Abstract
Omalizumab (Xolair®) is a recombinant humanized monoclonal antibody that selectively binds to human immunoglobulin E (IgE). Omalizumab is used to treat IgE-mediated diseases such as chronic idiopathic urticaria (CIU) and moderate to severe allergic asthma. In pre-marketing clinical trials in patients with asthma, anaphylaxis was reported in 3 of 3,507 (0.1%) patients. In post-marketing spontaneous reports, the frequency of anaphylaxis attributed to omalizumab use was estimated to be at least 0.2% of patients based on an estimated exposure of about 57,300 patients from June 2003 through December 2006. To better understand the risk of anaphylaxis in patients with allergic asthma receiving omalizumab, a post-marketing pharmacosurveillance study was initiated in 2009. As part of this study, an assay was developed to detect antibodies of IgE isotype to omalizumab. Serum samples from patients in the study were evaluated using this assay. Our results indicated that there was no observable correlation between either anaphylaxis or skin test reactivity and the presence of antibodies of IgE isotype to omalizumab. Here, we discuss the development of this assay as well as the results of the immunogenicity assessment.
Key Words: biotin-mutant omalizumab-AAA, DIG-FcεR1-IgG, IgE isotype, omalizumab (Xolair®), omalizumab/total IgE molar ratio
INTRODUCTION
Omalizumab (Xolair®) is a recombinant humanized monoclonal antibody (mAb) that was designed to treat allergic asthma and chronic idiopathic urticaria (CIU) by selectively binding to human immunoglobulin E (IgE). The United States Food and Drug Administration (FDA) approved omalizumab in June 2003; approval was granted in Europe by the European Medicines Agency (EMA) in 2005. Its use was initially limited to the treatment of moderate to severe persistent asthma in adults and adolescents aged 12 years or older who had a positive skin test or in vitro reactivity to a perennial aeroallergen and whose symptoms were inadequately controlled by inhaled corticosteroids. More recently, omalizumab received approval by the FDA for treatment of CIU (March 2014).
Type I hypersensitivity reactions to omalizumab administration have been reported at a frequency of 0.1% in clinical trials (n = 3/3,507). Similar observations were made through post-marketing pharmacosurveillance. Through the end of 2006, a total of 124 anaphylaxis cases associated with omalizumab administration had been reported in post-marketing spontaneous reports of anaphylactic reactions (1). The responses met the clinical definition of anaphylaxis established by Sampson et al. (2,3). The reporting rate of anaphylaxis based on the 124 cases was at least 0.2% from the time of initial marketing through 2006 (n = 124/57,269) (1). The risk factors for these anaphylactic reactions were uncertain given the limitations of spontaneous reports and the lack of control data (1). Due to this increase in the rate of anaphylaxis, a black box warning was added to the omalizumab label in 2007.
To understand the risk factors associated with anaphylaxis among omalizumab-treated patients, a pharmacosurveillance data repository (Q4458g, X-PAND) was initiated in early 2009 as a post-marketing commitment. The objective of this study was to collect data from patients treated with omalizumab who showed or lacked hypersensitivity reactions. Data from this study was intended to assess the risk factors for anaphylaxis among the patient population treated with omalizumab. Data collected included clinical histories, immunogenicity assessment, and an optional allergy skin test (unpublished data). As part of this study, an assay that could detect antibodies of IgE isotype to omalizumab was developed. The purpose was to evaluate the potential correlation of anaphylaxis to the presence of IgE antibodies to omalizumab. The study was completed with the final database locked in April 2014 (Genentech, Inc., data on file). In addition to the assay described above, all samples were also evaluated for antibodies of an immunoglobulin G (IgG) isotype to omalizumab. To ensure assay quality, the assay criteria were pre-specified based on available data at the time. The target sensitivity was set at 0.1 IU/mL to ensure high sensitivity based on data from food allergy reactions. The total IgE tolerance was required to be ≥700 IU/mL as 700 IU/mL is the highest total IgE treatment level on the omalizumab dosing table. The omalizumab tolerance was set to 50–100 ng/mL based on the doses that were given and the specified washout periods. To accomplish these criteria, we designed a novel strategy that required the use of custom designed reagents. Here, we describe the development and characterization of the assay system as well as the final antibody assay that was developed and qualified to detect the IgE anti-therapeutic antibody (ATA) directed against omalizumab.
MATERIALS AND METHODS
Construction of Triple Mutants of Omalizumab (Mutant Omalizumab-AAA)
Three alanine substitutions were made in the light chain complementarity-determining region 1 (CDR-1) region of omalizumab. The substitutions were introduced using Kunkel mutagenesis as described elsewhere (4). A single oligonucleotide was designed to encompass all the substitutions (5′-GATACCAGTTCATGTAGCTGGCACCAGCGTAGGCG ACGCTCTGACTGGCAC-3′) at positions D30A, D32A, and D34A (Genentech Oligonucleotide Synthesis Group). The light chain plasmid containing the three mutations was combined with the omalizumab heavy chain IgG1 plasmid and transfected into the 293s human embryonic kidney cell line (Genentech Cell Banking Group) for protein expression.
