Abstract
DNA methylation is one of the major epigenetic modifications and has been involved in a number of biological processes in mammalian cells. Yeast is widely used as a model organism for studying cell metabolism, cell cycle regulation, and signal transduction. However, it remains controversial whether methylated cytosine (5-methylcytosine, 5mC) exists in the yeast genome. In the current study, we developed a highly sensitive method based on gas chromatography/mass spectrometry (GC/MS) and systematically examined the incidence of 5mC in 19 yeast strains, which represent 16 yeast species. Our results showed that DNA methylation is widespread in yeast and the genome-wide DNA methylation of the studied yeast strains ranged from 0.014 to 0.364%, which were 1 to 2 orders of magnitude lower than that in mammalian cells (i.e., 3–8%). Furthermore, we found that the 5mC content in yeast varied considerably at different growth stages and DNA methylation inhibitor 5-azacytidine could induce a decrease in genome-wide DNA methylation as that in mammalian cells. The demonstration of the universal presence of DNA cytosine methylation in yeast constituted the first and essential step toward understanding the functions of this methylation in yeast.
Methylation at the 5-position of cytosine (C) residues in CpG dinucleotide is one of the major epigenetic modifications that plays crucial roles in embryogenesis, gene regulation, and genomic imprinting.1 In mammalian cells, 5-methylcytosine (5mC) is the only methylated base found in DNA, and many studies show a correlation between hypomethylation and gene reactivation while DNA hypermethylation represses transcription.2 Properly established and maintained DNA methylation patterns are vital for mammalian development and for the normal functions of the adult organism.3 Consistent with the important roles of DNA methylation, aberrant DNA methylation links to dysregulation of gene expression and leads to a growing number of human diseases.4
For many human diseases, including cancer, it remains challenging to develop efficacious treatment strategies due to the complexity of the regulatory networks that underlie the onset and progression of the diseases.5 In this respect, model organisms, which are easy to manipulate and allow detailed molecular studies, have been frequently employed for studying cell metabolism and signal transduction. Since a number of key regulatory pathways are highly conserved between yeast and humans, yeast has been extensively studied as a model organism for decades in the biological field due to the ease of genetic manipulation, the small genome, and the numerous collections of well-characterized mutants.6 Therefore, a better understanding of DNA methylation regulation in mammalian systems may be achieved by studying its role in genetically tractable yeasts.
Up to date, however, the existence of 5mC in yeast species, such as Schizosaccharomyces pombe and Saccharomyces cerevisiae, is still controversial. DNA methylation is normally considered to be absent in yeast, and no DNA methyltransferase was found in yeast.7–10 Nevertheless, a gene (pmt1+) encoding a protein sharing significant sequence identity with both prokaryotic and eukaryotic DNA (cytosine-5)-methyltransferase was found in Schizosaccharomyces pombe,9 which may indicate the existence of methylated cytosine in this species. Interestingly, a DNA methylation level of 0.3–1% in Saccharomyces cerevisiae was described more than 30 years ago,11 but this result remained in doubt because later studies were unable to reproduce the data.7,8 The inconsistent results on the existence and content of 5mC in yeast hamper DNA methylation studies in yeast.
5mC content in eukaryotic genomes varies considerably. In mammalian cells, the percentages of methylated cytosine are ~3–8% of total cytosine, whereas in plants the proportion of methylated cytosine can be as high as 30%.12 In the previous reports which showed no DNA methylation in yeast, high-performance liquid chromatography (HPLC) or restriction enzyme isoschizomer analyses were employed for the detection of 5mC.7,8 The most sensitive method used to analyze the 5mC content of DNA required that at least 0.075% of the total cytosine is methylated.13 Therefore, it is possible that the low level of 5mC in yeast genomes prevents its detection. However, the 0.3–1% DNA methylation level reported by Hattman and co-workers11 was much higher than the detection limit of the HPLC or restriction enzyme digestion methods.7,8 This suggested that, in theory, 5mC should be detectable with these methods if yeast genome indeed contains 0.3–1% 5-methylcytosine. The contradictory results on whether DNA methylation occurs in yeast reported by different groups warrants further investigation of this subject.
In this study, we developed a highly sensitive and reliable method based on gas chromatography/mass spectrometry (GC/MS) for the systematic study on the incidence of 5mC in yeast genomes. The genome-wide DNA methylation levels in 19 yeast strains representing 16 yeast species were examined. Our results demonstrated that 5mC was present in all the examined yeast species, with the levels ranging from 0.014 to 0.364% [5mC/(5mC + cytosine)]. Furthermore, we found that the 5mC content in yeast varied considerably between the exponential growth stage and the stationary stage and DNA methylation inhibitor of 5-azacytidine could induce a significant decrease of genome-wide DNA methylation level.
EXPERIMENTAL SECTION
Chemicals and Reagents
Five standard nucleobases (purity >99%), cytosine (C), adenine (A), thymine (T), guanine (G), and uracil (U), were purchased from Aladdin Reagent Co., Ltd. (Shanghai, China). 5-Methylcytosine (5mC, purity >99%) was purchased from Hanhong Chemical Co., Ltd. (Shanghai, China).
