Abstract
Porphyromonas gingivalis is a gram-negative anaerobic bacterium that is considered the key etiologic agent of chronic periodontitis. Arg- and Lys-gingipain cysteine proteinases produced by P. gingivalis are key virulence factors and are believed to be essential for significant tissue component degradation, leading to host tissue invasion by periodontopathogens. Two in vitro models were used to determine the extent to which P. gingivalis can reach connective tissue. The tissue penetration potential of P. gingivalis was first investigated by using an engineered human oral mucosa model composed of normal human epithelial cells and fibroblasts. Internalized bacteria were assessed by transmission electron microscopy. Bacteria were observed within multilayered gingival epithelial cells and in the space between the stratified epithelium and the lamina propria. A gingipain-null mutant strain of P. gingivalis was found to be less potent in penetrating tissue than the wild-type strain. Proinflammatory responses to P. gingivalis infection were evaluated. P. gingivalis increased the secretion of interleukin-1β, interleukin-6, interleukin-8, and tumor necrosis factor alpha. In the second part of the study, the contribution of P. gingivalis gingipains to tissue penetration was investigated by using a reconstituted basement membrane model (Matrigel). The penetration of 14C-labeled P. gingivalis cells through Matrigel was significantly reduced when leupeptin, a specific inhibitor of Arg-gingipain activity, was added or when a gingipain-null mutant was used. The results obtained with these two relevant models support the capacities of P. gingivalis to infiltrate periodontal tissue and to modulate the proinflammatory response and suggest a critical role of gingipains in tissue destruction.
Periodontitis is an inflammatory disorder of the periodontium initiated by specific bacterial species and characterized by the destruction of supporting connective tissue and, in severe cases, the exfoliation of teeth. Porphyromonas gingivalis, a gram-negative anaerobic bacterium, has been identified as a major etiologic agent of chronic periodontitis (44). This bacterial species produces cysteine proteinases (gingipains) that can be cell bound or secreted (15, 49). Increasing numbers of reports have stressed the potential roles of Arg- and Lys-gingipains produced by P. gingivalis in the pathogenesis of periodontitis, since they can degrade a large variety of host proteins, resulting in tissue destruction and perturbation of host defenses (8, 15, 49). The critical role of gingipains in P. gingivalis pathogenicity is also supported by the fact that immunization with purified Arg-gingipain A or Arg-gingipain B protects against colonization and invasion by P. gingivalis in a mouse chamber model (7).
Invasion of mammalian epithelial cells is an important strategy developed by pathogenic bacteria to evade the host immune system and cause tissue damage (6). P. gingivalis has been observed in deep gingival tissue biopsy specimens, suggesting that it may pass through the epithelial barrier (9, 29, 37). Furthermore, in vitro studies have shown that P. gingivalis can infiltrate human transformed and primary cultures of gingival epithelial cells (5, 22, 24, 52) as well as multilayered pocket epithelial cells (40). However, the in vitro infiltration of connective tissue by P. gingivalis has never been investigated. In addition, few studies have evaluated the contribution of P. gingivalis gingipains to tissue invasion and destruction processes. The degradation of extracellular matrix components, such as fibronectin and collagens, by gingipains has been reported (42, 45). Moreover, gingipains indirectly contribute to tissue damage through the activation of latent host matrix metalloproteinases (MMPs) (4) and the inactivation of host proteinase inhibitors (10, 12). MMP-1 and MMP-9 can be activated by Lys-gingipain, while MMP-3 can be activated by Arg-gingipain (4). Purified gingipains also enhance significantly the synthesis of latent MMPs in rat mucosal epithelial cells and human fibroblasts (4). Furthermore, gingipains have been demonstrated to affect cytokine signaling networks (14, 25, 48) and to modulate the production of proinflammatory mediators (interleukin-1 [IL-1], IL-6, IL-8, and tumor necrosis factor alpha [TNF-α]), two phenomena that may initiate tissue destruction and alveolar bone resorption (1, 3).
All of the above studies suggest that P. gingivalis gingipains can facilitate the penetration of bacteria across the basement membrane barrier and promote the destruction of underlying tissues, leading to bacteremia and focal infections. The present study was aimed at evaluating the potential of P. gingivalis for tissue penetration and destruction by using two in vitro models, an engineered human oral mucosa and a reconstituted basement membrane. The critical contribution of gingipains to these penetration processes was also investigated.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. gingivalis ATCC 33277 and the derivative gingipain-null mutant KDP128 (rgpA rgpB kgp) were grown anaerobically (N2-H2-CO2 [80:10:10]) at 37°C for 24 h in Todd-Hewitt broth (BBL Microbiology Systems, Cockeysville, Md.) supplemented with vitamin K (1 μg/ml) and hemin (10 μg/ml). Mutant KDP128, kindly provided by K. Nakayama (Nagasaki University, Nagasaki, Japan), was constructed by using suicide plasmids as described previously (28, 43). To prevent the appearance of revertants and to ensure the correct genotype, tetracycline (0.7 μg/ml) and erythromycin (10 μg/ml) were added to culture plates for growing mutant KDP128. The phenotype of the triple mutant was confirmed prior to each experiment by testing its ability to cleave the chromogenic substrates for Arg-gingipain (benzoyl-Arg-p-nitroanilide) and Lys-gingipain (N-p-tosyl-glycine-proline-lysine-p-nitroanilide) as previously described (2).
Invasion assay for EHOM.
The in vitro engineered human oral mucosa (EHOM) model was prepared with primary fibroblasts and epithelial cells isolated from human palatal biopsy specimens from healthy patients according to a previously described procedure (35, 36). Engineered lamina propria was produced by mixing bovine skin type III collagen (5 mg/ml) (Sigma-Aldrich Canada Ltd., Oakville, Ontario, Canada) with normal human oral fibroblasts (1.5 × 106 cells) in Dulbecco’s modified Eagle (DME) medium (Flow Laboratories, Mississauga, Ontario, Canada). The mixture was poured into a petri dish (35-mm diameter) containing an anchor to prevent collagen contraction. Tissues were grown in DME medium supplemented with 100 IU of penicillin/ml, 25 μg of streptomycin/ml, 0.5 μg of amphotericin B (Fungizone)/ml, and 10% fetal calf serum and incubated in a 5% CO2 atmosphere. Four days later, the engineered lamina propria was seeded with oral epithelial cells (9 × 104/cm2) to obtain the EHOM. Tissues were grown in DME medium-Ham F-12 medium (Flow Laboratories) (3:1) (DMEH medium) until the epithelial cells reached confluence. EHOM then was raised to the air-liquid interface for an additional 5 days to allow the epithelium to stratify (36) prior to infection.