Construction of Anti-Omalizumab IgE-Positive Controls
The anti-omalizumab IgE-positive control was constructed by ligating the cDNA encoding the variable heavy chain of a murine antibody specific for the CDR of omalizumab into a human IgE expression plasmid. In order to perform the ligation, cloning sites were added to the cDNA of the omalizumab-specific murine antibody. An EcoRI endonuclease restriction site was introduced by PCR using a 5′-forward oligonucleotide that encoded an EcoRI site. This step was followed by a mammalian signal sequence and a 20-base pair overhang with the N-terminal mature sequence of the omalizumab-specific murine antibody (5′-CTATCGATTGAATTCCACCATGGGATGGTCATGTATCATCCTTTTTCTAGTAGCAACTGC AACTGGAGTACATTCACAGGTTCAGCTGCAGCAGTC-3′) and a 3′-reverse oligonucleotide that introduced a Xho1 site at the 3′-end of the variable heavy region (5′-GATGGGGGTGTCGTTTTGGCACTCGAGACGGTGACTGTGGTTCC-3′). A similar format was used to introduce 5′-EcoR1 and 3′-Xho1 cloning ligation sites into the cDNA of the omalizumab-specific murine antibody variable light region cDNA (EcoR1-forward 5′-CTATCGATTGAATTCCACCATGGGATGGTCATGTATCATCCTTTTTCTAGTAGCAACTGCAACTGGAGTACATTCACAAATTGTTATCACCCAGTCTC-3′ and Xho1-reverse 5′-CCGTTTTATCTCGAGCTTTGTCCCCGAGCCGAAC-3′). The PCR products for both the heavy and light chains were digested with EcoR1 and XhoI and gel purified. The mammalian IgE expression vector was also digested with EcoR1 and XhoI and gel purified. Purified PCR fragments and vectors were combined and ligated overnight using T4 DNA ligase (New England Biolabs, Cat. No. 0202S). The plasmid for anti-omalizumab IgE was used for transient expression (Roche Diagnostics, Fugene 6) of anti-omalizumab IgE from transfected 293s cells.
Patient Samples
The objective was to collect samples from 30 cases (patients who experienced anaphylaxis upon treatment with omalizumab) with four controls for each case (120 controls) over 4 years. Case patients were to undergo a 16-week to 18-month washout period before sample collection, and at least one out of four control patients also needed to undergo a washout. Collection of samples with a proper washout period was a major logistical challenge limiting the number of samples that were eligible for analysis.
Screening Assay
The screening assay used biotin-conjugated mutant omalizumab-AAA diluted to 2 μg/mL to capture antibodies directed against omalizumab. The biotin-mutant omalizumab-AAA reagent was co-incubated overnight with an equal volume of each undiluted serum sample and anti-omalizumab IgE controls in a 96-well polypropylene microplate (Corning Life Sciences, Cat. No. 3365) at a 1:2 dilution in-well. The samples from the polypropylene microtiter plate were then transferred to a streptavidin-coated 96-well microtiter plate (StreptaWellTM High Bind; Roche Diagnostics, Cat. No. 11-989-685-001) and incubated for 1 h. Next, digoxigenin (DIG)-conjugated rhuFcεR1-IgG at 0.5 μg/mL was added and incubated for 1 h for detection of IgE isotype antibodies specific for omalizumab, followed by a 1-h incubation with a mouse anti-digoxigenin antibody conjugated with horseradish peroxidase (Jackson ImmunoResearch Laboratories Inc., Cat. No. 200-032-156) at 0.25 μg/mL. A peroxidase substrate (tetramethyl benzidine, two parts: TMB Peroxidase Substrate (A), Kirkegaard & Perry Laboratories Cat. No. 50-76-02; Peroxidase Solution; (B), Kirkegaard & Perry Laboratories Cat. No. 50-65-02) was added for color development, and the reaction was stopped by adding an equal volume of 1 M phosphoric acid. The absorbance values of the samples in the plate were evaluated at 450 nm, using 630 nm for reference absorbance (SpectraMax Plus 384 microplate reader, Molecular Devices, Sunnyvale, CA).
Screening Assay Cutpoint Factor Determination
The screening assay cutpoint is the threshold at which a sample with an assay signal at or above the cutpoint is determined to be positive for ATAs, while samples that fall below the cutpoint are determined to be negative for ATAs. The screening assay cutpoint was computed for each plate. A plate’s cutpoint is the product of the cutpoint factor (CPF) and the plate’s average negative control (NC).
The screen CPF was determined by assaying a panel of 100 individual therapeutically untreated human allergic asthma samples obtained from Bioreclamation (Hicksville, NY). The panel was screened in duplicate at a 1:2 dilution in two independent assays. A sample score (SS) was computed for each sample in each assay, and each assay was typically performed over several plates. The SS was calculated to be the ratio of the average sample assay signal to the average NC signal.
