Abstract
Streptococcus mutans is considered to be the major etiologic agent of human dental caries. Attachment of S. mutans to the tooth surface is required for the development of caries and is mediated, in part, by the 185-kDa surface protein variously known as antigen I/II, PAc, and P1. Such proteins are expressed by nearly all species of oral streptococci. Characteristics of P1 include an alanine-rich repeat region and a centrally located proline-rich repeat region. The proline-rich region of P1 has been shown to be important for the translational stability and translocation of P1 through the bacterial membrane. We show here that (i) several anti-P1 monoclonal antibodies require the simultaneous presence of the alanine-rich and proline-rich regions for binding, (ii) the proline-rich region of P1 interacts with the alanine-rich region, (iii) like the proline-rich region, the alanine-rich region is required for the stability and translocation of P1, (iv) both the proline-rich and alanine-rich regions are required for secretion of P1 in Escherichia coli, and (v) in E. coli, P1 is secreted in the absence of SecB.
Streptococcus mutans is considered to be the major etiological agent of human dental caries. Attachment to the tooth surface is mediated, in part, by the 185-kDa surface protein variously known as P1 (15), PAc (51), and antigen I/II (55), which is encoded by the spaP (38) or pac (51) gene. Researchers have identified P1 as a potential antigen for dental caries vaccine development (73). Studies have shown that active immunization with antigen I/II (28, 39) or passive immunization with an anti-P1 monoclonal antibody (43) can protect against dental caries caused by S. mutans. Major characteristics of P1 include an amino-terminal signal sequence, an alanine-rich repeat region (A region), a variable region, a proline-rich repeat region (P region), carboxy-terminal wall- and membrane-spanning regions, and an LPXTG wall anchor motif (Fig. 1).
FIG. 1.
Schematic representation of the linear structure of P1 showing salient features.
Initially identified in S. mutans (56), antigen I/II-like molecules have been found to be expressed in nearly all of the oral streptococci (44). Antigen I/II polypeptides are structurally complex and exhibit diverse binding properties, which mediate interactions with a variety of substrates including host salivary agglutinin, fibronectin, fibrinogen, and collagen (5, 58). Several regions have been implicated in the binding activities of antigen I/II polypeptides. Brady et al. (9) provided evidence that the interaction of P1 with salivary agglutinin likely involves complex conformational determinants and that different regions might be involved in binding to soluble or immobilized salivary agglutinin. Later, Scatchard analysis of antigen I/II binding to saliva-coated hydroxyapatite found the binding to be mediated by two sites (21). Investigators have shown that recombinant peptide fragments derived from the A region can bind salivary agglutinin (12) or salivary glycoproteins (47), and Senpuku et al. demonstrated that antibodies specific to a peptide fragment derived from PAc amino acid residues (aa) 200 to 481 inhibited the binding of fluid-phase salivary components to immobilized PAc (60). In addition, an antigen I/II peptide fragment consisting of aa 816 to 1213 blocked S. mutans cell adhesion to saliva-coated hydroxyapatite (50), and Kelly et al. found antigen I/II-derived peptides consisting of residues 1005 to 1044 and 1085 to 1114 to be inhibitory to S. mutans adhesion to salivary glycoproteins (30). Taken together, these results suggest that multiple regions within P1 contribute to the biological activity of the molecule, either as linear determinants or as components of more complex 3-dimensional, possibly discontinuous structures.
Previously, in an attempt to define a role for the P region in the adhesive function of P1, a deletion mutant, P1ΔP (Δ826-996), was constructed (7). The P region is highly conserved among the antigen I/II family of oral streptococcal proteins, and similar highly repetitive proline-rich sequences have been identified in a wide variety of bacterial proteins (10, 19, 22, 23, 25, 36, 53, 54, 62, 65). The deletion mutant P1ΔP was expressed both in Escherichia coli and in S. mutans PC3370, an isogenic spaP-negative mutant (11). Western blotting of P1ΔP expressed in E. coli revealed a loss in reactivity for 5 of 11 P1-specific monoclonal antibodies (MAbs), but these MAbs did not react to a subclone of the P region (aa 826 to 996), suggesting that they recognize a complex P1 epitope that is dependent on the presence of the P region (7). Although P1ΔP contains the N-terminal signal and C-terminal wall-anchoring sequences, it was not localized to the surface of S. mutans PC3370 (spaP-negative mutant). In comparison to full-length P1, only low levels of P1ΔP were detected in the cytoplasm of PC3370 despite equivalent mRNA levels, suggesting that the P region may be required for the stability of P1 and its subsequent translocation to the cell surface.
Given that the P region has been shown to be required for the native structure and surface localization of P1 (7) and that the P region has been shown to interact with a fragment of P1 containing the A region (70), the present study was undertaken to determine whether the A region may also play a role in the structure, stability, and translocation of P1.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The S. mutans serotype c strain NG8 and the spaP-negative mutant PC3370 and its derivatives (Table 1) were grown for 16 h at 37°C in Todd-Hewitt broth (BBL, Cockeysville, Md.) supplemented with 0.3% yeast extract. E. coli strains used in these experiments included DH5α, MC4100, and CK1953 (kindly provided by C. Kumamoto). E. coli DH5α and MC4100 were grown aerobically at 37°C with vigorous shaking in Luria-Bertani (LB) broth (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 1% [wt/vol] NaCl [pH 7.0]) supplemented with ampicillin (100 μg/ml) or kanamycin (50 μg/ml) as appropriate. E. coli CK1953 was grown aerobically at 37°C with vigorous shaking in M9 medium [0.625% (wt/vol) Na2HPO4, 0.075% (wt/vol) KH2PO4, 0.2% (wt/vol) NaCl, 0.028% (wt/vol) MgSO4, 0.1% (wt/vol) (NH4)2SO4, 1% glucose] supplemented with kanamycin (50 μg/ml) and ampicillin (100 μg/ml) as appropriate. Plasmids pUC18 and pDL289 (kindly provided by D. LeBlanc) (7) were used as cloning and expression vectors.
TABLE 1.
Strains and plasmids used in this study
| Plasmid or strain | Description (source or reference) |
|---|---|
| Plasmids | |
| pDL289 | E. coli-streptococcal shuttle vector (9a) |
| pMAD | pDL289-derived plasmid containing PCR-amplified spaP encoding full-length P1 (7) |
| pMal-p | Vector for expression of MBP fusions (NEB) |
| pMA3 | pMal-p derived plasmid containing PCR-amplified DNA encoding aa 819 to 1017 of P1 (7) |
| pMA41 | pMal-p-derived plasmid containing PCR-amplified DNA encoding aa 186 to 469 of P1 (7) |
| pDC9 | pUC18-derived plasmid containing internal deletions of spaP-encoding aa 1 to 824 and 998 to 1561 (7) |
| pDC20 | pUC18-derived plasmid containing spaP-encoding aa 1 to 1561 (7) |
| pTS20 | pUC18-derived plasmid containing internal deletions of spaP-encoding aa 1 to 178 and 465 to 1561 |
| pTS21 | pDL289-derived plasmid containing internal deletions of spaP-encoding aa 1 to 178 and 465 to 1561 |
| Strains | |
| PC3370 | spaP-negative mutant derived from S. mutans NG8 (12a) |
| PC3370A | PC3370 transformed with pDL289 (7) |
| PC3370C | PC3370 transformed with pMAD (7) |
| MC4100 | E. coli F−araD 139Δ (argF lac)U169 relA rspP thiA (34) |
| CK1953 | MC4100 secB::Tn5 (34) |
| TS20 | E. coli DH5α transformed with pTS20 |
Elimination of spaP DNA encoding the A region.
