M, Xu L, Biertumpfel C, Boyington JC, Moquin S, Shi W, Wu X, Yang Y, Yang ZY, et al. Outer Domain of HIV-1 gp120: Antigenic Optimization, Structural Malleability, and Crystal Structure with Antibody VRC-PG04. J Virol. 2013;87:2294–2306. doi: 10.1128/JVI.02717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Migueles SA, Welcher B, Svehla K, Phogat A, Louder MK, Wu X, Shaw GM, Connors M, Wyatt RT, et al. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med. 2007;13:1032–1034. doi: 10.1038/nm1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, O'Dell S, Wilson R, Wu X, Schmidt SD, Hogerkorp CM, Louder MK, Longo NS, Poulsen C, Guenaga J, et al. HIV-1 neutralizing antibodies display dual recognition of the primary and coreceptor binding sites and preferential binding to fully cleaved envelope glycoproteins. Journal of virology. 2012;86:11231–11241. doi: 10.1128/JVI.01543-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature's pathways. Immunological reviews. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, Cohen KW, Jardine J, Menis S, Scheid JF, West AP, et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J Exp Med. 2013;210:655–663. doi: 10.1084/jem.20122824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikell I, Sather DN, Kalams SA, Altfeld M, Alter G, Stamatatos L. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS pathogens. 2011;7:e1001251. doi: 10.1371/journal.ppat.1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pancera M, McLellan JS, Wu X, Zhu J, Changela A, Schmidt SD, Yang Y, Zhou T, Phogat S, Mascola JR, et al. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. Journal of virology. 2010;84:8098–8110. doi: 10.1128/JVI.00966-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R, Walker LM, Stanfield RL, Phogat SK, Koff WC, Poignard P, Burton DR, Wilson IA. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc Natl Acad Sci U S A. 2010;107:11483–11488. doi: 10.1073/pnas.1004600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci U S A. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- Vrancken B, Rambaut A, Suchard MA, Drummond A, Baele G, Derdelinckx I, Van Wijngaerden E, Vandamme AM, Van Laethem K, Lemey P. The genealogical population dynamics of HIV-1 in a large transmission chain: bridging within and among host evolutionary rates. PLoS computational biology. 2014;10:e1003505. doi: 10.1371/journal.pcbi.1003505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS pathogens. 2010;6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Ekiert DC, Ahmad I, Yu W, Zhang Y, Bazirgan O, Torkamani A, Raudsepp T, Mwangi W, Criscitiello MF, et al. Reshaping antibody diversity. Cell. 2013;153:1379–1393. doi: 10.1016/j.cell.2013.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- West AP, Jr., Diskin R, Nussenzweig MC, Bjorkman PJ. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2083–2090. doi: 10.1073/pnas.1208984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Yang ZY, Xu L, Welcher B, Winfrey S, Shao Y, Mascola JR, Nabel GJ. Cross-clade recognition and neutralization by the V3 region from clade C human immunodeficiency virus-1 envelope. Vaccine. 2006;24:4995–5002. doi: 10.1016/j.vaccine.2006.03.083. [DOI] [PubMed] [Google Scholar]
- Wu X, Wang C, O'Dell S, Li Y, Keele BF, Yang Z, Imamichi H, Doria-Rose N, Hoxie JA, Connors M, et al. Selection pressure on HIV-1 envelope by broadly neutralizing antibodies to the conserved CD4-binding site. J Virol. 2012;86:5844–5856. doi: 10.1128/JVI.07139-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Do Kwon Y, Scheid J, Shi W, Xu L, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev IS, Altae-Tran HR, Chuang GY, Joyce MG, et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity. 2013;39:245–258. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, O'Dell S, Ofek G, Pancera M, Wu X, Zhang B, Zhang Z, Program NCS, Mullikin JC, Simek M, et al. Somatic Populations of PGT135-137 HIV-1-Neutralizing Antibodies Identified by 454 Pyrosequencing and Bioinformatics. Frontiers in microbiology. 2012;3:315. doi: 10.3389/fmicb.2012.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Wu X, Zhang B, McKee K, O'Dell S, Soto C, Zhou T, Casazza JP, Program NCS, Mullikin JC, et al. De novo identification of VRC01 class HIV-1-neutralizing antibodies by next-generation sequencing of B-cell transcripts. Proc Natl Acad Sci U S A. 2013;110:E4088–4097. doi: 10.1073/pnas.1306262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.