To target a 1% untreated positive rate (UTPR), the estimated 99th percentile of the log transform of the SS was computed for each of two assays. The screen CPF was the average of the estimated 99th percentiles across both assays. The screen CPF was determined to be 1.71 for allergic asthma. The calculations for the screen CPF were performed using Microsoft Excel.
Confirmatory Assay
The confirmatory assay procedure was performed to characterize the specificity of a sample’s positive-screen antibody response. It is a competitive binding assay, comparing the assay signal of the sample co-incubated with a mixture of biotin-labeled and unlabeled therapeutic to the assay signal of the sample incubated with biotin-labeled therapeutic only. A specified decrease in the sample signal, upon incubation with the unlabeled therapeutic compared to the sample incubated with only biotin-labeled therapeutic, indicates that immune complexes have formed and the assay response is specific to the therapeutic. Both the labeled and unlabeled therapeutic-treated samples were assayed in duplicate on the same plate.
In this assay, the therapeutic was the mutant omalizumab-AAA rather than native omalizumab to ensure that only IgE antibodies specific to omalizumab were measured and to limit the interference of total IgE. The specificity of an IgE-positive antibody response to omalizumab in the screening assay was confirmed by co-incubating the sample overnight with an equal volume of a mixture of 2 μg/mL biotin-labeled mutant omalizumab-AAA with and without 25 μg/mL of unlabeled mutant omalizumab-AAA.
Confirmatory Assay Cutpoint Factor Determination
The assay confirmatory cutpoint (CCP) was determined by assaying the same panel of omalizumab-naïve individual samples used in the screening assay CPF determination. All samples were co-incubated overnight with an equal volume of a mixture of 2 μg/mL biotin-labeled mutant omalizumab-AAA (1 μg/mL in-well) with and without 25 μg/mL unlabeled mutant omalizumab-AAA in duplicate (12.5 μg/mL in-well). Confirmatory (inhibition) scores were computed for each sample. A sample’s confirmatory score (CS) is the sample signal without unlabeled therapeutic mAbs minus the sample signal with unlabeled therapeutic mAbs, divided by the sample signal without unlabeled therapeutic, multiplied by 100%. The assay CCP, which targets a 1% UTPR, is the estimated 99th percentile of the confirmatory scores. The calculations were performed using Microsoft Excel.
Measurement of Total Omalizumab
Omalizumab concentrations in human serum samples were measured by the omalizumab human enzyme-linked immunosorbent assay (ELISA) developed by Genentech, Inc., (South San Francisco, CA) (5).
Measurement of Total IgE Levels
Serum total IgE levels were measured using the Cobas IgE II assay (Roche Diagnostics, Cat. No. 04827031190).
RESULTS
Development of reagents was critical for the assay to detect antibodies of IgE isotype to omalizumab.
Three unique and critical reagents were used to develop a sensitive assay to detect antibodies of IgE isotype to omalizumab. The approach was based on a novel strategy to overcome interference from endogenous IgE (Fig. 1). The first reagent was an altered version of omalizumab with reduced binding affinity for IgE. We selected three amino acids in the CDR of omalizumab and mutated them to alanines (mutant omalizumab-AAA). This change significantly reduced omalizumab’s affinity for IgE, but it had a minimal impact on the overall structure of the molecule. The second critical reagent was a human FcεR1-receptor IgG fusion protein (rhuFcεR1-IgG) that bound to the same region on the IgE Fc as omalizumab. This reagent distinguishes between IgE anti-therapeutic antibody bound to omalizumab versus non-specific IgE that omalizumab bound to since once omalizumab is bound to IgE, the receptor could not bind to it. The third critical reagent was a recombinant chimeric human IgE where the variable heavy chain and the variable light chain in the human IgE were replaced with a murine monoclonal antibody variable heavy chain and variable light chain that was specific to the CDR of omalizumab. This chimeric human IgE reagent was used as the IgE-positive control for the assay.
Fig. 1.