Fragments of spaP both upstream and downstream of the A region were amplified by PCR and subsequently ligated together to create spaPΔA. The fidelity of the reactions was confirmed by restriction and sequence analysis. Primers TS9s (5′-GCGTCGACGTTGGATAAAGTGTGGAGTTTG-3′) and TS8 (5′-GCCATACTGTTCTTTAGTTGCCTG-3′) were used to amplify spaP DNA upstream of the A region, including the spaP promoter. Primers TS7 (5′-GCCGACTATCCAGTTAAGTTAAAGGC-3′) and TS10s were used to amplify spaP downstream of the A region. (Underlining in TS9s and TS10s indicates SalI restriction sites.) Reactions were carried out in a UNO thermoblock thermocycler (Biometra, Tampa, Fla.) with plasmid-encoded spaP, pDC20 (7) as the template, and VENT polymerase (New England Biolabs [NEB], Beverly, Mass.) under the following conditions: (i) 94°C for 2 min; (ii) 30 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 1 min or 3 min, 30 s; and (iii) 72°C for an additional 7 min. The resulting 727- and 3,568-bp gene fragments were ligated together and cloned into the SmaI site of pUC18, creating pTS20, which was introduced into E. coli DH5α by electroporation. PCR was then used to amplify spaPΔA from pTS20 by using primers TS9k (5′-GCGGTACCGTTGGATAAAGTGTGGAGTTTG-3′) and TS10k (5′-GCGGTACCGCAGTGCGAAGTACCTTATC-3′), which introduced KpnI restriction sites (underlined). The amplified SpaPΔA digested with KpnI was cloned into the KpnI site of the shuttle vector pDL289, creating pTS21, and introduced into the S. mutans spaP-negative mutant strain PC3370 by natural transformation. Transformants were selected for their ability to grow on Todd-Hewitt broth with 0.3% yeast extract containing 500 μg of kanamycin/ml. Sequences of all recombinant constructs were confirmed by the DNA sequencing core facility (University of Florida).
Evaluation of antibody binding to P1ΔA.
E. coli DH5α harboring pTS20 and pDC20 was grown for 16 h at 37°C, harvested by centrifugation, and lysed by boiling for 5 min in sodium dodecyl sulfate (SDS) sample buffer (4% [wt/vol] SDS, 2% [vol/vol] 2-mercaptoethanol, 20% [vol/vol] glycerol, 125 mM Tris-HCl [pH 6.8], 0.1 mg of bromophenol blue per ml). Proteins were separated by SDS-polyacrylamide gel electrophoresis on 7.5% acrylamide preparatory gels by the method of Laemmli (35). Proteins were electroblotted onto a nitrocellulose membrane (Schleicher & Schuell, Keene, N.H.) for 1 h at 100 V by the method of Towbin et al. (66). Immunoblots were blocked with phosphate-buffered saline (PBS) containing 0.03% Tween 20 (PBST) and cut into 0.5-cm strips. Strips were incubated with anti-P1 MAbs (3, 8) at dilutions of 1:1,000 in individual troughs of an Incutray (Schleicher & Schuell). After washing, strips were incubated in peroxidase-labeled goat anti-mouse immunoglobulin G (IgG) (Cappell) and developed with 4-chloro-1-naphthol solution (7 ml of PBS, 1 ml of 4-chloro-1-naphthol [3 mg/ml in ice-cold methanol; Sigma], and 8 μl of 30% hydrogen peroxide).
Purification of A-region-maltose-binding protein (MBP) and P-region-MBP fusion proteins.
Overnight cultures of E. coli harboring pMA3 (7) or pMA41 (12) (Table 1) were diluted 1:100 into fresh LB broth containing 100 μg of ampicillin/ml and were grown to an optical density at 600 nm (OD600) of 0.5. The medium was supplemented with 0.3 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and the culture was incubated for an additional 2 h at 37°C. Periplasmic contents were extracted by osmotic shock (2). The fusion proteins were affinity purified by passage of the periplasmic fractions through a column of amylose resin (Bio-Rad) and elution with 10 mM maltose by a standard protocol (2).
ELISA to detect interaction between the A region and the P region.
Binding of the A region to the P region was measured by enzyme-linked immunosorbent assays (ELISA). Sample wells of Costar High Binding plates (Corning Incorporated, Corning, N.Y.) were coated overnight at 4°C, in triplicate, with 100 μl of 0.1 M carbonate-bicarbonate buffer (pH 9.6) containing 0.02% sodium azide and 100 ng of either purified MBP, A-region-MBP, or P-region-MBP. The coating buffer and unbound antigens were removed from the ELISA plate wells, and unreacted sites were blocked with PBST and overnight incubation at 4°C. Plates were washed four times with PBST. Purified A-region-MBP, P-region-MBP, and MBP were twofold serially diluted in PBST and added to the wells, beginning at 1,000 ng/well. The plates were incubated overnight at 4°C and washed four times with PBST. The A-region-specific MAb 3-8D (12) or a rabbit anti-MBP antibody (NEB) was added to the wells at a 1:1,000 dilution. Plates were washed with PBST, and peroxidase-labeled goat anti-mouse IgG or goat anti-rabbit Ig (Cappell) was added to the wells at a 1:1,000 dilution. After a wash, 100 μl of 0.01 M phosphate citrate buffer (pH 5.0) containing 0.1 M o-phenylenediamine dihydrochloride and 0.012% hydrogen peroxide was added to each well. Plates were incubated for 30 min at room temperature, and the absorbance at 490 nm was recorded by using an MPM Titertek model 550 ELISA plate reader (Bio-Rad).
Dot blot analysis for detection of P1 surface expression by the mutant strain PC3370 and derivatives.
The S. mutans spaP-negative isogenic mutant PC3370 (11) and PC3370 derivatives harboring pDL289, pMAD, or pTS21 (Table 1) were grown for 16 h at 37°C. Then the cells were harvested by centrifugation and washed twice with PBS. Cells were resuspended in PBS to 160 Klett units. Twofold serial dilutions of cell suspensions were made in PBS, and 100 μl of each dilution was applied in duplicate to two nitrocellulose membranes by using a 96-well dot blot manifold (both from Schleicher & Schuell). Wells were washed twice with 200 ml of PBS, and the filters were removed from the apparatus and blocked with PBS containing 0.25% gelatin and 0.25% Tween 20. P1 on the cell surface was detected with rabbit antiserum 230 (50) or MAb 3-10E (3) as the primary antibody, peroxidase-conjugated goat anti-rabbit IgG or goat anti-mouse IgG as the secondary antibody, and development with 4-chloro-1-naphthol solution.
Western blot analysis of lysates of PC3370 and derivatives.
