Abstract
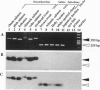
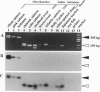
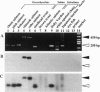
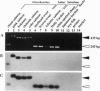
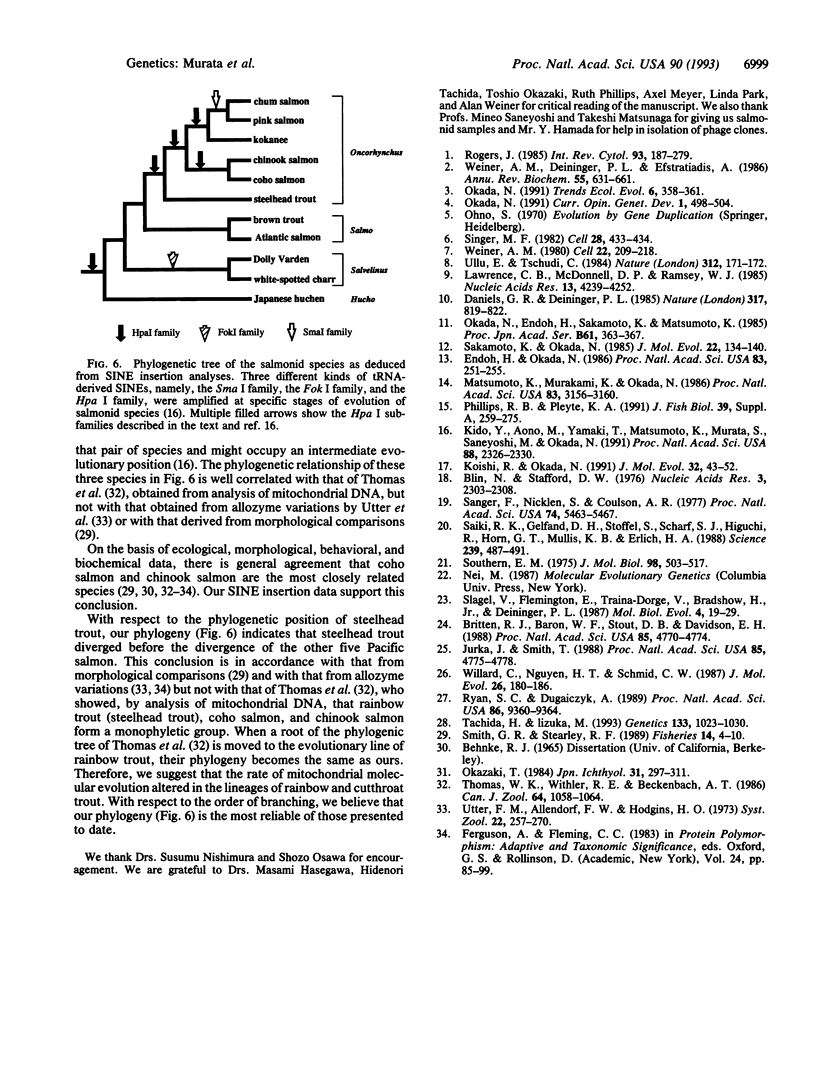
Several subfamilies of the salmonid Hpa I short interspersed element (SINE) family were isolated from salmonid genomes and were sequenced. For each genomic locus that represented the subfamily, amplification by PCR of the orthologous loci in the 12 fish allowed us to determine the order of branching of the Pacific salmonid species. The deduced phylogeny suggests three evolutionary lines, namely, a line of chum salmon, pink salmon, and kokanee; a line of coho salmon and chinook salmon; and a line of steelhead trout. Our data also support a change in the phylogenetic assignment of steelhead trout from Salmo to Oncorhynchus. We present here an extensive phylogenetic tree constructed from an analysis of differential insertion of SINEs, and we propose that SINE insertion analysis is one of the best available methods for clarifying the order of divergence of closely related species.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten R. J., Baron W. F., Stout D. B., Davidson E. H. Sources and evolution of human Alu repeated sequences. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4770–4774. doi: 10.1073/pnas.85.13.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels G. R., Deininger P. L. Repeat sequence families derived from mammalian tRNA genes. 1985 Oct 31-Nov 6Nature. 317(6040):819–822. doi: 10.1038/317819a0. [DOI] [PubMed] [Google Scholar]
- Endoh H., Okada N. Total DNA transcription in vitro: a procedure to detect highly repetitive and transcribable sequences with tRNA-like structures. Proc Natl Acad Sci U S A. 1986 Jan;83(2):251–255. doi: 10.1073/pnas.83.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J., Smith T. A fundamental division in the Alu family of repeated sequences. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4775–4778. doi: 10.1073/pnas.85.13.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kido Y., Aono M., Yamaki T., Matsumoto K., Murata S., Saneyoshi M., Okada N. Shaping and reshaping of salmonid genomes by amplification of tRNA-derived retroposons during evolution. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2326–2330. doi: 10.1073/pnas.88.6.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koishi R., Okada N. Distribution of the salmonid Hpa 1 family in the salmonid species demonstrated by in vitro runoff transcription assay of total genomic DNA: a procedure to estimate repetitive frequency and sequence divergence of a certain repetitive family with a few known sequences. J Mol Evol. 1991 Jan;32(1):43–52. doi: 10.1007/BF02099928. [DOI] [PubMed] [Google Scholar]