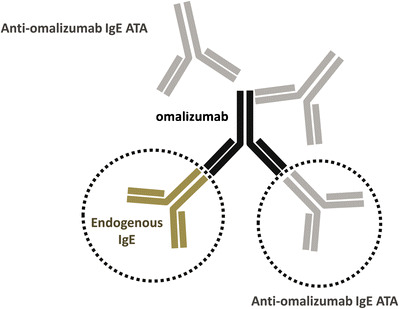
Omalizumab can bind to endogenous IgE as well as to anti-omalizumab IgE ATA
Characterization of Critical Reagents
Affinity Comparison of Omalizumab and the Omalizumab-AAA Mutant for Human IgE
Varying levels of human (Hu) IgE were captured on microtiter plates coated with either omalizumab or mutant omalizumab-AAA. The bound Hu IgE was detected with a horseradish peroxidase (HRP)-labeled goat anti-Hu IgE polyclonal antibody. The mutant omalizumab-AAA bound Hu IgE by approximately 100-fold less when directly compared to IgE binding to omalizumab (Fig. 2). The affinities of the anti-omalizumab IgE-positive control for omalizumab and mutant omalizumab-AAA were evaluated to ensure that the positive control generated could detect both omalizumab and mutant omalizumab-AAA equally. The assay was specifically designed to detect the binding of the positive control to omalizumab or the mutant omalizumab-AAA while eliminating the binding of omalizumab/mutant omalizumab-AAA to the Fc fragment of the positive control. Microtiter plates were first coated with rhuFcεR1-IgG to capture the Fc fragment of the positive control and thereby block the ability of omalizumab/mutant omalizumab-AAA to bind to the Fc fragment of the positive control. Varying levels of omalizumab or omalizumab-AAA mutant were then in turn captured by the rhuFcεR1-IgG-bound positive control, and the resulting IgE/omalizumab complexes of anti-omalizumab IgE-positive control specifically bound to omalizumab or mutant omalizumab-AAA were detected with an HRP-labeled goat anti-Hu IgG polyclonal antibody. The anti-omalizumab IgE-positive control demonstrated similar binding to both omalizumab and mutant omalizumab-AAA (Fig. 3).
Fig. 2.
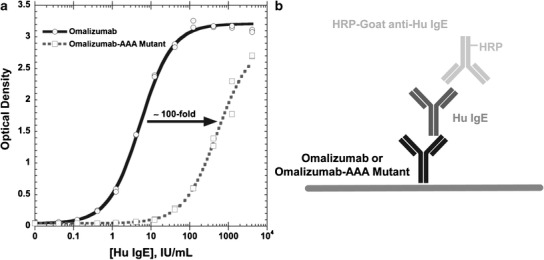
Omalizumab-AAA mutant demonstrates 100-fold lower affinity than omalizumab for Hu IgE
Fig. 3.
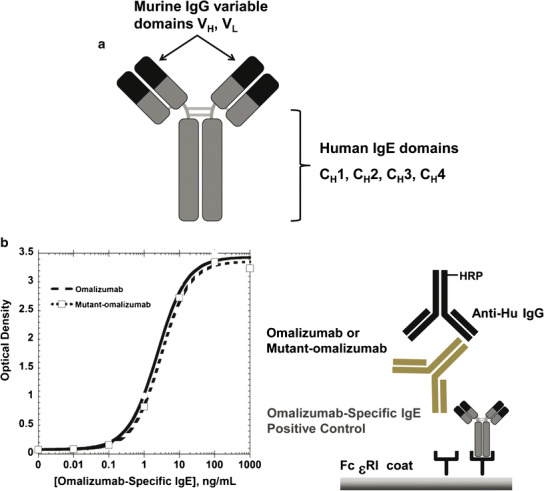
a An engineered chimeric human IgE antibody that consists of a human IgE constant domain (gray) with a murine IgG variable domain (black) containing a complementarity-determining region (CDR) that is specific for the CDR epitopes of omalizumab. The murine IgG variable domain was obtained from a Genentech proprietary monoclonal antibody. b Anti-Omalizumab IgE- positive control demonstrates comparable affinity for omalizumab and omalizumab-AAA mutant
Final Assay Format: Distinguishing Between Endogenous and Specific IgE
Biotin-labeled mutant omalizumab-AAA with ~100-fold reduced affinity for endogenous IgE was used as the capture reagent. Samples were incubated with biotin-labeled mutant omalizumab-AAA. The omalizumab-specific IgE antibody/biotin-omalizumab-AAA complexes were captured on a streptavidin-coated microtiter plate. The plate-bound complexes were detected with a recombinant human FcεRI IgG fusion protein that bound the Fc fragment of human IgE (Fig. 4). The FcεRI IgG reagent was not able to detect any non-specific human IgE bound by omalizumab-AAA. The combination of the mutant omalizumab for capture and the FcεRI IgG detection reagent ensured the selective detection of ATA consisting of omalizumab-specific IgE. Final assay validation parameters are summarized in Table I.
Fig. 4.
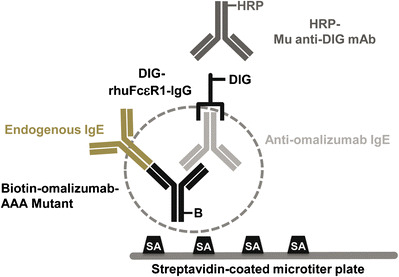
Anti-omalizumab IgE ATA assay. The assay is a semi-homogenous colorimetric enzyme-linked immunosorbent assay (ELISA). Serum samples are pre-incubated with biotin-labeled omalizumab-AAA mutant to form complexes of anti-omalizumab IgE and biotin-omalizumab-AAA mutant. The complex is then captured on a streptavidin-coated microtiter plate and detected using a combination of DIG-labeled rhuFceR1-IgG and HRP-labeled anti-DIG mAb. The FceR1 is not able to bind to the Fc fragment of human IgE if it is bound by omalizumab. Therefore, only anti-omalizumab IgE is detected by the DIG-rhuFceR1-IgG
Table I.