S. mutans PC3370, PC3370A, PC3370C (Table 1), and PC3370 harboring pTS21 were grown at 37°C to an OD600 of 1.0 to 1.1. The cells were harvested and mechanically homogenized in a Mini Beadbeater (Biospec Products, Inc., Bartlesville, Okla.) (33). Total protein concentrations of the recovered cell lysates were estimated by measurement of A280, and the samples were adjusted so that protein concentrations were equal. The glass beads and cell debris were resuspended in 1 ml of Tris-EDTA buffer. The glass beads quickly settled, and the cell debris remaining in suspension was removed by pipetting. The cell debris was centrifuged at 10,000 × g for 5 min and resuspended in 1 ml of SDS sample buffer. Cell lysates were diluted 1:1 in SDS sample buffer, and both cell lysates and cell debris were heated for 5 min at 100°C. Proteins were separated on SDS-7.5% polyacrylamide gels and transferred to nitrocellulose membranes for 1 h at 100 V. Immunoblots were blocked and developed as described above for the dot blot assay.
RNA isolation and RNA dot blotting.
By following the manufacturer's protocol, RNA was isolated from stationary-phase cultures of PC3370 harboring pDL289, pTS21, or pMAD by using the QIAGEN (Valencia, Calif.) RNeasy kit. Total RNA concentrations were measured by OD260/OD280 and standardized to ∼92 μg/ml by the addition of RNA dilution buffer (6× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]-20% formaldehyde). Samples were serially diluted twofold, and 50 μl of each was applied to a nylon membrane by using a 96-well dot blot manifold (Schleicher & Schuell). The membrane was baked for 30 min at 120°C and incubated in DIG Easy Hyb (Roche, Indianapolis, Ind.) for 2 h at 37°C. The membrane was probed overnight at 37°C with digoxigenin-labeled, PCR amplified DNA complementary to the 3′ end of spaP, nucleotides 3985 to 4125. The membrane was washed, blocked for 1 h at 25°C in Roche blocking buffer, and incubated with alkaline phosphatase-labeled anti-digoxigenin antibodies. After a wash in detection buffer, the chemiluminescence substrate disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2-(5-chloro)tricyclo[3.3.1.13,7]decan}-4-yl)phenyl phosphate was added and the membrane was exposed to X-ray film (Fuji, Tokyo, Japan).
Western blot analysis of E. coli periplasm extracts.
Periplasm contents of E. coli DH5α harboring pUC18, pDC20, pDC9, or pTS20 and of E. coli MC4100 and CK1953, a secB-negative mutant (34), harboring pUC18 or pDC20 were extracted by osmotic shock (2). Cells were grown for 16 h at 37°C, 0.3 mM IPTG was added, and the culture was incubated for an additional 2 h. Cells were harvested by centrifugation, resuspended in 30 mM Tris-HCl-20% sucrose-0.1 mM EDTA (pH 8.0), and incubated with shaking. Cells were again pelleted by centrifugation, resuspended in ice-cold 5 mM MgSO4, and incubated in an ice bath for 10 min. Lastly, cells were removed by centrifugation, and 1 M Tris-HCl (pH 7.4), to a final concentration of 20 mM, was added to the supernatant. The proteins in this periplasmic preparation were separated on SDS-7.5% polyacrylamide gels and transferred to nitrocellulose membranes. Corresponding cell pellets were boiled in SDS sample buffer and analyzed as well. Immunoblots were blocked and developed as described above for the dot blot assay.
RESULTS
Expression of recombinant P1ΔA and recognition by anti-P1 MAbs.
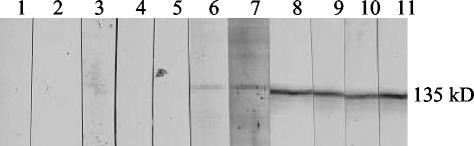
A spaP gene lacking DNA encoding the A region (nucleotides 537 to 1401) was constructed by PCR and cloned into pUC18, creating pTS20, as described in Materials and Methods. P1 lacking the A region (P1ΔA) was detectable by Western blotting in cell lysates of recombinant E. coli DH5α by using anti-P1 polyclonal antibodies (data not shown). P1ΔA migrates at its predicted molecular weight of 135,000. The effect of deleting the A region on the antigenicity of P1 was examined by Western blotting utilizing a panel of 11 anti-P1 MAbs (7). Deletion of the A region from P1 eliminated the reactivity of 5 of the 11 MAbs (Fig. 2). Three of the nonreactive MAbs, 4-9D, 5-5D, and 6-11A, are also not reactive with P1ΔP (7). The reactivity of MAbs 5-3E, 2-8G, 3-3B, and 6-8C, which are specific to the C-terminal end of P1 (8), confirmed that the deletion of the DNA encoding the A region did not disrupt the reading frame. The Western blot also shows that, like P1ΔP, P1ΔA is stably expressed and easily detectable in E. coli.
FIG. 2.
Western blot analysis of recombinant P1 lacking the A region. Lanes 1 through 11, cell lysates of TS20 reacted with anti-P1 MAbs 3-8D, 4-10A, 4-9D, 5-5D, 6-11A, 3-10E, 1-6F, 5-3E, 2-8G, 3-3B, and 6-8C, respectively.
Interaction of the A and P regions by ELISA.
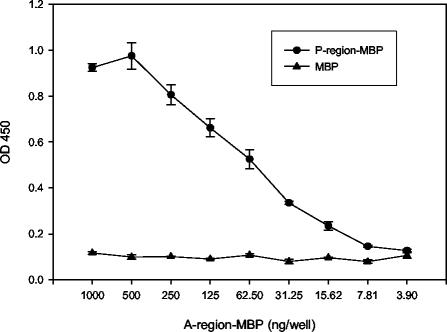
The dependence of several anti-P1 MAbs on the simultaneous presence of the A and P regions, and work by van Dolleweerd et al. (70) characterizing a complex epitope comprising the P region and a fragment of P1 containing the A region, suggests a specific interaction between these domains. To determine whether the isolated A region (aa 186 to 469) and P region (aa 819 to 1017) are capable of such an interaction, an ELISA was used to evaluate binding (Fig. 3). To facilitate protein purification, the A region and P region of P1 were expressed as fusions with MBP (pMA41 and pMA3, respectively [Table 1]). Purified P-region-MBP or MBP alone as a negative control was immobilized in ELISA plate wells. After the plates were washed and blocked, twofold serial dilutions of A-region-MBP were added to the wells. Binding of A-region-MBP to the immobilized proteins was detected by the A-region-specific MAb 3-8D. As shown, A-region-MBP binds to P-region-MBP in a dose-dependent manner, but not to MBP alone. MBP alone does not bind to MBP or to P-region-MBP (data not shown).
FIG. 3.
Demonstration of A-region and P-region interaction by ELISA. One hundred nanograms of the P-region-MBP fusion polypeptide (circles) or of MBP alone (triangles) was used to coat ELISA plate wells. Twofold serial dilutions of the purified A-region-MBP fusion polypeptide starting at 1,000 ng/well were added to the coated wells, and binding of the A region to the P region or to the MBP negative control was traced with the A-region-specific MAb 3-8D.