P. gingivalis was grown to late log phase, harvested by centrifugation (10,000 × g, 10 min), washed, and resuspended at final concentrations of 106 and 109 bacteria (ATCC 33277 or KPD128) per ml in DMEH medium without antibiotics, as determined by using a Petroff-Hausser counting chamber. Bacterial suspensions (100 μl) were placed on top of the EHOM in antibiotic-free DMEH medium containing growth factors and were incubated in an anaerobic chamber at 37°C. After 24 h of incubation, biopsy specimens were collected from infected and uninfected tissues and used for structural and ultrastructural analyses.
Optical microscopy.
Structural analyses of the EHOM tissues were performed as described by Rouabhia and Deslauriers (36). Briefly, EHOM biopsy specimens were fixed in Bouin's solution and then embedded in paraffin. Thin sections (5 μm) were prepared and stained with Masson trichome, mounted with 50% glycerol mounting medium, and viewed with a Nikon Elipse TS100 (Diagnostic Instruments Inc., Sterling Heights, Mich.) optical microscope.
Transmission electron microscopy.
Specimens were fixed with 4% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) for 20 min at room temperature. The samples were postfixed in 1% osmium tetraoxide prior to staining with 0.5% aqueous uranyl acetate at room temperature for 30 min. The samples then were dehydrated by using a graded ethanol series and were embedded in Epon resin. Ultrathin sections (1.5 μm) were stained with 3% citrate and examined with a JEOL 1200 transmission electron microscope.
Quantification of IL-1β, IL-6, IL-8, and TNF-α secreted by EHOM cells.
Culture media from 24-h infected or uninfected EHOM were collected and used to measure the levels of IL-1β, IL-6, IL-8, and TNF-α with enzyme-linked immunosorbent assay (ELISA) kits purchased from R&D Systems (Minneapolis, Minn.). Supernatants were filtered by using a 0.22-μm-pore-size filter and were used to quantify IL-1β, IL-6, IL-8, and TNF-α according to the manufacturer's instructions. Plates were read at 450 nm by using a microplate reader (model 680; Bio-Rad Laboratories Ltd., Mississauga, Ontario, Canada). The sensitivities of the commercial ELISA kits were 3.9 pg/ml for IL-1β, 4.5 pg/ml for IL-6, 7.8 pg/ml for IL-8, and 3.9 pg/ml for TNF-α, as specified by the manufacturer. Cytokine levels were compared by using the Student t test. Results presented as means and standard deviations (SDs) of three different experiments with two EHOM samples for each condition were considered significant when the P value was <0.05.
Radiolabeling of P. gingivalis cells.
Both strains of P. gingivalis (ATCC 33277 and KDP128) were radiolabeled by incubating mid-log-phase cultures (optical density at 660 nm, 0.5) with a mixture of 14C-labeled amino acids (Amersham Pharmacia Biotech, Baie d'Urfé, Quebec, Canada) at a final concentration of 20 μCi/ml at 37°C for 6 h in an anaerobic chamber. Cells were harvested by centrifugation at 10,000 × g for 10 min, washed three times in 100 mM phosphate-buffered saline (pH 7.2) (PBS), and suspended in PBS that had been reduced by overnight incubation in the anaerobic chamber. To determine the ratio of cells to disintegrations per minute, the bacterial suspensions (100 μl) were added to EcoLite scintillation liquid (ICN, Costa Mesa, Calif.) and counted by using a multipurpose scintillation counter (Beckman Coulter, Fullerton, Calif.). The concentration of each bacterial strain was evaluated by using a Petroff-Hausser counting chamber.
Basement membrane model (Matrigel).
Matrigel (Sigma-Aldrich Canada Ltd.) is a solubilized basement membrane preparation containing several proteins, including laminin, type IV collagen, and proteoglycans. Matrigel was diluted 1/3 in ice-cold PBS, and 100 μl was placed on 8-μm-pore-size filters in Transwell cell culture chamber inserts (Costar, Cambridge, Mass.). Matrigel was allowed to settle at 4°C for 30 min and then was gelled at 37°C for 24 h in an anaerobic chamber (N2-H2-CO2 [80:10:10]). Matrigel was rehydrated in 100 μl of sterile reduced PBS for 1 h at 37°C in the anaerobic chamber before the penetration assay was carried out, while 200 μl of PBS was placed in the lower chamber. Approximately 106 radiolabeled cells (ATCC 33277 or KDP128) in 100 μl of PBS were placed on top of the Matrigel in the double-chamber system, which was incubated in the anaerobic chamber at 37°C for 24 or 48 h. The migration of P. gingivalis cells through Matrigel was evaluated by measuring the radioactivity in the buffer recovered in the lower chamber.
The effect of selected proteinase inhibitors on the ability of P. gingivalis ATCC 33277 to migrate through Matrigel was investigated by adding the inhibitors to the bacterial cell suspensions 30 min before the penetration assay was carried out. N-α-p-Tosyl-l-lysine chloromethyl ketone (TLCK) (cysteine proteinase inhibitor), leupeptin (Arg-gingipain inhibitor), and cathepsin B inhibitor II (Lys-gingipain inhibitor) were used at 20 mM, 3.3 mM, and 30 μM, respectively. These concentrations were previously found to be effective (2). The assays were performed in triplicate, and the means and SDs were calculated. Statistical significance was evaluated by using the Student t test.
Degradation of basement membrane components.
Degradation of Matrigel was monitored by analyzing the degradation products by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) as described by Laemmli (21). P. gingivalis ATCC 33277 cells from a 10-h culture were harvested by centrifugation at 10,000 × g for 10 min, washed three times in reduced PBS, and suspended in the same buffer to an optical density at 660 nm of 0.5. Equal volumes of cell suspension and Matrigel (diluted 1/3 in PBS) were mixed together. Enzymatic digestions were carried out for 24 h at 37°C in the anaerobic chamber. Assays were also performed in the presence of the proteinase inhibitors listed above. Proteolysis was stopped by heating samples at 100°C for 10 min. Reaction mixtures were centrifuged (10,000 × g, 10 min) to remove cells, and equal volumes of supernatant and sampling buffer (0.125 M Tris-HCl buffer [pH 6.8], 2% SDS, 2% mercaptoethanol, 20% glycerol, 0.01% bromophenol blue) were mixed prior to heating at 100°C for 10 min. Samples (5 μl) then were subjected to electrophoresis with a 10% polyacrylamide resolving gel and a 4.5% polyacrylamide stacking gel. Protein bands were visualized by Coomassie brilliant blue R-250 staining. The gel was photographed, and the relative intensities of the bands were measured by using Scion Imaging Software (Scion Corporation).