Overview of Assay Validation Parameters for the Anti-Omalizumab IgE ATA Assay
| Assay parameters | Results |
|---|---|
| Relative sensitivity (IU/mL) | 0.041 IU/mL for allergic asthma (>target of 0.1 IU/mL) |
| Screen cutpoint factor | 1.71 (n = 100 allergic asthma) |
| Confirmatory cutpoint factor | −25.3% (n = 100 allergic asthma) |
| Total IgE tolerance | Up to ~5,000 IU/mL in allergic asthma |
| Omalizumab drug tolerance (for detection of 0.1 IU/mL PC) | ≤0.1 omalizumab/total IgE molar ratio for allergic asthma |
| Hook effect (high positive at 142 IU/mL) | No hook effect found |
| How positive results were reported | OD/cutpoint OD ratio (no titer due to limited sample volume) |
The anti-omalizumab IgE isotype assay demonstrated suitable tolerance for total IgE.
The potential cross-reactivity of total IgE in this assay was tested by analyzing 100 individual allergic asthma human serum samples with a broad range of endogenous total IgE levels (22–5,340 IU/mL). As part of the determination of the screening cutpoint factor (CPF) and the confirmatory cutpoint (CCP) threshold, each of the 100 individual serum samples was analyzed twice in the screening assay and once in the confirmatory assay. No correlation was observed between the sample total IgE levels and signal in either the screening or confirmatory assays (Fig. 5a, b). Total IgE levels up to 5,340 IU/mL were evaluated confirming the high endogenous IgE tolerance of the assay.
Fig. 5.
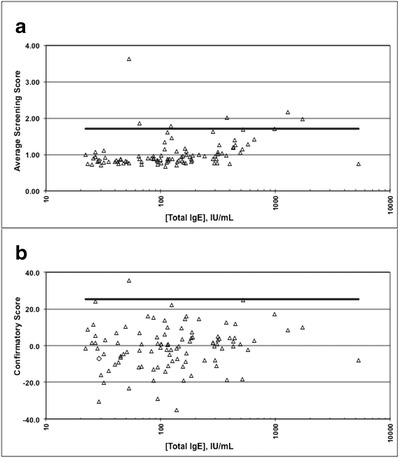
Total IgE tolerance of anti-omalizumab IgE ATA assay. One hundred individual human sera samples with 22–5,340 IU/mL of total IgE from patients diagnosed with allergic asthma were analyzed in the anti-omalizumab IgE ATA assay. Neither the screening score (a) or the confirmatory score (b) exhibited any correlation with the sample total IgE level indicating the assay could tolerate up to ~5,340 IU/mL of total IgE
Omalizumab tolerance in the IgE ATA assay was dependent on both omalizumab and total IgE.
The omalizumab tolerance in the assay was determined by assaying four levels of anti-omalizumab IgE-positive control antibody (0, 0.1, 0.4, and 1.6 IU/mL) in the presence of four levels of omalizumab (0, 20, 50, and 125 ng/mL) in four allergic asthma individual human serum samples and the human negative control (NC) serum (Fig. 6a). The omalizumab therapeutic levels chosen were based on the expected concentrations in the study population at the time of ATA sample collection after the washout period. The four individual allergic asthma human serum samples and NC serum had a wide range of endogenous total IgE levels (30, 91, 318, 982, and 177 IU/mL), providing samples with a wide range of omalizumab/total IgE molar ratios (0 and 0.01–2.2). The low-level IgE ATA concentration of 0.1 IU/mL was targeted at the desired sensitivity level of the assay. A sample with a mean OD/cutpoint OD ratio > 1.00 was considered positive in the assay.
Fig. 6.
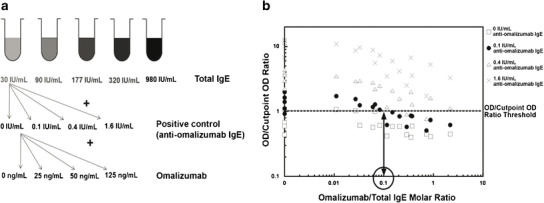
Drug (omalizumab) tolerance of anti-omalizumab IgE ATA assay. Omalizumab has the potential to create false-negative results in the anti-omalizumab IgE ATA assay by binding to the sample anti-omalizumab IgE and preventing its detection. To determine the omalizumab tolerance of the assay, a set of 80 human serum samples containing four levels of anti-omalizumab IgE (0, 0.1, 0.4, and 1.6 IU/mL) and a variety of omalizumab/total IgE molar ratios (0, 0.01–2.2) were prepared. Combinations of four levels of omalizumab (0, 20, 50, and 125 ng/mL) were added to five human sera with different levels of total IgE (30–982 IU/mL) and four levels of anti-omalizumab IgE. In order to detect low positive anti-omalizumab IgE (0.1 IU/mL), the omalizumab/total IgE molar ratio needed to be approximately less than 0.1 (see Fig. 6b)
As expected, samples with higher levels of IgE antibodies to omalizumab (positive control) tolerated higher levels of omalizumab compared to samples with low levels of IgE antibodies to omalizumab (Fig. 6b). These experiments confirmed that with the positive control antibody in this assay, low-level IgE ATA-positive samples (0.1 IU/mL) could only be detected when the omalizumab/total IgE molar ratio was <0.1. However, as shown in Fig. 6b, higher levels of specific IgE could easily be detected at molar ratios greater than 0.1.