Evaluation of surface expression of P1ΔA in S. mutans.
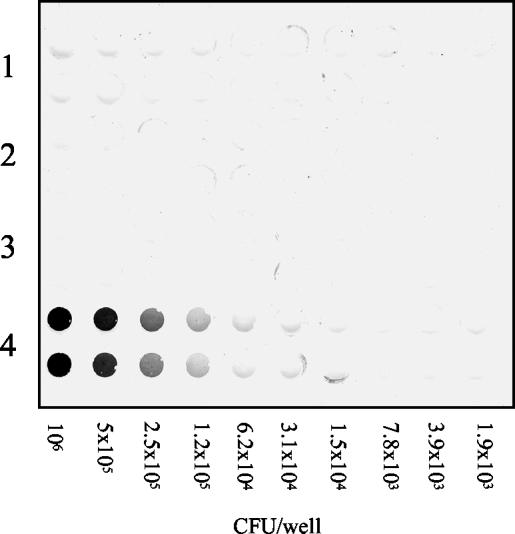
If an interaction between the P region and the A region of P1 is required for P1 structure, stability, and translocation, a similar phenotype should be observed for P1ΔA as that which was seen for P1ΔP (7). The spaP-negative mutant PC3370 was used as the host for plasmids pMAD and pTS21, encoding full-length P1 and P1ΔA, respectively. Whole-cell dot blot analysis was used to examine whether the A region, like the P region (7), is necessary for P1 surface expression in PC3370. These results are shown in Fig. 4. Twofold serial dilutions of the cells were applied to the nitrocellulose membrane in duplicate. The positive control PC3370C, expressing full-length P1, demonstrated the reactivity of the antiserum with surface-expressed P1. The negative controls, PC3370 and PC3370A (vector only), showed the lack of reactivity of the antiserum with cells lacking P1. PC3370 harboring pTS21, encoding P1ΔA, was not reactive with the polyclonal antiserum, indicating a lack of surface expression of P1. These results indicated that P1ΔA is not translocated to the surfaces of PC3370 cells. No P1ΔA was detected in spent culture liquor, although P1 is found in the spent culture liquor of PC3370C and NG8 (wild type) (data not shown). To determine if P1ΔA was detectable in S. mutans cell lysates, NG8, PC3370A, PC3370C, PC3370, and PC3370 harboring pTS21 were subjected to mechanical lysis in a Mini Beadbeater apparatus, and samples were analyzed by Western blotting (data not shown). Full-length P1 was present in both cell extracts and cell debris of NG8 and PC3370C. P1ΔA was not detected in either the cell extract or the cell debris of PC3370 harboring pTS21, and no P1 was observed in the negative controls, PC3370A and PC3370.
FIG. 4.
Dot blot analysis of P1 expression by the S. mutans spaP-negative mutant PC3370 and derivatives. Twofold serial dilutions of bacterial cells were applied in duplicate to the membrane. Rabbit antiserum 230, raised against purified P1 from S. mutans, was used as the primary antibody. Rows 1 through 4, the spaP-negative mutant PC3370, PC3370A (shuttle vector only), PC3370 harboring pTS21 (spaP with the A region deleted), and PC3370C (spaP encoding full-length P1), respectively.
Evaluation of spaP-specific mRNA in PC3370 harboring the deletion construct pTS21.
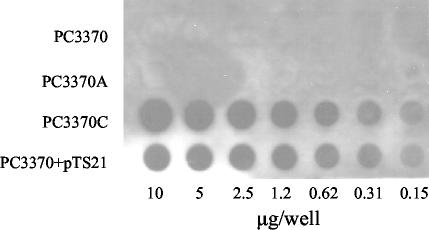
To confirm that spaPΔA was transcribed from the pDL289 shuttle vector in PC3370, an RNA dot blot was performed (Fig. 5). Dilutions of total cellular RNA were probed with a digoxigenin-labeled probe corresponding to the 3′ end of spaP. Rows 1 and 2 correspond to the negative controls, PC3370 and PC3370A (harboring the vector only). Row 3 contains RNA from PC3370 harboring pTS21, and row 4 contains RNA from the positive control, PC3370 harboring pMAD. The dot blot shows that the spaPΔA message is expressed at levels equivalent to those of the full-length spaP expressed from pMAD.
FIG. 5.
RNA dot blot analysis of spaP-specific mRNA levels in the spaP-negative mutant PC3370 and derivatives. Twofold serial dilutions of total cellular RNA, beginning with 5 mg, were probed with DNA encoding the C-terminal end of spaP. Rows 1 through 4, PC3370, PC3370A (shuttle vector only), PC3370C (full-length P1), and PC3370 harboring pTS21 (spaP with the A region deleted), respectively.
Evaluation of secretion of P1, P1ΔA, and P1ΔP in E. coli.
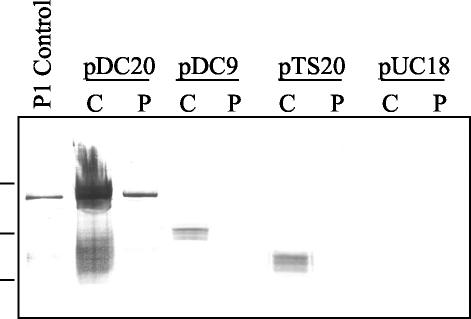
To analyze the secretion competency of P1, P1ΔA, and P1ΔP expressed in E. coli, periplasmic extracts of E. coli DH5α harboring pUC18, pDC20, pDC9, or pTS20 were prepared by osmotic shock, and the presence of P1 and derivatives was detected by Western blotting. These results are shown in Fig. 6. The leftmost lane contains a positive control for antibody reactivity, P1 extracted from S. mutans NG8 (8). The lanes containing cellular extracts from E. coli DH5α harboring pDC20 (full-length P1) clearly show that P1 is present in both the cytoplasm and the periplasm. The following lanes, containing cellular fractions from DH5α harboring pDC9 (spaP with the P region deleted), show that P1ΔP is present in the cytoplasm but absent from the periplasm. Cell fractions from E. coli harboring pTS20 show that, like P1ΔP, P1ΔA is present in the cytoplasm but is not translocated to the periplasm. The two rightmost lanes contain cellular lysates and periplasmic extracts from E. coli harboring pUC18 (vector only).
FIG. 6.
Western blot analysis of the cellular locations of P1, P1ΔA, and P1ΔP in E. coli DH5α. Leftmost lane, P1 extracted from S. mutans NG8, used as a positive control for antibody reactivity. As indicated, lanes contain cytoplasmic (C) or periplasmic (P) extracts of E. coli DH5α harboring either pDC20 (full-length P1), pDC9 (P1ΔP), pTS20 (P1ΔA), or pUC18. The membrane was reacted with a cocktail of MAbs including 5-3E, 2-8G, 3-3B, and 6-8C. Molecular weight standards, indicated on the left, correspond to Mrs of 250,000, 150,000, and 100,000.