Chromatographic grade acetonitrile was purchased from Honeywell Co., Ltd. (Shanghai, China). All other solvents and chemicals used were of analytical grade. Glucose, formic acid, aqueous ammonia (28%, by weight), sodium acetate, NaCl, ethylenediaminetetraacetic acid (EDTA), sodium dodecyl sulfate (SDS), and chloroform were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Triton X-100 was purchased from Aladdin Reagent Co., Ltd. Tris buffer-saturated phenol, Tris–HCl, and glass beads (425–600 μm) were purchased from Wenhan Technology Co., Ltd. (Wuhan, China). Yeast extract, peptone, and malt extract were purchased from Becton Dickinson Medical Devices Co., Ltd. (Shanghai, China). N,O-bis-(Trimethylsilyl)-trifluoroacetamide (BSTFA), cholorotrimethylsilane (TMCS), and 5-azacytidine were purchased from Sigma-Aldrich China Inc. (Beijing, China). The water used throughout the study was purified by a Milli-Q apparatus (Millipore, Bedford, MA, USA).
All the nucleobase stock solutions were prepared in 1.2% aqueous ammonia at a concentration of 400 μg/mL. Standard solutions for the construction of calibration curve were prepared with 5mC at 5 ng/mL and various concentrations of cytosine ranging from 0.5 to 50 μg/mL. The quality control samples (QCs) with the molar ratios of 5mC/(5mC + cytosine) ranging from 0.05% to 0.50% were prepared to validate the method by mixing a 27-mer ODN (5′-GAGTCGCACGCCAGGG XGCCGAATTC-3′, X indicates the 5mC) and a 17-mer ODN (5′-CCATGGGCGCCGAATTC-3′) at different amounts.
Yeast Culture and Drug Treatment
The Saccharomyces cerevisiae strains used in this study included the BY4741 (MATa his3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0) and YEF473A (MATa his3 leu2 lys2 trp1 ura3)14 in S288C background and the W1588-4C (MATa leu2-3,112 ura3-1 his3-11,15 trp1-1 ade2-1 can1-100)15 in W303 background. The ten yeast strains Debaryomyces hansenii (CGMCC 2.494), Debaryomyces polymorphus (CGMCC 2.1462), Cryptococcus laurentii (CGMCC 2.1491), Pachysolen tannophilus (CGMCC 2.1622), Schizosaccharomyces japonicus (CGMCC 2.1635), Schizosaccharomyces octosporus (CGMCC 2.1636), Yarrowia lipolytica (CGMCC 2.1718), Candida rugopelliculosa (CGMCC 2.1864), Pichia anomala (CGMCC 2.1886), and Candida magnolia (CGMCC 2.1919) were purchased from the China General Microbiological Culture Collection Center (CGMCC). The six yeast strains Kluyveromyces lactis (CICC 1773), Kluyveromyces marxianus (CICC 1953), Pichia pastoris (CICC 1958), Saccharomyces boulardii (CICC 1794), Schizosaccharomyces pombe (CICC 1056), and Schizosaccharomyces pombe (CICC 1372) were purchased from the China Center of Industrial Culture Collection (CICC).
All the yeast strains were cultured using similar procedures. Briefly, the yeast strains were first inoculated onto the solid medium and grew for 24–48 h at 25 °C. A colony from the plate was inoculated into 5 mL of liquid medium and grew at 25 °C on a rotary shaker (250 rpm) for 12 h. Two mL of the culture was subsequently added into 400 mL of medium, and the cells were cultured for another 12 h. The strain of K. latics (CICC 1773) was cultured in the YM media (yeast extract, 3 g/L; malt extract, 3 g/L; peptone, 5 g/L; glucose, 10 g/L). The strains of K. marxianus (CICC 1953), P. pastoris (CICC 1958), S. boulardii (CICC 1794), S. pombe (CICC 1056), and S. pombe (CICC 1372) were cultured in the 5° Brix malt extract medium (malt extract, 20 g/L; peptone, 1 g/L; glucose, 20 g/L). Three S. cerevisiae strains (BY4741, W1588-4C, and YEF473A) were cultured in the YPD medium (yeast extract, 10 g/L; peptone, 20 g/L; glucose, 20 g/L). The rest of the yeast strains were cultured in the 12° Brix malt extract medium (malt extract, 20 g/L; peptone, 1 g/L; glucose, 50 g/L). As for the drug treatment experiments, the cells of Y. lipolytica (CGMCC 2.1718) and S. cerevisiae (W1588-4C) were treated with 10 or 50 μM 5-azacytidine (DNA methylation inhibitor) at 25 °C for 16 h.