Immunogenicity and Skin Test Evaluation of the Study Subjects
Serum samples from all eligible patients (21/30 patient samples, see “MATERIALS AND METHODS”) were analyzed for antibodies of IgE and IgG isotype to omalizumab (Table II). There were no ATAs of IgG or IgE isotypes detected among the eligible cases or control subjects (21 cases; 10 controls). In addition, all 10 subjects enrolled in the skin test sub-study (three anaphylaxis case subjects and seven control subjects) were skin tested with both omalizumab and placebo. Nine of these subjects (three anaphylaxis case subjects and six control subjects) were eligible for ATA analysis based on the inclusion criteria. None of the eligible cases had a positive skin reaction to omalizumab or to excipient. Two of seven controls tested positive for both omalizumab and excipient but tested negative in the IgE and IgG antibody assays.
Table II.
Skin and Antibody of IgE Isotype Results for Eligible Patients in the Study
| CASE subjects Omalizumab treated Anaphylaxis |
Control subjects Omalizumab treated No anaphylaxis |
||||
|---|---|---|---|---|---|
| Main study | Skin test | Main study | Skin test | ||
| Enrolled (118) | 30 | 3 | 88 | 7 | |
| ATA eligible (31) | 21 | 3 | 10 | 6 | |
| IgE ATA confirmed positive (0) | 0 | 0 | |||
| Reported IgE ATA results | Negative | 21 (100%) | 3 (100%) | 10 (100%) | 6 (100%) |
| Positive | 0 (0%) | 0 (100%) | 0 (0%) | 0 (100%) | |
DISCUSSION
Anaphylaxis is a serious, potentially life-threatening reaction that involves multiple organs and is usually rapid in onset. It is an IgE-mediated type I hypersensitivity reaction that results in mast cell activation and release of multiple mediators such as histamine, leukotrienes, and cytokines including tumor necrosis factor (TNF) (6). Whereas anaphylactic reactions have been reported after treatment with several monoclonal antibodies, their rates have generally been less than 1% (7–14). To date, the only published case that has reported a high rate of hypersensitivity reactions has been with cetuximab. In that case, the risk was reported to be higher in particular areas of the southeastern USA (15,16) due to environmental factors leading to development of an anti-alpha-gal IgE, a carbohydrate known to be present on cetuximab as a result of the production cell line (15). These hypersensitivity reactions were immediate and severe in nature, primarily occurring during the initial cetuximab infusion, supporting the role of pre-existing IgE targeting cetuximab. In fact, pre-existing IgE against cetuximab has been detected in patients who experience hypersensitivity reactions (15–17).
It is important to note that the reactions that are observed with cetuximab are very different from the reactions observed with omalizumab. The anaphylactic cases reported for omalizumab patients range anywhere from immediate to delayed reactions that persist from hours to days after treatment (1). This delayed response has been previously reported in the literature for various allergenic triggers, as well as for cases of food-dependent exercise-induced anaphylaxis and its triggers are poorly understood, all of which make its prevention challenging (18). In addition, although hypersensitivity reactions have occurred after first infusion with omalizumab, the majority of the reactions have occurred after two or more infusions (1) suggesting that the reactions are not due to pre-existing antibodies to omalizumab.
To better understand the risk of anaphylaxis in patients with allergic asthma who received omalizumab, a post-marketing pharmacosurveillance study was initiated, and as part of this study, an assay to detect IgE antibody to omalizumab was developed.
Development of an assay that could detect antibodies of IgE isotype specific for omalizumab which is itself an anti-IgE antibody was a challenging task, especially in the allergic asthma population which has high and variable levels of total IgE. Significant technical challenges were overcome in developing an assay to detect antibodies of IgE isotype to omalizumab. One challenge was the high and variable levels of total IgE in the asthmatic population. The omalizumab dosing table is designed to treat patients with baseline total IgE levels as high as 700 IU/mL. However, these levels can be further elevated by binding of IgE to omalizumab which decreases its clearance rate and elevates total IgE levels in serum. Elevated total IgE in the samples can significantly interfere in an assay designed to detect IgE specific for omalizumab. In addition, levels of omalizumab are also expected to be variable in these patients due to a large washout period window (16 weeks–18 months) before sample collection. The study was designed to have a washout period of at least 4 months or ~4 half-life to minimize the drug interference in the assay. We do acknowledge however that the longer washout period may potentially lead to false-negative signal. This large washout window was based on logistical challenges of the study (discussed further below) and is also the reason for the ineligibility of a large number of patients.