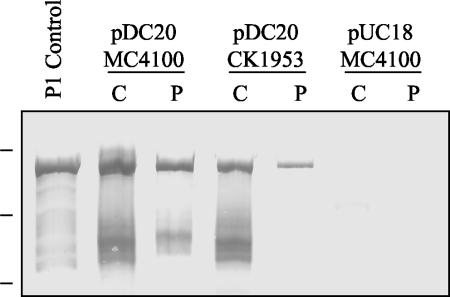
To determine whether P1 secretion is dependent on SecB in E. coli, periplasmic extracts of an E. coli SecB-negative mutant, CK1953, and the wild-type strain MC4100 (34) were prepared by osmotic shock, and the presence of P1 was detected by Western blotting. These results are shown in Fig. 7. Lane 1 contains a positive control for antibody reactivity, P1 extracted from S. mutans NG8 (8). The lanes containing cellular extracts from E. coli MC4100 harboring pDC20 (full-length P1) clearly show that P1 is detected in both the cytoplasm and the periplasm. The lanes containing cellular fractions from the SecB mutant CK1953 harboring pDC20 (full-length P1) show that, as in MC4100, P1 is clearly detected in both the cytoplasm and the periplasm. The two rightmost lanes correspond to cell fractions from MC4100 harboring pUC18 (vector only). These cell extracts were analyzed by Western blotting for β-galactosidase to confirm the integrity of the periplasmic extractions (data not shown). No β-galactosidase was detected in the periplasmic extracts. These results show that P1 translocation in E. coli is not dependent on SecB, the chaperone that is central to the general secretory pathway of E. coli.
FIG. 7.
Western blot analysis of P1 cellular location in the E. coli secB-negative mutant CK1953. Leftmost lane, P1 extracted from S. mutans NG8, used as a positive control for antibody reactivity. As indicated, lanes contain cytoplasmic (C) or periplasmic (P) extracts of E. coli MC4100 (wild type) or CK1953 (secB::Tn5) harboring pDC20 (full-length P1) or of MC4100 harboring pUC18. The membrane was reacted with a cocktail of MAbs including 5-3E, 2-8G, 3-3B, and 6-8C. Molecular weight standards, indicated on the left, correspond to Mrs of 250,000, 150,000, and 100,000.
DISCUSSION
In this paper, we report that the phenotype of the P1 polypeptide lacking the A region (P1ΔA) is the same as that of P1 lacking the P region (P1ΔP). While equivalent levels of mRNA are expressed, P1ΔA appears to be unstable in S. mutans and is not expressed on the cell surface. Like P1ΔP, P1ΔA is detectable in the cell lysates of recombinant E. coli. Deletion of the A region from P1 abolishes the reactivities of several P-region-dependent anti-P1 MAbs, and a direct interaction between the A region and the P region was demonstrated. In E. coli, P1 but not P1ΔA or P1ΔP is translocated to the periplasm, and P1 translocation is SecB independent.
An interaction between the P region and the A region of P1 that is required for P1 structure, stability, and translocation should impart a similar phenotype to P1ΔA as that which was seen for P1ΔP (7). To address the possibility of such an interaction, a spaP gene lacking the A region (nucleotides 537 to 1401) was constructed by PCR and cloned into pUC18, creating pTS20. The reactivities of MAbs 5-3E, 2-8G, 3-3B, and 6-8C, which are specific to the C-terminal end of P1, confirmed that the deletion of the DNA encoding the A region did not disrupt the reading frame. Western blotting also shows that, like P1ΔP, P1ΔA is stably expressed and easily detectable in E. coli.
Deletion of the P region of P1 resulted in a loss of reactivity for 5 of 11 anti-P1 MAbs. Deletion of the A region also results in a loss of reactivity for 5 of 11 of the anti-P1 MAbs, and 3 of the MAbs are dependent on both the A and the P region. This suggests that the epitopes are complex and that they may be nonlinear and composed of portions of both regions, or that an interaction between the regions may result in conformational epitopes being produced within one or both of the regions or perhaps elsewhere in the molecule. The requirement of both the A and the P region for MAb reactivity and the recent evidence presented by van Dolleweerd et al. (70) for the binding of the P region with an N-terminal fragment of P1 containing the A region led us to look into a specific interaction between the A and P regions. Binding of the A region (aa 186 to 469) with the P region (aa 819 to 1017) was analyzed by ELISA, and binding in a dose-dependent manner was observed.
In addition to the requirement for MAb reactivity, the simultaneous presence of both the A and the P region appears to be required for P1 stability in S. mutans. Analysis of mRNA encoding P1ΔA demonstrated that, as with P1ΔP (7), the internally deleted spaP gene was transcribed at levels equivalent to those of the wild-type spaP gene. However, no P1ΔA was detected in the cytoplasm, on the cell surface, or in the culture liquor. While P1ΔP contains a deletion of 170 residues and P1ΔA lacks 287 residues, there are examples of stable antigen I/II polypeptides that, compared to P1, are lacking large segments of the molecules. The antigen I/II protein expressed by Streptococcus intermedius, Pas, lacks ∼270 residues from the A region and ∼80 residues from the P region (64); Paa from Streptococcus cricetus possesses an additional ∼139 residues in the A region and ∼39 residues fewer in the P region (64); and S. mutans GS-5 expresses a PAc molecule lacking the C-terminal ∼400 residues (57). The A region of P1 consists of three 82-residue repeats, and the P region consists of three 39-residue repeats; both Paa and Pas retain repeats in both regions. This suggests that the A and P regions may contain inherent structural information and possible chaperone binding sites, or that perhaps they possess chaperone-like activities that are critical to P1 stability. Proline-rich regions are known to be involved in a variety of intra- and intermolecular protein-protein interactions (1, 4, 16, 18, 26, 29, 32, 45, 68, 72), including chaperone-like activities. Wang et al. identified a centrally located proline-rich region in the serine protease Limulus factor C that is required for the proper folding and secretion of the molecule (71). These investigators proposed that internal proline-rich domains may serve as intramolecular chaperones. It has also been proposed that many proteins may contain uncleaved intramolecular chaperone-like fragments that interact with adjoining regions and are essential for achieving native structure (42). Proline containing motifs similar to those found repeatedly within the P region of P1 are found in a wide variety of bacterial proteins (6, 31, 61, 63). These similarities and the conservation of the P region within oral streptococci suggest that it may play an important functional role in this regard. Considering the prevalence of proline-rich regions in protein-protein interactions, it is likely that the P region is involved in such an interaction. Recently, van Dolleweerd et al. demonstrated that the interaction of the P region of P1 with a peptide fragment of P1 containing the A region restored the reactivity of an antigen I/II-reactive MAb that was not reactive with either fragment alone (70). X-ray crystallography has also revealed that the variable region of P1 (aa 464 to 840) forms a beta-sandwich that would place the P region and A region in close proximity (67).