DNA Extraction
Yeast cell suspensions were centrifuged at 4600g for 5 min, and the cell pellet was collected. The pellet was washed with water to remove the residual medium. Yeast genomic DNA was extracted according to a previously described method with some modification.16 Briefly, yeast cells were suspended in 1 mL of cell lysis buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA, 100 mM NaCl, 1% SDS, and 2% Triton X-100) plus an equal volume of phenol/chloroform (1:1, v/v, saturated with 10 mM Tris, pH 8.0, 1 mM EDTA) and 1.5 g of glass beads (425–600 μm) and rigorously vortexed for 10 min. To the solution was added 1 mL of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). The resulting solution was centrifuged at 13 500g for 5 min, and the aqueous layer was transferred into a clean tube. An equal volume solvent mixture of phenol/chloroform was added; the sample solution was vortexed and centrifuged at 13 500g for 5 min. The aqueous layer was subsequently transferred into a clean tube, and genomic DNA was precipitated by isopropanol. RNA was removed from the extracted DNA samples by digestion with RNase A (5 μL, 10 μg/μL) and RNase T1 (0.5 μL, 1000 unit/μL). DNA purity and concentration were determined using a spectrophotometer (Shimadzu, Japan).
Derivatization
Prior to the real samples analysis, the derivatization conditions, including temperature and time, were optimized to obtain the best detection sensitivity. According to the previously described DNA hydrolysis method,17 5 μg of genomic DNA was dissolved in 500 μL of 88% formic acid and then hydrolyzed at 140 °C for 90 min in a glass vial. After cooling, the hydrolyzate was dried at 35 °C under vacuum. The derivatization was performed by adding 25 μL of BSTFA (with 1% TMCS) plus 25 μL of acetonitrile under optimized conditions (i.e., derivatized at 30 °C for 30 min). The derivatization mixture (2 μL) was directly injected into GC/MS for analysis.
Analysis of the Genome-Wide DNA Methylation in Yeast by GC/MS
The GC/MS analysis was performed on a Shimadzu GCMS-QP2010 Plus which was equipped with an AOC-20i+s autosampler (Kyoto, Japan). The GC separation was achieved on an Rxi-5 ms column (30 m × 0.25 mm ×0.25 μm) purchased from Restek (Bellefonte, USA). The oven temperature was held at 120 °C for 3 min and increased to 150 °C at a rate of 8 °C/min, then to 240 °C at a rate of 8 °C/min, and finally to 300 °C at a rate of 30 °C/min. The sample was injected in splitless mode. Helium (purity ≥ 99.999%) was used as the carrier gas at a flow rate of 1 mL/min. The injection port, transfer line, and ion source temperature were maintained at 250, 280, and 250 °C, respectively. The MS was operated in electron impact (EI) mode with an electron energy of 70 eV, and the spectra were scanned within the range of m/z 50–400. For quantitative analysis, selective ion monitoring (SIM) mode was adopted. The characteristic ions used for the quantification of cytosine and 5mC were described in Table 1.
Table 1.
GC/MS Parameters for the Nucleobases Analysis
| analytes | retention time (min) | qualitative ion 1 | qualitative ion 2 | qualitative ion 3 | quantitative ion |
|---|---|---|---|---|---|
| uracil | 5.87 | 256 | 255 | 241 | 241 |
| thymine | 6.90 | 270 | 255 | 255 | |
| cytosine | 9.43 | 255 | 254 | 240 | 240 |
| 5mC | 10.28 | 269 | 255 | 254 | 269 |
| adenine | 16.70 | 279 | 264 | 264 | |
| guanine | 20.34 | 367 | 352 | 352 |
Calculation of the Percentage of DNA Methylation
The genome-wide DNA methylation was calculated using the following formula:
where A(5mC) is the molar quantity of 5mC and A(cytosine) is the molar quantity of cytosine determined in the DNA sample.
Statistical Analysis
The statistical data were processed with SPSS 16.0 software (Chicago, USA). The one-way ANOVA was performed to evaluate the differences in genomic DNA methylation. The Z-scores method was used to normalize variances prior to one-way ANOVA, and a post hoc analysis was performed using Tukey’s test. The differences were considered to be significant when p < 0.05.