RESULTS
Penetration of P. gingivalis through the EHOM.
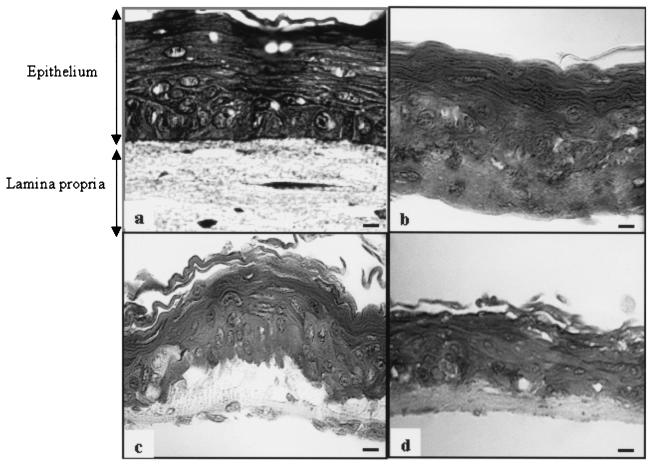
The infiltrative potential of P. gingivalis was first investigated by using an in vitro EHOM model composed of epithelial cells and fibroblasts obtained from a human biopsy specimen. Microscopic examination of the histological structures of the EHOM showed well-organized and stratified tissues with a structural organization similar to that of in vivo normal human oral mucosa (Fig. 1a). The EHOM incubated in the anaerobic chamber (80% N2- 10% CO2- 10% H2) for 24 h displayed no structural changes (Fig. 1b) and was visually identical to the EHOM kept in the incubator containing a 5% CO2 atmosphere. When the EHOM was infected with 109 P. gingivalis ATCC 33277 cells, significant structural modifications at the junction between the stratified epithelium and the lamina propria were observed after 24 h of incubation under anaerobic conditions (Fig. 1c). On the other hand, no visible histological changes were observed in the junction area of the EHOM infected with 106 P. gingivalis ATCC 33277 cells (data not shown) or with 109 P. gingivalis KDP128 cells (Fig. 1d). However, initiation of tissue desquamation, characterized by the presence of highly differentiated cells (large cells with faint nuclei and a large amount of cytoplasm), was observed in the basal layer of the EHOM infected with the gingipain-null mutant KDP128 (Fig. 1d).
FIG. 1.
Effect of P. gingivalis on EHOM structures revealed by optical microscopy. (a and b) Histological structures of sections of the control EHOM placed in a CO2 (a) or an anaerobic (b) atmosphere for 24 h. (c and d) Histological structures of sections of the EHOM infected with P. gingivalis ATCC 33277 (109 cells) (c) or P. gingivalis KDP128 (109 cells) (d) for 24 h. Biopsy specimens were collected and embedded in paraffin. Thin sections were stained with Masson trichome. Scale bars, 50 μm.
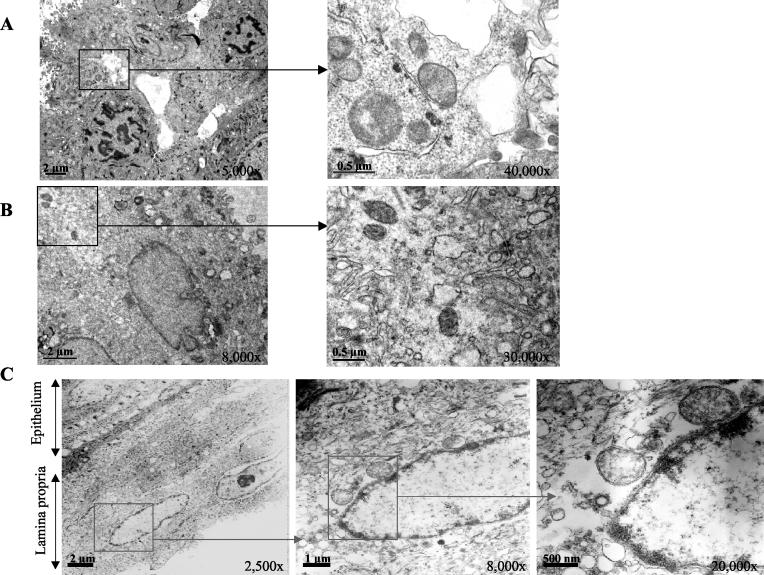
To confirm that cells of P. gingivalis migrated through the EHOM, ultrathin sections were examined by transmission electron microscopy (Fig. 2). Internalized P. gingivalis ATCC 33277 cells were observed within multilayered gingival epithelial cells as well as at the junction between the stratified epithelium and the lamina propria (Fig. 2A). As shown in Fig. 2C, infiltration of the basement membrane and connective tissue by P. gingivalis ATCC 33277 cells was also observed. Internalized P. gingivalis KPD128 cells were observed only in multilayered gingival epithelial cells (Fig. 2B), and no visible bacterial cells were detected in the basement membrane or in connective tissue.
FIG.2.
Effect of P. gingivalis on EHOM structures revealed by transmission electron microscopy. Shown are ultrathin sections of the EHOM infected with P. gingivalis ATCC 33277 (109 cells) or P. gingivalis KDP128 (109 cells). Cells of P. gingivalis ATCC 33277 were localized in multilayered epithelial cells (A) as well as in connective tissue (C), whereas cells of P. gingivalis KDP128 (B) were localized only in multilayered epithelial cells.
Secretion of cytokines by the EHOM in response to P. gingivalis infection.