To fully understand the roles of the A and P regions in P1 translocation, it is imperative to identify the molecule's route of translocation. There are no experimental data that identify the secretion pathway employed by P1 or antigen I/II-like proteins. Cell wall anchoring of P1 and PAc is mediated by the transpeptidase sortase (27, 37), and sortase-anchored proteins are believed to be translocated via the Sec translocase (46). The Sec-dependent secretion pathway has been well characterized and studied in E. coli and, to a lesser extent, in Bacillus subtilis. In E. coli, the Sec translocase consists of SecA, SecY, SecE, SecG, SecD, SecF, and YajC (13). Two major targeting pathways converge on the Sec translocase: the signal recognition particle (SRP) pathway and the SecB pathway. The E. coli SRP consists of a 4.5S RNA and the GTPase Ffh, both of which are required for cell viability in this organism (52). Signal peptides of nascent polypeptides are recognized by the SRP as they emerge from the ribosome (69). SRP binding stalls translation and targets the SRP-ribosome complex to the SRP receptor, FtsY (41, 59). The complex is then targeted to the Sec translocon, where the ribosome docks and translation is restored. The preprotein is cotranslationally translocated across the membrane via an integral membrane complex consisting of SecY, SecE, and SecG. The ATPase SecA provides energy for the translocation (13). In the case of posttranslational secretion, the cytoplasmic chaperone SecB targets preproteins to the Sec translocon. SecB binds to nascent and full-length preproteins as they emerge from the ribosome (17). SecB interaction prevents premature folding of the preprotein and delivers it to the Sec translocon in a secretion-competent state. Binding of the SecB-preprotein complex with SecA results in the transfer of the preprotein to SecA and the release of SecB (24). The preprotein is subsequently translocated across the membrane through the Sec translocon (14). Due to the faint expression of P1ΔP and the undetectable expression of P1ΔA in S. mutans, E. coli was used to examine P1 secretion. P1, P1ΔP, and P1ΔA are stable and easily detectable in whole-cell lysates of E. coli by Western blotting. Analysis of periplasmic extracts by Western blotting revealed that P1 was secreted into the periplasm but P1ΔP and P1ΔA were not. This suggests that while the A and P regions apparently are not required for stability in E. coli, they are required for secretion. If a lack of chaperone interaction with the recombinant deletion proteins results in a lack of secretion, perhaps a similar lack of interaction favors degradation of the molecules in S. mutans.
The SRP pathway is known to exist in both gram-negative and gram-positive bacteria. Identified homologs of the general secretory pathway components in B. subtilis include SecA, SecYEG, SecDF, YrbF, Ffh, and scRNA. As the genome sequences of gram-positive bacteria have become available, investigators have searched for homologs of SecB, to no avail. However, a B. subtilis complementation study of an E. coli SecB-null mutant revealed a functional ortholog, CsaA, with partially overlapping binding characteristics (40, 48, 49). As previously stated, the SRP is essential for viability in E. coli, and this was assumed to be the case in all organisms. However, an Ffh-null mutant of S. mutans is viable (20), and P1 is translocated and expressed on the cell surface (data not shown). This would suggest that if P1 secretion is Sec dependent, the targeting pathway should likely be SecB-like and require a functional SecB ortholog. To examine the possibility of a role for SecB in P1 secretion, P1 was expressed in a SecB-negative E. coli mutant, CK1953. P1 was shown to be stable and to be secreted into the periplasm in CK1953. This suggests that, in E. coli, if P1 is secreted via the Sec pathway, it is associating with an alternative chaperone to SecB, or it may be able to use the SRP pathway. Further research is currently under way to establish the route of P1 secretion in S. mutans and the chaperones involved.
Acknowledgments
We thank C. Kumamoto for providing the secB-negative mutant CK1953.
This work was supported by NIH grants DE13882 and DE08007 and training grant T32-DE07200.
Editor: J. D. Clements
REFERENCES
- 1.Aravind, L. 2001. The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem. Sci. 26:273-275. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, and K. Struhl. 1992. Expression and purification of maltose-binding protein fusions. (ed.), John Wiley and Sons, Inc., New York, N.Y.
- 3.Ayakawa, G. Y., L. W. Boushell, P. J. Crowley, G. W. Erdos, W. P. McArthur, and A. S. Bleiweis. 1987. Isolation and characterization of monoclonal antibodies specific for antigen P1, a major surface protein of mutans streptococci. Infect. Immun. 55:2759-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball, L. J., T. Jarchau, H. Oschkinat, and U. Walter. 2002. EVH1 domains: structure, function and interactions. FEBS Lett. 513:45-52. [DOI] [PubMed] [Google Scholar]
- 5.Beg, A. M., M. N. Jones, T. Miller-Torbert, and R. G. Holt. 2002. Binding of Streptococcus mutans to extracellular matrix molecules and fibrinogen. Biochem. Biophys. Res. Commun. 298:75-79. [DOI] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Brady, L. J., D. G. Cvitkovitch, C. M. Geric, M. N. Addison, J. C. Joyce, P. J. Crowley, and A. S. Bleiweis. 1998. Deletion of the central proline-rich repeat domain results in altered antigenicity and lack of surface expression of the Streptococcus mutans P1 adhesin molecule. Infect. Immun. 66:4274-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brady, L. J., D. A. Piacentini, P. J. Crowley, and A. S. Bleiweis. 1991. Identification of monoclonal antibody-binding domains within antigen P1 of Streptococcus mutans and cross-reactivity with related surface antigens of oral streptococci. Infect. Immun. 59:4425-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brady, L. J., D. A. Piacentini, P. J. Crowley, P. C. Oyston, and A. S. Bleiweis. 1992. Differentiation of salivary agglutinin-mediated adherence and aggregation of mutans streptococci by use of monoclonal antibodies against the major surface adhesin P1. Infect. Immun. 60:1008-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Buckley, N. D., I. N. Lee, and D. J. LeBlanc. 1995. Use of a novel mobilizable vector to inactivate the scrA gene of Streptococcus sobrinus by allelic replacement. J. Bacteriol. 177:5028-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, R., W. Schmidmayr, C. Kramer, U. Chen-Schmeisser, and U. Henning. 1980. Primary structure of major outer membrane protein II (ompA protein) of Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 77:4592-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowley, P. J., L. J. Brady, S. M. Michalek, and A. S. Bleiweis. 1999. Virulence of a spaP mutant of Streptococcus mutans in a gnotobiotic rat model. Infect. Immun. 67:1201-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crowley, P. J., L. J. Brady, D. A. Piacentini, and A. S. Bleiweis. 1993. Identification of a salivary agglutinin-binding domain within cell surface adhesin P1 of Streptococcus mutans. Infect. Immun. 61:1547-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Crowley, P. I., J. A. Gutierrez, J. D. Hillman, and A. S. Bleiweis. 1997. Genetic and physiologic analysis of a formyltetrahydrofolate synthetase mutant of Streptococcus mutans. J. Bacteriol. 179:1563-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Keyzer, J., C. van der Does, and A. J. Driessen. 2003. The bacterial translocase: a dynamic protein channel complex. Cell. Mol. Life Sci. 60:2034-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Driessen, A. J., P. Fekkes, and J. P. van der Wolk. 1998. The Sec system. Curr. Opin. Microbiol. 1:216-222. [DOI] [PubMed] [Google Scholar]