RESULTS AND DISCUSSION
GC/MS Procedure
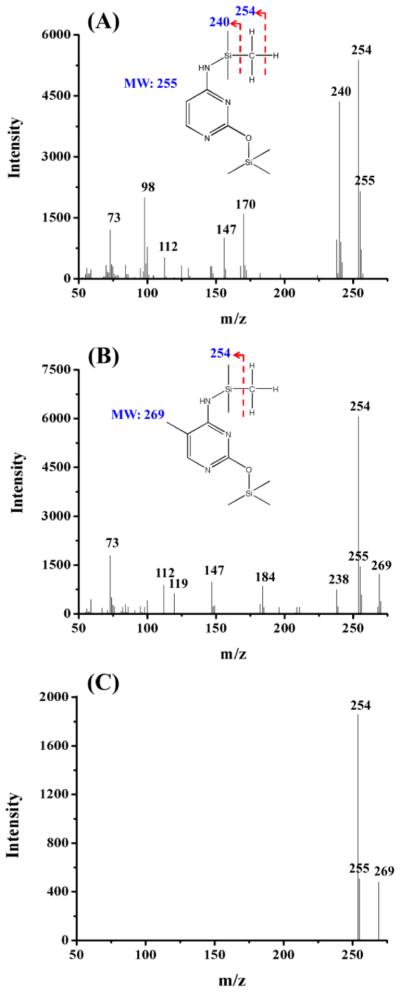
As shown in Figure 1A,B, cytosine and 5mC standards were successfully derivatized with BSTFA/TMCS. The molecular weights for the derivatized cytosine and 5mC were 255 and 269 Da, respectively (Figure 1A,B). The TMS derivative of cytosine yielded the major peaks at m/z 240, 254, and 255 after fragmentation, while the TMS derivative of 5mC produced the major peaks at m/z 254, 255, and 269. In addition, the TMS derivative of cytosine lost a hydrogen radical, whereas the corresponding fragmentation for the TMS derivative of 5mC was not observed (Figure 1A,B). Due to the presence of the fragment ions of m/z 254 and 255 in the mass spectra of both cytosine and 5mC, the fragment ions of m/z 240 and 269 were therefore chosen for specifically monitoring cytosine and 5mC, respectively. A previous study also suggested that the fragment ion at m/z 240 should be chosen for the quantitative cytosine analysis rather than the fragment ion at m/z 254 or 255 so that the interference by fragmentation products of 5mC can be avoided.18 A typical fragment ion profile of 5mC from Y. lipolytica (CGMCC 2.1718) obtained under selective ion monitoring (SIM) mode was displayed in Figure 1C. The GC/MS chromatogram of six standard nucleobases (U, T, C, 5mC, A, and G) showed that all six nucleobases can be well separated under optimized conditions (Figure 2A).
Figure 1.
Electron impact (EI) full-scan mass spectra of TMS derivatives of cytosine (A) and 5mC (B). (C) Selective ion monitoring (SIM) mass spectrum of 5mC from genomic DNA of yeast strain of Y. lipolytica (CGMCC 2.1718). The fragment ions of m/z 269, 255, and 254 were used for monitoring 5mC.
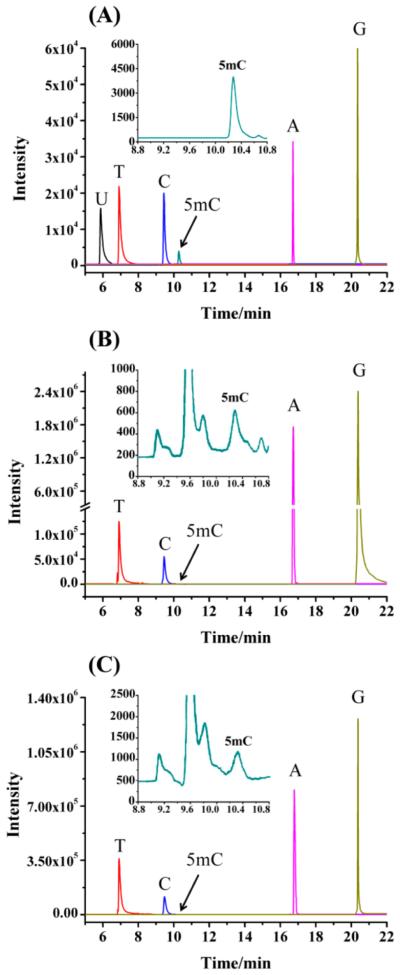
Figure 2.
Selective ion monitoring (SIM) chromatograms of nucleobases. (A) Chromatogram of 6 standard nucleobases. (B) Representative chromatogram for nucleobase mixture from genomic DNA of Y. lipolytica (CGMCC 2.1718). (C) Representative chromatogram for nucleobase mixture from genomic DNA of S. cerevisiae W1588-4C. U, uracil; T, thymine; C, cytosine; 5mC, 5-methylcytosine; A, adenine; G, guanine.
Cytosine and 5mC may undergo deamination to produce U or T under certain conditions,19 which affects the accuracy of the measurement. To evaluate the possibility of the deamination of cytosine and 5mC, we mimicked the whole process of acid hydrolysis and derivatization using the standard nucleobases. As shown in Figure 3, cytosine and 5mC did not artificially produce U or T by deamination under the aforementioned conditions of acid hydrolysis and derivatization, which was consistent with a previous report20 and suggested that the sample processing conditions were appropriate for the analysis of nucleobases from yeast DNA samples.
Figure 3.
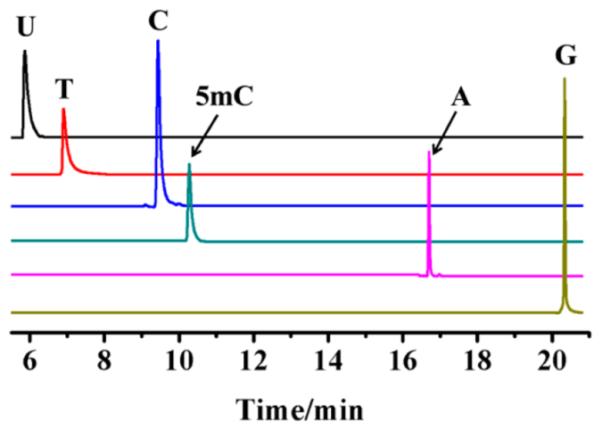
Selective ion monitoring (SIM) chromatogram of the TMS derivatives of individual nucleobase standards after acid hydrolysis. U, uracil; T, thymine; C, cytosine; 5mC, 5-methylcytosine; A, adenine; G, guanine.