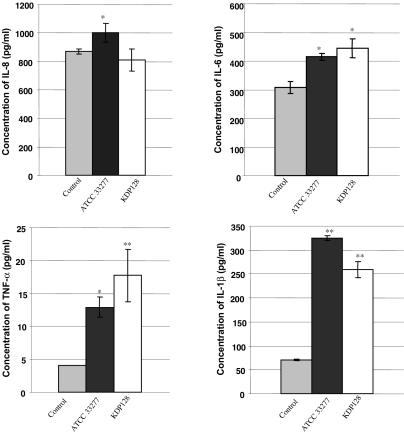
To evaluate the inflammatory response of the EHOM model to P. gingivalis infection, culture supernatants of the EHOM infected with 109 cells of P. gingivalis ATCC 33277 or KDP128 (gingipain-null mutant) were used to quantify four inflammatory cytokines: IL-1β, IL-6, IL-8, and TNF-α. Cytokine levels in supernatants from the EHOM infected with P. gingivalis ATCC 33277 were found to be higher than those in supernatants from uninfected EHOM (control) (Fig. 3). The same observations for IL-1β, IL-6, and TNF-α were noted for the EHOM infected with KDP128. For IL-8, the amount produced was comparable to that secreted by the control EHOM. P. gingivalis ATCC 33277 appeared to be more potent in inducing IL-1β secretion than the gingipain-null mutant KDP128 (324.4 ± 4.6 and 259 ± 16.7 pg/ml, respectively). In contrast, TNF-α levels tended to be higher in supernatants from the EHOM infected with KDP128 (17.7 ± 3.9 pg/ml) than in those from the EHOM infected with the wild-type strain (12.8 ± 1.5 pg/ml). Infection of EHOM with either P. gingivalis strain yielded comparable levels of IL-6 secretion.
FIG. 3.
Effect of P. gingivalis infection on IL-1β, IL-6, IL-8, and TNF-α secretion by the EHOM model. After 24 h of infection, culture supernatants were collected from EHOM samples, and cytokine levels were measured with ELISA kits. Results are reported as the means and SDs of three different EHOM experiments per condition. Single and double asterisks indicate P values of <0.05 and <0.01, respectively, for comparisons of infected and uninfected tissues.
Penetration of P. gingivalis through Matrigel and contribution of gingipains.
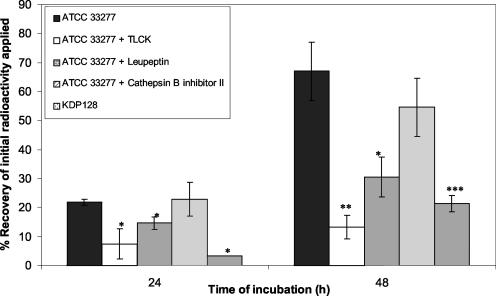
Matrigel was used to investigate the contribution of gingipains to the penetration of the basement membrane by P. gingivalis ATCC 33277. Bacterial cells were radiolabeled with 14C-labeled amino acids. Samples of approximately 50,000 dpm, corresponding to 106 14C-labeled bacteria, were initially applied on top of the Matrigel. As shown in Fig. 4, significant (P < 0.05) differences in penetration through Matrigel between untreated and treated (TLCK or leupeptin) 14C-labeled cells of P. gingivalis ATCC 33277 were observed after 24 or 48 h of incubation. However, the difference was more significant (P < 0.01) after 48 h of incubation with TLCK.
FIG. 4.
Comparative analysis of the penetration potential of P. gingivalis ATCC 33277 and its gingipain-null derivative mutant (KDP128) in the reconstituted basement membrane (Matrigel) model. 14C-labeled P. gingivalis cells were placed on top of the Matrigel, and incubation was carried out for 24 or 48 h. When the effect of proteinase inhibitors was tested, bacterial cells were preincubated in the presence of the inhibitors prior to initiation of the penetration assays. Radioactivity in the lower chamber was measured with a scintillation counter. The values represent the percent recovery of initial radioactivity added to the upper chamber and are reported as the means and SDs of nine assays. Single, double, and triple asterisks indicate significant differences at P values of <0.05, <0.01, and <0.002, respectively.
Phase-contrast microscopy observations of the contents of the lower chamber following P. gingivalis infection (48 h) revealed the presence of intact bacterial cells, whereas these were absent from assays with TLCK-pretreated P. gingivalis. TLCK, which inhibits gingipain activity, reduced the number of migrating bacteria recovered in the lower chamber by over 75% (48 h of incubation). The amount of radioactivity in the buffer recovered from the lower chamber following infection with P. gingivalis cells pretreated with TLCK may correspond to the leakage of radioactivity by 14C-labeled cells in the Matrigel during incubation. The small amount of buffer collected in the lower chamber in these experiments did not make it possible to accurately determine the proportions of free radioactivity versus P. gingivalis-associated radioactivity. A specific Arg-gingipain inhibitor (leupeptin) but not a Lys-gingipain inhibitor (cathepsin B inhibitor II) reduced the capacity of P. gingivalis ATCC 33277 to pass through Matrigel by 58%. Triple mutant KDP128, which is deficient in Arg-gingipains A and B and Lys-gingipain, exhibited a 3.5-fold-lower capacity to migrate through Matrigel than wild-type ATCC 33277 (Fig. 4).
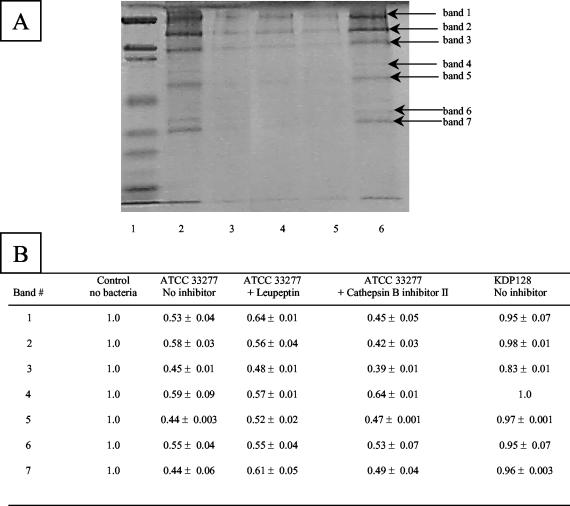
To establish a correlation between the abilities of P. gingivalis to degrade components of Matrigel and to migrate through Matrigel, ATCC 33277 and mutant KDP128 (rgpA rgpB kgp) cell suspensions were incubated with Matrigel. Protein degradation was monitored by SDS-PAGE and Coomassie blue staining. Wild-type ATCC 33277 completely hydrolyzed the protein constituents of Matrigel, whereas the gingipain-null mutant did not hydrolyze any of these proteins (Fig. 5). The contribution of each gingipain to the degradation of the proteins was further investigated with specific Arg- and Lys-gingipain inhibitors. Leupeptin (Arg-gingipain A and B inhibitor) was found to be more effective than cathepsin B inhibitor II (Lys-gingipain inhibitor) in inhibiting protein degradation (Fig. 5).