- 15.Forester, H., N. Hunter, and K. W. Knox. 1983. Characteristics of a high molecular weight extracellular protein of Streptococcus mutans. J. Gen. Microbiol. 129:2779-2788. [DOI] [PubMed] [Google Scholar]
- 16.Foucault, I., Y. C. Liu, A. Bernard, and M. Deckert. 2003. The chaperone protein 14-3-3 interacts with 3BP2/SH3BP2 and regulates its adapter function. J. Biol. Chem. 278:7146-7153. [DOI] [PubMed] [Google Scholar]
- 17.Francetic, O., M. P. Hanson, and C. A. Kumamoto. 1993. prlA suppression of defective export of maltose-binding protein in secB mutants of Escherichia coli. J. Bacteriol. 175:4036-4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freund, C., V. Dotsch, K. Nishizawa, E. L. Reinherz, and G. Wagner. 1999. The GYF domain is a novel structural fold that is involved in lymphoid signaling through proline-rich sequences. Nat. Struct. Biol. 6:656-660. [DOI] [PubMed] [Google Scholar]
- 19.Gilkes, N. R., B. Henrissat, D. G. Kilburn, R. C. Miller, Jr., and R. A. Warren. 1991. Domains in microbial β-1,4-glycanases: sequence conservation, function, and enzyme families. Microbiol. Rev. 55:303-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutierrez, J. A., P. J. Crowley, D. G. Cvitkovitch, L. J. Brady, I. R. Hamilton, J. D. Hillman, and A. S. Bleiweis. 1999. Streptococcus mutans ffh, a gene encoding a homologue of the 54 kDa subunit of the signal recognition particle, is involved in resistance to acid stress. Microbiology 145:357-366. [DOI] [PubMed] [Google Scholar]
- 21.Hajishengallis, G., T. Koga, and M. W. Russell. 1994. Affinity and specificity of the interactions between Streptococcus mutans antigen I/II and salivary components. J. Dent. Res. 73:1493-1502. [DOI] [PubMed] [Google Scholar]
- 22.Han, N., J. Whitlock, and A. Progulske-Fox. 1996. The hemagglutinin gene A (hagA) of Porphyromonas gingivalis 381 contains four large, contiguous, direct repeats. Infect. Immun. 64:4000-4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hannavy, K., G. C. Barr, C. J. Dorman, J. Adamson, L. R. Mazengera, M. P. Gallagher, J. S. Evans, B. A. Levine, I. P. Trayer, and C. F. Higgins. 1990. TonB protein of Salmonella typhimurium. A model for signal transduction between membranes. J. Mol. Biol. 216:897-910. [DOI] [PubMed] [Google Scholar]
- 24.Hartl, F. U., S. Lecker, E. Schiebel, J. P. Hendrick, and W. Wickner. 1990. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell 63:269-279. [DOI] [PubMed] [Google Scholar]
- 25.Heden, L. O., E. Frithz, and G. Lindahl. 1991. Molecular characterization of an IgA receptor from group B streptococci: sequence of the gene, identification of a proline-rich region with unique structure and isolation of N-terminal fragments with IgA-binding capacity. Eur. J. Immunol. 21:1481-1490. [DOI] [PubMed] [Google Scholar]
- 26.Hung, A. Y., and M. Sheng. 2002. PDZ domains: structural modules for protein complex assembly. J. Biol. Chem. 277:5699-5702. [DOI] [PubMed] [Google Scholar]
- 27.Igarashi, T., E. Asaga, and N. Goto. 2003. The sortase of Streptococcus mutans mediates cell wall anchoring of a surface protein antigen. Oral Microbiol. Immunol. 18:266-269. [DOI] [PubMed] [Google Scholar]
- 28.Katz, J., M. W. Russell, C. C. Harmon, G. P. Buckner, P. L. White, G. J. Richardson, and S. M. Michalek. 1995. Induction of salivary IgA responses to Streptococcus mutans antigen I/II after intranasal immunization. Adv. Exp. Med. Biol. 371B:1153-1156. [PubMed] [Google Scholar]
- 29.Kay, B. K., M. P. Williamson, and M. Sudol. 2000. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14:231-241. [PubMed] [Google Scholar]
- 30.Kelly, C. G., S. Todryk, H. L. Kendal, G. H. Munro, and T. Lehner. 1995. T-cell, adhesion, and B-cell epitopes of the cell surface Streptococcus mutans protein antigen I/II. Infect. Immun. 63:3649-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, Y. R., S. E. Lee, C. M. Kim, S. Y. Kim, E. K. Shin, D. H. Shin, S. S. Chung, H. E. Choy, A. Progulske-Fox, J. D. Hillman, M. Handfield, and J. H. Rhee. 2003. Characterization and pathogenic significance of Vibrio vulnificus antigens preferentially expressed in septicemic patients. Infect. Immun. 71:5461-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koepf, E. K., H. M. Petrassi, G. Ratnaswamy, M. E. Huff, M. Sudol, and J. W. Kelly. 1999. Characterization of the structure and function of W→ F WW domain variants: identification of a natively unfolded protein that folds upon ligand binding. Biochemistry 38:14338-14351. [DOI] [PubMed] [Google Scholar]
- 33.Kremer, B. H., M. van der Kraan, P. J. Crowley, I. R. Hamilton, L. J. Brady, and A. S. Bleiweis. 2001. Characterization of the sat operon in Streptococcus mutans: evidence for a role of Ffh in acid tolerance. J. Bacteriol. 183:2543-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumamoto, C. A., and J. Beckwith. 1985. Evidence for specificity at an early step in protein export in Escherichia coli. J. Bacteriol. 163:267-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 36.Larsen, R. A., G. E. Wood, and K. Postle. 1993. The conserved proline-rich motif is not essential for energy transduction by Escherichia coli TonB protein. Mol. Microbiol. 10:943-953. [DOI] [PubMed] [Google Scholar]
- 37.Lee, S. F., and T. L. Boran. 2003. Roles of sortase in surface expression of the major protein adhesin P1, saliva-induced aggregation and adherence, and cariogenicity of Streptococcus mutans. Infect. Immun. 71:676-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, S. F., A. Progulske-Fox, and A. S. Bleiweis. 1988. Molecular cloning and expression of a Streptococcus mutans major surface protein antigen, P1 (I/II), in Escherichia coli. Infect. Immun. 56:2114-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lehner, T., M. W. Russell, J. Caldwell, and R. Smith. 1981. Immunization with purified protein antigens from Streptococcus mutans against dental caries in rhesus monkeys. Infect. Immun. 34:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Linde, D., R. Volkmer-Engert, S. Schreiber, and J. P. Muller. 2003. Interaction of the Bacillus subtilis chaperone CsaA with the secretory protein YvaY. FEMS Microbiol. Lett. 226:93-100. [DOI] [PubMed] [Google Scholar]