Optimization of Derivatization Conditions
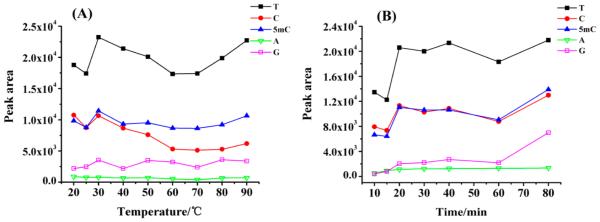
We also optimized the derivatization temperature and time to achieve the best sensitivity for detecting 5mC. The derivatization temperature ranging from 20 to 90 °C and derivatization time ranging from 20 to 80 min were investigated successively. In general, temperature above 30 °C did not provide additional benefit for the 5mC derivatization (Figure 4A). Therefore, 30 °C was chosen as the derivatization temperature for the following experiments. As for the derivatization time, a slight increase of the signal intensity of 5mC was observed from 10 to 20 min and no significant further increase was observed until 80 min (Figure 4B). To ensure sufficient derivatization, 30 min was chosen for the subsequent experiments. Therefore, the optimized derivatization conditions for 5mC detection were at 30 °C for 30 min.
Figure 4.
(A) Optimization of derivatization temperature. (B) Optimization of derivatization time.
As noted by Razin and Sedat,21 the derivatization of purines (A and G) is not efficient in pyridine solution at room temperature. Similarly, due to lack of derivatization in pyridine, guanine was also not detected even using an exceptionally strong silylation reagent, tert-butyldimethylchlorosilane, at 40 °C.17 Moreover, the presence of pyridine in the derivatization solution could cause a rapid decrease of MS performance due to the soiling of the system.17 To circumvent this, we chose acetonitrile as the derivatization medium. The results showed that nucleobases A and G can readily react with BSTFA to produce their TMS derivatives in acetonitrile (Figure 2A). We reason that the rapid keto–enol tautomerization of purines may occur in pyridine rather than in acetonitrile because pyridine is a weak base, and the TMS derivatives of A and G in pyridine are relatively unstable. Although the derivatization efficiency of 5mC in acetonitrile was approximately 40% of that in pyridine (data not shown), with the use of acetonitrile, one can avoid the soiling of the MS system caused by pyridine and therefore increase the reproducibility of the method. Thus, acetonitrile was selected as the derivatization medium in this study.
Method Validation
Under the optimized derivatization conditions, a series of experiments were performed to validate the method. As shown in Table S1, Supporting Information, the detection linearity of the method was investigated using 2 μg of cytosine standard spiked with 5mC at different amounts ranging from 0.2 to 20 ng. The calibration curve was constructed by plotting the mean peak area ratio of 5mC/(5mC + cytosine) versus the mean molar ratio of 5mC/(5mC + cytosine) based on data obtained from triplicate measurements. The results showed that good linearity within the range of 0.01–1% (molar ratio of 5mC/(5mC + cytosine)) was obtained with a coefficient value (R2) higher than 0.99 (Table S1, Supporting Information).
The limit of detection (LOD) and the limit of quantification (LOQ) were calculated as the amounts of the analytes at a signal-to-noise ratio (S/N) of 3 and 10, respectively. Our results showed that the LOD and LOQ values for 5mC were 0.8 pg (6.4 fmol) and 2.5 pg (20 fmol), respectively, which was much lower compared to previous GC/MS analysis for 5mC (LOQ, 56 fmol).17 To the best of our knowledge, the LOQ value obtained in the current study was the lowest in the GC/MS analysis for 5mC. Therefore, 100 ng of genomic DNA with even as low as 0.01% 5mC can be easily detected with our method.
The accuracy of the method was assessed using the synthesized 5mC-containing oligodeoxynucleotide by comparing the measured 5mC content to the theoretical 5mC content in 5mC-containing oligodeoxynucleotide. Three different molar ratios of 5mC/cytosine ranging from 0.05% to 0.5% were measured (Table 2). Our results showed that good reproducibility and accuracy could be achieved, which are manifested by relative standard deviations (RSDs) being less than 7.2% and relative errors being less than 14%, respectively.
Table 2.
Accuracy and Precision of the GC/MS Method for the Determination of Cytosine Methylation
| theoretical value (%) |
measured value (%) |
RSD (%) (n = 3) |
relative error (%) |
|
|---|---|---|---|---|
| 5mC/cytosine molar ratioa |
0.050 | 0.043 | 7.2 | −14.0 |
| 0.100 | 0.089 | 5.8 | −11.0 | |
| 0.500 | 0.442 | 5.5 | −11.6 |
The quality control samples (QCs) were prepared at three different molar ratios of 5mC/cytosine.