FIG. 5.
Degradation of Matrigel constituents by cells of P. gingivalis ATCC 33277 and KDP128, as determined by SDS-PAGE analysis and Coomassie blue staining. (A) Lanes: 1, molecular weight markers; 2, Matrigel alone; 3, Matrigel plus ATCC 33277; 4, Matrigel plus ATCC 33277 and leupeptin; 5, Matrigel plus ATCC 33277 and cathepsin B inhibitor II; 6, Matrigel plus KDP128 (rgpA rgpB kgp). (B) The relative intensities of bands were assessed by using Scion Imaging Software. A value of 1 was given to each band of the Matrigel control. Results are reported as means and SDs.
DISCUSSION
P. gingivalis, a bacterium associated with chronic periodontitis, has been observed within gingival tissues in vivo, suggesting that in addition to colonizing mucosal surfaces, it may also pass through the epithelial barrier (9, 37). While bacterial tissue invasion during periodontitis has not been extensively investigated, P. gingivalis can invade primary cultures of human gingival epithelial cells (24), cultured multilayered human pocket epithelial cells (40), and transformed human KB epithelial cells (5, 22). Most of these studies focused on bacterial-oral epithelial cell interactions and invasion. The lack of a relevant and reliable in vitro model has limited studies on the capacity of P. gingivalis to infiltrate tissues. In this study, we investigated tissue penetration by P. gingivalis by using a three-dimensional EHOM model composed of primary fibroblasts and epithelial cells isolated from normal human palatal biopsy specimens. This model has well-organized and stratified structures in which epithelial cells interact with fibroblasts in the lamina propria, leading to the secretion and deposition of basement membrane proteins (36). As the EHOM model exhibits the salient histological and functional features of oral mucosa, we used it to investigate bacterial tissue infiltration by P. gingivalis.
We demonstrated that P. gingivalis cells infiltrated multilayered epithelial cell structures. This finding is in agreement with previous observations reported by Sandros et al. (40), who used a multilayered epithelial cell model. Moreover, we showed that P. gingivalis could migrate through the basement membrane and reach the underlying connective tissue. This is the first in vitro demonstration of the infiltration of gingival tissue by P. gingivalis. Ultrastructural analyses showed that the infiltrating bacteria penetrated beneath the superficial cell layer and did not appear to be surrounded by an endosomal membrane, unlike previous observations reported by Sandros et al. (40) for gingival pocket epithelium. These findings suggest that the vacuolization of internalized P. gingivalis is likely restricted to the early steps of the invasion process. P. gingivalis, like a number of enteropathogenic bacteria, invades oral epithelial cells via endocytic pathways (26) and is capable of replicating intracellularly (22, 23, 26). In vitro studies on epithelial cell invasion involving the periodontal pathogen Actinobacillus actinomycetemcomitans and the enteropathogen Shigella flexneri indicated that bacteria can escape vacuoles by rapid efficient lysis of the phagocytic vacuole, a phenomenon that has a strong correlation with the ability of the bacteria to replicate freely in the cytoplasm (27, 41). Although internalized dividing P. gingivalis cells were not observed in epithelial cells, it is attempting to speculate that after 24 h of incubation in the anaerobic chamber, the replication of P. gingivalis is likely to occur because the EHOM model may provide nutrients for bacterial growth. This hypothesis is supported by previous studies which demonstrated the replication of P. gingivalis in a monolayered epithelial cell system (22, 26). Our electron microscopic observations showing internalized P. gingivalis not surrounded by an endosomal membrane would tend to support the notion that the intracellular survival of P. gingivalis is similar to that of other pathogens.
Histological analyses of EHOM sections following infection with P. gingivalis ATCC 33277 showed that the tissue had undergone structural modifications. The space between the stratified epithelium and the lamina propria may reflect detachment of cells from the basement membrane, suggesting possible degradation of the intercellular matrix by P. gingivalis. Cells of the stratified epithelium are normally bound to extracellular proteins, while basal cells are normally bound to proteins making up the basement membrane. In our EHOM model, epithelial cells interacted with fibroblasts in the lamina propria by secreting basement membrane proteins (laminins B1, B2, and 5) and by expressing β1 and α2β1 integrins (36). Recent studies demonstrated that P. gingivalis interferes with both cell-cell and cell-matrix adhesion involving oral keratinocytes by degrading junction proteins, such as cell-extracellular matrix junction transmembrane protein β1 integrin (13, 17). Moreover, the adhesion of oral keratinocytes to laminin 5 was found to decrease by more than 50% when cells were infected with P. gingivalis (13). The ability of certain pathogens to breach the epithelial cell barrier is well established as a virulence-associated mechanism that ensures a minimal exposure of bacteria to the extracellular environment (20).
Gingipains of P. gingivalis were recently implicated in the proteolysis of junction proteins and were suggested to contribute significantly to bacterial tissue penetration (17, 18). Gene inactivation of Arg- or Lys-gingipain result in a decreased ability of P. gingivalis to bind to epithelial cells and the extracellular matrix (28) as well as in a reduction in the colonization and pathogenic properties of the bacteria (8). We propose that the EHOM structural modifications observed following P. gingivalis ATCC 33277 infection can be attributed to gingipain-associated proteolysis of proteins involved in epithelial cell and extracellular matrix interactions, a phenomenon that may promote bacterial connective tissue infiltration. This hypothesis is supported by our histological observations of the EHOM infected with gingipain-null mutant KDP128, which revealed no deeper tissue structural changes. This finding is in accordance with the results of previous in vivo studies which demonstrated that an Arg- or Lys-gingipain-deficient mutant of P. gingivalis induced a significantly smaller abscess in a mouse chamber model than did the wild-type strain, whereas a gingipain-null mutant failed to induce lesion formation (54). Moreover, in our study, KDP128 was found to be ineffective in infiltrating connective tissue, in agreement with the results of Park and Lamont (32), who reported that an Arg- or Lys-gingipain-deficient mutant was 10-fold less invasive than the parent strain.