The second challenge was the need to distinguish between omalizumab bound by IgE (omalizumab-specific IgE) and IgE bound by omalizumab (non-omalizumab-specific IgE). Omalizumab binds to endogenous IgE with high affinity (Kd = 15.5 nM). In addition, omalizumab can also bind to IgE ATAs that are directed against it. The third challenge was the sensitivity requirements needed to be achieved in the current assay.
In order to increase sensitivity as well as specificity in our assay and detect IgE antibodies specific for omalizumab while minimizing the detection of IgE antibodies that were not specific for omalizumab (essentially total IgE), a mutant form of omalizumab (with a mutation in the CDR region) with an approximately 100-fold lower affinity for the Fc fragment of IgE was used as a capture reagent. Although the affinity of this mutated omalizumab was reduced ~100-fold, it could still potentially bind to endogenous IgE in the patient samples. In order to distinguish between specific antibodies of IgE isotype to omalizumab and non-specific IgE, FcεRI IgG was used for detection. Due to the IgE Fc fragment specificity of the detection reagent, it is unable to detect any omalizumab-bound human IgE. The combined use of the mutated omalizumab for capture and the FcεRI IgG detection reagent ensured the selective detection of omalizumab-specific IgE. This assay was able to reliably quantitate the omalizumab-specific IgE-positive control antibody with a sensitivity of less than 0.1 IU/mL. Samples that screened positive were confirmed in a competitive binding assay using unlabeled mutated omalizumab.
A tiered based strategy is typically used in immunogenicity assessments. In such a strategy, samples are first evaluated in a screening and confirmatory assay followed by further characterization including isotyping evaluations performed on the confirmed positive samples. However, this strategy is based on the new bridging assay formats where all antibody isotype responses to the drug can be detected. In the case of omalizumab, the screening assay consisted of two fragment assays: one for Fab and one for the Fc portion of the antibody and could only detect antibodies of IgG isotype. Therefore, in this instance, all eligible case and control samples from the study were tested in parallel in both the IgG and IgE ATA assays. Our data demonstrated no observable correlation between IgE or IgG ATAs and anaphylaxis under our assay conditions. It also demonstrated no apparent correlation between skin test positivity and anaphylaxis.
It is important to highlight that in addition to the technical challenges with this assay, there were also significant logistical challenges. The study was designed to collect samples from 30 anaphylactic patients, and it took 5 years to collect these samples. As mentioned earlier, only 21 of the 30 samples had the appropriate drug washout making them eligible for this analysis. We acknowledge that the limited sample size of this study would make it difficult to make definitive conclusions on correlation between anaphylaxis and presence of antibodies of IgE isotype to omalizumab, though the study itself highlights the challenges of design and execution of such studies even when a suitable assay is available.
One potential explanation for the observed incidence of anaphylaxis occurring during therapy with omalizumab could be the higher predisposition of asthmatics to anaphylaxis and a range of other allergy diagnoses (19). For example, a previous history of anaphylaxis increases the risk of developing a subsequent anaphylactic reaction with omalizumab by more than 19-fold (19). This study further reiterates the complexities associated with understanding the causes of anaphylactic reactions in the allergic asthma population with higher rates of baseline anaphylaxis. Clearly, many unknowns still remain in our understanding of the causes of these events, and therefore preventive measures have yet to be determined.
CONCLUSION
The post-marketing pharmacosurveillance study using a novel assay to detect IgE antibody to omalizumab showed no apparent correlation between either anaphylaxis or skin test reactivity and the presence of antibodies of IgE isotype to omalizumab.
ACKNOWLEDGMENTS
The authors would like to acknowledge Chris Lowe and Rich Erickson for providing the total IgE data. We would also like to thank Samantha Lien, Dorothea Reilly, Rod Keck, and Reed Harris for critical reagent generation and characterization.