Quantitative Determination of the Genome-Wide DNA Methylation in 16 Yeast Species
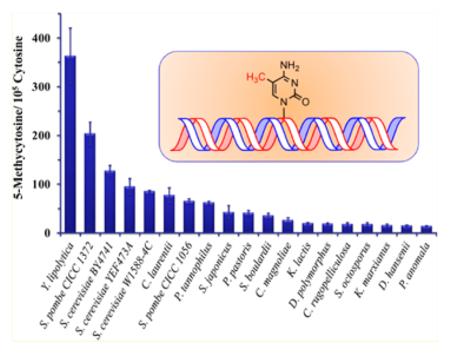
A total of 19 yeast strains from 16 different yeast species were chosen to investigate the genome-wide DNA methylation. The yeast cells were cultured at 30 °C for 12 h, and then, genomic DNA was extracted for the subsequent analysis. Figure 2B,C showed the typical chromatograms for Y. lipolytica (CGMCC 2.1718) and S. cerevisiae W1588–4C, respectively, and good separation of the aforementioned nucleobases was achieved in these two real samples as that in a standard nucleobase mixture (Figure 2A). The absence of the uracil peak in the chromatogram ruled out the presence of RNA contamination or cytosine deamination (Figure 2B,C). It turned out that 5mC was present in all the examined yeast strains with the contents ranging from 0.014 to 0.364% [5mC/(5mC + cytosine), Table 3]. In addition to the strains of S. cerevisiae and S. pombe, our study is the first one to quantify the genome-wide 5mC content in the other 14 yeast species. Y. lipolytica was widely used as the model organism for the studies of protein secretion as well as in industrial applications such as citric acid and peach flavor production.22 As shown in Table 3, the highest value of genome-wide DNA methylation was found in Y. lipolytica (CGMCC 2.1718), which may assist future studies toward understanding gene regulation in this species. Proffitt et al. failed to observe 5mC in genomic DNA of S. cerevisiae.8 However, in our work, the DNA methylation levels detected in three S. cerevisiae strains (BY4741, YEF473A, and W1588-4C) were 0.128%, 0.095%, and 0.085%, respectively (Table 3). In addition, the methylation levels in two S. pombe strains (CICC 1056 and CICC 1372) were found to be 0.065% and 0.205%, respectively. Therefore, the genome-wide DNA methylation levels varied considerably among different yeast strains and species. These findings suggest that cytosine methylation is widespread in yeast, but the overall DNA methylation levels in yeast are about 10- to 100-fold less than that observed in mammalian cells [3–8% of 5mC/(5mC + cytosine)].
Table 3.
Genome-Wide DNA Methylation Level in 19 Yeast Strains
| yeast strainsa | methylation %b
5mC/(5mC + cytosine) |
|---|---|
| Candida magnoliae (CGMCC 2.1919) | 0.026 ± 0.005 |
| Candida rugopelliculosa (CGMCC 2.1864) | 0.018 ± 0.003 |
| Cryptococcus laurentii (CGMCC 2.1491) | 0.079 ± 0.015 |
| Debaryomyces hansenii (CGMCC 2.494) | 0.015 ± 0.001 |
|
Debaryomyces polymorphus (CGMCC 2.1462) |
0.019 ± 0.002 |
| Kluyveromyces lactis (CICC 1773) | 0.019 ± 0.002 |
| Kluyveromyces marxianus (CICC 1953) | 0.015 ± 0.003 |
| Pachysolen tannophilus (CGMCC 2.1622) | 0.062 ± 0.003 |
| Pichia anomala (CGMCC 2.1886) | 0.014 ± 0.001 |
| Pichia pastoris (CICC 1958) | 0.041 ± 0.005 |
| Saccharomyces boulardii (CICC 1794) | 0.036 ± 0.005 |
| Saccharomyces cerevisiae BY4741 | 0.128 ± 0.011 |
| Saccharomyces cerevisiae W1588–4C | 0.085 ± 0.002 |
| Saccharomyces cerevisiae YEF473A | 0.095 ± 0.016 |
|
Schizosaccharomyces japonicus (CGMCC 2.1635) |
0.043 ± 0.014 |
|
Schizosaccharomyces octosporus (CGMCC 2.1636) |
0.018 ± 0.003 |
| Schizosaccharomyces pombe (CICC 1056) | 0.065 ± 0.005 |
| Schizosaccharomyces pombe (CICC 1372) | 0.205 ± 0.023 |
| Yarrowia lipolytica (CGMCC 2.1718) | 0.364 ± 0.057 |
All yeast cells were harvested from a 12 h culture.
The data represent the means and standard deviations of results from three independent measurements.