P. gingivalis cysteine proteinase gingipains are important virulence factors that contribute to the pathogenesis of periodontitis and degrade extracellular matrix proteins, such as laminin, fibronectin, and type IV collagen (11, 33, 51). We extended studies on the direct contribution of P. gingivalis gingipains to the degradation of extracellular matrix proteins by using an in vitro reconstituted basement membrane model (Matrigel). P. gingivalis ATCC 33277 degraded purified laminin, fibronectin, and type IV collagen as well as Matrigel protein constituents. Our study provides evidence of the importance and direct involvement of gingipain catalytic activities in the degradation of extracellular matrix proteins. Moreover, using a gingipain-null mutant and specific gingipain inhibitors, we demonstrated that gingipain activities were involved in the ability of P. gingivalis to penetrate an in vitro reconstituted basement membrane model. These results suggest that Arg-gingipain and, to a lesser extent, Lys-gingipain can promote the penetration of P. gingivalis through Matrigel by degrading extracellular matrix components.
The host response to bacterial infections can also lead to tissue damage. Proinflammatory cytokines (IL-1β, IL-6, IL-8, and TNF-α) can be induced by several components of periodontopathogens (53). Excessive production of these cytokines in inflamed periodontal tissues was proposed to be responsible for the progression of periodontitis (31). Indeed, high levels of IL-1β, IL-6, IL-8, and TNF-α are detected in gingival crevicular fluid from patients with periodontitis (34, 46, 50). Cytokines and inflammatory mediators in turn promote the release of tissue-derived enzymes, the MMPs, which are destructive for the extracellular matrix and bone (30). To assess the inflammatory response to P. gingivalis infection, we measured IL-1β, IL-6, IL-8, and TNF-α secretion in culture supernatants of EHOM infected with P. gingivalis ATCC 33277 or KDP128. Our results revealed that P. gingivalis induces cytokine responses in our model, in accordance with the results reported by Sandros et al. (39), who demonstrated that P. gingivalis can induce a strong cytokine response in primary cultures of pocket epithelium. The induction of cytokine expression following P. gingivalis infection was also reported for a mouse model (19).
Our results also showed that a gingipain-null mutant induced quantitavely different levels of proinflammatory cytokines than did wild-type strain ATCC 33277. A variety of bacterial components, including lipopolysaccharides and fimbriae, are known to be potent inducers of cytokine synthesis in human epithelial cells (47). Inactivation of gingipain genes has been reported to modulate the pathophysiological properties of P. gingivalis strains, since these enzymes contribute to the processing and/or maturation of various cell surface proteins of P. gingivalis (16). For example, gene inactivation of Arg-gingipain results in attenuation of the expression of major fimbriae, which ultimately affects the ability of P. gingivalis to bind to epithelial cells as well as extracellular matrix proteins (21). Thus, the differences observed between P. gingivalis ATCC 33277 and KDP128 in their abilities to induce cytokine release in the EHOM model may be related to their different phenotypic properties rather than simply to the presence or absence of gingipain molecules. This hypothesis is supported by the study of Sandros et al. (39), which suggested that the adhesive or invasive ability of P. gingivalis used to infect epithelial cells was positively correlated with the cytokine response. However, one should not exclude the possibility that the high level of proteolytic activity exhibited by the parent strain (ATCC 33277) might be responsible for the differences observed in cytokine responses. Gingipains have been implicated not only in the induction of cytokine synthesis locally but also in the degradation of exogenous cytokines (1, 3). Further studies will investigate the kinetics of cytokine production following infection of the EHOM with P. gingivalis.
Our results confirm and extend previous investigations demonstrating that P. gingivalis can infiltrate multilayered epithelial cells. Moreover, this is the first study reporting the ability of P. gingivalis to reach the underlying connective tissue. These observations are in agreement with previous in vivo observations of P. gingivalis in the junctional epithelium and connective tissue surrounding periodontal lesions (38). Our results also suggest the critical contribution of gingipains in gingival tissue destruction and penetration by P. gingivalis. Proteolytic activities presumably affect the epithelial barrier by damaging epithelial cells and loosening the epithelial tissue from the basement membrane. These factors may be important in facilitating the penetration of the space between epithelial cells and the lamina propria by P. gingivalis. However, the degradation of extracellular matrix proteins involves complex interactions between bacterial and host cells, and a contribution of host proteinases to tissue destruction cannot be ruled out. Future studies with the EHOM model, which allows host responses to bacterial infiltration to be taken into consideration, will address the contribution of P. gingivalis to MMP production and activation.
Acknowledgments
We thank Yakout Mostefaoui (Université Laval) for expert technical assistance. We also thank Koji Nakayama (Nagasaki University) for providing the gingipain-null mutant of P. gingivalis.
This work was supported by the Canadian Institutes of Health Research and the Fonds de la Recherche en Santé du Québec.
Editor: V. J. DiRita
REFERENCES
- 1.Banbula, A., M. Bugno, A. Kuster, P. C. Heinrich, J. Travis, and J. Potempa. 1999. Rapid and efficient inactivation of IL-6 gingipains, lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Biochem. Biophys. Res. Commun. 261:598-602. [DOI] [PubMed] [Google Scholar]
- 2.Brochu, V., D. Grenier, K. Nakayama, and D. Mayrand. 2001. Acquisition of iron from human transferrin by Porphyromonas gingivalis: a role for Arg- and Lys-gingipain activities. Oral Microbiol. Immunol. 16:79-87. [DOI] [PubMed] [Google Scholar]
- 3.Calkins, C. C., K. Platt, J. Potempa, and J. Travis. 1998. Inactivation of tumor necrosis factor-alpha by proteinases (gingipains) from the periodontal pathogen, Porphyromonas gingivalis. Implications of immune evasion. J. Biol. Chem. 273:6611-6614. [DOI] [PubMed] [Google Scholar]
- 4.DeCarlo, A. A., Jr., L. J. Windsor, M. K. Bodden, G. J. Harber, B. Birkedal-Hansen, and H. Birkedal-Hansen. 1997. Activation and novel processing of matrix metalloproteinases by a thiol-proteinase from the oral anaerobe Porphyromonas gingivalis. J. Dent. Res. 76:1260-1270. [DOI] [PubMed] [Google Scholar]
- 5.Duncan, M. J., S. Nakao, Z. Skobe, and H. Xie. 1993. Interactions of Porphyromonas gingivalis with epithelial cells. Infect. Immun. 61:2260-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falkow, S. 1991. Bacterial entry into eukaryotic cells. Cell 65:1099-1102. [DOI] [PubMed] [Google Scholar]
- 7.Genco, C. A., B. M. Odusanya, J. Potempa, J. Mikolajczyk-Pawlinska, and J. Travis. 1998. A peptide domain on gingipain R which confers immunity against Porphyromonas gingivalis infection in mice. Infect. Immun. 66:4108-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genco, C. A., J. Potempa, J. Mikolajczyk-Pawlinska, and J. Travis. 1999. Role of gingipain R in the pathogenesis of Porphyromonas gingivalis-mediated periodontal disease. Clin. Infect. Dis. 28:456-465. [DOI] [PubMed] [Google Scholar]
- 9.Gibson, W. A., and I. L. Shannon. 1965. Simulation with carbon particles of bacterial infiltration of human gingival tissues. Periodontics 59:57-59. [PubMed] [Google Scholar]
- 10.Grenier, D. 1996. Degradation of host protease inhibitors and activation of plasminogen by proteolytic enzymes from Porphyromonas gingivalis and Treponema denticola. Microbiology 142:955-961. [DOI] [PubMed] [Google Scholar]
- 11.Grenier, D., and D. Mayrand. 1993. Proteinases, p. 227-243. In H. N Shah, D. Mayrand, and R. J. Genco (ed.), Biology of the species Porphyromonas gingivalis. CRC Press, Inc., Boca Raton, Fla.