- 41.Luirink, J., C. M. ten Hagen-Jongman, C. C. van der Weijden, B. Oudega, S. High, B. Dobberstein, and R. Kusters. 1994. An alternative protein targeting pathway in Escherichia coli: studies on the role of FtsY. EMBO J. 13:2289-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma, B., C. J. Tsai, and R. Nussinov. 2000. Binding and folding: in search of intramolecular chaperone-like building block fragments. Protein Eng. 13:617-627. [DOI] [PubMed] [Google Scholar]
- 43.Ma, J. K., M. Hunjan, R. Smith, C. Kelly, and T. Lehner. 1990. An investigation into the mechanism of protection by local passive immunization with monoclonal antibodies against Streptococcus mutans. Infect. Immun. 58:3407-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma, J. K., C. G. Kelly, G. Munro, R. A. Whiley, and T. Lehner. 1991. Conservation of the gene encoding streptococcal antigen I/II in oral streptococci. Infect. Immun. 59:2686-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macias, M. J., S. Wiesner, and M. Sudol. 2002. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 513:30-37. [DOI] [PubMed] [Google Scholar]
- 46.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40:1049-1057. [DOI] [PubMed] [Google Scholar]
- 47.Moisset, A., N. Schatz, Y. Lepoivre, S. Amadio, D. Wachsmann, M. Scholler, and J. P. Klein. 1994. Conservation of salivary glycoprotein-interacting and human immunoglobulin G-cross-reactive domains of antigen I/II in oral streptococci. Infect. Immun. 62:184-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muller, J. P., S. Bron, G. Venema, and J. M. van Dijl. 2000. Chaperone-like activities of the CsaA protein of Bacillus subtilis. Microbiology 146:77-88. [DOI] [PubMed] [Google Scholar]
- 49.Muller, J. P., J. Ozegowski, S. Vettermann, J. Swaving, K. H. Van Wely, and A. J. Driessen. 2000. Interaction of Bacillus subtilis CsaA with SecA and precursor proteins. Biochem. J. 348:367-373. [PMC free article] [PubMed] [Google Scholar]
- 50.Munro, G. H., P. Evans, S. Todryk, P. Buckett, C. G. Kelly, and T. Lehner. 1993. A protein fragment of streptococcal cell surface antigen I/II which prevents adhesion of Streptococcus mutans. Infect. Immun. 61:4590-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okahashi, N., C. Sasakawa, M. Yoshikawa, S. Hamada, and T. Koga. 1989. Cloning of a surface protein antigen gene from serotype c Streptococcus mutans. Mol. Microbiol. 3:221-228. [DOI] [PubMed] [Google Scholar]
- 52.Phillips, G. J., and T. J. Silhavy. 1992. The E. coli ffh gene is necessary for viability and efficient protein export. Nature 359:744-746. [DOI] [PubMed] [Google Scholar]
- 53.Pistor, S., T. Chakraborty, K. Niebuhr, E. Domann, and J. Wehland. 1994. The ActA protein of Listeria monocytogenes acts as a nucleator inducing reorganization of the actin cytoskeleton. EMBO J. 13:758-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Proft, T., H. Hilbert, G. Layh-Schmitt, and R. Herrmann. 1995. The proline-rich P65 protein of Mycoplasma pneumoniae is a component of the Triton X-100-insoluble fraction and exhibits size polymorphism in the strains M129 and FH. J. Bacteriol. 177:3370-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Russell, M. W., L. A. Bergmeier, E. D. Zanders, and T. Lehner. 1980. Protein antigens of Streptococcus mutans: purification and properties of a double antigen and its protease-resistant component. Infect. Immun. 28:486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Russell, M. W., and T. Lehner. 1978. Characterisation of antigens extracted from cells and culture fluids of Streptococcus mutans serotype c. Arch. Oral Biol. 23:7-15. [DOI] [PubMed] [Google Scholar]
- 57.Sato, Y., K. Okamoto, and H. Kizaki. 2002. gbpC and pac gene mutations detected in Streptococcus mutans strain GS-5. Oral Microbiol. Immunol. 17:263-266. [DOI] [PubMed] [Google Scholar]
- 58.Sciotti, M. A., I. Yamodo, J. P. Klein, and J. A. Ogier. 1997. The N-terminal half part of the oral streptococcal antigen I/IIf contains two distinct binding domains. FEMS Microbiol. Lett. 153:439-445. [DOI] [PubMed] [Google Scholar]
- 59.Seluanov, A., and E. Bibi. 1997. FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J. Biol. Chem. 272:2053-2055. [DOI] [PubMed] [Google Scholar]
- 60.Senpuku, H., T. Miyauchi, N. Hanada, and T. Nisizawa. 1995. An antigenic peptide inducing cross-reacting antibodies inhibiting the interaction of Streptococcus mutans PAc with human salivary components. Infect. Immun. 63:4695-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Signas, C., G. Raucci, K. Jonsson, P. E. Lindgren, G. M. Anantharamaiah, M. Hook, and M. Lindberg. 1989. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc. Natl. Acad. Sci. USA 86:699-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Southwick, F. S., and D. L. Purich. 1996. Intracellular pathogenesis of listeriosis. N. Engl. J. Med. 334:770-776. [DOI] [PubMed] [Google Scholar]
- 63.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 64.Tamura, H., T. Kikuchi, R. Shirato, and H. Kato. 2001. Cloning and DNA sequencing of the surface protein antigen I/II (PAa) of Streptococcus cricetus. FEMS Microbiol. Lett. 196:251-256. [DOI] [PubMed] [Google Scholar]
- 65.Timoney, J. F., J. Walker, M. Zhou, and J. Ding. 1995. Cloning and sequence analysis of a protective M-like protein gene from Streptococcus equi subsp. zooepidemicus. Infect. Immun. 63:1440-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Troffer-Charlier, N., J. Ogier, D. Moras, and J. Cavarelli. 2002. Crystal structure of the V-region of Streptococcus mutans antigen I/II at 2.4 Å resolution suggests a sugar preformed binding site. J. Mol. Biol. 318:179-188. [DOI] [PubMed] [Google Scholar]
- 68.Tzivion, G., Y. H. Shen, and J. Zhu. 2001. 14-3-3 proteins; bringing new definitions to scaffolding. Oncogene 20:6331-6338. [DOI] [PubMed] [Google Scholar]
- 69.Valent, Q. A., J. W. de Gier, G. von Heijne, D. A. Kendall, C. M. ten Hagen-Jongman, B. Oudega, and J. Luirink. 1997. Nascent membrane and presecretory proteins synthesized in Escherichia coli associate with signal recognition particle and trigger factor. Mol. Microbiol. 25:53-64. [DOI] [PubMed] [Google Scholar]
- 70.van Dolleweerd, C. J., D. Chargelegue, and J. K. Ma. 2003. Characterization of the conformational epitope of Guy's 13, a monoclonal antibody that prevents Streptococcus mutans colonization in humans. Infect. Immun. 71:754-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang, J., N. S. Tan, B. Ho, and J. L. Ding. 2002. Modular arrangement and secretion of a multidomain serine protease. Evidence for involvement of proline-rich region and N-glycans in the secretion pathway. J. Biol. Chem. 277:36363-36372. [DOI] [PubMed] [Google Scholar]
- 72.Yaffe, M. B., and A. E. Elia. 2001. Phosphoserine/threonine-binding domains. Curr. Opin. Cell Biol. 13:131-138. [DOI] [PubMed] [Google Scholar]
- 73.Zhang, P., C. Jespersgaard, L. Lamberty-Mallory, J. Katz, Y. Huang, G. Hajishengallis, and S. M. Michalek. 2002. Enhanced immunogenicity of a genetic chimeric protein consisting of two virulence antigens of Streptococcus mutans and protection against infection. Infect. Immun. 70:6779-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]