Genome-Wide DNA Methylation of Yeast at Different Growth Stages
We further evaluated the genome-wide DNA methylation of yeast at different growth stages. The exponential growth stage of yeast occurred between 8 and 16 h in both Y. lipolytica (CGMCC 2.1718) and S. cerevisiae W1588-4C, and the growth rate slowed down significantly and then entered into the stationary growth stage after 16 h (Figure S1, Supporting Information). Shown in Figure 5 was the dynamic change of genome-wide DNA methylation in the aforementioned two yeast strains from the exponential growth stage (12 h) to the stationary growth stage (20 h). The results demonstrated that the cytosine methylation decreased significantly from 0.360% to 0.163% in Y. lipolytica (CGMCC 2.1718) and from 0.086% to 0.040% in S. cerevisiae (W1588-4C) from 12 to 16 h, respectively. In addition, the values of cytosine methylation in the yeast strains of Y. lipolytica (CGMCC 2.1718) and S. cerevisiae W1588-4C did not change too much in the stationary growth stage (from 16 to 20 h, Figure 5). The approximately 60% decrease of genome-wide DNA methylation from exponential growth stage to the stationary stage (p < 0.05) suggested that 5mC may be involved in yeast cell development.
Figure 5.
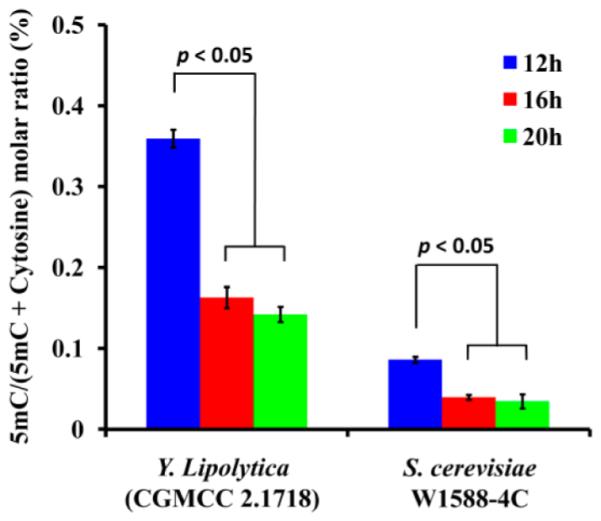
Genome-wide DNA methylation levels in Y. lipolytica (CGMCC 2.1718) and S. cerevisiae W1588-4C at different growth stages. The DNA methylation level was shown as average molar ratio of 5mC/(5mC + cytosine) ± SD (n = 3); p values were derived from Tukey’s test.
Effect of 5-Azacytidine Treatment on the Genome-Wide DNA Methylation
To investigate whether the cytosine methylation can be inhibited in yeast as that in mammalian cells, we treated yeast cells with 5-azacytidine, a cytosine methylation inhibitor.23 It turned out that culturing of Y. lipolytica (CGMCC 2.1718) cells in a medium containing 10 or 50 μM of 5-azacytidine for 16 h led to an appreciable decrease in genome-wide DNA methylation (p < 0.05), i.e., the percentage of cytosine methylation decreased from 0.163% in untreated cells to 0.112% and 0.088% in the cells treated with 10 and 50 μM 5-azacytidine, respectively (Table 4). In addition, a similar treatment of S. cerevisiae (W1588-4C) cells with 10 and 50 μM 5-azacytidine resulted in a significant drop of the cytosine methylation level from 0.039% to 0.017% and 0.018%, respectively (p < 0.05) (Table 4). These results also indicated that 10 μM of 5-azacytidine was sufficient to induce the decrease of genomic DNA methylation since 50 μM of 5-azacytidine gave the similar effect as 10 μM of 5-azacytidine did. Since studies in cultured mammalian cells have shown that the incorporation of 5-azacytosine into DNA led to rapid loss of DNA methyltransferase activity through irreversible binding of the enzyme to the modified cytosine residues in DNA,23 our results may suggest the possible presence of active DNA methyltransferase in yeast.
Table 4.
5-Azacytidine Induced the Decrease of Genome-Wide DNA Methylation in Yeast Strains
| yeast strains | drug concentration |
methylation (%)a
5mC/(5mC + cytosine) |
|---|---|---|
|
Y. lipolytica (CGMCC 2.1718) |
control | 0.163 ± 0.013 |
| 10 μM | 0.112 ± 0.012 | |
| 50 μM | 0.088 ± 0.012 | |
|
S. cerevisiae (W1588– 4C |
control | 0.039 ± 0.003 |
| 10 μM | 0.017 ± 0.002 | |
| 50 μM | 0.018 ± 0.002 |
The data represent the means and standard deviations of results from three independent treatments. The control and drug-treated yeast cells were harvested at 16 h after culturing.