- 12.Grenier, D., and D. Mayrand. 2001. Inactivation of tissue inhibitor of metalloproteinases-1 (TIMP-1) by Porphyromonas gingivalis. FEMS Microbiol. Lett. 203:161-164. [DOI] [PubMed] [Google Scholar]
- 13.Hintermann, E., S. K. Haake, U. Christen, A. Sharabi, and V. Quaranta. 2002. Discrete proteolysis of focal contact and adherence junction components in Porphyromonas gingivalis-infected oral keratinocytes: a strategy for cell adhesion and migration disabling. Infect. Immun. 70:5846-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, G. T., D. Kim, J. K. Lee, H. K. Kuramitsu, and S. K. Haake. 2001. Interleukin-8 and intercellular adhesion molecule 1 regulation in oral epithelial cells by selected periodontal bacteria: multiple effects of Porphyromonas gingivalis via antagonistic mechanisms. Infect. Immun. 69:1364-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadowaki, T., K. Nakayama, K. Okamoto, N. Abe, A. Baba, Y. Shi, D. B. Ratnayake, and K. Yamamoto. 2000. Porphyromonas gingivalis proteinases as virulence determinants in progression of periodontal diseases. J. Biochem. 128:153-159. [DOI] [PubMed] [Google Scholar]
- 16.Kadowaki, T., K. Nakayama, F. Yoshimura, K. Okamoto, N. Abe, and K. Yamamoto. 1998. Arg-gingipain acts as a major processing enzyme for various cell surface proteins in Porphyromonas gingivalis. J. Biol. Chem. 273:29072-29076. [DOI] [PubMed] [Google Scholar]
- 17.Katz, J., V. Sambandam, J. H. Wu, S. M. Michalek, and D. F. Balkovetz. 2000. Characterization of Porphyromonas gingivalis-induced degradation of epithelial cell junctional complexes. Infect. Immun. 68:1441-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katz, J., Q. B. Yang, P. Zhang, J. Potempa, J. Travis, S. M. Michalek, and D. F. Balkovetz. 2002. Hydrolysis of epithelial junctional proteins by Porphyromonas gingivalis gingipains. Infect. Immun. 70:2512-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kesavalu, L., B. Chandrasekar, and J. L. Ebersole. 2002. In vivo induction of proinflammatory cytokines in mouse tissue by Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans. Oral Microbiol. Immunol. 17:177-180. [DOI] [PubMed] [Google Scholar]
- 20.Labrec, E. H., H. Schneider, T. J. Magnani, and S. B. Formal. 1964. Epithelial cell penetration is an essential step in the pathogenesis of bacterial dysentery. J. Bacteriol. 88:1503-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lamont, R. J., A. Chan, C. M. Belton, K. T. Izutsu, D. Vasel, and A. Weinberg. 1995. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 63:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamont, R. J., D. Oda, R. E. Persson, and G. R. Persson. 1992. Interaction of Porphyromonas gingivalis with gingival epithelial cells maintained in culture. Oral Microbiol. Immunol. 7:364-367. [DOI] [PubMed] [Google Scholar]
- 25.Lourbakos, A., J. Potempa, J. Travis, M. R. D'Andrea, P. Andrade-Gordon, R. Santulli, E. J. Mackie, and R. N. Pike. 2001. Arginine-specific protease from Porphyromonas gingivalis activates protease-activated receptors on human oral epithelial cells and induces interleukin-6 secretion. Infect. Immun. 69:5121-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madianos, P. N., P. N. Papapanou, U. Nannmark, G. Dahlen, and J. Sandros. 1996. Porphyromonas gingivalis FDC381 multiplies and persists within human oral epithelial cells in vitro. Infect. Immun. 64:660-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer, D. H., J. E. Lippmann, and P. M. Fives-Taylor. 1996. Invasion of epithelial cells by Actinobacillus actinomycetemcomitans: a dynamic, multistep process. Infect. Immun. 64:2988-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama, K., T. Kadowaki, K. Okamoto, and K. Yamamoto. 1995. Construction and characterization of arginine-specific cysteine proteinase (Arg-gingipain)-deficient mutants of Porphyromonas gingivalis. Evidence for significant contribution of Arg-gingipain to virulence. J. Biol. Chem. 270:23619-23626. [DOI] [PubMed] [Google Scholar]
- 29.Njoroge, T., R. J. Genco, H. T. Sojar, N. Hamada, and C. A. Genco. 1997. A role for fimbriae in Porphyromonas gingivalis invasion of oral epithelial cells. Infect. Immun. 65:1980-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Offenbacher, S. 1996. Periodontal diseases: pathogenesis. Ann. Periodontol. 1:821-878. [DOI] [PubMed] [Google Scholar]
- 31.Okada, H., and S. Murakami. 1998. Cytokine expression in periodontal health and disease. Crit. Rev. Oral Biol. Med. 9:248-266. [DOI] [PubMed] [Google Scholar]
- 32.Park, Y., and R. J. Lamont. 1998. Contact-dependent protein secretion in Porphyromonas gingivalis. Infect. Immun. 66:4777-4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potempa, J., A. Banbula, and J. Travis. 2000. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontology 24:153-192. [DOI] [PubMed] [Google Scholar]
- 34.Reinhardt, R. A., M. P. Masada, W. B. Kaldahl, L. M. DuBois, K. S. Kornman, J. I. Choi, K. L. Kalkwarf, and A. C. Allison. 1993. Gingival fluid IL-1 and IL-6 levels in refractory periodontitis. J. Clin. Periodontol. 20:225-231. [DOI] [PubMed] [Google Scholar]