ABBREVIATIONS
- ATA
Anti-therapeutic antibody
- CIU
Chronic idiopathic urticaria
- CDR
Complementarity-determining region
- CCP
Confirmatory cutpoint
- CPF
Cutpoint factor
- CS
Confirmatory score
- ELISA
Enzyme-linked immunosorbent assay
- FDA
Food and Drug Administration
- HRP
Horseradish peroxidase
- Hu
Human
- mAb
Monoclonal antibody
- PC
Positive control
- NC
Negative control
- SS
Sample score
- UTPR
Untreated positive rate
- DIG
Digoxigenin
- IgG
Immunoglobulin G
- IgE
Immunoglobulin E
REFERENCES
- 1.Limb S, Starke P, Lee C, Chowdhury BA. Delayed onset and protracted progression of anaphylaxis after omalizumab administration in patients with asthma. J Allergy Clin Immunol. 2007;120:1378–81. doi: 10.1016/j.jaci.2007.09.022. [DOI] [PubMed] [Google Scholar]
- 2.Sampson H, Munoz-Furlong A, Bock SA, Schmitt C, Bass R, Chowdhury BA, et al. Symposium on the definition and management of anaphylaxis: summary report. J Allergy Clin Immunol. 2005;115:584–91. doi: 10.1016/j.jaci.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Sampson H, Munoz-Furlong A, Campbell RL, Adkinson NF, Jr, Bock SA, Branum A, et al. Second symposium on the definition and management of anaphylaxis: summary report—Second National Institute of Allergy and Infectious Disease/Food Allergy and Anaphylaxis Network symposium. J Allergy Clin Immunol. 2006;117:391–7. doi: 10.1016/j.jaci.2005.12.1303. [DOI] [PubMed] [Google Scholar]
- 4.Kelley RF, Meng YG. Methods to engineer and identify IgG1 variants with improved FcRn binding or effector function G. In: Proetzel H, Ebersbach H, editors. Methods in molecular biology: antibody methods and protocols. New York, NY: 901 Humana Press; 2012. pp. 277–293. [DOI] [PubMed] [Google Scholar]
- 5.Lowe PJ, Tannenbaum S, Gautier A, Jimenez P. Relationship between omalizumab pharmacokinetics, IgE pharmacodynamics and symptoms in patients with severe persistent allergic (IgE-mediated) asthma. Br J Clin Pharmacol. 2009;68:61–76. doi: 10.1111/j.1365-2125.2009.03401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phil Leiberman. Definition and criteria for the diagnoses of anaphylaxis. Mariana C. Castells, editor. Anaphylaxis and hypersensitivity reactions. Humana Press, Springer New York; 2010. P. 1–13.
- 7.Edwards JC, Szczepanski L, Szechinski J, Filipowicz-Sosnowka A, Emery P, Close DR, et al. Efficacy of B-cell-targeted therapy with rituximab in patients with rheumatoid arthritis. N Engl J Med. 2004;350:2572–81. doi: 10.1056/NEJMoa032534. [DOI] [PubMed] [Google Scholar]
- 8.Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–78. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 9.Holgate S, Casale T, Wenzel S, Bousquet J, Deniz Y, Reisner C. The anti-inflammatory effects of omalizumab confirm the central role in IgE in allergic inflammation. J Allergy Clin Immunol. 2005;115:459–65. doi: 10.1016/j.jaci.2004.11.053. [DOI] [PubMed] [Google Scholar]
- 10.Cheifetz A, Smedley M, Martin S, Reiter M, Leone G, Mayer L, et al. The incidence and management of infusion reactions to infliximab: a large center experience. Am J Gastroenterol. 2003;98:1315–24. doi: 10.1111/j.1572-0241.2003.07457.x. [DOI] [PubMed] [Google Scholar]
- 11.Cook-Bruns N. Retrospective analysis of the safety of Herceptin immunotherapy in metastatic breast cancer. Oncology. 2001;61(Suppl 2):58–66. doi: 10.1159/000055403. [DOI] [PubMed] [Google Scholar]
- 12.Omalizumab (marketed as Xolair®) Information. Rockville, MD: Food and Drug Administration, Center for Drug Evaluation and Research, 2007. (Accessed February 19, 2008, at http://www.fda.gov/cder/drug/infopage/omalizumab/default.htm.)
- 13.Erbitux (cetuximab). New York: Im-Clone Systems Incorporated and Bristol-Myers Squibb, 2007 (drug product label).
- 14.Cheifetz A, Mayer L. Monoclonal anti-bodies, immunogenicity, and associated infusion reactions. Mt Sinai J Med. 2005;72:250–6. [PubMed] [Google Scholar]
- 15.Chung CH, Mirakhur B, Chan E, Le QT, Berlin J, Morse M, et al. Cetuximab-induced anaphylaxis and IgE specific for galactose-α-1,3-galactose. N Engl J Med. 2008;358:1109–1117. doi: 10.1056/NEJMoa074943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maier S, Chung CH, Morse M, Platts-Mills T, Townes L, Mukhopadhyay P, et al. A retrospective analysis of cross-reacting cetuximab IgE antibody and its association with severe infusion reactions. Cancer Med. 2015;4(1):36–42. doi: 10.1002/cam4.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Neil BH, Allen R, Spigel DR, Stinchcombe TE, Moore DT, Berlin JD, et al. High incidence of cetuximab-related infusion reactions in Tennessee and North Carolina and the association with atopic history. J Clin Oncol. 2007;25:3644–3648. doi: 10.1200/JCO.2007.11.7812. [DOI] [PubMed] [Google Scholar]
- 18.Oyefara BI, Bahna SL. Delayed food-dependent, exercise-induced anaphylaxis. Allergy Asthma Proc. 2007;28:64–6. doi: 10.2500/aap.2007.28.2951. [DOI] [PubMed] [Google Scholar]
- 19.Iribarren C, Tolstykh IV, Miller MK, Eisner MD. Asthma and the prospective risk of anaphylactic shock and other allergy diagnoses in a large integrated health care delivery system. Ann Allergy Asthma Immunol. 2010;104:371–7. doi: 10.1016/j.anai.2010.03.004. [DOI] [PubMed] [Google Scholar]