CONCLUSION
In the current study, we developed a highly sensitive and accurate GC/MS method for the determination of genome-wide DNA methylation. Under optimized derivatization conditions, 5mC was detected in all the yeast strains examined, which was the first systematic and conclusive report for the confirmation of the widespread existence of DNA methylation in yeast. Our studies showed that the genome-wide DNA methylation of the studied yeast strains ranged from 0.014 to 0.364%, which were 1 to 2 orders of magnitude lower than that in mammalian cells (i.e., 3–8%). Furthermore, the 5mC percentage in yeast varied considerably at different growth stages, and treatment with a DNA methylation inhibitor, i.e., 5-azacytidine, can induce the decrease of genome-wide DNA methylation as that in mammalian cells, suggesting the possible existence of DNA methyltransferase in yeast. Given the critical role of 5mC in epigenetic regulation in mammalian cells, the widespread existence of 5mC in yeast should not be neglected. Although the function of 5mC in yeast DNA remains unclear, we believe that the mystery of the 5mC in yeast will be unveiled by further investigations. Along this line, experiments are under way to ascertain whether DNA methylation is indeed developmentally modulated in yeast.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the financial support from the National Basic Research Program of China (973 Program) (2012CB720600, 2012CB720601), the National Natural Science Foundation of China (91017013, 31070327, 30871347), the Natural Science Fund for Creative Research Groups (No. 20921062, NSFC), the Fundamental Research Funds for the Central Universities (1104001, 203271204, 203274612), the Chinese 111 Project (B06018), and the Natural Science Foundation of Hubei Province (2011CDB440).
Footnotes
Supporting Information
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Jones PA, Baylin SB. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Gupta R, Nagarajan A, Wajapeyee N. Biotechniques. 2010;49:3–11. doi: 10.2144/000113493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Robertson KD. Nat. Rev. Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- (4).Esteller M. Annu. Rev. Pharmacol. Toxicol. 2005;45:629–656. doi: 10.1146/annurev.pharmtox.45.120403.095832. [DOI] [PubMed] [Google Scholar]
- (5).Petranovic D, Tyo K, Vemuri GN, Nielsen J. FEMS Yeast Res. 2010;10:1046–1059. doi: 10.1111/j.1567-1364.2010.00689.x. [DOI] [PubMed] [Google Scholar]
- (6).Bolotin-Fukuhara M, Dumas B, Gaillardin C. FEMS Yeast Res. 2010;10:959–960. doi: 10.1111/j.1567-1364.2010.00693.x. [DOI] [PubMed] [Google Scholar]
- (7).Antequera F, Tamame M, Villanueva JR, Santos TJ. Biol. Chem. 1984;259:8033–8036. [PubMed] [Google Scholar]
- (8).Proffitt JH, Davie JR, Swinton D, Hattman S. Mol. Cell. Biol. 1984;4:985–988. doi: 10.1128/mcb.4.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Wilkinson CRM, Bartlett R, Nurse P, Bird AP. Nucleic Acids Res. 1995;23:203–210. doi: 10.1093/nar/23.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Bulkowska U, Ishikawa T, Kurlandzka A, Trzcinska-Danielewicz J, Derlacz R, Fronk J. Yeast. 2007;24:871–882. doi: 10.1002/yea.1538. [DOI] [PubMed] [Google Scholar]
- (11).Hattman S, Kenny C, Berger L, Pratt KJ. Bacteriol. 1978;135:1156–1157. doi: 10.1128/jb.135.3.1156-1157.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Zhang M, Kimatu JN, Xu K, Liu BJ. Genet. Genomics. 2010;37:1–12. doi: 10.1016/S1673-8527(09)60020-5. [DOI] [PubMed] [Google Scholar]
- (13).Crain PF, McCloskey JA. Anal. Biochem. 1983;132:124–131. doi: 10.1016/0003-2697(83)90434-7. [DOI] [PubMed] [Google Scholar]
- (14).Bi E, Pringle JR. Mol. Cell. Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ko N, Nishihama R, Tully GH, Ostapenko D, Solomon MJ, Morgan DO, Pringle JR. Mol. Biol. Cell. 2007;18:5139–5153. doi: 10.1091/mbc.E07-05-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Hoffman CS, Winston F. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- (17).Rossella F, Polledri E, Bollati V, Baccarelli A, Fustinoni S. Rapid Commun. Mass Spectrom. 2009;23:2637–2646. doi: 10.1002/rcm.4166. [DOI] [PubMed] [Google Scholar]
- (18).Singer J, Schnute WC, Jr, Shively JE, Todd CW, Riggs AD. Anal. Biochem. 1979;94:297–301. doi: 10.1016/0003-2697(79)90363-4. [DOI] [PubMed] [Google Scholar]
- (19).Duncan BK, Miller JH. Nature. 1980;287:560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- (20).San Romerio A, Fiorillo G, Terruzzi I, Senesi P, Testolin G, Battezzati A. Anal. Biochem. 2005;336:158–163. doi: 10.1016/j.ab.2004.09.034. [DOI] [PubMed] [Google Scholar]
- (21).Razin A, Sedat J. Anal. Biochem. 1977;77:370–377. doi: 10.1016/0003-2697(77)90250-0. [DOI] [PubMed] [Google Scholar]
- (22).Beckerich J-M, Boisrame-Baudevin A, Gaillardin C. Int. Microbiol. 1998;1:123–130. [PubMed] [Google Scholar]
- (23).Christman JK. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.