- 35.Rouabhia, M. 1996. In vitro production and transplantation of immunologically active skin equivalents. Lab. Investig. 75:503-517. [PubMed] [Google Scholar]
- 36.Rouabhia, M., and N. Deslauriers. 2002. Production and characterization of an in vitro engineered human oral mucosa. Biochem. Cell Biol. 80:189-195. [DOI] [PubMed] [Google Scholar]
- 37.Saglie, F. R., M. T. Resende, F. Carranza, Jr., and M. G. Newman. 1988. Intragingival presence of microorganisms associated with grade I active destructive periodontal lesions. Av. Odontoestomatol. 4:385-390. (In Spanish.) [PubMed] [Google Scholar]
- 38.Saglie, F. R., C. T. Smith, M. G. Newman, F. A. Carranza, Jr., J. H. Pertuiset, L. Cheng, E. Auil, and R. J. Nisengard. 1986. The presence of bacteria in the oral epithelium in periodontal disease. II. Immunohistochemical identification of bacteria. J. Periodontol. 57:492-500. [DOI] [PubMed] [Google Scholar]
- 39.Sandros, J., C. Karlsson, D. F. Lappin, P. N. Madianos, D. F. Kinane, and P. N. Papapanou. 2000. Cytokine responses of oral epithelial cells to Porphyromonas gingivalis infection. J. Dent. Res. 79:1808-1814. [DOI] [PubMed] [Google Scholar]
- 40.Sandros, J., P. N. Papapanou, U. Nannmark, and G. Dahlen. 1994. Porphyromonas gingivalis invades human pocket epithelium in vitro. J. Periodontal Res. 29:62-69. [DOI] [PubMed] [Google Scholar]
- 41.Sansonetti, P. J., A. Ryter, P. Clerc, A. T. Maurelli, and J. Mounier. 1986. Multiplication of Shigella flexneri within HeLa cells: lysis of the phagocytic vacuole and plasmid-mediated contact hemolysis. Infect. Immun. 51:461-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scragg, M. A., S. J. Cannon, M. Rangarajan, D. M. Williams, and M. A. Curtis. 1999. Targeted disruption of fibronectin-integrin interactions in human gingival fibroblasts by the RI protease of Porphyromonas gingivalis W50. Infect. Immun. 67:1837-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi, Y., D. B. Ratnayake, K. Okamoto, N. Abe, K. Yamamoto, and K. Nakayama. 1999. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J. Biol. Chem. 274:17955-17960. [DOI] [PubMed] [Google Scholar]
- 44.Slots, J., and M. Ting. 1999. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in human periodontal disease: occurrence and treatment. Periodontol. 2000. 20:82-121. [DOI] [PubMed] [Google Scholar]
- 45.Smalley, J. W., A. J. Birss, and C. A. Shuttleworth. 1988. The degradation of type I collagen and human plasma fibronectin by the trypsin-like enzyme and extracellular membrane vesicles of Bacteroides gingivalis W50. Arch. Oral Biol. 33:323-329. [DOI] [PubMed] [Google Scholar]
- 46.Stashenko, P., J. J. Jandinski, P. Fujiyoshi, J. Rynar, and S. S. Socransky. 1991. Tissue levels of bone resorptive cytokines in periodontal disease. J. Periodontol. 62:504-509. [DOI] [PubMed] [Google Scholar]
- 47.Sugiyama, A., A. Uehara, K. Iki, K. Matsushita, R. Nakamura, T. Ogawa, S. Sugawara, and H. Takada. 2002. Activation of human gingival epithelial cells by cell-surface components of black-pigmented bacteria: augmentation of production of interleukin-8, granulocyte colony-stimulating factor and granulocyte-macrophage colony-stimulating factor and expression of intercellular adhesion molecule 1. J. Med. Microbiol. 51:27-33. [DOI] [PubMed] [Google Scholar]
- 48.Tada, H., S. Sugawara, E. Nemoto, N. Takahashi, T. Imamura, J. Potempa, J. Travis, H. Shimauchi, and H. Takada. 2002. Proteolysis of CD14 on human gingival fibroblasts by arginine-specific cysteine proteinases from Porphyromonas gingivalis leading to down-regulation of lipopolysaccharide-induced interleukin-8 production. Infect. Immun. 70:3304-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Travis, J., R. Pike, T. Imamura, and J. Potempa. 1997. Porphyromonas gingivalis proteinases as virulence factors in the development of periodontitis. J. Periodontal Res. 32:120-125. [DOI] [PubMed] [Google Scholar]
- 50.Tsai, C. C., Y. P. Ho, and C. C. Chen. 1995. Levels of interleukin-1 beta and interleukin-8 in gingival crevicular fluids in adult periodontitis. J. Periodontol. 66:852-859. [DOI] [PubMed] [Google Scholar]
- 51.Uitto, V. J., M. Haapasalo, T. Laakso, and T. Salo. 1988. Degradation of basement membrane collagen by proteases from some anaerobic oral micro-organisms. Oral Microbiol. Immunol. 3:97-102. [DOI] [PubMed] [Google Scholar]
- 52.Weinberg, A., C. M. Belton, Y. Park, and R. J. Lamont. 1997. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 65:313-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson, M., K. Reddi, and B. Henderson. 1996. Cytokine-inducing components of periodontopathogenic bacteria. J. Periodontal Res. 31:393-407. [DOI] [PubMed] [Google Scholar]
- 54.Yoneda, M., T. Hirofuji, H. Anan, A. Matsumoto, T. Hamachi, K. Nakayama, and K. Maeda. 2001. Mixed infection of Porphyromonas gingivalis and Bacteroides forsythus in a murine abscess model: involvement of gingipains in a synergistic effect. J. Periodontal Res. 36:237-243. [DOI] [PubMed] [Google Scholar]