- Lawrence C. B., McDonnell D. P., Ramsey W. J. Analysis of repetitive sequence elements containing tRNA-like sequences. Nucleic Acids Res. 1985 Jun 25;13(12):4239–4252. doi: 10.1093/nar/13.12.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Murakami K., Okada N. Gene for lysine tRNA1 may be a progenitor of the highly repetitive and transcribable sequences present in the salmon genome. Proc Natl Acad Sci U S A. 1986 May;83(10):3156–3160. doi: 10.1073/pnas.83.10.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada N. SINEs. Curr Opin Genet Dev. 1991 Dec;1(4):498–504. doi: 10.1016/s0959-437x(05)80198-4. [DOI] [PubMed] [Google Scholar]
- Rogers J. H. The origin and evolution of retroposons. Int Rev Cytol. 1985;93:187–279. doi: 10.1016/s0074-7696(08)61375-3. [DOI] [PubMed] [Google Scholar]
- Ryan S. C., Dugaiczyk A. Newly arisen DNA repeats in primate phylogeny. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9360–9364. doi: 10.1073/pnas.86.23.9360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sakamoto K., Okada N. Rodent type 2 Alu family, rat identifier sequence, rabbit C family, and bovine or goat 73-bp repeat may have evolved from tRNA genes. J Mol Evol. 1985;22(2):134–140. doi: 10.1007/BF02101691. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M. F. SINEs and LINEs: highly repeated short and long interspersed sequences in mammalian genomes. Cell. 1982 Mar;28(3):433–434. doi: 10.1016/0092-8674(82)90194-5. [DOI] [PubMed] [Google Scholar]
- Slagel V., Flemington E., Traina-Dorge V., Bradshaw H., Deininger P. Clustering and subfamily relationships of the Alu family in the human genome. Mol Biol Evol. 1987 Jan;4(1):19–29. doi: 10.1093/oxfordjournals.molbev.a040422. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tachida H., Iizuka M. A population genetic study of the evolution of SINEs. I. Polymorphism with regard to the presence or absence of an element. Genetics. 1993 Apr;133(4):1023–1030. doi: 10.1093/genetics/133.4.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984 Nov 8;312(5990):171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Weiner A. M. An abundant cytoplasmic 7S RNA is complementary to the dominant interspersed middle repetitive DNA sequence family in the human genome. Cell. 1980 Nov;22(1 Pt 1):209–218. doi: 10.1016/0092-8674(80)90169-5. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Willard C., Nguyen H. T., Schmid C. W. Existence of at least three distinct Alu subfamilies. J Mol Evol. 1987;26(3):180–186. doi: 10.1007/BF02099850. [DOI] [PubMed] [Google Scholar]